The ATR–CHK1 axis stabilizes stalled replication forks and prevents their collapse into DNA double-strand breaks (DSBs). Here, Ragland et al. show that fork collapse in Atr-deleted cells is mediated through the combined effects of the sumo targeted E3-ubiquitin ligase RNF4 and activation of the AURKA–PLK1 pathway. DSB generation is also shown to be dependent on the SLX4–endonuclease complex. The results indicate that replication fork collapse following ATR inhibition is an active and multistep process necessary for the cleavage of the replication fork into DSBs.
Keywords: ATR, PLK1, RNF4, replication fork collapse, SLX4, SUMO
Abstract
The ATR–CHK1 axis stabilizes stalled replication forks and prevents their collapse into DNA double-strand breaks (DSBs). Here, we show that fork collapse in Atr-deleted cells is mediated through the combined effects the sumo targeted E3-ubiquitin ligase RNF4 and activation of the AURKA–PLK1 pathway. As indicated previously, Atr-deleted cells exhibited a decreased ability to restart DNA replication following fork stalling in comparison with control cells. However, suppression of RNF4, AURKA, or PLK1 returned the reinitiation of replication in Atr-deleted cells to near wild-type levels. In RNF4-depleted cells, this rescue directly correlated with the persistence of sumoylation of chromatin-bound factors. Notably, RNF4 repression substantially suppressed the accumulation of DSBs in ATR-deficient cells, and this decrease in breaks was enhanced by concomitant inhibition of PLK1. DSBs resulting from ATR inhibition were also observed to be dependent on the endonuclease scaffold protein SLX4, suggesting that RNF4 and PLK1 either help activate the SLX4 complex or make DNA replication fork structures accessible for subsequent SLX4-dependent cleavage. Thus, replication fork collapse following ATR inhibition is a multistep process that disrupts replisome function and permits cleavage of the replication fork.
DNA replication is a significant source of double-strand breaks (DSBs) that promote cancer and age-associated pathologies. Accordingly, increased cancer risk and stem cell exhaustion directly correlate with conditions that increase replicative demand in various tissues. These occurrences have been more recently characterized as forms of replication stress, which at the molecular level is defined by an increased frequency of the uncoupling of DNA unwinding and synthesis. The ssDNA that results from this uncoupling activates the ATR–CHK1 pathway as well as other proreplicative responses (Cimprich and Cortez 2008; Branzei and Foiani 2010).
In addition to regulating cell cycle phase progression, the ATR–CHK1 (Mec1–Rad53 in Saccharomyces cerevisiae) pathway has been shown to prevent replication fork collapse, which is generally defined as the inability to reinitiate DNA replication after fork stalling (Lopes et al. 2001; Tercero and Diffley 2001; Cimprich and Cortez 2008; Branzei and Foiani 2010). The generation of replication fork-associated recombination structures (e.g., reversed replication forks) and DSBs is increased in ATR/Mec1 pathway-deficient metazoan and yeast cells (Brown and Baltimore 2000; Lopes et al. 2001; Myung et al. 2001; Tercero and Diffley 2001; Cimprich and Cortez 2008; Branzei and Foiani 2010). However, substantial defects in replication restart appear to take place earlier than the generation of recombinatorial structures in S. cerevisiae mec1 mutants (Lopes et al. 2001; Tercero and Diffley 2001; Cobb et al. 2003; Tercero et al. 2003; Raveendranathan et al. 2006). Along these lines, it has been shown in yeast that loss of replication reinitiation coincides with the loss of replisome components from chromatin in mec1 cells (Cobb et al. 2003; Raveendranathan et al. 2006). These findings suggest an association between replisome disassembly and the recombinatorial processes that lead to DSB generation. Indeed, remodeling of the replication apparatus has been proposed to be necessary for replication fork repair and damage bypass in Escherichia coli (Goldfless et al. 2006).
In support of the existence of such a mechanism, several replisome components in yeast and metazoans are known to be targeted for protein degradation after lengthy fork stalling. Mrc1 (S. cerevisiae) and CLSPN (metazoans) are orthologous replication factors associated with the MCM2–7 helicase complex that have additional functions in the transmission of Mec1–Rad53 and ATR–CHK1 signaling in response to replication stress (Cimprich and Cortez 2008). Degradation of these proteins is stimulated by their phosphorylation by the Cdc5 (S. cerevisiae) and PLK1 (metazoans) kinases, an event that promotes K48-linked polyubiquitination of CLSPN via recruitment of the β-TRCP ubiquitin ligase in metazoan cells (Yoo et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006). Notably, because CLSPN is required as a docking site to recruit CHK1 for phosphorylation by ATR, the degradation of CLSPN causes a downward spiral driven by the combined effects of replication factor removal and the loss of fork stabilization by the ATR–CHK1 axis (Yoo et al. 2004; Cimprich and Cortez 2008). The same PLK1–β-TRCP pathway regulates the degradation of FANCM, another replisome-associated factor that functions as part of the Fanconi anemia group of repair proteins (Kee et al. 2009).
PLK1 and orthologs in yeast (S. cerevisiae Cdc5) have also been shown to regulate the activation of MUS81–EME1 and stimulate its association with the SLX4–endonuclease complex (Matos et al. 2011; Gallo-Fernandez et al. 2012; Munoz-Galvan et al. 2012; Schwartz et al. 2012; Szakal and Branzei 2013). Although it is not clearly essential for fork collapse in all cell types, the Mus81–Eme1 complex in Schizosaccharomyces pombe plays a crucial role in DSB generation at stalled replication forks (Froget et al. 2008), and a similar relationship has been observed in mammalian cells when CHK1 is inhibited (Forment et al. 2011). These findings in aggregate suggest that PLK1 could play an active facilitative role in replication fork collapse at the level of both the replisome and cleaving aberrant fork structures.
The S. cerevisiae ubiquitin ligase complex Slx5–Slx8 has also been implicated in replication restart after prolonged stalling (Mullen et al. 2001; Prudden et al. 2007; Nagai et al. 2008). These studies indicated that yeast Slx5–Slx8 and RNF4 in mammalian cells play facilitative roles in the reformation of the replication fork structure by homologous recombination (HR) (Nagai et al. 2008; Galanty et al. 2012; Yin et al. 2012). However, in aggregate, these and other studies also suggest that this complex may affect replication restart through regulation of replication factor stability (Branzei et al. 2006; Bruderer et al. 2011; Plechanovova et al. 2011; Cremona et al. 2012). Slx5–Slx8 and the orthologous mammalian RNF4 dimer are targeted to sites of polysumoylation through four contiguous SUMO-interacting motifs (Bruderer et al. 2011; Plechanovova et al. 2011). In S. cerevisiae, polysumoylation of replisome components is stimulated after fork stalling and occurs initially in a Mec1-independent manner (Branzei et al. 2006; Cremona et al. 2012), although loss of Mec1 activity increases sumoylation in the absence of fork stalling (Cremona et al. 2012). Thus, the known targeting of RNF4 to polySUMO2/3-conjugated proteins suggests that RNF4 is localized to replisome components after fork stalling and participates in replication fork collapse when ATR/Mec1 is absent.
Although these advances imply that PLK1 and RNF4 may regulate replication fork collapse in ATR-deficient cells, this hypothesis has not been tested. Here, we show that suppression of either the AURKA–PLK1 pathway or RNF4 permits replication restart following collapse in Atr-deleted cells. Replication recovery in Atr-deleted cells is not associated with dormant origin firing but instead takes place at formerly stalled replication forks. Notably, combined suppression of RNF4 or the AURKA–PLK1 pathway almost completely prevents replication fork collapse into DSBs, with suppression of RNF4 alone having a more pronounced rescuing effect than inhibition of PLK1. These data indicate that replication fork collapse in mammalian cells is an active and cell-dominant signaling process that is necessary for the cleavage of replication forks into DSBs.
Results
Failure to restart replication in ATR-deficient cells coincides with the loss of replisome components from chromatin and polysumoylation
Previous studies suggest that prolonged replication stress leads to a decrease in the association of replication factors with chromatin (Cobb et al. 2003; Raveendranathan et al. 2006) and the elevated degradation of specific replisome components (Yoo et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006; Kee et al. 2009). Indeed, a more recent study has identified additional novel pathways that lead to replication factor degradation in response to replisome instability (Roseaulin et al. 2013). These effects on replisome integrity suggest that replication fork collapse in cells deficient in the Mec1–Rad53 (S. cerevisiae), Rad3–Cds1 (S. pombe), or ATR–CHK1 (metazoans) pathways may be caused initially by the targeted disengagement of replisome components. Although attractive, this model is still under investigation in yeast (De Piccoli et al. 2012).
