Abstract
F-box proteins (FBPs) are substrate-recruiting subunits of Skp1-cullin1-FBP (SCF)-type E3 ubiquitin ligases. To date, 69 FBPs have been identified in humans, but ubiquitinated substrates have only been identified for a few, with the majority of FBPs remaining ‘orphans’. In recent years, a growing body of work has identified non-canonical, SCF-independent roles for about 12% of the human FBPs. These atypical FBPs affect processes as diverse as transcription, cell cycle regulation, mitochondrial dynamics and intracellular trafficking. Here, we provide a general review of FBPs, with a particular emphasis on these expanded functions. We review Fbxo7 as an exemplar of this special group as it has well-defined roles in both SCF and non-SCF complexes. We review its function as a cell cycle regulator, via its ability to stabilize p27 protein and Cdk6 complexes, and as a proteasome regulator, owing to its high affinity binding to PI31. We also highlight recent advances in our understanding of Fbxo7 function in Parkinson's disease, where it functions in the regulation of mitophagy with PINK1 and Parkin. We postulate that a few extraordinary FBPs act as platforms that seamlessly segue their canonical and non-canonical functions to integrate different cellular pathways and link their regulation.
Keywords: Fbxo7/PARK15, F-box protein, ubiquitin, E3 ligase, Parkinson's disease, mitophagy
2. Introduction
As with actors on a stage, the timely exit of cellular proteins is as important as their entrance; they must play their part at the appropriate moment and then depart on command. The ‘exit’ or destruction of proteins within the cell goes beyond the simple removal of proteins; it provides a means to achieve rapid activation, by the degradation of an inhibitor, or conversely, the inactivation of a given process faster than could be achieved by the synthesis of new inhibitor proteins. Furthermore, degradation is irreversible, imparting a unidirectionality that is absolutely fundamental to basic processes like the cell cycle, which requires the coordinated degradation of kinase inhibitors and activating cyclins, helping to ensure a single and complete round of replication of the genome [1,2].
The degradation of cellular proteins is not random but is directed by signalling pathways and carried out by the ubiquitin proteasome system (UPS) [3–5]. Proteins are marked for degradation by an initial post-translational modification or a series of modifications, such as phosphorylation, creating a ‘degron’ to which ubiquitin ligases are recruited to label targeted proteins with ubiquitin. These small, 8.5 kDa proteins, when attached as polymer chains, can direct proteins to proteasomes, large multi-subunit complexes with multiple proteolytic activities. Proteasomes essentially ‘recycle’ proteins by breaking them into short seven to eight amino acid polypeptides that are further broken down to their composite amino acids to be re-used or catabolized by the cell. The ubiquitination reaction itself is a well-defined process mediated by a cascade of ubiquitin-handling enzymes and has been reviewed in detail elsewhere [3,6–8]. The final step in this process is orchestrated by an E3 ubiquitin ligase. Its role is to recognize and bind degrons within a target protein and bring it into proximity with a ubiquitin-charged E2 protein, stimulating transfer of a ubiquitin moiety to recipient lysine residues within the target.
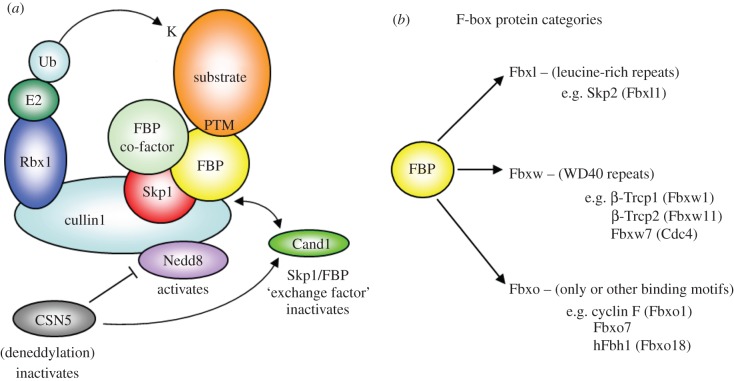
The array of targets regulated by E3 ubiquitin ligases is as broad as the proteome itself. One strategy the cell employs to handle the magnitude of this task is to express a large number of different E3 ligases. Indeed, the human genome encodes over 500 distinct E3s [9], roughly separated across two main families; the homologous to E6-associated protein (E6-AP) C-terminus (HECT) domain and the really interesting new gene (RING) finger domain E3s [3]. Another strategy that goes beyond merely increasing E3 numbers is to employ adaptor proteins that change the substrate specificity of the E3, which may enable a tailoring of substrate engagement with a ligase as per the changing needs of the cell. This strategy is typified by the Skp1-cullin1-F-box protein (SCF)-type E3 ubiquitin ligases, the largest group of multi-subunit E3 ligases within the RING finger domain family [5,10,11]. The F-box protein (FBP) family is fundamental to this flexible substrate-recognition, as they act as interchangeable docking sites for the ligase. The SCF holoenzyme is formed around a central cullin (Cul1) backbone, which provides a rigid scaffold, holding the E2 binding subunit, Rbx1, at a distance of approximately 50 Å from the substrate docking site (figure 1a) [8]. Substrate recruitment is the role of the FBP, and its tethering to cullin is mediated by Skp1.
