The Nrf2–Keap1 signaling pathway is key to cell defense and survival pathways. Nrf2 can protect cells and tissues from toxicants and carcinogens, and several Nrf2 activators are currently being tested as chemopreventive compounds. However, several studies also suggest that Nrf2 may protect cancer cells from chemotherapeutic agents and promote cancer cell proliferation. Here, Jaramillo and Zhang provide an overview of the Nrf2–Keap1 signaling pathway in cancer. They discuss the dual role of Nrf2 in cancer and the challenges in developing Nrf2-based drugs for chemoprevention and therapy.
Keywords: Nrf2, Keap1, chemoprevention, chemoresistance, ARE, oxidative stress
Abstract
The Nrf2 (nuclear factor erythroid 2 [NF-E2]-related factor 2 [Nrf2])–Keap1 (Kelch-like erythroid cell-derived protein with CNC homology [ECH]-associated protein 1) signaling pathway is one of the most important cell defense and survival pathways. Nrf2 can protect cells and tissues from a variety of toxicants and carcinogens by increasing the expression of a number of cytoprotective genes. As a result, several Nrf2 activators are currently being tested as chemopreventive compounds in clinical trials. Just as Nrf2 protects normal cells, studies have shown that Nrf2 may also protect cancer cells from chemotherapeutic agents and facilitate cancer progression. Nrf2 is aberrantly accumulated in many types of cancer, and its expression is associated with a poor prognosis in patients. In addition, Nrf2 expression is induced during the course of drug resistance. Collectively, these studies suggest that Nrf2 contributes to both intrinsic and acquired chemoresistance. This discovery has opened up a broad spectrum of research geared toward a better understanding of the role of Nrf2 in cancer. This review provides an overview of (1) the Nrf2–Keap1 signaling pathway, (2) the dual role of Nrf2 in cancer, (3) the molecular basis of Nrf2 activation in cancer cells, and (4) the challenges in the development of Nrf2-based drugs for chemoprevention and chemotherapy.
The transcription factor nuclear factor erythroid 2 (NF-E2)-related factor 2 (Nrf2) belongs to the Cap ‘N’ Collar (CNC) family that contains a conserved basic leucine zipper (bZIP) structure. The main function of Nrf2 is to activate the cellular antioxidant response by inducing the transcription of a wide array of genes that are able to combat the harmful effects of extrinsic and intrinsic insults, such as xenobiotics and oxidative stress. As a result, Nrf2 has traditionally been regarded as the cell's main defense mechanism and a major regulator of cell survival. Activation of the Nrf2 defense response has been shown to protect against neurodegenerative diseases, aging, diabetes, photo-oxidative stress, cardiovascular disease, inflammation, pulmonary fibrosis, acute pulmonary injury, and cancer (for review, see Motohashi and Yamamoto 2004; Jeong et al. 2006; Zhang 2006; Kensler et al. 2007; Lau et al. 2008).
Recent studies, however, have demonstrated that Nrf2 promotes the survival of not only normal cells but also cancer cells. Accumulation of Nrf2 in cancer cells creates an environment conducive for cell growth and protects against oxidative stress, chemotherapeutic agents, and radiotherapy. This phenomenon has been termed the “dark side of Nrf2” (Lau et al. 2008; Wang et al. 2008b). This discovery has opened up a broad spectrum of research geared toward a better understanding of the role of Nrf2 signaling in cancer and has set a new paradigm for the development of pharmacological reagents targeting Nrf2 for cancer prevention and treatment.
The Nrf2–Keap1 (Kelch-like erythroid cell-derived protein with CNC homology [ECH]-associated protein 1)–ARE (antioxidant response element) signaling pathway
Nrf2
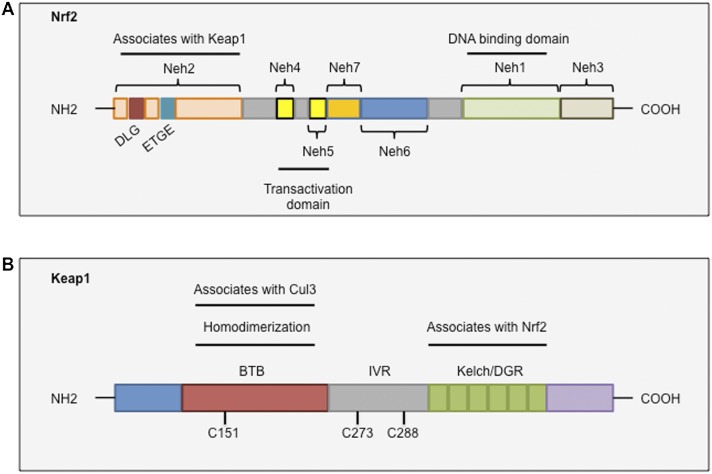
Nrf2 contains seven functional domains, known as Neh1–Neh7 (Fig. 1A). Of these, the Neh2 domain, located in the N terminus of Nrf2, is the major regulatory domain. Neh2 contains seven lysine residues that are responsible for ubiquitin conjugation (Zhang et al. 2004) as well as two binding sites (termed ETGE and DLG motifs) that help regulate Nrf2 stability (McMahon et al. 2006; Tong et al. 2006b). The ETGE and DLG motifs interact with Keap1, which is a substrate adaptor protein for the Cullin 3 (Cul3)-dependent E3 ubiquitin ligase complex that represses Nrf2 by promoting its ubiquitination and subsequent proteasomal degradation (Cullinan et al. 2004; Kobayashi et al. 2004; Zhang et al. 2004; Furukawa and Xiong 2005).
Figure 1.
Conserved domains of Nrf2 and Keap1. (A) Nrf2 contains seven domains, known as Neh1–Neh7. The Neh2 domain contains two binding motifs, DLG and ETGE, which interact with Keap1. The Neh4, Neh5, and Neh3 domains are important for the transactivation activity of Nrf2. The Neh6 domain is a serine-rich region that regulates Nrf2 stability. The Neh1 domain is a basic region leucine zipper motif that is important for its stability, DNA binding, and dimerization with Maf. (B) Keap1 contains three major domains. The BTB domain mediates Keap1 homodimerization and associates with Cul3. The IVR domain contains critical cysteine residues and connects the BTB domain with the C terminus Kelch/DGR domain. The Kelch/DGR domain mediates binding with the Neh2 domain of Nrf2.
The Neh1 and Neh6 domains have also been reported to regulate the stability of Nrf2. Neh1 contains a CNC-type bZIP DNA-binding motif that allows Nrf2 to bind DNA and dimerize with other transcription factors (Moi et al. 1994). Additionally, the Neh1 domain has been shown to interact with UbcM2, a ubiquitin-conjugating enzyme, to regulate the stability of Nrf2 (Plafker et al. 2010). The Neh6 domain contains two binding sites (DSGIS and DSAPGS motifs) for the β-transducin repeat-containing protein (β-TrCP). β-TrCP acts as a substrate adaptor for the Skp1–Cul1–Rbx1/Roc1 ubiquitin ligase complex. Phosphorylation of the DSGIS motif by GSK-3 increases the ability of β-TrCP to ubiquitinate Nrf2 and promote its rapid turnover (McMahon et al. 2004; Rada et al. 2011; Chowdhry et al. 2013).
The Neh3, Neh4, and Neh5 domains interact with coactivators to enable the transactivation of Nrf2 target genes. The Neh3 domain binds to the chromo-ATPase/helicase DNA-binding protein family member CHD6, which functions as an Nrf2 transcriptional coactivator (Nioi et al. 2005). The Neh4 and Neh5 domains have been shown to interact with the CH3 domains of CBP (CREB-binding protein) to facilitate the transactivation of Nrf2 target genes (Katoh et al. 2001; Zhu and Fahl 2001). Recently, however, a seventh Neh domain (Neh7, amino acids 209–316) was identified and shown to interact with the retinoic X receptor α, an Nrf2 repressor, and repress Nrf2 target gene transcription (Wang et al. 2013).