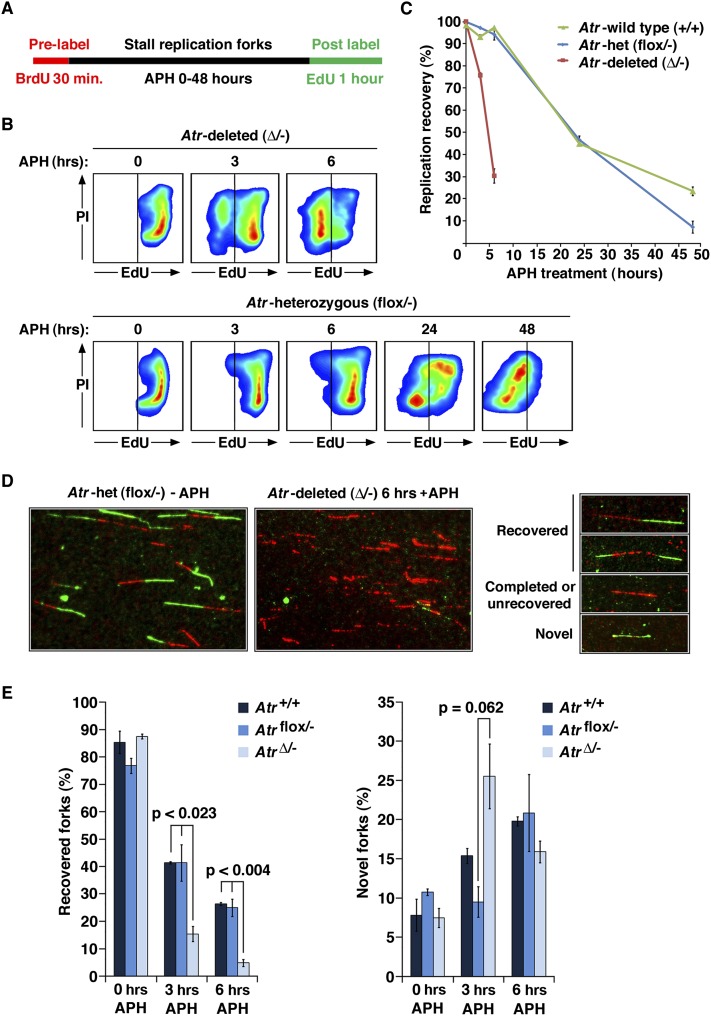
To examine replication fork collapse in mammalian cells, immortalized murine embryonic fibroblasts (MEFs) containing both null and Cre-lox conditional alleles (Atrflox/−) were treated to delete the Atrflox allele (Brown and Baltimore 2003; Ruzankina et al. 2007). After ATR protein depletion, S-phase cells were labeled with BrdU and then pulse-treated with the replicative polymerase inhibitor aphidicolin (APH) for varying lengths of time. These BrdU-tagged S-phase cells were then assayed for the ability to restart replication by washing out APH and allowing cells to recover replication in the presence of EdU (Fig. 1A; Supplemental Fig. 1A). Notably, on a population basis, Atr-deleted cells were bimodally distributed in their ability to restart replication after 3 or 6 h of APH treatment and demonstrated a time-dependent decrease in the frequency of cells capable of recovering DNA replication (Fig. 1A,B). This bimodal distribution was also observed in Atrflox/− tail fibroblasts and following small molecule inhibition of ATR (Supplemental Fig. 1B). This bimodal outcome per cell is inconsistent with replication fork collapse being stochastically degenerative at the level of the replication fork; rather, it indicates that fork collapse is a dominant process that occurs on a per-cell basis.
Figure 1.
Replication fork collapse in wild-type and ATR-deficient cells. (A) Dual-labeling method to measure replication restart at the cellular level by flow cytometry. Pulse BrdU labeling before APH-mediated stalling served as a tag for S-phase cells; these cells are monitored subsequently for replication recovery by EdU pulse labeling. (B) Representative examples of replication recovery after 5 μM APH treatment of Atrflox/− and AtrΔ/− cells. EdU labeling (X-axis) of BrdU-positive cells is shown. An example of BrdU gating for EdU detection is shown in Supplemental Figure 1A. (C) Quantification of the frequency of replication restart in AtrΔ/− and control cells. AtrΔ/− cells lose the ability to recover replication approximately six times faster than either Atrflox/− or Atr+/+ cells treated with APH. (D) Examples of DNA-combing results and the categories used for analysis. (E) Quantification of the frequency of replication reinitiation after release from 6 h of APH treatment as determined by DNA combing. Replication restart was quantified as the percentage of BrdU and EdU double-labeled strands (recovered) divided by the total frequency of the three categories shown in D (recovered, completed/unrecovered, and novel). Novel fork representation was calculated similarly (novel forks divided by recovered + completed/unrecovered + novel). Note that the increase in novel forks observed at 3 h in AtrΔ/− is consistent with ATR's known role in preventing late origin firing. The modest general increase in novel origins in ATR-expressing cells over time is consistent with the passage of cells from G1 into S during APH treatment.
The mixed populations of replication-competent and replication-incompetent Atr-deleted cells observed after 6 h of APH treatment (Fig. 1A,B) were then examined for the frequency of replication reinitiation on a per-fork basis, as assayed by DNA combing (Fig. 1D,E). Using this method, Atr-deleted cells again demonstrated an inability to restart replication, consistent with prior studies. The loss of replication restart in Atr-deleted cells on both a population level (Fig. 1B, C) and a per-fork basis (Fig. 1D,E) was particularly penetrant after 6 h of APH treatment. At this time point, the firing of novel origins observed after 3 h of APH treatment in Atr-deleted cells appeared to be exhausted (Fig. 1E), suggesting that the forks that emerge from these origins ultimately succumb to collapse by 6 h of fork stalling. These data indicate that the inability to recover replication in Atr-deleted cells is attributable to the loss of replication activity at previously active replication forks. The inability to restart replication persisted for up to 48 h post-APH treatment in Atr-deleted cells and correlated with the accumulation and maintenance of phospho-S139 H2AX (Supplemental Fig. 1C), a marker of DSBs (Rogakou et al. 1998).
Although wild-type and ATR heterozygous controls were able to restart replication after short periods of stalling at the cellular level (3–6 h), replication restart was compromised in these cells when detected by DNA combing (Fig. 1D,E). Indeed, even at the cellular level, APH treatment for extensive periods (24–48 h) was capable of preventing ATR-expressing cells from reinitiating DNA replication (Fig. 1C,B), consistent with previous findings (Borel et al. 2002). These results indicate that prolonged fork stalling compromises replication fork stability even in ATR-competent cells, albeit with significantly delayed kinetics in comparison with Atr-deleted cells. Thus, ATR deficiency appears to accelerate a response to replication stress that ultimately occurs in wild-type cells and culminates in replication fork collapse.
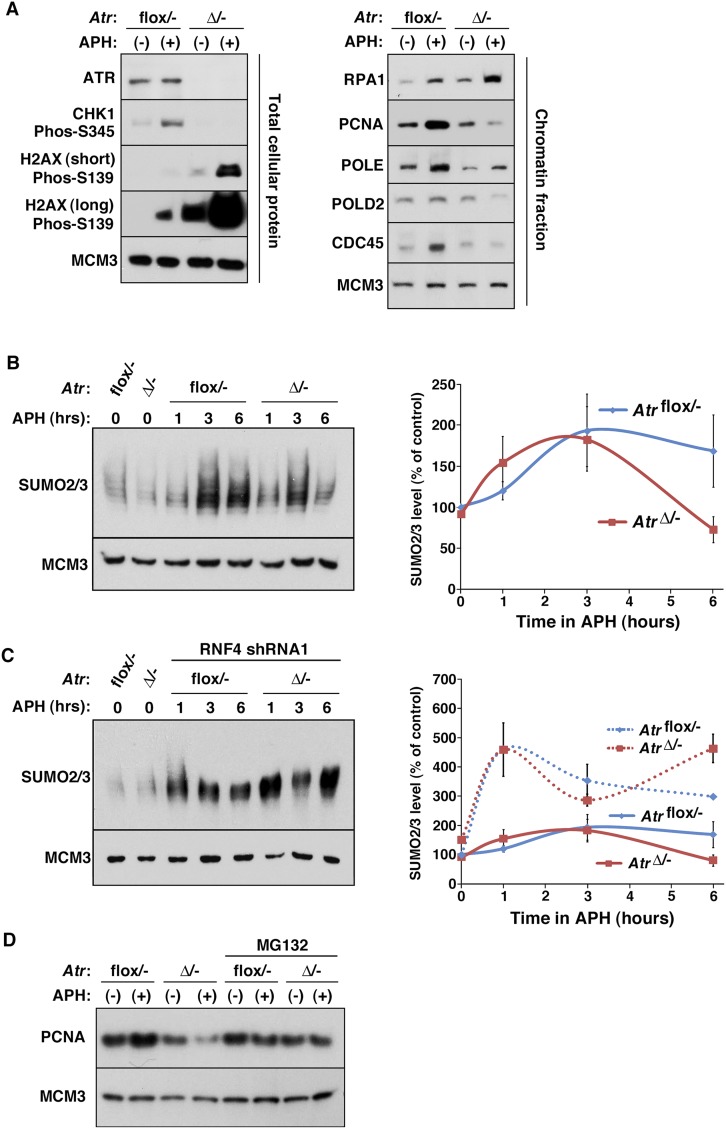
To determine whether replication fork collapse in Atr-deleted cells correlates with the disassociation of replisome components from chromatin, nuclear-insoluble fractions were prepared (Mendez and Stillman 2000) and examined for the abundance of a variety of replication factors. As expected, the levels of many replisome components on chromatin increased in ATR-expressing control cells during the 6-h APH treatment period due to the continued accumulation of cells in S phase from G1 (Fig. 2A). However, with the exception of most MCM components, which are associated with chromatin in excess (Ibarra et al. 2008), a reduced abundance of several replication factors was observed in ATR-depleted cells following APH treatment (Fig. 2A). The levels of CDC45, PCNA, POLD2 (50-kDa POLδ subunit), and POLE on chromatin in Atr-deleted cells were consistently below those observed in similarly treated control cells (Fig. 2A). In accord with increased rates of fork collapse into DSBs and 5′ end resection in ATR/Mec1-deficient cells (Cotta-Ramusino et al. 2005; Cimprich and Cortez 2008; Segurado and Diffley 2008), H2AX phosphorylation and RPA1 (70-kDa RPA subunit) abundance on chromatin increased in Atr-deleted cells compared with ATR-expressing controls following APH treatment, indicating a concomitant increase in both DSBs and ssDNA (Fig. 2A). These data demonstrate that the association of several replication factors with chromatin is compromised upon replication fork stalling in the absence of ATR.
