Abstract
Neuroendocrine ACTH secretion responds to peripheral inflammatory and stress signals. We previously demonstrated that the proinflammatory cytokine, leukemia inhibitory factor (LIF), affects the hypothalamo-pituitary-adrenal axis (HPA) by stimulating in vitro and in vivo pituitary proopiomelanocortin (POMC) gene expression and ACTH secretion and by potentiating the action of hypothalamic corticotropin releasing hormone (CRH). Whereas pathways shown thus far to regulate POMC expression exclusively involve cAMP or calcium, we here describe a direct and indirect STAT3-dependent regulation of POMC transcription by LIF. Using progressive 5′-deletions of POMC promoter, we identified a LIF-responsive –407/–301 region that contains two juxtaposed sequences within –399/–379 related to a STAT3 DNA-binding motif. Each sequence within –399/–379 separately corresponds to a low-affinity and direct binding site for STAT3, but, in combination, these sequences bind STAT3 cooperatively and with high affinity. Moreover, LIF-activated STAT3 indirectly mediates LIF corticotroph action by inducing and potentiating CRH-induced c-fos and JunB expression and binding to the POMC AP-1 element. We therefore conclude that both a direct and indirect route mediate LIF-induced STAT3 activation of POMC transcription. Demonstration of STAT3-dependent regulation of the POMC gene represents a powerful mechanism for immuno-neuroendocrine interfacing and implies a direct stimulation of ACTH secretion by inflammatory and stress-derived STAT3-inducing cytokines.
Introduction
Leukemia inhibitory factor (LIF) is a pleiotropic IL-6 family-related cytokine (1). We have shown LIF to regulate the hypothalamo-pituitary-adrenal axis (HPA). LIF and LIF receptor are expressed in murine hypothalamus and pituitary and in human fetal and adult pituitary (2, 3). Both peripheral, as well as hypothalamic and pituitary, LIF, and LIFR expression are upregulated in response to inflammatory stimuli (2). LIF contributes to HPA axis activation in inflammatory states, as evidenced by systemic LIF injection inducing ACTH in vivo (4, 5), and increased circulating levels of LIF are observed in septic patients and in a variety of acute and chronic inflammatory conditions (6). Lipopolysaccharide endotoxin injection also results in a concordant increase of plasma LIF and ACTH levels in mice (6). Models of LIF–/– knockout or pituitary-directed LIF transgenic mice have yielded interesting insights into the integrative function of LIF in pituitary function and development. LIF–/– knockout mice showed attenuated ACTH responses after 36 hours of fasting (7) or immobilization stress for 30 or 45 minutes (5). Furthermore, poststress pituitary proopiomelanocortin (POMC) mRNA content was significantly decreased in LIF–/– knockout mice (5). Early transgenic pituitary LIF overexpression results in features consistent with Cushing’s disease, including corticotroph hyperplasia and hypercortisolism (8), confirming a key role for LIF in pituitary development as well as for mature pituitary function.
In vitro, LIF stimulates ACTH secretion in primary human fetal pituitary cultures (3) or in murine AtT20 cells (9). Interestingly, LIF synergizes with corticotropin-releasing hormone (CRH) (10) to stimulate in vivo and in vitro POMC transcription and ACTH secretion (3, 11). In AtT20 cells transfected with a –706/+64 rat POMC promoter-luciferase construct, LIF-induced luciferase activity was robustly potentiated by CRH (∼15-fold) (9, 11).
CRH-induced POMC expression is mediated through the cAMP/PKA pathway and through calcium-mediated events (12, 13). Three elements on the POMC promoter bind CRH-induced transcription factors. The AP-1 site in exon-1 (+41/+47) binds CRH-induced CREB (12) and c-fos and JunB (14). Two elements within POMC promoter also bind CRH-induced Nurr family transcription factors: The –71/–64 element corresponds to a pivotal responsive sequence for positive or negative POMC regulation by Nurr and the glucocorticoid receptor, respectively (15), and the –404/–383 element recognizes Nur77 homo- or heterodimers (16, 17). In contrast, LIF action on POMC is independent of cAMP production (9) but is transduced through stimulation of STAT3 (18). LIF binding to its receptor (LIFRα) indeed induces LIFRα and gp130 subunit dimerization. Consequently, JAK2 is activated to phosphorylate the gp130 subunit on tyrosine residues, providing docking sites for STAT3. STAT3 is tyrosine phosphorylated on Y705 by JAKs, dimerizes by interaction between the SH2 domain of a STAT3 molecule and the tyrosine phosphorylated Y705 residue of the other STAT3 molecule. The STAT3/STAT3 dimer is then translocated to the nucleus, where it binds to specific STAT3 DNA-binding motifs (19). By using two dominant negative forms of STAT3 (20), we previously showed LIF corticotroph action to depend on STAT3 (18). These STAT3 mutants are either mutated on the Y705 residue or on the DNA-binding domain and, consequently, are unable to dimerize or to bind DNA, respectively. By binding to endogenous wild-type STAT3, they exert a dominant negative action on the corticotrope and exhibit abrogated LIF signaling to POMC (18).
Despite the STAT3-dependency for corticotroph LIF action, no STAT3 binding site has heretofore been identified on the POMC promoter. To localize a LIF-responsive STAT3 DNA-binding element on this promoter, we constructed 5′ progressive deletions of the POMC promoter, and identify a STAT3 specific region, which accounts for significant LIF- and LIF and CRH–induced POMC promoter activity. We also show that LIF potently induces, in a STAT3-dependent manner, c-fos and JunB expression in corticotroph cells, and potentiates CRH-induced c-fos and JunB expression and binding to the POMC exon-1 AP-1 element. Therefore, we propose a model for LIF and CRH synergy, which combines independent and cooperative effects of both peptides on corticotroph function.
Methods
Culture of AtT20 cells.
AtT20 cells were grown in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml streptomycin, 100 U/ml penicillin, and 0.25 μg/ml amphotericin B (Life Technologies Inc., Gaithersburg, Maryland, USA).
Transient transfection and luciferase assay.