Keap1
Nrf2 is primarily regulated by Keap1, a substrate adaptor for a Cul3-containing E3 ubiquitin ligase. Keap1 possesses three functional domains, including a broad complex/tramtrack/bric-a-brac (BTB) domain, an intervening region (IVR), and a Kelch domain, also known as the double glycine repeat (DGR) domain (Fig. 1B). The BTB domain binds Cul3 and is required for Keap1 dimerization (Zipper and Mulcahy 2002; Lo et al. 2006). The Kelch/DGR domain is critical for maintaining the interaction between Nrf2 and Keap1 by interacting with the Neh2 domain of Nrf2 (Itoh et al. 1999; McMahon et al. 2004). The IVR links the BTB and Kelch/DGR domains and contains several cysteine residues that have been proposed to regulate Keap1 activity (Kobayashi et al. 2004; Ogura et al. 2010). Thus, each of the three domains is thought to play a unique role in mediating Nrf2 ubiquitination and repression.
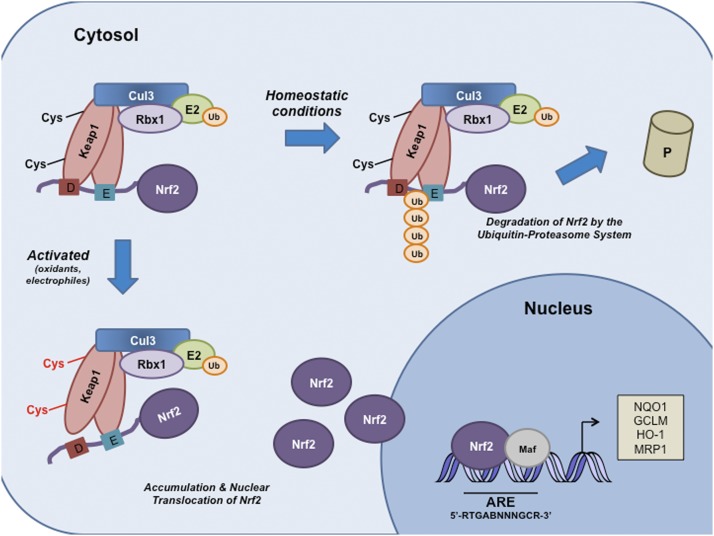
Under basal conditions, Nrf2 is primarily localized in a complex with Keap1 via direct protein–protein interactions between the Keap1 Kelch domain and the ETGE and DLG motifs on the Neh2 domain of Nrf2 (Fig. 2). Keap1 has been shown to bind to the ETGE motif with a higher affinity than to the DLG motif (McMahon et al. 2006; Tong et al. 2006a). Based on these observations, a two-site substrate recognition hinge-and-latch model describing the interaction between Nrf2 and Keap1 was developed (McMahon et al. 2006; Tong et al. 2006a,b). The model suggests that Keap1 recruits Nrf2 via the ETGE motif (hinge), and once this interaction has been established, the DLG motif (latch) docks onto an adjacent unoccupied Kelch repeat domain on Keap1. Two Keap1 molecules position the seven ubiquitin-accepting lysine residues that are located between the DLG and ETGE motifs of Nrf2 in a favorable position and promote Nrf2 polyubiquitination and its subsequent proteasomal degradation by the 26S proteasome (Zhang et al. 2004; McMahon et al. 2006; Tong et al. 2007). Therefore, the Keap1–Cul3–E3 ubiquitin ligase complex tightly regulates Nrf2 protein to maintain it at a low level. Conversely, recent evidence from our laboratory demonstrated that USP15 (ubiquitin-specific peptidase 15), a deubiquitinating enzyme, also plays an important role in mediating the ubiquitination and degradation of Nrf2. USP15 deubiquitinates Keap1, stabilizes the Keap1–Cul3–E3 ligase complex, and enhances its E3 ligase activity, which ultimately leads to the degradation of Nrf2 (Villeneuve et al. 2013).
Figure 2.
Schematic model of the Nrf2–Keap1 signaling pathway. Under basal conditions, Keap1 binds to the ETGE and DLG motifs on Nrf2 and brings Nrf2 into Keap1–Cul3–E3 ubiquitin ligase complex, leading to ubiquitination and subsequent degradation of Nrf2. Oxidative stress or electrophiles can cause a conformational change in the Keap1–Cul3–E3 ubiquitin ligase by acting on specific cysteine residues in Keap1. These changes disrupt Nrf2–Keap1 binding at the DLG domain. Nrf2 is stabilized, and free Nrf2 translocates to the nucleus, where it dimerizes with members of the small Maf family and binds to AREs (5′-RTGABNNNGCR-3′) within regulatory regions of a wide variety of cell defense genes, including NQO1, GCLM, HO-1, and MRP1. (E) ETGE; (D) DLG.
In response to a diverse array of stimuli, it has been proposed that critical cysteine residues, especially Cys151, within Keap1 can be covalently modified, allowing Nrf2 to evade Keap1-mediated ubiquitination (Fig. 2). The human Keap1 protein contains 27 cysteine residues that can be oxidized to sulfenic acid, form disulfides, or be covalently adducted by electrophiles (Dinkova-Kostova et al. 2002; Zhang and Hannink 2003; McMahon et al. 2010). The modification of thiols on Keap1 is thought to alter its conformation and results in the release of Nrf2 from the low-affinity binding site (DLG motif); however, Nrf2 remains attached to Keap1 by the ETGE motif. These changes are thought to prevent Nrf2 ubiquitination (McMahon et al. 2006; Tong et al. 2006a,b). Consequently, Keap1 molecules become saturated with Nrf2 that is no longer targeted for degradation, and newly synthesized, free Nrf2 translocates to the nucleus. In the nucleus, Nrf2 dimerizes with members of the masculoaponeurotic fibrosarcoma (Maf) protein family that have been shown to facilitate the binding of Nrf2 to AREs located within the regulatory regions of a wide variety of genes involved in cytoprotection and metabolism (Itoh et al. 1997; Nguyen et al. 2000; Motohashi et al. 2004; Hirotsu et al. 2012). The ARE is a cis-acting DNA enhancer sequence with the consensus sequence 5′-RTGABnnnGCR-3′, where conserved nucleotides are in capitals, and the “n” represents any nucleotide (Rushmore et al. 1991; Wasserman and Fahl 1997; Hayes and McMahon 2001). The Nrf2–Maf heterodimer recruits transcriptional coactivators that promote the transcription of genes involved in (1) regulating the synthesis and metabolism of glutathione, such as glutamate–cysteine ligase catalytic subunit (GCS); (2) antioxidant proteins specializing in neutralizing reactive species such as glutathione peroxidase (GPX); (3) drug-metabolizing enzymes like UDP-glucuronosyl-transferase 1A1; (4) xenobiotic transporters, including multidrug resistance protein 1 (MRP1); and (5) numerous other stress response proteins (for review, see Zhang and Gordon 2004; Hayes et al. 2010). By inducing the expression of this battery of genes, Nrf2 is able to augment a wide range of cell defense processes, thereby enhancing the overall capacity of cells to detoxify potentially harmful entities. As such, the Nrf2–Keap1 pathway is generally considered a major cellular defense pathway.
The dual role of Nrf2 in cancer
Tumor suppressor functions of Nrf2: ‘the good side of Nrf2’
Several studies using Nrf2 knockout mice (Nrf2−/−) show that Nrf2 protects against chemical carcinogen-induced tumor formation in the stomach, bladder, and skin. For example, Nrf2-null mice are more likely to develop gastric neoplasia after exposure to benzo(a)pyerene compared with wild-type mice (Ramos-Gomez et al. 2001). Higher tumor burdens were reported in the intestines of Nrf2-deficient mice challenged with azoxymethane followed by dextran sodium sulfate compared with wild-type mice (Osburn et al. 2007; Khor et al. 2008). In addition, Nrf2-deficient mice had a higher incidence of bladder tumors following exposure to N-nitrosobutyl(4-hydroxybutyl)amine (Fahey et al. 2002) as well as an increased incidence of skin tumors after exposure to 7,12-dimethylbenz(a)anthracene or 12-O-tetradecanoylphorbol-13-acetate, two potent carcinogens (Xu et al. 2006). The mechanism by which Nrf2 protects against chemical-induced carcinogenesis may be due in part to its ability to reduce the amount of reactive oxygen species (ROS) and DNA damage in cells (Hirayama et al. 2003; Morito et al. 2003).