Figure 1.
F-box proteins. (a) Schematic of active SCF complexes, which are neddylated (Nedd8 moiety attached to cullin1). FBPs can bind to the SCF holoenzyme alone, with accessory cofactors, or as a homo- or heterodimer allowing for dimeric SCF complex formation. The SCF complex is orientated so the lysine residue (K) in the substrate, usually recruited by FBPs after being PTM, is in close proximity to the ubiquitin moiety on the E2 enzyme. SCF complexes can be inactivated by deneddylation, at which point Cand1 can compete with Skp1/FBP for binding to cullin1, allowing for exchange of SCF subunits. (b) There are three classes of FBP, which are listed, along with examples from each group.
In this review, we present a brief outline of the function and diversity of the FBP family, describing how they operate within the UPS system as conventional components of SCF-type E3 ubiquitin ligases. We will then discuss an array of alternative functions displayed by FBPs, as many are being reported to lead ‘double-lives’, operating outside the UPS system and independently of the SCF to regulate a diverse range of cellular processes. Lastly, we will focus on Fbxo7, an FBP implicated in cancer and neurodegenerative disease, whose atypical functions include regulating cell cycle, differentiation, proteasomal function and mitochondrial quality control. As illustrated by the exemplary case of Fbxo7, we propose that having both ubiquitination functions and non-canonical regulatory roles endows a subset of special FBPs with the potential to act as hubs to link the UPS to other cellular signalling networks.
3. F-box proteins
FBPs are defined by a 40–50 amino acid F-box domain that binds Skp1 [12]. After the discovery of the first FBP, cyclin F (Fbxo1) [13], further family members were identified using a yeast-two-hybrid screen for Skp1-interacting proteins and bioinformatics analysis for homologous sequences [14,15]. Without taking into account the various isoforms that may be produced, there are 69 human FBPs [16], and the number is significantly higher in other organisms, such as Caenorhabditis elegans and plants. FBPs are subdivided into three separate classes, Fbxw, Fbxl and Fbxo, based on their individual complement of protein interaction domains: WD40, leucine-rich repeats (LRR) and ‘other’, respectively (figure 1b) [17]. These three classes did not diverge from a single common ancestral gene, as might be expected. Instead the F-box phylogenetic tree is made up of two main groups with FBPs from all classes found in both, suggesting frequent swapping of protein interaction domains during their evolution [17]. The WD40 and LRR domains of Fbxw and Fbxl family members fold to create large surface areas for protein–protein interaction. WD40 repeats form a circularized propeller-like structure [18], while the successive repeats of LRR domains stack in a horseshoe formation [19]. The Fbxo class, on the other hand, features a broad array of different interaction domains, including in between-ring domains (IBR), TRAF-domain like motifs and proline-rich regions (PRR), among others. Although these domains from all classes of FBPs directly recruit substrates, in some cases an additional cofactor is essential or can change the substrate specificity of the FBP. For example, Fbxl1 (Skp2) requires Cks1 to bind the Cdk inhibitor p27 or the pRb-related protein, p130 for ubiquitination [20,21]. However, Cks1 is not needed for the recruitment of other Skp2 substrates.