AtT20 cells were plated in 2-ml dishes (100,000 cells per well) and allowed to adhere for 16 hours. Cells were transiently transfected as described (9) with 0.5 μg per dish of each rat POMC promoter fragment fused to the luciferase reporter gene, and 0.5 μg of pSV-lacZ (Promega Corp., Madison, Wisconsin, USA) expressing β-galactosidase, as an internal transfection control. Twenty-four hours after transfection, cells were treated with 1 nM LIF and/or 10 nM CRH in triplicate for 7 hours. Cells were then washed once in PBS (pH 7), lysed in reporter lysis buffer, and subjected to assay for luciferase (9) or β-galactosidase activity (Promega Corp.).
Construction of POMC promoter deleted fragments fused to the luciferase reporter gene.
Deletions of rat POMC promoter were performed using a PCR technique (ExSite PCR-based site-directed mutagenesis; Stratagene, La Jolla, California, USA). All constructs were verified by sequencing.
Electromobility shift assay.
For nuclear extract preparations, AtT20-treated cells were harvested in cold PBS buffer. Nuclear extracts were prepared as described previously (21). Final concentrations were typically 3 μg/μl as determined by Bio-Rad protein assay (Bio-Rad Laboratories Inc., Hercules, California, USA). For electromobility shift assay (EMSA), 15 μg nuclear extracts and 10–20 pg 32P end-labeled double stranded DNA probe (100,000 cpm) were used per reaction. Nuclear extracts were preincubated for 15 minutes at room temperature in 20 μl of binding buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 5% glycerol, 1 mg/ml BSA [pH 7.5]) with 1 μg of poly(dI-dC). 32P-labeled probe was added and the binding reaction was left at room temperature for 20 minutes. In competition experiments, a 25- to 100-fold molar excess of unlabeled competitor oligonucleotide was added to the preincubation reaction. Antibody competitions were performed using 1 μg anti-STAT3, JunB or c-fos antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) that were added to the preincubation reaction for 1 hour at 4°C. The protein-DNA complexes were resolved on a 6% polyacrylamide gel in 0.5 X Tris borate/EDTA. Gels were dried and autoradiographed.
Western blot.
For whole cell extract preparations, cells were lysed in 20 mM Tris-HCl, 20% glycerol, 2 mM DTT, 400 mM KCl, 20 μM leupeptin, 1 mM PMSF, and 2 μg/μl aprotinin, with repeated freezing at –70°C and thawing at 37°C. Protein concentration was measured after removal of membrane debris by centrifuging at 4°C for 10 minutes at 12,000 g. Western blot was performed as described previously (22) using 30 μg of protein per lane and 1 μg/ml STAT3 antibody (Santa Cruz Biotechnology Inc.).
Northern blot.
Total RNA were prepared from AtT20 cells treated with LIF, CRH, or both, using Trizol reagent (Life Technologies Inc). A total of 10 μg RNA per lane were size-fractionated under denaturing conditions using formaldehyde-agarose (1%) gel, transferred to nitrocellulose, and hybridized using 32P-labeled probe corresponding to c-fos, JunB, c-jun, JunD, Nurr1, Nur77, or β-actin cDNA.
Results
Identification of an LIF-responsive element on the POMC promoter.
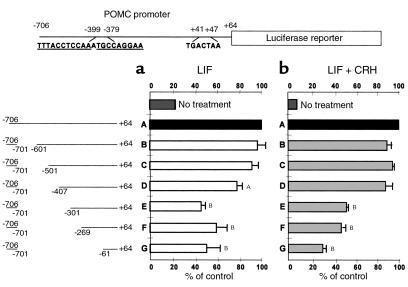
LIF-induced POMC promoter activity is STAT3 mediated (18). As no direct STAT3 action on the POMC promoter has been shown, progressive 5′ deletions of the POMC promoter were transiently transfected in AtT20 cells (Figure 1, a and b). LIF stimulated full-length rat POMC promoter activity (–706/+64) 4.5 ± 0.5–fold. Progressive promoter deletions from nucleotide –701 revealed a marked attenuation of full-length promoter activity (100%) with construct E versus construct D (77 ± 6% vs. 46 ± 3%, P = 0.001). Further POMC promoter deletions to nucleotide –269 and to nucleotide –61 (constructs F and G) did not affect LIF induction of promoter activity. This result indicated the presence of an LIF-responsive region located within –407/–301 region of the POMC promoter.
Figure 1.
Identification of a LIF-responsive region on the POMC promoter. Progressive 5′ deletions of the POMC promoter were fused to the luciferase reporter gene and transiently transfected in AtT20 cells (9). Cells were stimulated with 1 nM LIF (a) or with 1 nM LIF + 10 nM CRH (b) for 6 hours. Luciferase activity was measured in cell lysates in the presence of the luciferin substrate. Results are expressed as percentage of control (n ≥ 3; control = 100% = full-length –706/+64 POMC promoter). AP < 0.05 and BP < 0.01 for deleted POMC promoter constructs (constructs B–G) versus full-length POMC promoter (construct A).
To explore whether this –407/–301 region is also critical for known LIF and CRH synergy on the POMC promoter, transiently transfected AtT20 cells were stimulated with LIF and CRH (Figure 1b). Under these conditions, full-length rat POMC promoter activity (–706/+64) was stimulated by 15.1 ± 2.1–fold. Furthermore, significant attenuation of LIF+CRH–induced POMC promoter activity was also observed for construct E versus construct D (88 ± 7% vs. 52 ± 3%, P = 0.01), which further emphasized the importance of the –407/–301 region for LIF action. A significant decrease in LIF+CRH–induced POMC promoter activity was also observed for construct G versus construct F (48 ± 3% versus 30 ± 4%, P = 0.02), implying a role for the rat POMC –269/–61 region in LIF and CRH synergy, as described previously (11).
LIF-induced STAT3 binds to the rat POMC promoter –399/–379 region.