Further evidence supporting the protective role of Nrf2 comes from studies with mice harboring a single-nucleotide polymorphism (SNP) in the promoter region of the mouse Nrf2 gene. Mice with this SNP have reduced expression of Nrf2 and are more susceptible to hyperoxia-induced lung damage (Cho et al. 2002). The human NRF2 gene also harbors a SNP in its promoter region (rs6721961) (Yamamoto et al. 2004; Marzec et al. 2007). Individuals with this SNP have significantly lower NRF2 messenger RNA (mRNA) levels and an increased risk of developing non-small-cell lung cancer (NSCLC) (Suzuki et al. 2013).
Oncogenic functions of Nrf2: ‘the dark side of Nrf2’
Although a wide body of evidence indicates that activation of Nrf2 protects against a variety of toxicants and diseases, the prolonged activation of Nrf2 has been shown to favor the progression of several types of cancers. Nrf2 has been shown to be constitutively elevated in lung, breast, head and neck, ovarian, and endometrial carcinomas (Singh et al. 2006; Shibata et al. 2008a,b; Wang et al. 2008b; Jiang et al. 2010; Kim et al. 2010; Solis et al. 2010; Zhang et al. 2010). The prognosis of patients with tumors expressing high levels of Nrf2 in the clinic is poor (Shibata et al. 2008a; Solis et al. 2010; Sasaki et al. 2012) partly due to Nrf2's ability to enhance cancer cell proliferation and promote chemoresistance and radioresistance. In addition, Nrf2 expression is induced during the course of drug resistance. Collectively, these studies suggest that Nrf2 contributes to both intrinsic and acquired chemoresistance (Shibata et al. 2008a,b; Singh et al. 2008; Solis et al. 2010; Zhang et al. 2010; Inoue et al. 2012).
Nrf2 and cancer cell proliferation
The elevated levels of Nrf2 in cancer cells have been shown to promote cancer cell proliferation. Recently, Mitsuishi et al. (2012) demonstrated that Nrf2 is indeed important for the ability of A549 lung cancer cells to proliferate. They performed microarray analysis to identify Nrf2 target genes involved in cancer cell proliferation and identified several genes involved in the pentose phosphate pathway, including glucose-6-phosphate dehydrogenase (G6PD), phosphogluconate dehydrogenase (PGD), transketolase (TKT), and transaldolase 1 (TALDO1), which are responsible for nicotinamide adenine dinucleotide phosphate (NADPH) regeneration. In addition, other metabolic genes, including malic enzyme 1 (ME1), phosphoribosyl pyrophosphate amidotransferase (PPAT), methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), and isocitrate dehydrogenase 1 (IDH1), were also identified as transcription targets of Nrf2. Nrf2 directly activated G6PD, PGD, TKT, TALDO1, ME1, and IDH1 by binding to their respective AREs. These proteins support glucose flux and generate purines, which are the building blocks of DNA and RNA and are important for accelerating proliferation in cancer cells.
Nrf2 may also promote cancer cell proliferation and tumorigenesis by maintaining the redox balance and generating antioxidants in cancer cells. Recent studies have revealed that glutathione is critical for cell proliferation (Reddy et al. 2007a,b; Ishimoto et al. 2011). Mitsuishi et al. (2012) also found that, in A549 cells, the cellular levels of glutamine are elevated and that a substantial amount of the glutamine was used for the generation of glutathione. Therefore, the enhancement of glutathione synthesis is another important effect of Nrf2 in accelerating cancer cell proliferation.
Nrf2 and chemoresistance and radioresistance
Several studies have demonstrated that cancer cells with elevated levels of Nrf2 are less sensitive to common chemotherapeutic agents such as etoposide, carboplatin, cisplatin, 5-fluorouracil, and doxorubicin (Singh et al. 2006; Ohta et al. 2008; Shibata et al. 2008a; Wang et al. 2008b; Homma et al. 2009; Jiang et al. 2010; Lister et al. 2011). Consistent with this notion, ectopic expression of Nrf2 in cancer cell lines that have low basal levels of Nrf2 renders them more resistant to a variety of anti-cancer agents, whereas siRNA-mediated inhibition of Nrf2 in cells with high levels of Nrf2 has been shown to reverse drug resistance (Shibata et al. 2008a; Wang et al. 2008b; Homma et al. 2009; Jiang et al. 2010; Lister et al. 2011). Furthermore, cells that have developed resistance to chemotherapeutic agents have also been shown to express high levels of Nrf2. For example, human ovarian cancer cells selected for resistance to doxorubicin have elevated levels of Nrf2 compared with wild-type cells, and depletion of Nrf2 restores drug sensitivity in the resistant cells (Shim et al. 2009). In vivo xenografts derived from Nrf2 silenced lung cancer cells were more susceptible to platin-based chemotherapeutics compared with xenografts with control cells (Singh et al. 2008; Jiang et al. 2010).
Constitutively high levels of Nrf2 have also been shown to protect cancer cells against ionizing radiation and confers radioresistance in NSCLC cells (Singh et al. 2010). A549 and H460 NSCLC cells transfected with Nrf2 siRNA were more susceptible to ionizing radiation, which was measured by an increase in protein oxidation and the total protein carbonyl content, than cells transfected with a control siRNA. Taken together, these findings suggest that Nrf2 plays a role in not only intrinsic chemoresistance but also acquired resistance in cancer cells and that selectively inhibiting Nrf2 may improve the clinical outcome of patients receiving chemotherapy or radiation therapy.
All of these findings suggest that Nrf2 has a dual role in cancer. Based on numerous studies, it appears that transient activation of Nrf2 in normal cells (where the Nrf2–Keap1 axis is intact) is protective; however, constitutive activation of Nrf2, as seen in cancer, enhances the survival and progression of cancer cells (Lau et al. 2008). A recent study supports this hypothesis. Satoh et al. (2013) found that Nrf2-deficient mice exposed to the carcinogen urethane exhibited a relative increase in tumor foci 8 wk after urethane administration. However, after 16 wk, tumors in Nrf2-null mice showed less advanced malignancy (Satoh et al. 2013).
Molecular basis of Nrf2 activation in cancer cells
Several mechanisms have been reported for the increased activity of Nrf2 in cancers, including (1) somatic mutations in KEAP1, CUL3, or NRF2; (2) epigenetic silencing of Keap1; (3) aberrant accumulation of proteins that disrupt the interaction between Nrf2 and Keap1; (4) transcriptional up-regulation of NRF2 through oncogene-dependent signaling; and (5) modification of Keap1 by metabolic intermediates (for review, see Mitsuishi et al. 2013). A detailed description of each mechanism is discussed below.
Somatic mutations in KEAP1, NRF2, or CUL3
Gain-of-function mutations in NRF2 and loss-of-function mutations in KEAP1 and CUL3 have been identified in several human cancers (Padmanabhan et al. 2006; Singh et al. 2006; Nioi and Nguyen 2007; Ohta et al. 2008; Shibata et al. 2008a; Li et al. 2011; Ooi et al. 2013; Sato et al. 2013). Mutations in the KEAP1 gene were initially identified in two human lung adenocarcinoma cell lines (Padmanabhan et al. 2006). The mutations involved a glycine-to-cysteine substitution in the Kelch/DGR domain of Keap1. The mutant Keap1 exhibited reduced affinity to Nrf2, and, consequently, Nrf2 was constitutively activated in these cells. Since then, multiple somatic mutations have also been identified in the Kelch or IVR domains of the Keap1 protein in NSCLC cell lines and human tissues from NSCLC patients (Singh et al. 2006; Ohta et al. 2008). The decreased Nrf2/Keap1 interaction resulted in the constitutive activation of Nrf2 and induction of ARE-containing genes. KEAP1 gene mutations have also been identified in breast cancer cells (Sjoblom et al. 2006) and patients with gallbladder (Shibata et al. 2008a), liver (Shibata et al. 2008b), ovarian (Konstantinopoulos et al. 2008), endometrial (Wong et al. 2008), and lung papillary (Li et al. 2011) cancers. As more studies accessing the status of KEAP1 in different cancers are undertaken, it is likely that these studies will reveal that KEAP1 mutations occur frequently in many cancer types.