Figure 2.
Loss of replisome proteins from chromatin in ATR-deficient cells. (A) Western blot detection of total cellular and chromatin-bound proteins as indicated. (B) Western blot detection of SUMO2/3 in chromatin lysates and quantification by optical density. APH was added to the cells as indicated, and chromatin-containing fractions were Western-blotted and detected for SUMO2/3. Note that AtrΔ/− cells consistently exhibited reduced SUMO2/3-conjugated proteins in chromatin lysates after 6 h of APH treatment. (C) Western blot detection of SUMO2/3 in chromatin lysates from RNF4-suppressed cells. This experiment was performed similarly to that described in B, with the exception that RNF4 levels were suppressed by shRNA targeting. Note that while the amplitude of SUMO2/3 presence was variable using crude nuclear-insoluble preparations, the changes shown represent the average effects of the experiments (n = 14). RNF4 suppression prevents the loss of SUMO2/3 conjugates on chromatin in AtrΔ/− cells. (D) Western blot detection of PCNA in chromatin lysates from the indicated cells treated with MG132 or left untreated. Loss of PCNA in AtrΔ/− cells was consistently mitigated by the addition of MG132.
Studies in yeast have indicated that replisome components are sumoylated in a Mec1-independent manner (Cremona et al. 2012). Consistent with these findings, a twofold to threefold increase in SUMO2/3 conjugates was observed after 3 h of APH treatment in both ATR-expressing controls and Atr-deleted cells (Fig. 2B). However, after 6 h of treatment, chromatin-bound sumoylated proteins declined significantly, particularly in ATR-depleted cells. Notably, the decline in SUMO conjugates on chromatin after 6 h of replication arrest was suppressed by the targeted reduction of RNF4 expression (Fig. 2C) or treatment with MG132 (data not shown). Finally, addition of the proteasome inhibitor MG132 during APH treatment preserved unmodified PCNA on chromatin in Atr-deleted cells (Fig. 2D), suggesting that protein degradation facilitates fork collapse. Although immunoprecipitation of hypermodified forms of replisome subunits was not possible due to the insoluble nature of these complexes (data not shown), these data indicate that replication stalling leads to the polysumoylation of chromatin-bound proteins and that suppression of RNF4 prevents their removal (Fig. 2A–D). These data demonstrate that irrecoverable replication fork collapse in ATR-deficient cells correlates with the APH-induced polysumoylation and loss of replisome components from chromatin (Figs. 1, 2).
Inhibition of RNF4 or the AURKA–Plk1 pathway promotes replication restart after fork collapse in Atr-deleted cells
As described above, replisome disengagement may play a key role in replication fork collapse. The two main pathways implied to regulate this process are RNF4 and PLK1. However, the effect of inhibiting these pathways on replication restart has not been tested. To do so, shRNA reduction of RNF4 expression and small molecule inhibition of the CDK1–AURKA–PLK1 circuit (Lens et al. 2010) were examined for their ability to promote replication restart in ATR-deficient cells, as assayed on both a population and per-fork basis.
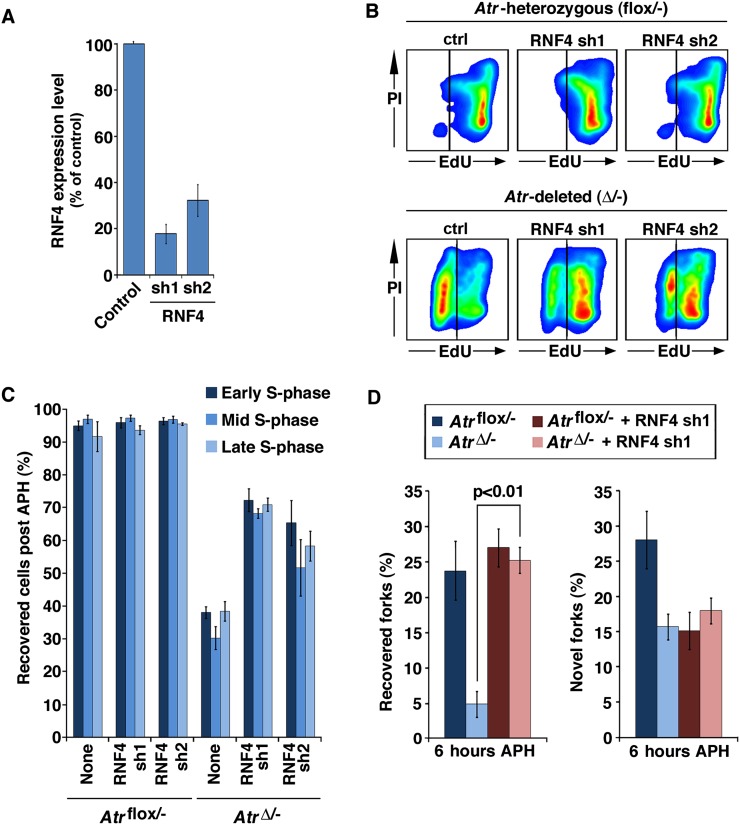
Although suppression of RNF4 expression did not affect replication restart in ATR-expressing control cells after APH treatment, this suppression did permit a majority of Atr-deleted cells to restart DNA replication following 6 h of APH treatment (Fig. 3A–C). The level of restart afforded by RNF4 reduction in ATR-deleted cells correlated with the degree of RNF4 suppression, with greater reduction in RNF4 levels leading to a higher frequency of replication restart in Atr-deleted cells (Fig. 3A–C). Once again, the rescue of Atr-deleted cells from irrecoverable replication failure was bimodal, leading to either near full recovery or none at all within the same time point (Fig. 3A–C).
Figure 3.
RNF4 suppression prevents replication fork collapse in ATR-deficient cells. (A) Quantitative real-time PCR detection of RNF4 mRNA expression 48 h after lentivirus-mediated transduction. The sequences and regions of RNF4 targeted by these shRNAs are distinct (Supplemental Fig. 5). (B) Representative replication restart following RNF4 suppression, as detected by flow cytometry. (C) Quantification of replication restart results in B. (D) Quantification of replication reinitiation on a per-fork basis in RNF4-suppressed cells, as determined by DNA combing. Note that RNF4 suppression permitted significant recovery of replication in previously unrecoverable AtrΔ/− cells (B,C); this reinitiation occurs at previously active replication forks but not at novel origins (D).
Similar effects of RNF4 suppression were observed at individual replication forks by DNA combing (Fig. 3D). The fivefold reduction in replication fork restart in Atr-deleted cells (Figs. 1D,E, 3D) was fully suppressed by reduction of RNF4 levels (Fig. 3D). Notably, the effect of RNF4 suppression on restart occurred almost exclusively at previously active replication forks, indicating that replication recovery on a per-cell basis was due to bona fide reinitiation of replication at previously active replication forks, not de novo activation of dormant origins. These data demonstrate that replication fork collapse attributable to ATR deletion can be overcome by suppressing RNF4 and, by extension, that RNF4 plays an active role in fork collapse in ATR-deficient cells.