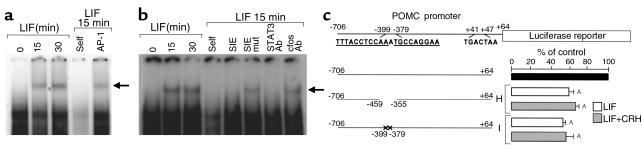
We had previously shown that corticotroph LIF function was disrupted by dominant negative STAT3 forms (18). However, no classic STAT3 consensus DNA-binding sequence is apparent in the POMC promoter. Nevertheless, the previously identified LIF- and LIF+CRH–responsive region –407/–301 on the POMC promoter (Figure 1, a and b) contains two sequences (–399- TTTACCTCCAAATGCCAGGAA –379) distantly related to the STAT3 consensus DNA-binding element (TTCCA/TGGAA). To explore STAT3 binding to this element, EMSAs were performed using sequence –407/–374 as a probe and nuclear extracts from LIF-stimulated AtT20 cells (Figure 2a). Stimulation of cells with 1 nM LIF for 15 or 30 minutes induced formation of a single specific complex, which was not detectable in nonstimulated cells. This complex was effectively competed by 100-fold molar excess of cold homologous oligonucleotide (self), but not by a heterologous nonspecific (AP-1) oligonucleotide. We show direct STAT3 binding to this POMC promoter sequence; preincubation of LIF-stimulated nuclear extracts with an anti-STAT3 antibody completely abrogated the complex formation, unlike the retained complex observed with an anti–c-fos antibody (Figure 2b). Furthermore, 100-fold molar excess of a STAT-inducible element (SIE) oligonucleotide which contains a STAT3 DNA-binding motif (18) efficiently competed for STAT3 with the –407/–374 probe, unlike a mutated SIE oligonucleotide (SIE mut) (Figure 2b). We therefore conclude that the observed STAT3 containing complex binds directly through a STAT3 DNA-binding motif to the POMC probe. These results indicate a novel specific and direct binding of STAT3 on the –407/–374 region of the pituitary POMC promoter.
Figure 2.
Characterization of a STAT3 binding site on the POMC promoter. (a and b) A 32P-labeled double-stranded oligonucleotide corresponding to region –407/–374 of the POMC promoter (5′- –407TAGTGATATTTACCTCCAAATGCCAGGAAGGCA G–374 -3′; in bold are the two STAT3 DNA-binding motifs) was used as probe. Cells were either untreated or treated with 1 nM LIF for 15 or 30 minutes. Using 15 μg nuclear extracts from 15-minute LIF-treated AtT20 cells, competition was performed with either 100-fold molar excess of (a) cold –407/–374 double-stranded oligonucleotide (self), of cold nonspecific POMC exon-1 AP-1 double-stranded oligonucleotide (AP-1) (5′- +33AGGAGCAGTGACTAAGAGAGGC+54 -3′; in bold is the consensus AP-1 motif), of (b) cold SIE oligonucleotide (5′- CGCGTCATTTCCCGTAAATCA -3′; in bold is the consensus SIE motif) (SIE), of cold mutated SIE oligonucleotide (5′- CGCGTCAGATCCCGTCATTCA -3′; the bold underlined italic letters correspond to the mutated bases from the SIE motif) (SIE mut), or with 1 μg anti-STAT3 (STAT3 Ab) or c-fos antibody (c-fos Ab). (c) POMC promoter deleted from –459/–355 (construct H) or mutated in the –399/–379 region (construct I) (5′- –399GTGACATCCAGAGGCTAGTG–379 -3′; the bold underlined italic letters correspond to the mutated bases from the POMCSTAT3 motifs) were fused to the luciferase reporter gene and transiently transfected in AtT20 cells (9). Cells were stimulated with 1 nM LIF or with 1 nM LIF + 10 nM CRH for 6 hours. Luciferase activity was measured in cell lysates in the presence of the luciferin substrate. Results are expressed as percentage of control (n ≥ 3; control = 100% = full-length –706/+64 POMC promoter). AP < 0.01 for deleted or mutated POMC promoter constructs (constructs H and I) versus full-length POMC promoter (black bar).
The functional relevance of LIF-induced STAT3 binding to the –407/–374 region was confirmed by using POMC promoter constructs either deleted within the region –459/–355 (construct H) or mutated at both STAT3-related binding sequences (construct I) (Figure 2c). Indeed, LIF- and LIF+CRH–stimulated transfectants showed markedly decreased POMC promoter activity (59.2 ± 4% for LIF, P < 0.001, and 65.2 ± 2% for LIF+CRH, P = 0.01, for construct H; 53.2 ± 4% for LIF, P < 0.001, and 56.5 ± 12% for LIF+CRH, P = 0.01, for construct I) (Figure 2c), compared with wild-type promoter transfectants.
High-affinity STAT3 binding to the POMC promoter –399/–379 region requires cooperation between both STAT3-related binding elements.
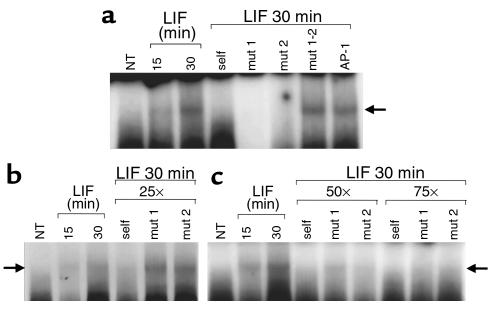
To explore whether each STAT3-related DNA-binding sequence, contained within the POMC promoter –399/–379 region, independently binds STAT3, we performed EMSA using the same oligonucleotide (–407/–374) as in Figure 2a as a probe. Competition was then assessed with 100-fold molar excess of this same cold probe mutated at either the first putative STAT3 DNA-binding site (mut1) or at the second putative STAT3 DNA-binding site (mut2) (Figure 3a). Similar to competition observed with wild-type cold –407/–374 oligonucleotide (self), competitions with mut1 or mut2 oligonucleotide disrupted STAT3 complex formation (Figure 3a). However, competition with a cold oligonucleotide mutated at both STAT3 DNA-binding sites (mut1-2), nor an heterologous oligonucleotide AP-1, was inefficient in abrogating complex formation, indicating that each STAT3 DNA-binding site might independently bind STAT3.
Figure 3.
Characterization of two cooperative low-affinity STAT3 binding sites on the POMC promoter. (a) This experiment was performed as in Figure 2a, except that competition was performed with 100-fold molar excess of cold mut1 (5′- –407 TAGTGATAGTGACATCCAGATGCCAGGAAGGCAG–374 -3′), mut2 (5′- –407TAGTGATATTTACCTCCAAAGGCTAGTGAGGCAG–374 -3′), mut1-2 (5′- –407TAGTGATAGTGACATCCAGAGGCTAGTGAGGCAG–374 -3′), or nonspecific POMC exon-1 AP-1 oligonucleotide. (b and c) Competition was performed as in a with 25-fold, 50-fold, or 75-fold molar excess of wild-type –407/–374 (self) or mut1 or mut2 cold double-stranded oligonucleotide. NT, vehicle-treated control.