Quite how the most appropriate FBPs are ‘selected’ and loaded onto a cullin scaffold is not fully understood, but it is thought to be regulated by two processes: availability and exchange. The pool of available FBPs is likely to be controlled by a combination of transcriptional regulation, targeted stabilization/degradation and auto-ubiquitination in the absence of their substrates [22,23]. For example, the muscle catabolism regulator, Atrogin-1 (Fbxo32) is controlled transcriptionally. Its expression is enhanced under starvation conditions, promoting muscle atrophy, but is suppressed by testosterone, stimulating increased muscle mass [24]. On the other hand, the putative tumour suppressor, Fbxo31, is regulated post-translationally. It is stabilized in response to DNA damage and targets ATM-phosphorylated cyclin D1 for UPS-dependent degradation, thus bringing about a G1 cell cycle arrest [25]. Auto-ubiquitination in the absence of a suitable substrate is a process thought to be very common among FBPs. It provides a means by which ‘idle’ FBPs target themselves for UPS-dependent degradation. This is typified by β-Trcp2 (Fbxw11), a regulator of the Wnt and NF-κB signalling pathways, which is intrinsically unstable under basal conditions but has been shown to be stabilized by the induction of one of its substrates, the phosphorylated form of inhibitor of NF-κB alpha (IκBα) [26]. The variety of these mechanisms provides the means to upregulate required FBPs in response to intrinsic and extrinsic signals, while continually clearing those that are no longer required, ensuring that the available pool addresses the needs of the cell at any given time. The mechanism by which one FBP is ‘switched-out’ and replaced by another has recently been reported and is controlled by the ‘exchange factor’ Cand1 [27,28]. By increasing the dissociation rate of Skp1 : FBP complexes from the cullin scaffold, without affecting the kinetics of its reassembly, Cand1 keeps the spectrum of SCF activity within the cell dynamic and capable of responding to a changing cellular environment.
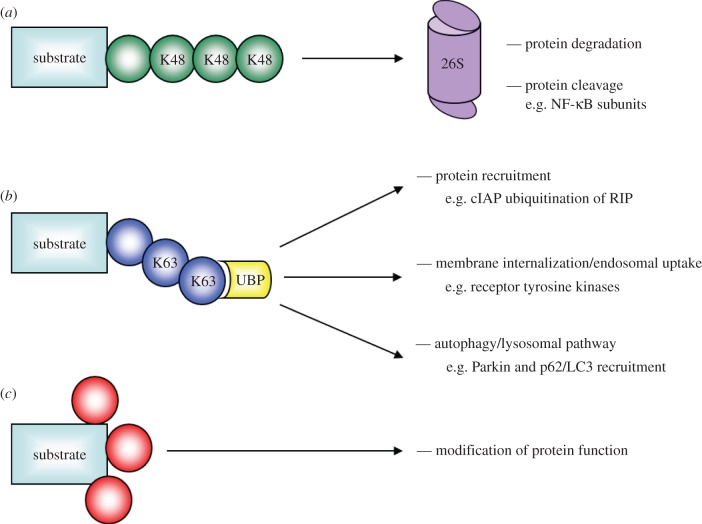
FBPs interact with and ubiquitinate their own particular panel of substrates, usually showing a preference for post-translationally modified (PTM), often phosphorylated, proteins. In this way, signal transduction networks that use protein kinases (e.g. GSK3β, Cdks, IKK) can engage a UPS response. Recognition by an FBP can be via a single PTM or cumulative PTMs of the substrate on multiple sites. This latter scenario, which creates a switch that senses and responds to a threshold of modifications, is used by the yeast Cdk inhibitor, Sic1. Phosphorylation on any six of its nine phospho-acceptor sites stimulates its ubiquitination by SCFCdc4 as part of the transition from G1 to S phase [29]. Thus as part of an SCF, FBPs ‘translate’ a PTM from upstream signalling pathways into a ubiquitination response, and that signal can be further acted upon and/or stratified. Ubiquitin can be conjugated as individual moieties to single or multiple sites within a protein (mono and multi-monoubiquitination, respectively) or attached as a polyubiquitin chain (figure 2). Such chains can differ in both length and topology, which forms the basis of the ‘ubiquitin code’ (reviewed in [30]). Ubiquitin contains eight residues (Met1, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) that are capable of forming an isopeptide bond with the C-terminal glycine residue (Gly76) of the preceding ubiquitin molecule in a nascent chain. Chains can be constructed using either a single type of linkage (homogeneous), several types (mixed) or be branched. The ‘deciphering’ of this code is due to the fact that different ubiquitin chain topologies recruit distinct ubiquitin binding proteins (UBPs) with activities that bring about changes to protein activity, location and/or levels [30]. The two best understood types of polyubiquitin chain linkage are those assembled on Lys48 or Lys63. Lys48 polyubiquitination is the most common linkage used in mammalian cells and is almost exclusively associated with the targeting of proteins for proteasomal degradation. It is the only essential lysine residue in yeast ubiquitin [31]. By contrast, Lys63 chains are generally not associated with UPS-based proteolysis but do play a major role in regulating the destruction of proteins and organelles by the lysosomal/autophagy pathway [32]. They are also used in other signalling networks, including the DNA damage response and the NF-κB signal transduction pathway [33,34]. In these instances, they are used to build scaffolds or ‘platforms’ for protein recruitment via UBPs.