Although Nrf2 mutations occur less frequently than Keap1 mutations, mutations in the NRF2 gene have also been identified in several cancers, including lung, head and neck, and esophageal carcinoma (Shibata et al. 2008b, 2011; Kim et al. 2010). Shibata et al. (2008b) recently identified NRF2 somatic mutations in 11 of 103 patients with lung cancer and in three of 12 patients with head and neck cancers. Of the 14 NRF2 mutations described in the study, six were within the DLG motif and eight were within the ETGE motif (Shibata et al. 2008b). Mutations in this region impair the two-site substrate recognition of Keap1, resulting in the stabilization of Nrf2, increased nuclear translocation of Nrf2, and activation of Nrf2 target genes (for review, see Mitsuishi et al. 2013).
Recently, somatic mutations in CUL3 were identified in hereditary type 2 papillary renal cell carcinoma (Ooi et al. 2013). While CUL3 has a role in regulating the Nrf2/Keap1 pathway, CUL3 also has many other targets. Thus, mutation of CUL3 likely has many downstream effects, one of which is up-regulation of Nrf2. Interestingly, all of the KEAP1, NRF2, and CUL3 mutations identified thus far are somatic. No genomic amplification or deletion of NRF2 has been reported in cancer thus far.
Epigenetic silencing of Keap1
Epigenetic modifications in KEAP1 have also been shown to promote the accumulation of Nrf2. The promoter region of the KEAP1 gene is hypermethylated in lung (Wang et al. 2008a; Muscarella et al. 2011), prostate (Zhang et al. 2010), malignant glioma (Muscarella et al. 2011), and colorectal cancers (Hanada et al. 2012). The methylation of specific CpG sites within the promoter region of KEAP1 affects KEAP1 expression by inducing local chromatin remodeling and restricting the ability of the transcriptional machinery to bind to the necessary DNA sequences (for review, see Copple 2012). Therefore, hypermethylation inhibits KEAP1 gene expression, which results in the accumulation of Nrf2. These epigenetic modifications in KEAP1 confer a growth advantage in cancer cells. In fact, the epigenetic abnormalities in the KEAP1 gene in lung cancers and malignant gliomas have been associated with poor clinical prognoses in patients (Muscarella et al. 2011).
Several microRNAs (miRNAs) have been shown to be involved in the regulation of NRF2, including miR-144, miR-28, and miR-200a. miRNAs are short, single-stranded, noncoding RNAs that regulate gene expression by sequence-specific binding to mRNA that either inhibits translation or causes degradation of mRNA (Bartel 2004). miR-144 was the first miRNA to be fully characterized as a negative regulator of Nrf2 expression (Sangokoya et al. 2010). miR-144 decreased the expression of NRF2 and its target genes by targeting two distinct sites at positions 265–271 and 370–377 of the 3′ untranslated region of NRF2 (Sangokoya et al. 2010). Similarly, MCF-7 breast cancer cells ectopically expressing miR-28 exhibited lower levels of NRF2 mRNA and protein (Yang et al. 2011). miR-200a was shown to negatively regulate the stability of KEAP1 mRNA and the level of Keap1 protein, suggesting that miR-200a indirectly up-regulates Nrf2 by suppressing Keap1 levels (Eades et al. 2011). Supporting a role for these miRNAs in cancer, aberrant expression of miR-28 and miR-144 has been reported in various cancers, including lymphoma, glioma, and squamous cell carcinoma (Bryan et al. 2012).
Interactions between Nrf2 and other signaling pathways
A growing body of evidence suggests that in tumors, increased levels of Nrf2 can occur in the absence of genomic alterations in the NRF2 and KEAP1 genes. Numerous studies have revealed that a wide variety of proteins can activate the Nrf2–Keap1 pathway by altering Nrf2–Keap1 binding (for review, see Yates et al. 2009; Wakabayashi et al. 2010). In this section, we focus on recent evidence highlighting the interactions between the Nrf2–Keap1 complex and p21, p62, and other proteins (Fig. 3).
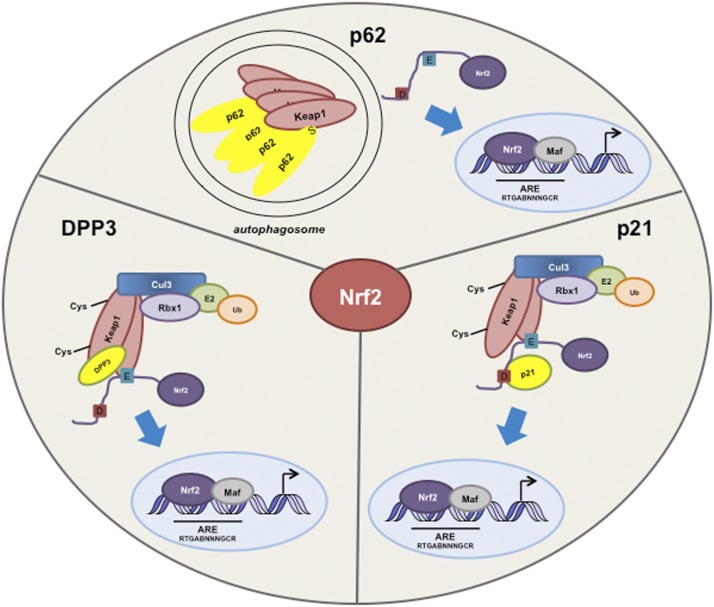
Figure 3.
Cross-talk between Nrf2 and other proteins. The substrate adaptor sequestosome 1 protein (p62) modulates the Nrf2–Keap1 signaling pathway by directly interacting with Keap1. p62 sequesters Keap1 in autophagosomes, which results in a decrease in Nrf2 ubiquitination, an increase in Nrf2 stability, and activation of Nrf2 target genes. Nrf2 is stabilized in response to p21 up-regulation. p21 associates with the DLG motif on Nrf2, thereby disrupting the ability of Keap1 to properly bind and ubiquitinate Nrf2. DPP3 binds to Keap1, inhibits Nrf2 ubiquitination, and drives Nrf2-dependent transcription in cancer cells. (E) ETGE; (S) STGE; (D) DLG.
Nrf2 and p21
Studies have demonstrated that p53 negatively regulates Nrf2-mediated gene transcription (Faraonio et al. 2006). Specifically, p53 has been shown to suppress the transcription of x-ct, NQO1, and GST1, three Nrf2 target genes (Faraonio et al. 2006). In this study, various cell lines were transfected with expression vectors containing Nrf2 or p53. Cotransfection with Nrf2 and p53 resulted in decreased expression of the three target genes compared with cells transfected with Nrf2 alone. A report from our laboratory examining the relationship between p21 (a direct downstream target of p53) and Nrf2, however, demonstrated that the basal and inducible activity of Nrf2 is subject to positive regulation by p21 (Chen et al. 2009). p21 associates with the DLG motif within Nrf2, thereby disrupting the ability of Keap1 to properly bind and promote the ubiquitination of Nrf2. As a result, Nrf2 is stabilized in response to p21 up-regulation. Conversely, in the absence of p21, the basal and inducible expression of Nrf2 target genes is reduced in vitro and in vivo (Fig. 3). These findings may suggest that under unfavorable conditions, strong induction of p53 inhibits Nrf2 by an undefined mechanism, which reduces the antioxidant defense and cell survival pathways in order to promote cell death. On the other hand, under mild stress conditions, when p53 is only weakly induced, p21 is increased, stalling the cell cycle at the G1/S-phase checkpoint and eliciting the cytoprotective response to promote cell survival through (1) the induction of DNA damage repair functions of p53 and (2) activation of the Nrf2-mediated cytoprotective response.