Accordingly, we explored this hypothesis by using the mut1 or mut2 oligonucleotide as hybridization probe in two independent EMSAs (data not shown). Under these conditions, no STAT3 complex was observed. We therefore reasoned that STAT3 binds independently to both single STAT3 DNA-binding elements, but at a lower affinity than it does to both STAT3 DNA-binding elements combined. This could explain the efficient competition with 100-fold excess of mut1 and mut2 oligonucleotide for STAT3 DNA-binding to the –407/–374 probe (Figure 3a), and why no direct STAT3 binding was detected when each individual STAT3 DNA-binding element (mut1 and mut2) was used separately as a probe (data not shown).
Binding affinity of STAT3 to wild-type, mut1, and mut2 –407/–374 elements was therefore subsequently explored (Figure 3, b and c). Interestingly, cold wild-type –407/–374 oligonucleotide (self) totally disrupted STAT3 DNA-binding to the –407/–374 probe at concentrations as low as 25-fold molar excess (Figure 3b). However, mut1 and mut2 were only fully effective at concentrations higher than 50-fold molar excess, as a reminiscent band was still observed at this concentration, but not at 75-fold molar excess (Figure 3c).
We therefore conclude that both STAT3 DNA-binding elements are required for cooperative STAT3 high affinity binding to the –407/–374 probe.
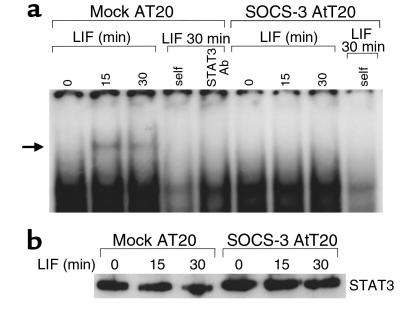
STAT3 binding to the POMC promoter –399/–379 region is abrogated in SOCS-3 overexpressing AtT20 cells.
We previously demonstrated in AtT20 cells that blockade of LIF-induced STAT3 tyrosine phosphorylation is mediated by SOCS-3, a member of the inhibitors of cytokine signaling family (23, 24). SOCS-3 overexpression in AtT20 cells results in abrogation of LIF-induced POMC gene expression, POMC promoter activity and ACTH secretion (23). We therefore explored whether LIF-induced STAT3 binding to the –407/–374 probe is affected in SOCS-3 overexpressing cells. Using nuclear extracts from LIF-stimulated mock- or SOCS-3–overexpressing AtT20 cells, formation of the STAT3 complex with the –407/–374 probe was abrogated when SOCS-3 is overexpressed (Figure 4a). However, STAT3 protein content was similar in whole cell protein extracts from mock or SOCS-3–overexpressing transfectants, as confirmed with an anti-STAT3 immunoblot (Figure 4b). This result confirms that STAT3 DNA-binding activity to the POMC promoter is directly mediated through the LIF-induced JAK-STAT3 pathway.
Figure 4.
Inhibition of LIF-induced STAT3 in SOCS-3 overexpressing AtT20 cells disrupts STAT3 binding to the POMC –407/–374 probe. (a) EMSA was performed as in Figure 2a with 15 μg nuclear extracts derived from LIF-treated mock- or SOCS-3–overexpressing cells for 15 or 30 minutes. A 100-fold molar excess of cold POMC –407/–374 double-stranded oligonucleotide (self) or 1 μg anti-STAT3 antibody (STAT3 Ab) was used as competitor. (b) To show that STAT3 concentration was identical in both mock- and SOCS-3–overexpressing cells and at each reaction time point, LIF-treated whole cell extracts were prepared from the same cell aliquot as the one used for preparation of nuclear extracts and used for Western blotting with an anti-STAT3 antibody.
LIF and CRH additively induce c-fos and JunB transcription.
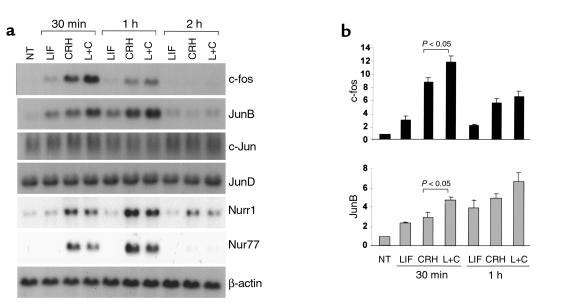
The IL-6 cytokine family (25), to which LIF belongs, also stimulate AP-1 transcription factor expression, including c-fos or JunB (26, 27). Interestingly, a functional AP-1 binding site in the POMC exon-1 promoter binds and transduces transcriptional activity both of CRH-induced CREB (12) and of CRH-induced c-fos and JunB (13, 14). Although LIF does not affect CREB activity (11), we investigated whether the cytokine might affect c-fos and JunB gene expression (Figure 5, a and b). Treatment with 1 nM LIF for 30 minutes stimulated c-fos and JunB expression approximately 3.1 ± 0.5–fold and approximately 2.3 ± 0.1 –fold, respectively, whereas 10 nM CRH induced gene expression approximately 9 ± 0.7–fold and approximately 2.9 ± 0.5 –fold, respectively. Interestingly, treatment with LIF+CRH for 30 minutes was additive for c-fos (∼12 ± 0.9–fold) and JunB transcription (4.9 ± 0.3–fold) (Figure 5, a and b). However, neither LIF nor CRH stimulated the transcription of c-Jun or JunD, which demonstrated LIF and CRH specificity for c-fos and JunB. This result indicates that LIF stimulates c-fos and JunB mRNA expression both alone and additively in combination with CRH, suggesting that the POMC promoter AP-1 binding site might transduce a component of the LIF-and LIF+CRH- induced POMC promoter activity through binding of the c-fos/JunB complex.
Figure 5.