Figure 2.
Common ubiquitin linkages and their consequences. (a) Polyubiquitin chains linked via lysine 48 (K48) to the preceding ubiquitin molecule usually target proteins for degradation via the 26S proteasome. Poly-K48 chains can also result in proteasome-mediated protein cleavage such as in the case of the NF-κB subunits, p100 and p105, being processed into p52 and p50, respectively. (b) Polyubiquitination via lysine 63 (K63) is also common. This type of ubiquitination usually acts as a scaffold for UBPs that recruit other protein complexes, and also mediates alternate outcomes such as changes to cell signalling (e.g. ubiquitinated RIP can recruit NF-κB signalling pathway components), activation of the endosomal pathway (e.g. resulting in internalization of membrane proteins such as receptor tyrosine kinases) and activation of the autophagic/lysosomal pathway (e.g. resulting in degradation of target proteins and organelles independently of the proteasome). (c) Monoubiquitination, or multi-monoubiquitination, can result in changes to substrate function, localization or protein binding.
The ability of an E3 enzyme to conjugate one type of polyubiquitin chain or another on its substrate is largely dictated by the spectrum of E2 enzymes with which it interacts [30,35,36]. With few exceptions, this appears to limit SCF-type E3 complexes to participating almost exclusively in the production of Lys48 polyubiquitin chains, thus promoting degradation. However, in some cases F-box-dependent Lys48 modification does not lead to complete proteolysis. SCFβ-Trcp-induced poly-Lys48 ubiquitination can stimulate the 26S proteasome-dependent processing of the NF-κB transcription factors p100 and p105 to p52 and p50, respectively [37–39]. Interestingly, a growing number of reports have shown that FBPs are capable of participating in a broader range of ubiquitin–conjugation reactions. For example, Chen et al. [40] have shown that monoubiquitination of CTP : phosphocholine cytidylyltransferase (CCTα) by Fbxl2 targets it for endosome-lysosomal degradation. Another study has also suggested that β-Trcp has the capacity to direct Lys63 as well as Lys48 polyubiquitination of the interferon α/β receptor 1, when paired with an appropriate E2 enzyme in vitro [41].
In sum, through recognition of PTMs in its substrates, FBPs can use the SCF machinery to codify a ubiquitin-based response thus relaying and diversifying a signal into downstream cellular pathways.
4. SCF-independent functions of F-box proteins
The study of FBPs to date has focused on their SCF-dependent functions, even though the majority of FBPs in humans and other species remain ‘orphans’. The identification of substrates for these orphans remains a major endeavour for the field. However, an additional, perhaps under-appreciated consideration is that the many FBPs within a cell must compete for binding to the cullin scaffold and consequently may not be able to participate readily in ubiquitination reactions. It is therefore conceivable that these unengaged subunits may be free to participate in other reactions. In yeast, several FBPs have been found bound to Skp1, but not as part of an SCF complex. Instead FBP–Skp1 dimers participate in processes like centromere complex assembly and the recycling of endosome components (reviewed in [42]). Although the first descriptions of SCF-independent functions were in yeast, such roles for mammalian FBPs have also been reported [43]. For example, Emi1 (Fbxo5) functions as an SCF-independent suppressor of APC/C activity. Emi1 negatively regulates APC/C activity by binding to its activators, Cdc20 and Cdh1, which recruit APC/C substrates [44,45]. Thus, Emi1 prevents DNA re-replication and helps to link DNA replication to mitosis (reviewed in [44]).