CDK1–CYCB, AURKA, and PLK1 form a positive feedback circuit that leads ultimately to maximal activation of each kinase (Lens et al. 2010). This circuit activates PLK1 through AURKA phosphorylation of T210 on PLK1 and the localization of PLK1 through its Polo box domain to CDK-phosphorylated S/TP sites (Elia et al. 2003; Macurek et al. 2008). Therefore, inhibition of CDK1, AURKA, or PLK1 should each limit the activation of PLK1. Furthermore, if PLK1 does indeed participate in replication fork collapse, then inhibition of any of these kinases should foster replication recovery in ATR-deficient cells.
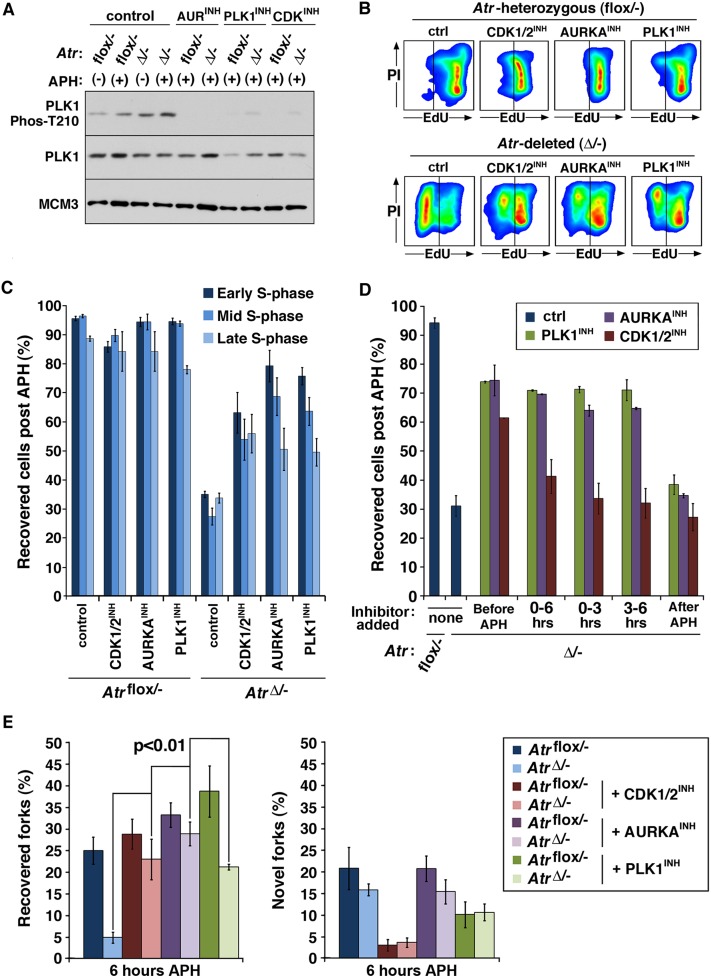
Small molecule inhibitors of CDK1, AURKA, and PLK1 were first examined for their effects on PLK1 regulation. As expected, inhibition of CDK1/2, AURKA, or PLK1 resulted in a significant decrease in phospho-T210 PLK1 on chromatin. In addition, we found that ATR deletion alone was sufficient to increase levels of chromatin-bound phospho-T210 PLK1 (Fig. 4A; data not shown), consistent with ATR absence leading to inappropriate activation of the CDK1–AURKA–PLK1 circuit (van Vugt et al. 2001; Lens et al. 2010).
Figure 4.
The AURKA–PLK1 pathway suppresses replication restart in ATR-deficient cells. (A) Western blot detection of the indicated proteins in chromatin fraction lysates from cells conditionally treated with APH and inhibitors of CDK1/2 (Purvalanol A), AURKA, or PLK1 (BI-2536) for 6 h. Note that the effect of PLK1 inhibition on AURKA-mediated phosphorylation of T210 is likely the result of suppressing the PLK1-driven feedback activation of AURKA (Lens et al. 2010). (B) Representative replication recovery from 6 h of APH treatment of the indicated cells concurrently treated with AURKA, CDK1/2, or PLK1 inhibitors. (C) Quantification of flow-cytometric data shown in B. (D) Quantification of replication restart by flow cytometry following CDK1/2, AURKA, or PLK1 inhibition at varying times after APH treatment. Inhibitors were added either (1) 1 h before APH, (2) at the same time as APH and continued for the entire 6-h treatment (0–6), (3) at the time of APH and removed 3 h into APH treatment (0–3), (4) 3 h after APH and continued onward, or (5) only for the hour of recovery after APH was removed (after APH). Note that whereas AURKA or PLK1 inhibition permitted replication restart in AtrΔ/− cells even when added 3 h after the addition of APH, CDK1/2 inhibitor treatment only permitted restart when added before APH addition. (E) Quantification of replication restart by DNA combing. Inhibitors of CDK1/2, AURKA, or PLK1 added just prior to APH-induced stalling for 6 h permitted replication restart at previously active replication forks but not at novel origins.
We then tested the effects of individually inhibiting CDK1/2, AURKA, and PLK1 on replication restart in Atr-deleted cells. In total, four distinct CDK1/2 inhibitors, one AURKA inhibitor, and two PLK1 inhibitors were tested (Fig. 4B–E; Supplemental Fig. 2). In each case, application of CDK1/2, AURKA, or PLK1 inhibitors led a significant increase in replication restart in Atr-deleted cells (Fig. 4B,C). The ability of CDK1/2, AURKA, or PLK1 inhibition to promote replication reinitiation in Atr-deleted cells was consistently more robust in early to mid-S-phase populations (Fig. 4B), where active replication forks are more numerous than in late S phase. This apparent selectivity may result from the normal activation of AURKA–PLK1 in late S phase, after which time inhibition of PLK1 may be insufficient to prevent fork collapse in such cells. Notably, CDK1/2 inhibitors only promoted replication restart in ATR-depleted cells when added at or before APH treatment (Fig. 4D). In contrast, AURKA or PLK1 inhibitors rescued replication reinitiation in ATR-deleted cells even when added 3 h after APH addition. These findings indicate that inhibition of AURKA and PLK1 have more immediate effects on replication fork stability than CDK1 inhibition.
As determined by DNA combing, inhibition of the CDK1–AURKA–PLK1 pathway in Atr-deleted cells permitted a high frequency of replication reinitiation at previously active replication forks (Fig. 4E). The percentage of stalled forks that restarted replication in Atr-deleted cells that were treated with CDK, AURKA, or PLK1 inhibitors was similar to that observed in ATR-expressing controls either treated or left untreated with these inhibitors (Fig. 4E). Again, reinitiation of replication was not attributable to the de novo firing of dormant origins. Consistent with this finding, CDK1/2 inhibition prevented the firing of novel origins, as expected, and yet was still capable of promoting the recovery of replication at previously active forks in Atr-deleted cells. Therefore, similar to RNF4, these findings indicate that the CDK1–AURKA–PLK1 pathway participates in preventing replication reinitiation after fork stalling in ATR-deficient cells.