LIF and CRH are additive for c-fos and JunB mRNA expression. (a) A total of 10 μg total RNA extracted from 1 nM LIF–, 10 nM CRH–, or LIF+CRH–treated AtT20 cells for 30 minutes or 1 hour were separated on a 1% agarose/formaldehyde gel and transferred to nitrocellulose membrane. Hybridization was performed with a 32P-labeled double-stranded DNA probe corresponding to the c-fos, JunB, c-Jun, JunD, Nurr1, Nur77, or β-actin coding sequence, as described previously (23). (b) Northern blot signals for c-fos and JunB mRNA were analyzed by quantitative densitometry and normalized for β-actin. The relative increase of LIF–, CRH–, and LIF+CRH–induced c-fos and JunB mRNA was calculated from three independent experiments (n = 3). NT, vehicle-treated control.
LIF does not induce transcription of Nurr-family transcription factors.
CRH functions through the secondary messengers cAMP or calcium to potently stimulate POMC gene transcription within corticotroph cells. Beside induction of CREB, c-fos and JunB by CRH, Nurr1 and Nur77 mRNA expression are also rapidly and robustly increased by CRH (12–17). Nurr1 and Nur77 then bind to two specific sequences (–70/–63 or –404/–383) of the POMC promoter to induce POMC gene expression (15–17). We therefore investigated whether LIF corticotroph action is partially mediated through Nurr transcription factors. In a Northern blot representing AtT20 cell-extracted total RNA after cell stimulation with LIF, CRH, or LIF+CRH for 30 minutes or 1 hour, CRH indeed rapidly stimulated Nurr1 and Nur77 mRNA expression, as described previously (Figure 5a) (15–17). However, expression of Nurr-family transcription factors assessed by Northern blot were not regulated by LIF, minimizing the probability of this pathway as being important for LIF corticotroph signaling.
LIF and CRH additively induce c-fos and JunB binding to the POMC AP-1 exon-1 promoter binding element.
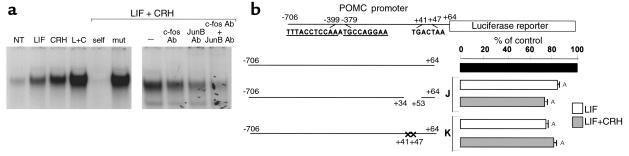
To ascribe a functional importance for LIF-induced c-fos and JunB mRNA expression, and for the additive effects of both LIF and CRH on their transcription, we investigated whether, in a similar manner as CRH (13, 14), LIF could also stimulate AP-1 binding activity to the well-described AP-1 element on POMC promoter exon-1. Nuclear extracts derived from cells stimulated for 1 hour with LIF, CRH, or LIF+CRH were used in an EMSA with a 32P-labeled probe comprising the POMC AP-1 site (Figure 6a). Formation of a specific AP-1 complex, which was completely competed by 100-fold molar excess of cold AP-1 oligonucleotide (self), but not by a mutated AP-1 oligonucleotide (mut), was induced by both LIF and CRH, and additively stimulated by combined LIF+CRH treatment. Both c-fos and JunB were shown to be components of this LIF- and CRH-inducible complex, as its formation was reduced after preaddition of anti–c-fos or anti-JunB antibodies to LIF+CRH–treated nuclear extracts. Modest residual binding, which probably reflects CRH-induced CREB binding activity (12), was still observed when both anti–c-fos and anti-JunB antibodies were added. This result indicates that the LIF-induced c-fos/JunB binding to the POMC promoter AP-1 site might be involved in LIF corticotroph action, in addition to the previously identified STAT3 DNA-binding site on the POMC promoter. Consequently, we deleted the POMC promoter region +34/+53 (construct J), or mutated this AP-1 site on the POMC promoter (construct K), and used these constructs to drive luciferase gene expression in transiently transfected AtT20 cells (Figure 6b). Deletion or mutation of the POMC exon-1 AP-1 binding site significantly (P < 0.001) altered LIF- and LIF+CRH–induced POMC promoter activity (83 ± 1% and 74 ± 4%, respectively, for LIF; 72 ± 0.4% and 80 ± 4%, respectively for LIF+CRH) as compared with full-length wild-type construct (Figure 6b), which confirmed functional involvement of this binding element in mediating LIF and CRH induction of POMC promoter activity.
Figure 6.
LIF and CRH have additive effects on c-fos and JunB binding to the POMC exon-1 AP-1 element. (a) A 32P-labeled double-stranded oligonucleotide corresponding to region +33/+54 AP-1 of POMC promoter (5′- +33AGGAGCAGTGACTAAGAGAGGC+54 -3′) was used as probe. Cells were either untreated or treated for 1 hour with 1 nM LIF and/or 10 nM CRH. By using 15 μg nuclear extracts from 1-hour LIF+CRH–treated AtT20 cells, competition was performed with either 100-fold molar excess of cold +33/+54 AP-1 double-stranded oligonucleotide (self), mutated AP-1 double-stranded oligonucleotide (mut) (5′- +33AGGAGCAGCGTACGAGAGAGGC+54 -3′), with 1 μg anti–c-fos antibody (c-fos Ab), with 1 μg anti-JunB antibody (JunB Ab), or with 1 μg anti–c-fos and 1 μg anti-JunB antibody (c-fos+JunB Ab). (b) POMC promoter deleted from +34/+53 (construct J) or mutated in the +41/+47 region (construct K) (5′- +41CGTACGA+47 -3′) were fused to the luciferase reporter gene and transiently transfected in AtT20 cells (9). Cells were stimulated with 1 nM LIF or with 1 nM LIF + 10 nM CRH for 6 hours. Luciferase activity was measured in cell lysates in the presence of the luciferin substrate. Results are expressed as percentage of control (n ≥ 3; control = 100% = full-length –706/+64 POMC promoter). AP < 0.01 for deleted or mutated POMC promoter constructs (constructs J and K) versus full-length POMC promoter (black bar). NT, vehicle-treated control.
LIF-induced c-fos and JunB transcription is mediated through STAT3.
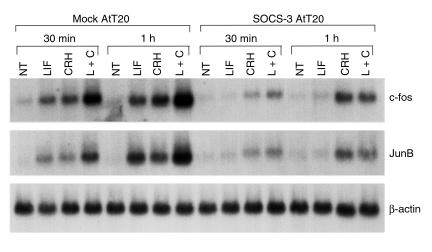
Because the JAK2-STAT3 pathway was shown to be critical for corticotroph LIF function (18), and as c-fos and JunB promoters also contain STAT3 binding sites (28, 29), we explored whether LIF-induced c-fos and JunB transcription was STAT3 dependent (Figure 7). Indeed, disruption of LIF-induced STAT3 activation in SOCS-3 overexpressing cells resulted in abrogation of LIF-induced, but not of CRH-induced, c-fos and JunB mRNA expression, whereas LIF effects in mock-transfected cells were similar to those observed in wild-type nontransfected cells (Figure 7).