Nrf2 and p62
Several laboratories, including ours, have demonstrated that sequestosome 1 protein (p62/SQSTM) can modulate the activity of Nrf2 (Copple et al. 2010; Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010). p62 is a scaffold protein that binds to polyubiquitinated proteins and targets protein aggregates and damaged organelles for degradation via the autophagy pathway. p62 contains an STGE-binding motif that is similar to the Nrf2 ETGE motif between amino acids 349 and 354 (Jain et al. 2010). p62 directly interacts with the Kelch domain of Keap1 via its STGE motif, thereby disrupting the proper confirmation of the Keap1–Nrf2 complex (Copple et al. 2010; Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010). p62 is also capable of binding to LC3, which is associated with the autophagosome membrane, thereby providing a link between the Nrf2–Keap1 pathway and the autophagy pathway (Komatsu et al. 2010). Our laboratory found that ectopic overexpression of p62 or blockage of autophagosomal flux sequestered Keap1 in autophagosomes via direct interaction between Keap1 and p62. The sequestration of Keap1 into autophagosomes resulted in a decrease in Nrf2 ubiquitination, an increase in Nrf2 stability, and, ultimately, the enhanced expression of ARE-bearing genes (Fig. 3). This aberrant accumulation of Nrf2 by autophagy deregulation is p62-dependent and Keap1-Cys151-independent, which we named the noncanonical mechanism of Nrf2 activation. The pathological importance of prolonged Nrf2 activation through the noncanonical mechanism is exemplified in studies with knockout mice showing that accumulation of Nrf2 is the major cause of liver damage in autophagy-deficient mice (Taguchi et al. 2012). Consistent with the notion that Keap1 is degraded through the autophagy–lysosome pathway (Taguchi et al. 2012), siRNA-mediated knockdown of p62 increased the half-life of Keap1 twofold, increased the levels of Keap1, lowered the level of Nrf2, and reduced the expression of Nrf2 target genes (Copple et al. 2010; Lau et al. 2010, 2013). This noncanonical mechanism of Nrf2 activation caused by autophagy deregulation adds another dimension to the regulation of the Nrf2–Keap1 pathway.
Recently identified proteins that interact with Keap1
The list of proteins that interact with Nrf2 or Keap1 and therefore modulate Nrf2 levels is continuously expanding. Recent studies revealed that the Wilms tumor gene on the X chromosome (WTX) tumor suppressor protein physically binds to Keap1 to competitively inhibit Nrf2 ubiquitination (Camp et al. 2012). PALB2 has also been shown to associate with Keap1 and sterically inhibit Nrf2 ubiquitination (Ma et al. 2012). Very recently, Hast et al. (2013) identified the protein dipeptidyl peptidase 3 (DPP3) that binds to Keap1, inhibits Nrf2 ubiquitination, and drives Nrf2-dependent transcription in cancer cells (Fig. 3). All of these proteins contain an ETGE motif, suggesting that certain ETGE-containing proteins are capable of up-regulating Nrf2 by competing with Nrf2 for Keap1 binding and suppressing Keap1-mediated ubiquitination of Nrf2. Therefore, the functional roles of these proteins may derive at least partially from activation of the Nrf2 pathway.
Transcriptional up-regulation of Nrf2 by oncogene-dependent signaling
Until recently, it was believed that Nrf2 activity is primarily regulated at the protein level through ubiquitination and degradation, based on the observations that a vast majority of Nrf2 activators enhance Nrf2 at the protein level without affecting the mRNA levels of NRF2. A recent study, however, demonstrated that Nrf2 expression might be constitutively elevated by oncogenic activation in cancer cells via transcriptional up-regulation of NRF2 (DeNicola et al. 2011). Activation of oncogenic alleles of KRAS, BRAF, and C-MYC (KRASG12D, BRAFV619E, and C-MYCERT12) increased the mRNA level of NRF2 and its target genes 60% and resulted in a more reduced intracellular redox environment. Furthermore, results from studies employing Jun and myc siRNA indicate that KRASG12D and BRAFV619E may up-regulate the transcription of NRF2 via Jun and Myc. However, more concrete mechanistic studies are needed to dissect the molecular mechanisms underlying the enhanced expression of NRF2 in response to oncogenic activation of KRAS, BRAF, and C-MYC.
Metabolic activation of Nrf2
Fumarate, a Kreb cycle metabolite, was recently shown to form adducts with Keap1 and activate the Nrf2 signaling pathway in the hereditary form of type 2 papillary renal cell cancer (Ooi et al. 2011). In the Krebs cycle, fumarate is normally metabolized to malate by the enzyme fumarate hydratase (FH). Homozygous FH loss-of-function mutations have been shown to cause the hereditary form of type 2 papillary renal carcinoma in humans (for review, see Eng et al. 2003). The loss of the FH enzyme activity causes the accumulation of high levels of fumarate in the affected cells. Mechanistic analysis revealed that fumarate modifies cysteine residues within Keap1, abrogating its ability to ubiquitinate Nrf2 and causing prolonged activation of Nrf2, which is likely to promote the progression of hereditary type 2 papillary renal cell carcinoma (Adam et al. 2011; Ooi et al. 2011).
Pharmacological modulators of the Nrf2–Keap1 signaling pathway
The discovery of mechanisms underlying the regulation of the Nrf2/Keap1 pathway has led to the development of agents that can manipulate Nrf2 in order to obtain therapeutic benefit. Compounds that increase the activity of Nrf2 are being tested for disease prevention, whereas Nrf2 inhibitors are being developed as an adjuvant therapy to enhance the efficacy of chemotherapeutic drugs.
Nrf2 activators
Many Nrf2 activators are naturally occurring, plant-derived phytochemicals. Some examples of natural Nrf2 activators include sulforaphane (SF), curcumin, epigallocatechin-3-gallate, resveratrol, cafestol, kahweol, cinnamonyl-based compounds, zerumbone, garlic oganosulfur compounds, lycopene, and carnosol (for review, see Jeong et al. 2006; Lau et al. 2008; Kensler and Wakabayashi 2010). These chemopreventive compounds exert their effects by inducing the Nrf2-mediated defense response, including activation of phase II detoxification enzymes, antioxidants, and transporters that protect cells from subsequent carcinogenic insults.
One of the most extensively investigated natural products that target the Nrf2–Keap1 signaling pathway is SF, an isothiocyanate present in cruciferous vegetables such as broccoli (Kensler et al. 2000). In a mouse cancer model, topical application of SF protected mice from developing skin tumors after they were exposed to several different carcinogens (Xu et al. 2006). SF also inhibited tobacco-induced lung carcinogenesis (Conaway et al. 2005) and reduced the formation of aberrant crypt foci and pancreatic carcinogenesis following carcinogen exposure (Chung et al. 2000; Kuroiwa et al. 2006).
In humans, several cohort and case control studies have shown that there is an inverse correlation between broccoli consumption and the risk of developing colon, lung, breast, liver, and prostate cancer (Kensler et al. 1987, 2000, 2005; Spitz et al. 2000; Seow et al. 2002; Ambrosone et al. 2004; Joseph et al. 2004). Broccoli sprouts contain glucosinolates, which are the precursors of isothiocyanates such as SF. The most abundant glucosinolate found in broccoli is glucoraphanin, the precursor to SF. A phase II clinical trial study in China evaluating the chemopreventive properties of broccoli sprout preparations containing >60% glucoraphanin determined that SF reduced aflatoxin–DNA adducts in humans (Kensler et al. 2005). The success of these studies has led to several phase II clinical trials to study SF and broccoli sprouts as chemopreventive agents (http://www.clinicaltrials.gov).
Curcumin is another well-investigated chemopreventive natural product that is capable of activating Nrf2 (Balogun et al. 2003). Dietary administration of curcumin enhanced the expression of Nrf2 and blocked DNA adduct formation, oxidative stress, and inflammation in the livers and lungs of mice treated with benzo(a)pyrene (Garg et al. 2008; Thimmulappa et al. 2008).
Several synthetic Nrf2 activators have also been developed. Oltipraz (4-methyl-5-[2-pyrazinyl]-1,2-dithiole-3-thione) is a synthetic Nrf2 activator that has been shown to inhibit chemical-induced carcinogenesis in a variety of organs in rodents, including the colon, kidney, liver, stomach, and bladder (Kensler et al. 1987; Roebuck et al. 1991; Rao et al. 1993, 1996; Moon et al. 1994; Clapper et al. 1995; Nishikawa et al. 1998; Iida et al. 2004; Sharma et al. 2006). Oltipraz reduced the number of pulmonary adenomas, gastric cancer, and forestomach tumors in Nrf2 wild-type mice treated with benzo(a)pyrene but not in Nrf2-null mice, suggesting that Nrf2 is in fact important for its chemopreventive effects (Rao et al. 1993; Ramos-Gomez et al. 2003; Sharma et al. 2006). Oltipraz also inhibited N-nitrosobutyl (4-hydroxybutyl) amine (BBN)-induced urinary bladder cancer in an Nrf2-dependent manner (Iida et al. 2004). These promising preclinical studies led to phase I and II clinical trials in humans; however, the results are inconclusive. In a randomized, double-blind phase II clinical trial in China, 1 mo of weekly administration of 500 mg of oltipraz reduced the levels of a toxic hydroxylated metabolite of aflatoxin (Kensler et al. 1998a,b; Wang et al. 1999). However, another randomized, double-blind trial in smokers did not show significant differences in the amount of polyaromatic hydrocarbon DNA adduct levels in lung epithelial cells and blood among trial groups (Kelley et al. 2005). Side effects such as flatulence, gastrointestinal irritation, and paresthesia in fingertips stopped the evaluation of this agent.