RNF4 and PLK1 are required for SLX4-dependent DSB formation in ATR-deficient cells upon fork stalling
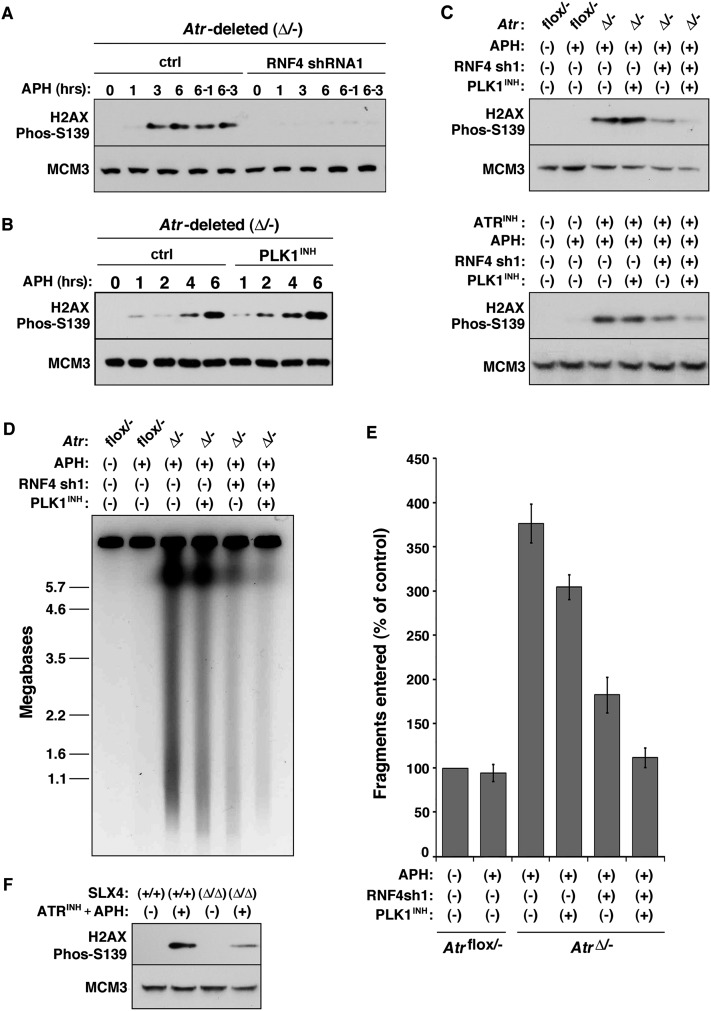
Previously, we demonstrated that inhibition of DNA replication in Atr-deleted cells leads to DSB formation and the phosphorylation of H2AX in S phase (Brown and Baltimore 2003; Chanoux et al. 2009). To examine the roles of RNF4 and PLK1 in DSB generation following replication fork stalling, RNF4 and PLK1 were inhibited singly or together in Atr-deleted and control cells treated with APH for 1–6 h. As described (Chanoux et al. 2009), H2AX phosphorylation on S139 is mediated by ATM and DNA-PK in response to fork collapse into DSBs. Indeed, H2AX phosphorylation was visible within 1 h of APH treatment in Atr-deleted cells and continued to increase subsequently (Fig. 5A,B). In contrast, APH treatment of ATR-expressing controls for up to 6 h led to comparably little phospho-H2AX (Fig. 2A; Chanoux et al. 2009). Strikingly, the increase in phospho-H2AX in Atr-deleted cells was largely suppressed by the targeted repression of RNF4. Although this phosphorylation was not affected by PLK1 inhibition alone, the phosphorylation of H2AX that remained in RNF4-depleted cells was further reduced by addition of PLK1 inhibitor (Fig. 5A–C). These effects were observed both following ATR deletion and upon ATR kinase inhibition using a highly specific small molecule inhibitor (Fig. 5C; Supplemental Fig. 3).
Figure 5.
Generation of DSBs in ATR-deficient cells requires RNF4. (A,B) Western blot detection of phospho-S139 H2AX in AtrΔ/− cells following 5 μM APH treatment with and without RNF4 suppression (A) or PLK1 inhibition (B). Cells were treated with 5 μM APH for the times indicated. Some cells were treated with APH for 6 h followed by a 1-h (6/1) or 3-h (6/3) period after APH removal. Note that PLK1 inhibition does not significantly change the rate or degree of H2AX phosphorylation in AtrΔ/− cells. Phosphorylation of H2AX on S139 is mediated by ATM and DNA-PK in response to fork collapse into DSBs (Chanoux et al. 2009). (C) Western blot detection of phospho-S139 H2AX in ATR-inhibited cells following suppression of RNF4 and PLK1. The separate and combined effects of RNF4 suppression and PLK1 inhibition on H2AX phosphorylation following 6 h of APH treatment were tested in AtrΔ/− cells and Atrflox/− cells treated with ATR inhibitor (ATR-45) (Charrier et al. 2011). ATR inhibitor was added 1 h prior to the 6-h APH treatment. Note that combined suppression of RNF4 and PLK1 together decreases the phosphorylation of H2AX more than either treatment alone. (D) PFGE analysis of the effects of separate and combined suppression of RNF4 and PLK1. The experiment was performed as described in C. (E) Quantification of PFGE results shown in D. The total amount of fragmented DNA divided by the total DNA (DNA in the well plus fragmented DNA) was calculated by fluorescence density and area. These fractions were normalized to untreated Atrflox/− controls. Values represent five to eight independent experiments. Note that a further reduction of DSBs was observed upon dual suppression of RNF4 and PLK1. (F) Detection of H2AX phosphorylation following ATR inhibition in the presence or absence of SLX4. Slx4 was deleted in immortalized Slx4flox/flox MEFs by tamoxifen-mediated activation of Cre-ERT2 for 48 h followed by culturing cells for an additional 2 d to deplete the SLX4 protein. Cells were then left untreated or treated with ATR kinase inhibitor (2 μM ATR-45) and APH (5 μM) for 6 h to promote replication fork collapse.
Although H2AX phosphorylation is frequently used as a surrogate for DSB formation, it is conceivable that some chromatin modifications could alter its levels in a manner unrelated to breakage of the parental DNA strands at replication forks. With this in mind, pulsed field gel electrophoresis (PFGE) was implemented to further quantify the abundance of DSBs in cells exposed to the conditions described above. Once again, these results indicated that a majority fraction of breaks following ATR deletion required expression of RNF4 (Fig. 5D,E) and that PLK1 inhibition further reduces DSB formation in Atr-deleted cells. Interestingly, a small but significant decrease in mobilized DNA fragments was observed with PLK1 inhibition alone using this method (Fig. 5D,E), again indicating a modest role for PLK1 in DSB generation. In aggregate, these results demonstrate that the vast majority of DSBs produced following replication fork stalling in ATR-deficient cells occurs in an RNF4-dependent manner and that PLK1 appears to play a secondary or compensatory role in this process.
The SLX4–endonuclease complex has previously been implicated as a mediator of replication fork collapse, particularly in cooperation with the MUS81–EME1 complex (Froget et al. 2008; Forment et al. 2011; Matos et al. 2011; Gallo-Fernandez et al. 2012; Munoz-Galvan et al. 2012; Schwartz et al. 2012; Szakal and Branzei 2013). To examine the involvement of this complex in the DSBs generated upon fork stalling in ATR-deficient cells, Cre-lox conditional SLX4 cells were used to delete Slx4 and determine the effect of its absence on DSB generation following ATR inhibition and fork stalling. As expected, ATR inhibition combined with inhibition of DNA replication led to a robust increase in H2AX phosphorylation. However, this increase was substantially diminished by Slx4 deletion (Fig. 5F). A similar effect of SLX4 deficiency was observed on the generation of DNA fragments in ATR-inhibited cells, as resolved by PFGE (data not shown). These data demonstrate that SLX4 is also required for DSB generation upon ATR inhibition. Collectively, these findings suggest that SLX4 and/or endonucleases dimers known to associate with it are directly responsible for the DSBs generated in ATR-deficient cells upon fork stalling. The effect of Slx4 deletion on DSBs in ATR-inhibited cells phenocopies the decrease in DSBs upon suppressing RNF4 and PLK1, suggesting some form of linkage between these processes.
RNF4 suppression does not foster long-range replication processivity
Our findings are consistent with a model in which RNF4 and the AURKA–PLK1 pathway work in concert to prevent replication restart and promote fork collapse into DSBs (Figs. 3–5). In the context of RNF4 suppression, the substantial reduction in DSBs and the near wild-type frequency of replication restart in Atr-deleted cells suggested that RNF4 reduction might rescue the essential function of ATR in cell viability. In contrast, a time-dependent decrease in the ability of RNF4 suppression to foster DNA synthesis in AtrΔ/− cells was observed after APH removal (Fig. 6A). Paradoxically, these results indicate that RNF4-suppressed cells lack the ability to complete DNA replication after efficient restart. Therefore, while reinitiation of replication was made possible by RNF4 suppression (Fig. 3), longer-range processivity after restart was inefficient.
Figure 6.
RNF4 and PLK1 function in processive replication after restart. (A) Replication restart at differing time points after APH removal. Short pulses of EdU from 0 to 30, 30 to 60, and 150 to 180 min after APH removal were applied to cells to quantify the frequency of replication recovery by flow cytometry. Note that the majority of replication recovery in RNF4-suppressed AtrΔ/− cells occurs within the first hour after APH removal, which is the total length of replication recovery assayed in all other experiments described (D–F and Figs. 1, 3, 4). (B) Representative examples of pre-APH BrdU (red) and post-APH EdU (green) labeling of combed DNA in Atrflox/− cells and AtrΔ/− cells with RNF4 suppression. Cells were BrdU-labeled for 15 min, treated with 5 μM APH for 6 h, and then EdU-labeled for 20 min immediately after APH removal. (C) Quantification of EdU-labeled replication tracks 20, 30, and 45 min after APH removal in Atrflox/− cells and AtrΔ/− cells with RNF4 suppression. The experiment was conducted as described in B, with the 20-min time point representing the earliest replication tracks after APH removal. Note that track lengths after APH removal in RNF4-suppressed AtrΔ/− cells fail to extend further than that observed in the initial 20 min of labeling. (D) Quantification of replication restart by DNA combing in cells subjected to combined suppression of RNF4 and PLK1. ATR deletion, RNF4 suppression, PLK1 inhibition, and subsequent DNA combing were performed and scored as described in Figures 3 and 4. (E) Replication recovery on a population basis in cells subjected to combined or separate suppression of RNF4 and PLK1. APH treatment was for 6 h before removal and subsequent EdU labeling; replication recovery was quantified by flow cytometry. Note that although suppression of RNF4 or PLK1 singly permits replication recovery, suppression of both together does not. (F) Quantification of replication restart shown in E.