The presence of an F-box domain itself is no guarantee that a protein will function as part of an SCF. Fbxo38 (MoKA) uses its F-box domain to interact with Kruppel-like transcription factor 7 (Klf7) [46]. Klf7 plays a key role in the development of the mammalian central nervous system by regulating differentiation, and maintaining cell cycle arrest, of post-mitotic neuro-progenitor cells [47]. Fbxo38 supports Klf7 in this role by acting as a transcriptional cofactor at the promoter of the cell cycle inhibitor p21WAF1/Cip1 [46]. Although it can bind to Skp1, to date an SCF-dependent role for Fbxo38 has yet to be identified. Moreover, this study raises the possibility that other FBPs might also use their F-box domains in transcriptional regulation.
Alternative functions for FBPs are not limited to simple binding interactions with other proteins, as they may also possess distinct and intrinsic enzymatic activities. hFbh1 (Fbxo18) has been shown to operate as a DNA helicase and is important for the maintenance of genomic stability through regulating homologous recombination [48,49]. The purified hFbh1 protein has DNA helicase activity, and strikingly this capacity is maintained when it is part of an SCF ligase [50]. However, as hFbh1 is an orphan FBP, the biological significance of linking ubiquitination activity to a helicase remains to be determined [49].
These examples illustrate that several FBPs, to date about 12% and usually of the Fbxo class, have activities beyond ubiquitination (table 1). We speculate that this class of special FBPs provides a means to link modification with ubiquitin to other enzymatic or functional interactions. Below, we will expand on Fbxo7 as a case in point for such FBPs, as it has well-defined SCF-dependent and independent activities; and furthermore, it is important in human health, having been linked to two diseases, cancer and Parkinson's disease (PD), and to alterations in red blood cell parameters.
Table 1.
FBPs with SCF-independent functions (abbreviations are Saccharomyces cerevisiae, Caenorhabditis elegans, Schizosaccharomyces pombe and Fusarium oxysporum).
| FBP | organism | function | references | additional SCF function |
|---|---|---|---|---|
| cell cycle regulation | ||||
| Fbxo7 | mammals | cell cycle, proteasome and mitophagy regulator | [51,52] | yes |
| Emi1 (Fbxo5) | mammals | suppressor of APC/C activity | [44,45,53] | possible |
| Emi2 (Fbxo43) | mammals | suppressor of APC/C activity | [54] | unknown |
| Cyclin F (Fbxo1) | mammals | promotes nuclear localization of cyclin B1 | [55] | yes |
| Ctf13p | S. cerevisiae | structural component of the CBF3 kinetochore complex | [56] | no |
| transcription/translation | ||||
| MoKA (Fbxo38) | mammals | transcriptional cofactor for KLF7 | [46,47] | unknown |
| hFbh1 (Fbxo18) | mammals | DNA helicase | [48–50] | yes |
| KDM2B (Fbxl10) | mammals | histone demethylase | [57] | unknown |
| KDM2A (Fbxl11) | mammals | histone demethylase, inhibitor of NF-κB | [58] | unknown |
| Elongin A | mammals | translation elongation activator | [59,60] | no, but E3 activity via Cul5 |
| FOG-2 | C. elegans | translational repressor, binds GLD-1 | [61] | no |
| intracellular trafficking | ||||
| Roy1/Ymr258c | S. cerevisiae | inhibits Ypt52 and consequently intracellular trafficking | [62] | no |