Dimethyl fumarate (DMF) is another synthetic Nrf2 activator that has been shown to alkylate critical cysteine residues on Keap1, prevent Nrf2 ubiquitination, and promote Nrf2 stabilization and subsequent activation of Nrf2 target genes (Phillips and Fox 2013). In phase III clinical trials for multiple sclerosis (MS), BG-12, an oral preparation of DMF, led to a reduction in the annual relapse rate for MS (Gold et al. 2012). In addition, DMF protected heart tissues from ischemia–reperfusion injury (Ashrafian et al. 2013). As a result, BG-12 (Tecfidera) has been approved by the Food and Drug Administraion for the treatment of MS in March 2013 (http://www.fda.gov). Although the cancer-preventive properties of DMF in humans have not been tested, recent studies have also shown that DMF may have potential as an anti-cancer agent in several different types of cancer cells, including melanoma, head and neck carcinoma, glioblastoma, and colon cancer (for review, see Chen and Kirsner 2011).
Nrf2 inhibitors
Given the role of constitutive Nrf2 activation in promoting cancer progression and mediating resistance to cancer therapy, the pharmacological inhibition of Nrf2 signaling has recently emerged as a promising approach for cancer therapy, especially for cancers with elevated levels of Nrf2. Recently, our laboratory screened many plant extracts to identify Nrf2 inhibitors and found that brusatol, a component of Brucea javanica seeds, is able to inhibit the Nrf2 pathway (Ren et al. 2011). Brusatol decreased the protein levels of Nrf2 across a panel of mammalian cells at nanomolar concentrations and decreased the expression of Nrf2 target genes in cancer cells. Consequently, brusatol decreased the intracellular levels of glutathione by suppressing the expression of GCLM and GCLC, which encode for GCS, a glutathione synthesis enzyme. Additionally, brusatol decreased the intracellular concentration of cisplatin, presumably through enhanced expression of Mrp1, a drug efflux protein highly expressed in cancer cells. As a result, brusatol enhanced the cytotoxic effects of several chemotherapeutic agents both in vitro and in vivo. Currently, the molecular mechanism by which brusatol inhibits Nrf2 is under investigation.
Several other small molecules have been found to suppress the Nrf2 pathway, including ascorbic acid, luteolin, ochratoxin A, trigonelline, and all-trans retinoic acid (ATRA) (for review, see Magesh et al. 2012). ATRA and other RA receptor α (RARα) agonists have been shown to inhibit the basal and inducible activity of Nrf2 in vitro and in vivo (Wang et al. 2007). Mechanistically, in the presence of ATRA, Nrf2 forms a complex with RARα. The Nrf2:RARα complex is not able to bind to the ARE and thus decreases the ability of Nrf2 to activate ARE-driven genes. Intriguingly, although these compounds were reported to inhibit Nrf2, they also have been shown to act as Nrf2 activators in other studies. Thus, additional research is needed to establish the specificity and mechanism of action of these and other putative Nrf2 inhibitors before they can be used as therapeutics for cancer patients.
Future directions in the development of Nrf2 modulators
Given the dual role of Nrf2 in cancer, it is natural to wonder whether Nrf2 activation can lead to cancer. There have been a substantial number of studies assessing the ability of Nrf2 activators to promote cancer. There is no evidence to date suggesting that the agents used to activate the Nrf2 pathway have adverse impacts on tumor growth. Administration of oltipraz in rats, for example, did not affect hepatic tumor yield or burden, and similar results were seen with other Nrf2 activators (Maxuitenko et al. 1993). Therefore, transient activation of Nrf2 in cells with an intact Nrf2–Keap1 axis by pharmacological activators is safe for the purpose of chemoprevention.
One of the major concerns with the Nrf2 activators that are currently being used as pharmacological agents, however, is cytotoxicity due to off-target effects. Given the increased tendency for these chemopreventive agents to react with cysteines, multiple signaling pathways can conceivably be modulated. For example, chemopreventive agents such as CDDO-Im, SF, and dithiolethione have been shown to affect multiple pathways. Bardoxolone methyl, a CDDO derivative, was recently withdrawn from a phase III clinical trial in end stage renal disease patients with type II diabetes following an excess of unspecified serious adverse effects and mortality among the treated patients (Tayek and Kalantar-Zadeh 2013; Zhang 2013). The reactive site on bardoxolone can undergo Michael addition reactions readily with a range of nucleophiles. Thus, it is possible that the adverse events associated with bardoxolone are due to off-target events and unlikely to be due to the activation of the Nrf2–Keap1 signaling pathway.
Several studies have been published that will help improve the specificity of Nrf2-based therapies. The identification of the cocrystal structure of Keap1 in complex with the Neh2 domain of Nrf2, for example, will provide opportunities to design molecules that specifically and selectively interfere with the binding of Keap1 and Nrf2 (McMahon et al. 2006; Tong et al. 2006a). Small mimetics of the ETGE or DLG domains of Nrf2, for example, may serve this purpose. The demonstration that the expression of NRF2 and KEAP1 is also controlled by specific miRNAs provides additional targets to manipulate the Nrf2–Keap1 pathway. Furthermore, mounting evidence suggests that Nrf2 can cross-talk with other pathways important for cell survival (for review, see Wakabayashi et al. 2010). These converging points may offer great opportunities for pharmacological intervention to control the level of Nrf2 and its downstream effects.
Acknowledgments
We thank Dr. Aikseng Ooi for critical reading of the manuscript. We apologize to colleagues whose work was not cited due to space limitations or oversight. This work was supported by the following grants: NCI R01 CA154377 and NIEHS ES015010 to D.D.Z., and center grant P30ES006694.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.225680.113.