To test this model, replication track lengths of RNF4-suppressed cells were quantified after restart by DNA combing. Replication tracks in the first 20 min after APH washout revealed that replication rates in RNF4-suppressed cells were subtly but significantly decreased in Atr-deleted cells in comparison with those in control cells (Fig. 6B,C). To test whether this decrease reflected an eventual loss of processivity during the 20-min labeling period, EdU-labeling times were lengthened, and track lengths were measured. Although replication tracks continued to extend in control cells for up to 45 min after APH washout, RNF4-suppressed ATR-null cells failed to show any further increase in track length after the initial 20 min of EdU labeling (Fig. 6C). These results indicate that RNF4 suppression is sufficient to promote efficient replication reinitiation after stalling in Atr-deleted cells (Fig. 3); however, these forks lack long-range processivity and fail to progress beyond an initial replicative stretch.
We then reasoned that the decreased rate of replication observed in RNF4-suppressed Atr-deleted cells might be caused by continued activity of the AURKA–PLK1 pathway. To test this hypothesis, RNF4 levels were reduced together with PLK1 inhibition, a condition that brings DSB generation in Atr-deleted cells down to near wild-type levels (Fig. 5). Surprisingly, despite the near complete elimination of DSBs, RNF4 and PLK1 dual suppression did not rescue replication restart in Atr-deleted cells (Fig. 6D–F). Replication reinitiation at previously active replication forks was abrogated in Atr-deleted cells when both RNF4 and PLK1 were suppressed (Fig. 6D). This finding is in stark contrast to the effect of inhibiting either RNF4 or PLK1 alone on replication reinitiation (Figs. 3, 4, 6E,F). ATR-expressing controls reinitiated replication normally following combined suppression of RNF4 and PLK1 inhibition (Fig. 6E). Interestingly, the pleiomorphic quality of limited replication reinitiation in Atr-deleted cells upon RNF4 and PLK1 suppression using the cell-based assay (Fig. 6E) may result from the moderate increase in novel origin firing under this condition that was observable with DNA combing (Fig. 6D).
These results indicate that, while suppression of either PLK1 or RNF4 can promote replication restart in Atr-deleted cells (Figs. 3, 4), the restart permitted in these contexts is only afforded by the continued activity of the unimpeded pathway. Therefore, although replication fork collapse is caused by the combined activity of PLK1 and RNF4, these proteins appear to play additional supportive roles in DNA replication after reinitiation. In addition, these findings further distinguish the collapse of replication forks into DSBs from the inability to re-engage the DNA replication machinery, as combined suppression of PLK1 and RNF4 substantially suppresses DSB formation (Fig. 5C–E), but this outcome does not recover replication after stalling (Fig. 6D–F).
Discussion
The results here are the first to demonstrate a requirement for RNF4 and the CDK1–AURKA–PLK1 pathway in replication fork collapse. Our findings indicate that replication fork collapse into DSBs is not a stochastic degenerative process but rather an active one that employs the concerted functions of RNF4 and AURKA–PLK1. As discussed below, the role of RNF4 and PLK1 in promoting DSB generation may be linked to both the activation of endonucleases and the disengagement of replisome components from the replication fork. Thus, RNF4 and AURKA–PLK1 actively suppress replication restart in instances of prolonged fork stalling or ATR pathway failure. Furthermore, these findings indicate that proficiency or deficiency in replication reinitiation is not necessarily predictable by the formation of DSBs, as discussed below.
RNF4 functions in replication fork collapse
Replisome component disengagement from chromatin is at least one component of fork collapse in mec1-mutant yeast strains (Cobb et al. 2003; Raveendranathan et al. 2006). Our results in mammalian cells are consistent with these findings (Figs. 1, 2). Furthermore, according to our studies and those in yeast, both mammalian RNF4 and S. cerevisiae Slx5–Slx8 have vital roles in the recovery from replication stress; however, it is unclear whether their respective functions in this process are entirely similar. In yeast, the Slx5–Slx8 complex was shown to be necessary for cellular viability after DSB induction from prolonged replication stress (Nagai et al. 2008). Although reinitiation of DNA replication was not directly examined, the investigators proposed that Slx5–Slx8 facilitates the recovery of collapsed replication fork structures through DNA repair (Nagai et al. 2008). In contrast, our results demonstrate that RNF4 prevents the reinitiation of replication after stalling and promotes the formation of DSBs in ATR-depleted cells (Figs. 3, 5).
Although these findings are seemingly contradictory, the apparent differences may simply reflect distinct time-dependent functions of RNF4 and Slx5–Slx8 during and after fork collapse. Our results indicate that RNF4 promotes the collapse of replication forks into DSBs and suppresses replication restart. This function of RNF4 would logically precede the requirement for repair-mediated recovery of the replication fork structure. Therefore, our findings are not mutually exclusive with supportive roles for Slx5–Slx8 in HR-mediated and non-HR-mediated repair of DSBs (Burgess et al. 2007; Nagai et al. 2008; Galanty et al. 2012; Yin et al. 2012). In addition, these time-dependent functions may vary with the status of ATR and Mec1. Currently, the function of RNF4 in promoting DSB formation is not clear, but two attractive possibilities are (1) the recruitment and activation of SLX4-associated endonuclease complexes or (2) replisome remodeling, whereby the replication fork is made accessible to DNA-processing enzymes and SLX4-dependent endonucleolytic cleavage. For either model, RNF4 maintains a key function in promoting replication fork collapse into DSBs, and its suppression facilitates replication restart in Atr-deleted cells.
In addition, our findings highlight the differences between the structural collapse of stalled replication forks into DSBs and the reactivation of the DNA synthesis machinery. According to our studies, the dual inhibition of PLK1 and RNF4 suppresses DSB formation (Fig. 5), but this outcome does not afford efficient reinitiation of replication after stalling (Fig. 6D–F). Furthermore, PLK1 inhibition promotes replication recovery in ATR-depleted cells but only modestly affects the formation of DSBs. Indeed, the reinitiation of replication in PLK1-inhibited cells is dependent on RAD51 (Supplemental Fig. 4), demonstrating that an HR-dependent step is still needed in this recovery. Collectively, these data indicate that the relevant functions of PLK1 and RNF4 in replication restart at least partly lie outside of their roles in promoting DSBs.
In accordance with Cobb et al. (2003), we propose that the roles of RNF4 and PLK1 in preventing replication restart are linked to the regulation of replisome integrity. Specifically, we argue that RNF4 and PLK1 participate in the recycling of replisome components. After lengthy stalling, this function can make the reinitiation of replication impossible, particularly when the activities of RNF4 and PLK1 are dysregulated by ATR deficiency. Notably, because suppression of both pathways abrogates reinitiation (Fig. 6D–F), it follows that the restart achieved through single-pathway suppression (Fig. 3, 4) is dependent on the continued activity of the uninhibited pathway, driven by either RNF4 or PLK1. Thus, only the repression of either RNF4 or PLK1 alone strikes the appropriate balance that permits replication reinitiation in Atr-deleted cells, albeit with defects in replication processivity. A role for RNF4 in processive replication is indicated by the inability of reinitiated replication forks to continue replicating beyond 20 min in RNF4-suppressed cells (Fig. 6B,C). Indeed, it is tempting to speculate that sumoylation and RNF4-dependent turnover of replisome components are part of a normal process of replisome maintenance following encounters with natural impediments to replication.
AURKA–PLK1 functions in replication fork collapse
Our results indicate that AURKA–PLK1 suppresses replication restart in ATR-deficient cells (Fig. 4). However, in contrast to RNF4 suppression, PLK1 inhibition has only minor effects on the appearance of DSBs in Atr-deleted cells (Fig. 5). These results indicate that PLK1 inhibition permits effective restart despite the processing of a majority of these forks through a DSB intermediate (Fig. 5; Supplemental Fig. 4). As discussed above, these results suggest that some mechanism other than limiting DSB generation is at play in rescuing replication by PLK1 inhibition.
A role for the degradation of replisome components in this outcome is consistent with the established functions of PLK1 in CLSPN and FANCM degradation (Yoo et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006; Kee et al. 2009). Indeed, it is conceivable that degradation of these two proteins is the sole means by which PLK1 prevents replication reinitiation (Schwab et al. 2010). However, it warrants noting that, similar to RNF4 suppression, PLK1 inhibition also led to the preservation of high-molecular-weight SUMO-conjugated proteins on chromatin, and this accumulation was further elevated by RNF4 suppression (data not shown). Because PLK1 inhibition only modestly affected the appearance of DSBs in Atr-deleted cells, the simplest explanation for the effect of PLK1 activity on replication restart is the disruption of replisome complexes.