Figure 7.
Blockade of LIF-induced STAT3 in SOCS-3–overexpressing AtT20 cells abrogates LIF stimulation of c-fos and JunB transcription. A total of 10 μg total RNA extracted from 1 nM LIF–, 10 nM CRH–, or LIF+CRH–treated mock or SOCS-3–transfected AtT20 cells for 30 minutes or 1 hour were separated on a 1% agarose/formaldehyde gel and transferred to nitrocellulose membrane. Hybridization was performed with a 32P-labeled double-stranded DNA probe corresponding to the c-fos, JunB, or β-actin coding sequence, as described previously (23). NT, vehicle-treated control.
Discussion
Our previous observations demonstrated a critical role for STAT3 in LIF corticotroph action (18). Indeed, overexpression of dominant negative STAT3 forms abrogated LIF-induced POMC promoter activity, POMC mRNA expression and ACTH secretion (18). Interestingly, all studies on POMC gene regulation have demonstrated regulation through the exon-1 AP-1 site via cAMP/CREB and c-fos pathways (12–14), or through the Nurr nuclear receptor superfamily of transcription factors (15–17). Therefore, POMC gene regulation was ascribed to activation of cAMP- or calcium-dependent transcription factors. Thus, the novel regulation of POMC gene expression through a STAT3-dependent mechanism shown here provides new mechanistic insights for HPA axis stimulation by inflammatory cytokines, such as LIF, which act through the JAK-STAT3 pathway. STAT3 regulation of POMC promoter activity is shown here to depend on both direct STAT3 binding to specific POMC promoter sequences and on indirect STAT3 stimulation of c-fos and JunB that bind to the exon-1 AP-1 binding element. Identification of a STAT3 DNA-binding sequence on the POMC promoter is of particular interest because no classical consensus STAT3 DNA-binding element is apparent on this promoter. Progressive deletions of POMC promoter proved useful for delineation of this LIF-responsive region. Interestingly, two juxtaposed STAT3-related DNA-binding elements were identified and shown to bind cooperatively STAT3 with high affinity. However, no STAT3 DNA-binding activity could be observed when each sequence was independently used as a probe in EMSA. In competition experiments, we show that each single sequence was not as powerful as the combination of both sequences for inhibition of LIF-induced STAT3 complex formation to the –407/–374 probe. We therefore conclude that these STAT3-related DNA-binding elements correspond to low-affinity binding sites for STAT3 when isolated or separate, but to a high-affinity binding site for STAT3 when combined. Interestingly, previous studies have shown that STAT3 binds to two juxtaposed DNA-binding elements as a tetramer (dimer-dimer formation), with each dimer binding symmetrically to each element (30, 31). Cooperative binding on clustered low-affinity nonconsensus STAT boxes have indeed been described for STAT5 binding as tetramers on CIS and IL-2 receptor α promoters (30, 31), resulting in high-affinity and high-specificity binding of STAT proteins with DNA. STAT dimer-dimer formation (tetramerization) on tandem STAT DNA binding sites is mediated through interactions between the NH2-terminal coiled-coil domain of each STAT dimer (32). The three-dimensional structure of these cooperative interactions between a STAT tetramer and adjacent STAT DNA-binding sites has been described with crystallization of the tyrosine phosphorylated STAT3 dimer bound to DNA (33).
Mutation of the POMC promoter STAT3 DNA-binding sequence –399/–379 significantly attenuated (∼50%) LIF-induced POMC promoter activity. However, our previous results showed that inactivation of STAT3 completely abrogated LIF corticotroph effects (18). We therefore investigated whether another POMC promoter sequence could account for those STAT3-mediated LIF effects. The IL-6 cytokine family (19), to which LIF belongs, also stimulates AP-1 transcription factor expression, including c-fos or JunB (26–29). Furthermore, c-fos and JunB promoters contain functional STAT3 binding sites (28, 29). Interestingly, the functional AP-1 binding site in the POMC exon-1 promoter binds and transduces transcriptional activity not only of CRH-induced CREB (12), but also of CRH-induced c-fos and JunB (13, 14). Although we previously showed that LIF does not affect CREB activity (11), we demonstrated here that LIF, in a STAT3-dependent manner, induces c-fos and JunB mRNA expression and binding to the POMC promoter exon-1 AP-1 site. Deletion or mutation of this AP-1 site showed this site to be functionally relevant and to account for approximately 20% of LIF corticotroph function. Interestingly, LIF and CRH have additive effects on c-fos and JunB mRNA expression and binding to the POMC promoter exon-1 AP-1 site. This result suggests that two different and independent routes transduce the action of both peptides. Overexpression of SOCS-3 in AtT20 cells, which, as we described previously (22, 23), directly inhibits LIF-induced JAK2 kinase activity and therefore LIF-induced gp130 and STAT3 tyrosine phosphorylation, resulted in abrogation of LIF induction of c-fos and JunB mRNA expression. We therefore conclude that independent mechanisms for LIF and CRH induction of these genes involve a STAT3-mediated pathway for LIF and cAMP or calcium-dependent mechanisms for CRH (12, 13). Our previous findings that LIF did not significantly induce c-fos mRNA were based on very short term cell treatment with LIF (15 minutes) (11). Our current data confirm that significant induction of c-fos is consistent only after 30 minutes of LIF stimulation whereas CRH-mediated action is already significant at 15 minutes (data not shown).