References
- Adam J, Hatipoglu E, O'Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. 2011. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosone CB, McCann SE, Freudenheim JL, Marshall JR, Zhang Y, Shields PG 2004. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J Nutr 134: 1134–1138 [DOI] [PubMed] [Google Scholar]
- Ashrafian H, Czibik G, Bellahcene M, Aksentijevic D, Smith AC, Mitchell SJ, Dodd MS, Kirwan J, Byrne JJ, Ludwig C, et al. 2013. Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway. Cell Metab 15: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, Alam J, Motterlini R 2003. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J 371: 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bryan HK, Olayanju A, Goldring CE, Park BK 2012. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717 [DOI] [PubMed] [Google Scholar]
- Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, Tang X, Zheng N, Major MB, Moon RT 2012. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J Biol Chem 287: 6539–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AF, Kirsner RS 2011. Mechanisms of drug action: The potential of dimethylfumarate for the treatment of neoplasms. J Invest Dermatol 131: 1181. [DOI] [PubMed] [Google Scholar]
- Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD 2009. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 34: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR 2002. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 26: 175–182 [DOI] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD 2013. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung FL, Conaway CC, Rao CV, Reddy BS 2000. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis 21: 2287–2291 [DOI] [PubMed] [Google Scholar]
- Clapper ML, Wood M, Leahy K, Lang D, Miknyoczki S, Ruggeri BA 1995. Chemopreventive activity of Oltipraz against N-nitrosobis(2-oxopropyl)amine (BOP)-induced ductal pancreatic carcinoma development and effects on survival of Syrian golden hamsters. Carcinogenesis 16: 2159–2165 [DOI] [PubMed] [Google Scholar]
- Conaway CC, Wang CX, Pittman B, Yang YM, Schwartz JE, Tian D, McIntee EJ, Hecht SS, Chung FL 2005. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Res 65: 8548–8557 [DOI] [PubMed] [Google Scholar]
- Copple IM 2012. The Keap1–Nrf2 cell defense pathway—a promising therapeutic target? Adv Pharmacol 63: 43–79 [DOI] [PubMed] [Google Scholar]
- Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK 2010. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1–Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci 99: 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eades G, Yang M, Yao Y, Zhang Y, Zhou Q 2011. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286: 40725–40733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C, Kiuru M, Fernandez MJ, Aaltonen LA 2003. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer 3: 193–202 [DOI] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A 2002. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci 99: 7610–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, Cimino F 2006. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem 281: 39776–39784 [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol 25: 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Gupta S, Maru GB 2008. Dietary curcumin modulates transcriptional regulators of phase I and phase II enzymes in benzo(a)pyrene-treated mice: Mechanism of its anti-initiating action. Carcinogenesis 29: 1022–1032 [DOI] [PubMed] [Google Scholar]
- Gold R, Kappos L, Arnold DL, Bar-Or A, Giovannoni G, Selmaj K, Tornatore C, Sweetser MT, Yang M, Sheikh SI, et al. 2012. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N Engl J Med 367: 1098–1107 [DOI] [PubMed] [Google Scholar]
- Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, et al. 2012. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN, Major MB 2013. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res 73: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M 2001. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett 174: 103–113 [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT 2010. Cancer chemoprevention mechanisms mediated through the keap1-nrf2 pathway. Antioxid Redox Signal 13: 1713–1748 [DOI] [PubMed] [Google Scholar]
- Hirayama A, Yoh K, Nagase S, Ueda A, Itoh K, Morito N, Hirayama K, Takahashi S, Yamamoto M, Koyama A 2003. EPR imaging of reducing activity in Nrf2 transcriptional factor-deficient mice. Free Radic Biol Med 34: 1236–1242 [DOI] [PubMed] [Google Scholar]
- Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M 2012. Nrf2–MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40: 10228–10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N, et al. 2009. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res 15: 3423–3432 [DOI] [PubMed] [Google Scholar]
- Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, Yamamoto M 2004. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res 64: 6424–6431 [DOI] [PubMed] [Google Scholar]
- Inoue D, Suzuki T, Mitsuishi Y, Miki Y, Suzuki S, Sugawara S, Watanabe M, Sakurada A, Endo C, Uruno A, et al. 2012. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci 103: 760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al. 2011. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc− and thereby promotes tumor growth. Cancer Cell 19: 387–400 [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322 [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285: 22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WS, Jun M, Kong AN 2006. Nrf2: A potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal 8: 99–106 [DOI] [PubMed] [Google Scholar]
- Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, Zhang DD 2010. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res 70: 5486–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph MA, Moysich KB, Freudenheim JL, Shields PG, Bowman ED, Zhang Y, Marshall JR, Ambrosone CB 2004. Cruciferous vegetables, genetic polymorphisms in glutathione S-transferases M1 and T1, and prostate cancer risk. Nutr Cancer 50: 206–213 [DOI] [PubMed] [Google Scholar]
- Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6: 857–868 [DOI] [PubMed] [Google Scholar]
- Kelley MJ, Glaser EM, Herndon JE 2nd, Becker F, Bhagat R, Zhang YJ, Santella RM, Carmella SG, Hecht SS, Gallot L, et al. 2005. Safety and efficacy of weekly oral oltipraz in chronic smokers. Cancer Epidemiol Biomarkers Prev 14: 892–899 [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N 2010. Nrf2: Friend or foe for chemoprevention? Carcinogenesis 31: 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Egner PA, Dolan PM, Groopman JD, Roebuck BD 1987. Mechanism of protection against aflatoxin tumorigenicity in rats fed 5-(2-pyrazinyl)-4-methyl-1,2-dithiol-3-thione (oltipraz) and related 1,2-dithiol-3-thiones and 1,2-dithiol-3-ones. Cancer Res 47: 4271–4277 [PubMed] [Google Scholar]
- Kensler TW, Groopman JD, Roebuck BD 1998a. Use of aflatoxin adducts as intermediate endpoints to assess the efficacy of chemopreventive interventions in animals and man. Mutat Res 402: 165–172 [DOI] [PubMed] [Google Scholar]
- Kensler TW, He X, Otieno M, Egner PA, Jacobson LP, Chen B, Wang JS, Zhu YR, Zhang BC, Wang JB, et al. 1998b. Oltipraz chemoprevention trial in Qidong, People's Republic of China: Modulation of serum aflatoxin albumin adduct biomarkers. Cancer Epidemiol Biomarkers Prev 7: 127–134 [PubMed] [Google Scholar]
- Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD 2000. Chemoprotection by organosulfur inducers of phase 2 enzymes: Dithiolethiones and dithiins. Drug Metabol Drug Interact 17: 3–22 [DOI] [PubMed] [Google Scholar]
- Kensler TW, Chen JG, Egner PA, Fahey JW, Jacobson LP, Stephenson KK, Ye L, Coady JL, Wang JB, Wu Y, et al. 2005. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin–DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People's Republic of China. Cancer Epidemiol Biomarkers Prev 14: 2605–2613 [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S 2007. Cell survival responses to environmental stresses via the Keap1–Nrf2–ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116 [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, et al. 2008. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res 1: 187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, Lee SH 2010. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 220: 446–451 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223 [DOI] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Fountzilas E, Pillay K, Zerbini LF, Libermann TA, Cannistra SA, Spentzos D 2008. Carboplatin-induced gene expression changes in vitro are prognostic of survival in epithelial ovarian cancer. BMC Med Genomics 1: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa Y, Nishikawa A, Kitamura Y, Kanki K, Ishii Y, Umemura T, Hirose M 2006. Protective effects of benzyl isothiocyanate and sulforaphane but not resveratrol against initiation of pancreatic carcinogenesis in hamsters. Cancer Lett 241: 275–280 [DOI] [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD 2008. Dual roles of Nrf2 in cancer. Pharmacol Res 58: 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD 2013. Arsenic inhibits autophagic flux, activating the Nrf2–Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QK, Singh A, Biswal S, Askin F, Gabrielson E 2011. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet 56: 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister A, Nedjadi T, Kitteringham NR, Campbell F, Costello E, Lloyd B, Copple IM, Williams S, Owen A, Neoptolemos JP, et al. 2011. Nrf2 is overexpressed in pancreatic cancer: Implications for cell proliferation and therapy. Mol Cancer 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M 2006. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J 25: 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, Mehta M, Cheung KL, Ganesan S, Kong AN, et al. 2012. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol 32: 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magesh S, Chen Y, Hu L 2012. Small molecule modulators of Keap1–Nrf2–ARE pathway as potential preventive and therapeutic agents. Med Res Rev 32: 687–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, et al. 2007. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21: 2237–2246 [DOI] [PubMed] [Google Scholar]
- Maxuitenko YY, MacMillan DL, Kensler TW, Roebuck BD 1993. Evaluation of the post-initiation effects of oltipraz on aflatoxin B1-induced preneoplastic foci in a rat model of hepatic tumorigenesis. Carcinogenesis 14: 2423–2425 [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279: 31556–31567 [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a ‘tethering' mechanism: A two-site interaction model for the Nrf2–Keap1 complex. J Biol Chem 281: 24756–24768 [DOI] [PubMed] [Google Scholar]
- McMahon M, Lamont DJ, Beattie KA, Hayes JD 2010. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci 107: 18838–18843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79 [DOI] [PubMed] [Google Scholar]