A modest compensatory role for PLK1 in DSB formation following ATR suppression would appear, on the surface, to conflict with recent reports of its involvement in MUS81–EME1 activation and association with SLX4 (Matos et al. 2011; Gallo-Fernandez et al. 2012; Munoz-Galvan et al. 2012; Schwartz et al. 2012; Szakal and Branzei 2013). However, these observations are likely distinct based on the unique upstream role that ATR plays in checkpoint activation and replication fork stability. Checkpoint failure in ATR-deficient cells allows the CDK1–AURKA–PLK1 pathway to become activated while stalled replication forks are still present. We propose that this inappropriate activation leads to replisome disassembly at stalled replication forks. A similar effect can be achieved through dysregulation of CDK1 and CDK2 activity (Neelsen et al. 2013). However, in this context, ATR remains active, and it is possible that this activity suppresses replication fork collapse into DSBs through the phosphorylation of targets locally. Consistent with this model, Couch et al. (2013) have demonstrated a direct role for ATR in inhibiting fork reversal by SMARCAL1. Thus, in the absence of ATR, forks are more prone to collapse through not only the inability to limit CDK1–AURKA–PLK1 activity but also permitting fork regression to occur at a higher frequency, leading to SLX4–SLX1-dependent cleavage. In this scenario, DSBs that occur in CDK1- and CDK2-dysregulated cells appear to be dependent only on PLK1 (Neelsen et al. 2013). In contrast, ATR inhibition leads to a concert of effects that promotes replication fork collapse at multiple levels.
Proposed model and implications
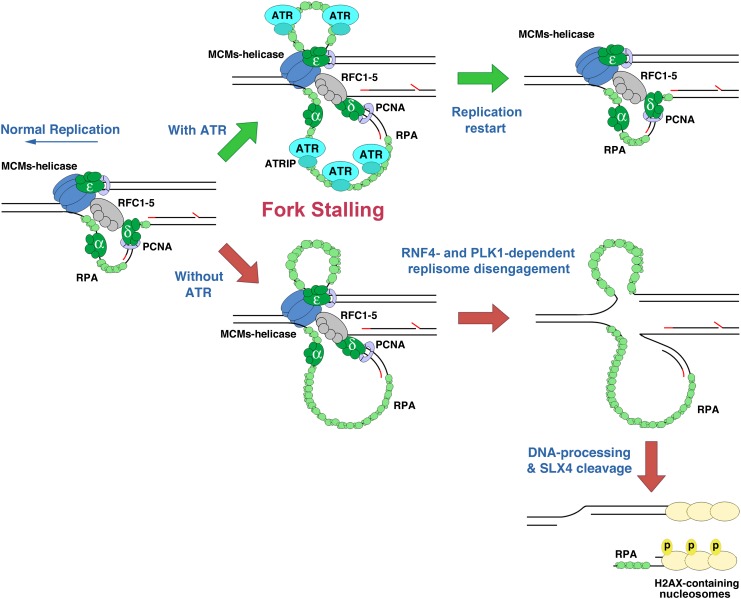
Previous studies and those here can be assembled into an overall model of the processes that promote replication fork collapse in ATR-deficient cells. We propose that fork collapse begins with sumoylation of replisome components and the premature activation of the CDK1–AURKA–PLK1 pathway. Localization of PLK1 to the replisome and the polySUMO2/3-mediated recruitment of RNF4 cause the ubiquitination and disengagement of replisome components. This process may normally function at the G2–M-phase transition to remove the MCM2–7 helicase and associated protein complexes from sister chromatids at replication termination sites. Under long-term replication stress, replisome removal may inadvertently promote checkpoint adaptation by eliminating components of the ATR signaling pathway, such as CLSPN.
We speculate that the premature activation of replisome disengagement from active replication forks in ATR-deficient cells makes these forks accessible to DNA-modifying enzymes, permitting the formation of altered fork structures, such as Holliday junctions. In light of Couch et al. (2013), at least one of these DNA-modifying enzymes includes SMARCAL1, an annealing helicase that reverses replication forks into structures cleavable by the SLX4–endonuclease complex when ATR signaling is absent. The dependence of DSB formation on RNF4, PLK1, and SLX4 (Fig. 5) is consistent with this model. Therefore, replication fork collapse requires several independent networks that are inappropriately engaged by ATR deficiency (Fig. 7).
Figure 7.
Model depicting replisome disassembly by RNF4 and PLK1 in the absence of ATR. In the absence of sufficient ATR expression, RNF4 and PLK1 become aberrantly active and function in the disengagement of replisome components from the fork. Replisome ubiquitination and removal may foster DNA processing, such as fork reversal and SLX4-dependent cleavage into DSBs.
Notably, >90% of cancers overexpress or overactivate AURKA at some level, and this outcome can be mediated directly by oncogene expression (den Hollander et al. 2010; Lens et al. 2010). According to our studies, accentuated AURKA–PLK1 pathway activity may promote replication fork collapse and an increased dependence on ATR function. In this light, we and others recently demonstrated that oncogene expression increases reliance on ATR activity for genome stability and cancer cell viability (Gilad et al. 2010; Murga et al. 2011; Schoppy et al. 2012). By extension, it is possible that AURKA overactivation could both elevate the genomic instability of cancer cells through elevated fork collapse and increase the sensitivity of these cells to ATR/CHK1 inhibitors.
Materials and methods
Atr and Sxl4 deletion
ATR was deleted in passage-immortalized (3T3 protocol) Atrflox/−CRE-ERT2+ MEFs by the addition of 0.5 μM 4-hydroxytamoxifen (4-OHT; EMD) to the culture medium, as described (Brown and Baltimore 2003; Ruzankina et al. 2007; Smith et al. 2009). The 4-OHT was added at the time of plating and washed out 24 h later. Cells were then cultured for an additional 24 h after 4-OHT washout to afford effective depletion of the ATR protein, at which time APH treatments were initiated. SV40 large T immortalized Slx4+/+ and Slx4flox/flox MEFs (Holloway et al. 2011) were generously provided by Dr. Paula Cohen, Dr. Robert Weiss, and Dr. Marcus Smolka (Cornell University). Cre-ERT2 was introduced into Slx4+/+ and Slx4flox/flox cells through lentivirus transduction, as previously described (Brown and Baltimore 2003).
Chemical inhibitors used
All chemical inhibitors were added to the cell culture medium 1 h prior to the addition of APH and remained in the medium both during APH and after APH removal unless otherwise noted. The following chemical inhibitors were used: 10 μM purvalanol (CalbioChem), 100 μM olomoucine (CalbioChem), 25 μM roscovitine (EMD Chemicals), and 5 μM olomoucine II (CalbioChem) for CDK1/2 inhibition; 10 μM BI 2536 (SelleckChem) and 1 μM GSK461364A (SelleckChem) for PLK1 inhibition; and 5 μM Aurora A Inhibitor I (SelleckChem) and ATR-45 (Charrier et al. 2011) for AURKA and ATR inhibition, respectively.
Western blot analysis
Total cell lysates were prepared in Laemmli sample buffer without bromophenol blue and boiled for 5 min prior to protein quantification. Total proteins were quantified by cell counting prior to collection and by the bicinchoninic acid protein assay (Thermo Fisher Scientific). The total protein quantities were normalized, separated by SDS-PAGE gel, and transferred to 0.45 μM polyvinylidene fluoride (PVDF) membranes. Blots were then detected for the following primary antibodies: PCNA (Santa Cruz Biotechnology, sc-56), phospho-PLK1 (Cell Signaling Technology, 5472S), PLK1 (Abcam, ab17056), ATR (Santa Cruz Biotechnology, sc-1887), phospho-S345 Chk1 (Cell Signaling Technology, 2348s), MCM3 (Bethyl Laboratories, A300-124A), phosphor-H2AX (Millipore, 05-636), RFC1 (Abcam, ab3566), RPA70 (Bethyl Laboratories, A300-241A), Sumo2/3 (Millipore, 07-2167), and ubiquitin (Santa Cruz Biotechnology, sc-8017).