In the anterior pituitary, the stimulatory effect of CRH on POMC gene transcription appears to be mediated also through cAMP or calcium-dependent activation of Nur77, Nurr1, and NOR-1 (15–17). These proteins correspond to orphan members of the nuclear receptor superfamily that bind as monomers to the cis-acting sequence AAAGGTCA. Nurr subfamily members are also implicated at multiple levels of the HPA axis. Indeed, Nur77 expression is strongly induced by a variety of stress stimuli in CRH-producing neurons of the hypothalamic paraventricular (34). Stress-induced signals also stimulate Nurr factor expression in the pituitary and the adrenal cortex (35, 36). Interestingly, Nurr1/Nur77 bind to the –70/–63 region of the POMC promoter, which also recognizes the glucocorticoid receptor. Thus, negative-feedback regulation of POMC transcription by glucocorticoids appears to be, at least in part, exerted on the Nurr signaling pathway by direct competition between Nurr transcription factors and glucocorticoid receptor for the –70/–63 element of the POMC promoter (37). Surprisingly, Nurr nuclear receptors also bind as Nur77 homo- or heterodimers to the POMC promoter sequence –404/–383 (16, 17), which partially overlaps with the –399/–379 POMC promoter STAT3 DNA-binding element identified here. It was therefore of particular interest to investigate whether Nurr nuclear receptors constituted potential targets for LIF action on the POMC promoter. However, we did not observe Nurr transcriptional activation by LIF. Nevertheless, as CRH-induced Nurr and LIF-induced STAT3 bind to overlapping sequences on the POMC promoter, an interaction between these transcription factors on these sequences remains to be explored. LIF-induced STAT3 binding to the POMC promoter –399/–379 region might also interfere with CRH-induced Nurr binding to the –404/–383 element or CRH-induced Nurr binding to the –404/–383 element might interfere with LIF-induced STAT3 binding to the –339/–379 region. This might result in privileging STAT3 and Nurr to use their respective alternative pathways for inducing POMC transcription, i.e., STAT3-mediated induction of c-fos and JunB expression, and Nurr binding to the POMC promoter –70/–63 element. These hypotheses are currently being tested.
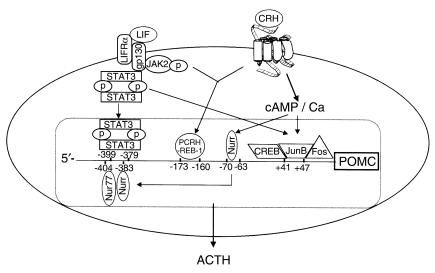
On the basis of these and previous results, we propose a model (Figure 8) in which the molecular basis for LIF and CRH synergy on the POMC promoter is defined as an integration of both independent and cooperative effects of both peptides on corticotroph function: first, LIF directly induces STAT3 binding to the POMC promoter, predominantly accounting for LIF-induced POMC promoter activity. Second, CRH induces CREB binding on the exon-1 AP-1 site, and Nurr to the glucocorticoid receptor responsive element (–70/–63). Third, two common elements on the POMC promoter mediate LIF and CRH synergy: the exon-1 AP-1 site, which additively binds both LIF and CRH-induced c-fos and JunB, and the –173/–160 element that we and others previously reported to correspond to a metallothionein responsive element (11, 38). The pro-opiomelanocortin corticotropin-releasing hormone responsive element binding protein 1 (PCRH-REB-1) was shown to bind to this sequence (38). Finally, the overlapping POMC promoter DNA-binding regions for Nurr and STAT3 might also correspond to common elements transducing LIF and CRH synergy. Further studies are required to explore putative dynamic interactions between Nurr nuclear receptors and STAT3, and their respective binding on overlapping DNA-binding elements within the –404/–383 and –399/–379 regions of the POMC promoter.
Figure 8.
Model for LIF and CRH synergistic induction of POMC promoter activity. The molecular basis for LIF and CRH synergy on the POMC promoter is defined as an integration of both independent and cooperative effects of both peptides on corticotroph function: First, LIF directly induces STAT3 binding to the POMC promoter on the –399/–379 region. Second, CRH induces CREB binding on the exon-1 AP-1 site (+41/+47), and Nurr binding to the glucocorticoid receptor responsive element (–70/–63) or to the –404/–383 element. Third, two common elements on the POMC promoter mediate LIF and CRH synergy: the exon-1 AP-1 site, which additively binds both LIF and CRH-induced c-fos and JunB, and the –173/–160 element, which corresponds to a metallothionein responsive element and binds PCRH-REB-1.
Demonstration of direct STAT3 binding to the POMC promoter represents a novel and powerful mechanism for regulation of pituitary corticotroph function, especially for proinflammatory systemic, hypothalamic or hypophyseal cytokines that stimulate the JAK-STAT pathway. As LIF injections induce pituitary POMC and stimulate circulating ACTH levels in vivo, and these responses are abrogated in the stressed LIF–/– knock-out mouse (5), the results shown here provide a molecular mechanism for cytokine-mediated HPA axis activation. Thus, the immuno-neuroendocrine interface is mediated by powerful synergistic interactions of both CRH and LIF occurring directly at the level of POMC transcription.
Acknowledgments
This study was supported by NIH grant R01-DK50238 and the Doris Factor Molecular Endocrinology Laboratory.