- Mitsuishi Y, Motohashi H, Yamamoto M 2013. The Keap1–Nrf2 system in cancers: Stress response and anabolic metabolism. Front Oncol 2: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc Natl Acad Sci 91: 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R, Rao K, Detrisac C, Kelloff G, Steele V, Doody L 1994. Chemoprevention of respiratory-tract neoplasia in the hamster by oltipraz, alone and in combination. Int J Oncol 4: 661–667 [DOI] [PubMed] [Google Scholar]
- Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S 2003. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene 22: 9275–9281 [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M 2004. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol Med 10: 549–557 [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M 2004. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1–Nrf2 regulatory pathway. Proc Natl Acad Sci 101: 6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscarella LA, Parrella P, D'Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M et al. 2011. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 6: 710–719 [DOI] [PubMed] [Google Scholar]
- Nguyen T, Huang HC, Pickett CB 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem 275: 15466–15473 [DOI] [PubMed] [Google Scholar]
- Nioi P, Nguyen T 2007. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 362: 816–821 [DOI] [PubMed] [Google Scholar]
- Nioi P, Nguyen T, Sherratt PJ, Pickett CB 2005. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol 25: 10895–10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa A, Tanakamura Z, Furukawa F, Lee IS, Kasahara K, Ikezaki S, Takahashi M 1998. Chemopreventive activity of oltipraz against induction of glandular stomach carcinogenesis in rats by N-methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis 19: 365–368 [DOI] [PubMed] [Google Scholar]
- Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M 2010. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci 107: 2842–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. 2008. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 68: 1303–1309 [DOI] [PubMed] [Google Scholar]
- Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BW, Tan MH, Zhang Z, Yang XJ, et al. 2011. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20: 511–523 [DOI] [PubMed] [Google Scholar]
- Ooi A, Dykema K, Ansari A, Petillo D, Snider J, Kahnoski R, Anema J, Craig D, Carpten J, Teh BT, et al. 2013. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res 73: 2044–2051 [DOI] [PubMed] [Google Scholar]
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW 2007. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer 121: 1883–1891 [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21: 689–700 [DOI] [PubMed] [Google Scholar]
- Phillips JT, Fox RJ 2013. BG-12 in multiple sclerosis. Semin Neurol 33: 56–65 [DOI] [PubMed] [Google Scholar]
- Plafker KS, Nguyen L, Barneche M, Mirza S, Crawford D, Plafker SM 2010. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J Biol Chem 285: 23064–23074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A 2011. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci 98: 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW 2003. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis 24: 461–467 [DOI] [PubMed] [Google Scholar]
- Rao CV, Rivenson A, Katiwalla M, Kelloff GJ, Reddy BS 1993. Chemopreventive effect of oltipraz during different stages of experimental colon carcinogenesis induced by azoxymethane in male F344 rats. Cancer Res 53: 2502–2506 [PubMed] [Google Scholar]
- Rao CV, Rivenson A, Zang E, Steele V, Kelloff G, Reddy BS 1996. Inhibition of 2-amino-1-methyl-6-phenylimidazo[4,5]pyridine-induced lymphoma formation by oltipraz. Cancer Res 56: 3395–3398 [PubMed] [Google Scholar]
- Reddy NM, Kleeberger SR, Cho HY, Yamamoto M, Kensler TW, Biswal S, Reddy SP 2007a. Deficiency in Nrf2–GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol 37: 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy NM, Kleeberger SR, Yamamoto M, Kensler TW, Scollick C, Biswal S, Reddy SP 2007b. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol Genomics 32: 74–81 [DOI] [PubMed] [Google Scholar]
- Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD 2011. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci 108: 1433–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck BD, Liu YL, Rogers AE, Groopman JD, Kensler TW 1991. Protection against aflatoxin B1-induced hepatocarcinogenesis in F344 rats by 5-(2-pyrazinyl)-4-methyl-1,2-dithiole-3-thione (oltipraz): Predictive role for short-term molecular dosimetry. Cancer Res 51: 5501–5506 [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266: 11632–11639 [PubMed] [Google Scholar]
- Sangokoya C, Telen MJ, Chi JT 2010. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 116: 4338–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Suzuki A, Shitara M, Hikosaka Y, Okuda K, Moriyama S, Yano M, Fujii Y 2012. Genotype analysis of the NRF2 gene mutation in lung cancer. Int J Mol Med 31: 1135–1138 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, et al. 2013. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 45: 860–867 [DOI] [PubMed] [Google Scholar]
- Satoh H, Moriguchi T, Takai J, Ebina M, Yamamoto M 2013. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res 73: 4158–4168 [DOI] [PubMed] [Google Scholar]
- Seow A, Yuan JM, Sun CL, Van Den Berg D, Lee HP, Yu MC 2002. Dietary isothiocyanates, glutathione S-transferase polymorphisms and colorectal cancer risk in the Singapore Chinese Health Study. Carcinogenesis 23: 2055–2061 [DOI] [PubMed] [Google Scholar]
- Sharma S, Gao P, Steele VE 2006. The chemopreventive efficacy of inhaled oltipraz particulates in the B[a]P-induced A/J mouse lung adenoma model. Carcinogenesis 27: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S 2008a. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135: 1358- 1368 [DOI] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S 2008b. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci 105: 13568–13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, Yoshimatsu Y, Tachimori Y, Kushima R, Kiyono T, et al. 2011. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 13: 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim GS, Manandhar S, Shin DH, Kim TH, Kwak MK 2009. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic Biol Med 47: 1619–1631 [DOI] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. 2006. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, et al. 2008. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res 68: 7975–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S 2010. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal 13: 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, et al. 2010. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 16: 3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz MR, Duphorne CM, Detry MA, Pillow PC, Amos CI, Lei L, de Andrade M, Gu X, Hong WK, Wu X 2000. Dietary intake of isothiocyanates: Evidence of a joint effect with glutathione S-transferase polymorphisms in lung cancer risk. Cancer Epidemiol Biomarkers Prev 9: 1017–1020 [PubMed] [Google Scholar]
- Suzuki T, Shibata T, Takaya K, Shiraishi K, Kohno T, Kunitoh H, Tsuta K, Furuta K, Goto K, Hosoda F, et al. 2013. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol Cell Biol 33: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M 2012. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci 109: 13561–13566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayek JA, Kalantar-Zadeh K 2013. The extinguished BEACON of bardoxolone: Not a Monday morning quarterback story. Am J Nephrol 37: 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Rangasamy T, Alam J, Biswal S 2008. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Med Chem 4: 473–481 [DOI] [PubMed] [Google Scholar]
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M 2006a. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol Cell Biol 26: 2887–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Kobayashi A, Katsuoka F, Yamamoto M 2006b. Two-site substrate recognition model for the Keap1–Nrf2 system: A hinge and latch mechanism. Biol Chem 387: 1311–1320 [DOI] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M 2007. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27: 7511–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve NF, Tian W, Wu T, Sun Z, Lau A, Chapman E, Fang D, Zhang DD 2013. USP15 negatively regulates Nrf2 through deubiquitination of Keap1. Mol Cell 51: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW 2010. When NRF2 talks, who's listening? Antioxid Redox Signal 13: 1649–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS, Shen X, He X, Zhu YR, Zhang BC, Wang JB, Qian GS, Kuang SY, Zarba A, Egner PA, et al. 1999. Protective alterations in phase 1 and 2 metabolism of aflatoxin B1 by oltipraz in residents of Qidong, People's Republic of China. J Natl Cancer Inst 91: 347–354 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Hayes JD, Henderson CJ, Wolf CR 2007. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor α. Proc Natl Acad Sci 104: 19589–19594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, An J, Ji F, Jiao H, Sun H, Zhou D 2008a. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 373: 151–154 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. 2008b. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29: 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, et al. 2013. RXRα inhibits the NRF2–ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res 73: 3097–3108 [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Fahl WE 1997. Functional antioxidant responsive elements. Proc Natl Acad Sci 94: 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TF, Yoshinaga K, Monma Y, Ito K, Niikura H, Nagase S, Yamamoto M, Yaegashi N 2008. Association of keap1 and nrf2 genetic mutations and polymorphisms with endometrioid endometrial adenocarcinoma survival. Int J Gynecol Cancer 21: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN 2006. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 66: 8293–8296 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Yoh K, Kobayashi A, Ishii Y, Kure S, Koyama A, Sakamoto T, Sekizawa K, Motohashi H, Yamamoto M 2004. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun 321: 72–79 [DOI] [PubMed] [Google Scholar]
- Yang M, Yao Y, Eades G, Zhang Y, Zhou Q 2011. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat 129: 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, Silkworth JB, Taguchi K, Yamamoto M, Williams CR, et al. 2009. Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 30: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD 2006. Mechanistic studies of the Nrf2–Keap1 signaling pathway. Drug Metab Rev 38: 769–789 [DOI] [PubMed] [Google Scholar]
- Zhang DD 2013. Bardoxolone brings Nrf2-based therapies to light. Antioxid Redox Signal 19: 517–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gordon GB 2004. A strategy for cancer prevention: Stimulation of the Nrf2–ARE signaling pathway. Mol Cancer Ther 3: 885–893 [PubMed] [Google Scholar]
- Zhang DD, Hannink M 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23: 8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S 2010. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Fahl WE 2001. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun 289: 212–219 [DOI] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem 277: 36544–36552 [DOI] [PubMed] [Google Scholar]