Extraction of nuclear-insoluble proteins
Extractions were performed as described by Mendez and Stillman (2000). Briefly, cells were collected by trypsin treatment and lysed in buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol) that was supplemented with protease inhibitors (Roche) and 0.1% Triton-X for 5 min at 4°C. Lysates were then centrifuged at 1300g for 5 min, and the supernatant was mixed 1:1 with 2× SDS sample buffer and collected as the cytoplasmic fraction. Pellets were then washed in buffer A once and incubated in buffer B (3 mM EDTA, 0.2mM EGTA) supplemented with protease inhibitors (Roche) for 30 min at 4°C. Samples were centrifuged at 1700g for 5 min, and the supernatant was collected and mixed 1:1 with 2× Laemmli sample buffer to generate the nuclear-soluble fraction. Pellets from this extraction were washed in buffer B twice prior to a final centrifugation at 1700g for 5 min. The pellets were resuspended in buffer A and mixed 1:1 with 2× Laemmli sample buffer as the nuclear-insoluble fraction. All samples were boiled for 5 min prior to protein quantification as described above.
Replication restart assay by flow cytometry
Cells were labeled with 10 μM BrdU for 30 min prior to the addition of 5 μM APH. After APH treatment for the times indicated, APH was removed by three washes of PBS followed by an additional wash of medium at 37°C and three final washes with PBS. After APH wash-off, cells were incubated in 50 μM EdU for 1 h. Cells were collected for flow cytometric analysis by trypsin treatment and fixed in 70% EtOH overnight. BrdU incorporation was detected by acid denaturaturing (3N HCl containing 0.5% Tween 20) and neutralizing (0.1 M sodium borate at pH 8.5) treatments of cellular DNA followed by the addition of anti-BrdU primary (Invitrogen, MoBu-1) and FITC-conjugated secondary (Jackson ImmunoResearch Laboratories) antibodies. EdU incorporation was detected using the Alexa Fluor-647 kit for flow cytometry per the manufacturer's instructions (Invitrogen). BrdU- and EdU-detected cells were stained with PI analyzed by flow cytometry using a FACScalibur (BD) and imaged using FlowJo (Tree Star) software.
DNA combing
Cells were labeled with 10 μM BrdU for 15 min prior to the addition of 10 μM APH. Cells were incubated at 37°C in APH containing medium for the indicated times and then washed three times with PBS followed by an additional wash of medium and a final three washes in PBS. After APH removal, cells were incubated in 50 μM EdU for 20 min. Cells were then collected by trypsinization, washed in PBS, counted, and embedded in 0.5% agarose plugs (2 × 105 cells per plug) in L buffer (100 mM EDTA, 20 mM NaCl, 10 mM Tris at pH 7.5). Plugs were then incubated in lysis buffer (L buffer with 1% SDS, 1 mg/mL proteinase K) for a total of 48 h at 50°C with a change of buffer after 24 h. Subsequently, plugs were washed in TE* (10 mM EDTA, 10 mM Tris-Cl at pH 8.0) and incubated for 24 h. Prior to DNA combing, plugs were digested in combing buffer (100mM MES at pH 6.5) supplemented with β-agarose overnight at 42°C. The DNA solution was poured into custom-made Teflon blocks, and silanized slides were dipped into the DNA solution via a KSV Nima single-vessel dip coater. DNA-combed slides were denatured (3N HCl containing 0.5% Tween 20), neutralized (0.1 M sodium borate at pH 8.5), and then detected for BrdU using anti-BrdU primary (Invitrogen MoBu-1) and FITC-conjugated secondary (Jackson ImmunoResearch Laboratories) antibodies. EdU was detected using the Alexa Fluor-647 kit as described above (Invitrogen). Slides were subsequently scored using a Nikon Eclipse 80i florescent microscope and a 100× lens.
Lentiviral shRNA infections
Lentiviral vectors expressing RNF4 or RAD51 shRNA (pLKO.1) were obtained from Thermo Fisher Scientific, modified to coexpress GFP, and used as previously described (Urtishak et al. 2009). Lentiviral virus was produced by transfecting 293T cells with shRNA-expressing vectors together with lentiviral packaging vectors (pMDLg/pRRE, CMV-VSVG, and RSV-REV) via calcium phosphate precipitation. After titration, experimental cells were transduced with a multiplicity of infection of 5–10 and assayed 48 h later.
PFGE
PFGE was performed as previously described (Smith et al. 2009). Following treatment as described, cells were collected by trypsinization, washed in PBS, counted, and embedded into agarose plugs (1 × 106 cells per plug) and L buffer (100 mM EDTA, 20 mM NaCl, 10 mM Tris at pH 7.5). Plugs were then incubated in lysis buffer (L buffer with 1% SDS, 1 mg/mL proteinase K) for a total of 48 h at 50°C with a change of buffer after 24 h. Subsequently, plugs were washed in TE* (10 mM Tris-Cl, 10 mM EDTA at pH 8.0) for 24 h and then TAE (40 mM Tris-acetate, 20 mM sodium acetate, 1 mM EDTA at pH 8.0) for either an additional 24 h or several days at 4°C. Electrophoresis in 0.8% PFGE-certified agarose gels was performed using a pulsed field electrophoresis system (CHEF-DR II, Bio-Rad Laboratories) in TAE at 2.8 V/cm2 with a 400- to 1800-sec switch time for a total of 60 h at 4°C.
Replicates and statistical tests
Unless otherwise noted, all data represented in figures represent three to five independent experiments. Error bars represent the standard error of the mean, and P-values were calculated using the Student's t-test.
Acknowledgments
We are grateful to David W. Schoppy for his kind assistance in these studies. We are also indebted to Dr. Paula Cohen, Dr. Robert Weiss, and Dr. Marcus Smolka (Cornell University) for generously providing immortalized Slxflox/flox MEFs. Funds for this research were provided by National Institute on Aging (R01AG027376), the Department of Defense Breast Cancer Research Program (W81XWH-09-1-0669), the Abramson Family Cancer Research Institute, and the Mary Kay Ash Charitable Trust. A.K.B. is a scholar of the Leukemia and Lymphoma Society and was supported by NIH GM74917.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.223180.113.
References
- Borel F, Lacroix FB, Margolis RL 2002. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J Cell Sci 115: 2829–2838 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev 17: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT 2011. Purification and identification of endogenous polySUMO conjugates. EMBO Rep 12: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X 2007. The Slx5–Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27: 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem 284: 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, Mortimore M, O'Donnell ME, Pinder JL, Reaper PM, et al. 2011. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54: 2320–2330 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, Kremer M, Graf N, Scheerer M, Hall MA, et al. 2010. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 116: 1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K 2012. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704 [DOI] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB 2003. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Forment JV, Blasius M, Guerini I, Jackson SP 2011. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE 6: e23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froget B, Blaisonneau J, Lambert S, Baldacci G 2008. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell 19: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26: 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Fernandez M, Saugar I, Ortiz-Bazan MA, Vazquez MV, Tercero JA 2012. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res 40: 8325–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, Brown EJ 2010. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res 70: 9693–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless SJ, Morag AS, Belisle KA, Sutera VA Jr, Lovett ST 2006. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell 21: 595–604 [DOI] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE 2011. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Kim JM, D'Andrea AD 2009. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev 23: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH 2010. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer 10: 825–841 [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455: 119–123 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Bartek J, Lukas J 2006. Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell 23: 307–318 [DOI] [PubMed] [Google Scholar]
- Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, Medema RH, Freire R 2006. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol 16: 1950–1955 [DOI] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A 2012. Distinct roles of Mus81, Yen1, Slx1–Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol 32: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, D'Artista L, Schleker T, Guerra C, Garcia E, et al. 2011. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 18: 1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IM, Herrador R, Lopes M 2013. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol 200: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M 2006. SCFβTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 23: 319–329 [DOI] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, Hay RT 2011. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol 18: 1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendranathan M, Chattopadhyay S, Bolon YT, Haworth J, Clarke DJ, Bielinsky AK 2006. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J 25: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Roseaulin LC, Noguchi C, Martinez E, Ziegler MA, Toda T, Noguchi E 2013. Coordinated degradation of replisome components ensures genome stability upon replication stress in the absence of the replication fork protection complex. PLoS Genet 9: e1003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ 2012. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest 122: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W 2010. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 29: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Wright WD, Ehmsen KT, Evans JE, Stahlberg H, Heyer WD 2012. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol Cell Biol 32: 3065–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Diffley JF 2008. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Fu MA, Brown EJ 2009. Tim-Tipin dysfunction creates an indispensible reliance on the ATR–Chk1 pathway for continued DNA synthesis. J Cell Biol 187: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakal B, Branzei D 2013. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J 32: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF 2003. A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Urtishak KA, Smith KD, Chanoux RA, Greenberg RA, Johnson FB, Brown EJ 2009. Timeless maintains genomic stability and suppresses sister chromatid exchange during unperturbed DNA replication. J Biol Chem 284: 8777–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MA, Smits VA, Klompmaker R, Medema RH 2001. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J Biol Chem 276: 41656–41660 [DOI] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 26: 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG 2004. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117: 575–588 [DOI] [PubMed] [Google Scholar]