References
- 1.Auernhammer CJ, Melmed S. Leukemia-inhibitory factor-neuroimmune modulator of endocrine function. Endocr Rev. 2000;21:313–345. doi: 10.1210/edrv.21.3.0400. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Ren SG, Melmed S. Hypothalamic and pituitary LIF gene expression in-vivo: a novel endotoxin-inducible neuroendocrine interface. Endocrinology. 1996;137:2947–2953. doi: 10.1210/endo.137.7.8770918. [DOI] [PubMed] [Google Scholar]
- 3.Shimon I, Yan X, Ray DW, Melmed S. Cytokine-dependent gp130 receptor subunit regulates human fetal pituitary adrenocorticotropin hormone and growth hormone secretion. J Clin Invest. 1997;100:357–363. doi: 10.1172/JCI119541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akita S, et al. Human and murine pituitary expression of LIF: novel intrapituitary regulation of adrenocorticotropin synthesis and secretion. J Clin Invest. 1995;95:1288–1298. doi: 10.1172/JCI117779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesnokova V, Auernhammer CJ, Melmed S. Murine LIF gene disruption attenuates the hypothalamo-pituitary-adrenal axis stress response. Endocrinology. 1998;139:2209–2216. doi: 10.1210/endo.139.5.6016. [DOI] [PubMed] [Google Scholar]
- 6.Ren SG, Seliktar J, Li X, Braunstein GD, Melmed S. Measurement of leukemia inhibitory factor in biological fluids by radioimmunoassay. J Clin Endocrinol Metab. 1998;83:1275–1283. doi: 10.1210/jcem.83.4.4702. [DOI] [PubMed] [Google Scholar]
- 7.Akita S, Conn PM, Melmed S. Leukemia inhibitory factor (LIF) induces acute adrenocorticotrophic hormone (ACTH) secretion in fetal rhesus macaque primates: a novel dynamic test of pituitary function. J Clin Endocrinol Metab. 1996;81:4170–4178. doi: 10.1210/jcem.81.11.8923879. [DOI] [PubMed] [Google Scholar]
- 8.Yano H, Readhead C, Nakashima M, Ren SG, Melmed S. Pituitary-directed leukemia inhibitory factor transgene causes Cushing’s syndrome: neuro-immune endocrine modulation of pituitary development. Mol Endocrinol. 1998;12:1708–1720. doi: 10.1210/mend.12.11.0200. [DOI] [PubMed] [Google Scholar]
- 9.Ray DW, Ren SG, Melmed S. Leukemia inhibitory factor (LIF) stimulates proopiomelanocortin (POMC) expression in a corticotroph cell line. Role of STAT pathway. J Clin Invest. 1996;97:1852–1859. doi: 10.1172/JCI118615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 11.Bousquet C, Ray DW, Melmed S. A common proopiomelanocortin element mediates LIF and CRH transcriptional synergy. J Biol Chem. 1997;272:10551–10557. doi: 10.1074/jbc.272.16.10551. [DOI] [PubMed] [Google Scholar]
- 12.Boutillier AL, Gaiddon C, Lorang D, Roberts JL, Loeffler JP. Transcriptional activation of the proopiomelanocortin gene by cyclic AMP-responsive element binding protein. Pituitary. 1998;1:33–43. doi: 10.1023/a:1009966808106. [DOI] [PubMed] [Google Scholar]
- 13.Boutillier AL, et al. Corticotropin-releasing hormone stimulates proopiomelanocortin transcription by cfos-dependent and -independent pathways: characterization of an AP1 site in Exon 1. Mol Endocrinol. 1995;9:745–755. doi: 10.1210/mend.9.6.8592520. [DOI] [PubMed] [Google Scholar]
- 14.Autelitano DJ, Cohen DR. CRF stimulates expression of multiple fos and jun related genes in the AtT-20 corticotroph cell. Mol Cell Endocrinol. 1996;119:25–35. doi: 10.1016/0303-7207(96)03791-4. [DOI] [PubMed] [Google Scholar]
- 15.Murphy EP, Conneely OM. Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol Endocrinol. 1997;11:39–47. doi: 10.1210/mend.11.1.9874. [DOI] [PubMed] [Google Scholar]
- 16.Philips A, et al. Novel dimeric Nurr77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the nurr subfamily of orphan receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bousquet C, Melmed S. Critical role for STAT3 in murine pituitary ACTH leukemia inhibitory factor (LIF) signaling. J Biol Chem. 1999;274:10723–10730. doi: 10.1074/jbc.274.16.10723. [DOI] [PubMed] [Google Scholar]
- 19.Hirano T, Nakajima K, Hibi M. Signaling mechanisms through gp130: a model of the cytokine system. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima K, et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bousquet C, Susini C, Melmed S. Inhibitory roles for SHP-1 and SOCS-3 following pituitary proopiomelanocortin by leukemia inhibitory factor. J Clin Invest. 1999;104:1277–1285. doi: 10.1172/JCI7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auernhammer CJ, Chesnokova V, Bousquet C, Melmed S. Pituitary corticotroph SOCS-3: novel intracellular regulation of leukemia-inhibitory factor-mediated proopiomelanocortin gene expression and adrenocorticotropin secretion. Mol Endocrinol. 1998;12:954–961. doi: 10.1210/mend.12.7.0140. [DOI] [PubMed] [Google Scholar]
- 24.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci USA. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 26.Botelho FM, Edwards DR, Richards CD. Oncostatin M stimulates c-Fos to bind a transcriptionally responsive AP-1 element within the tissue inhibitor of metalloproteinase-1 promoter. J Biol Chem. 1998;273:5211–5218. doi: 10.1074/jbc.273.9.5211. [DOI] [PubMed] [Google Scholar]
- 27.Symes A, Gearan T, Eby J, Fink S. Integration of Jak-Stat and AP-1 signaling pathways at the vasoactive intestinal peptide cytokine response element regulates ciliary neurotrophic factor-dependent transcription. J Biol Chem. 1997;272:9648–9654. doi: 10.1074/jbc.272.15.9648. [DOI] [PubMed] [Google Scholar]
- 28.Kojima H, Nakajima K, Hirano T. IL-6-inducible complexes on an IL-6 response element on the JunB promoter contain STAT3 and 36 kDa CRE-like site binding protein(s) Oncogene. 1996;12:547–554. [PubMed] [Google Scholar]
- 29.Rajotte D, et al. Contribution of both STAT and SRF/TCF to c-fos promoter activation by granulocyte-macrophage colony-stimulating factor. Blood. 1996;88:2906–2916. [PubMed] [Google Scholar]
- 30.Verdier F, et al. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer WK, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor alpha gene transcription. J Biol Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 32.Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 33.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 34.Parkes D, Rives S, Lee S, Rivier C, Vale W. Corticotropin-releasing factor activates c-fos, NGFI-B, and corticotropin-releasing factor gene expression within the paraventricular nucleus of the rat hypothalamus. Mol Endocrinol. 1993;7:1357–1367. doi: 10.1210/mend.7.10.8264665. [DOI] [PubMed] [Google Scholar]
- 35.Honkaniemi J, Kononen J, Kainu T, Pyykonen I, Pelto-Huikko H. Induction of multiple immediate early genes in rat hypothalamic paraventricular nucleus after stress. Brain Res. 1994;25:234–241. doi: 10.1016/0169-328x(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 36.Chan RKW, Brown ER, Ericsson A, Kovacs KJ, Sawchenko PE. A comparison of two immediate early genes, c-fos and NGFI-B, as markers for functional activation in stress-related neuroendocrine circuitry. J Neurosci. 1993;13:5126–5138. doi: 10.1523/JNEUROSCI.13-12-05126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philips A, et al. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol Cell Biol. 1997;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin WD, et al. Characterization of a corticotropin-releasing hormone-responsive element in the rat proopiomelanocortin gene promoter and molecular cloning of its binding protein. Mol Endocrinol. 1994;8:1377–1388. doi: 10.1210/mend.8.10.7854355. [DOI] [PubMed] [Google Scholar]