| Rcy1 | S. cerevisiae | v-SNARE recycling | [63] | no |
| Pof6 | S. pombe | endocytosis, cytokinesis and cell division (Rcy1 homolog) | [64] | yes |
| other | ||||
| Pof14 | S. pombe | inhibits Erg9, a squalene synthase involved in ergosterol synthesis | [65] | yes |
| Mfb1 | S. cerevisiae | mitochondrial morphogenesis - promotes fission | [66] | unknown |
| Mdm30 | S. cerevisiae | mitochondrial morphogenesis - prevents fission | [67] | yes |
| Frp1 | F. oxysporum | FBP required for pathogenicity in tomato wilt disease | [42,68,69] | yes |
5. Fbxo7: gene and protein structure
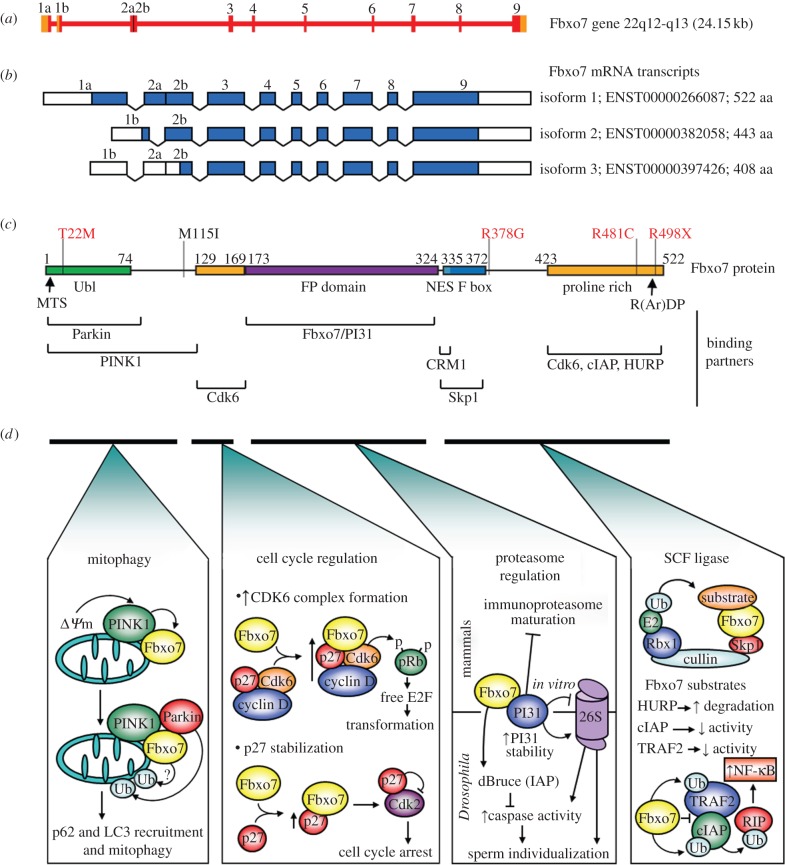
Fbxo7 was first identified as an FBP in 1999 [14,15], and the first report on its function was its canonical ubiquitination of HURP in 2004 [70]. However, a facilitating role for Fbxo7 in promoting cell cycle progression (discussed in §7) was discovered a year later and was the first clue that it had expanded activities [51]. This study also reported the basic domain structure of isoform 1 of Fbxo7 (figure 3c). In addition to the signature F-box domain, it contains a ubiquitin-like (Ubl) domain at its N-terminus and an unstructured PRR, used to recruit substrates, at its C-terminus [51]. While the F-box domain of Fbxo7 is most closely homologous to that of fellow FBPs, Fbxo9 and Fbxo11, its closest relative, proteasome inhibitor 31 (PI31), is in fact, not an FBP at all [71]. PI31 is a regulator of immunoproteasome maturation and an inhibitor of 20S proteasomes in vitro [72,73], and shares a common domain organization with Fbxo7 having a C-terminal PRR, containing a conserved R(Ar)DP motif, which is present in all orthologues of the two proteins [71]. Perhaps more significantly, both proteins share a core globular domain, designated the Fbxo7/PI31 (FP) domain that provides two distinct interaction surfaces that can mediate their homo- or heterodimerization [71]. Their relationship will be discussed in §8, but their common organization and shared domain structure raise the intriguing possibility that Fbxo7 was first a proteasome regulator that later gained the ability to ubiquitinate proteins.
Figure 3.
Human Fbxo7 gene and protein structure. (a) FBXO7 gene organization. Orange and red boxes are untranslated and translated portions of exons, respectively, with exon numbers, as indicated. (b) Protein coding Fbxo7 mRNA transcripts, with Ensembl accession numbers and protein size. (c) Fbxo7 isoform 1 protein structure. Protein binding partners are listed below the domains with which they interact. Pathogenic mutations associated with PD are in red, and SNPs associated with red blood cell parameters in black. MTS, mitochondrial targeting signal; Ubl, ubiquitin-like domain; FP, Fbxo7/PI31 interacting domain; R(Ar)DP motif, where Ar is an aromatic amino acid. (d) Fbxo7 functions described for each domain relative to the protein schematic above. They are subdivided into canonical (SCF ligase) and non-canonical functions (mitophagy, cell cycle regulation and proteasome regulation). See text for details.
The FBXO7 gene is located on chromosome 22q12-q13 in humans and is composed of nine exons spanning a region of approximately 24.15 kb (figure 3a). From this, 10 transcripts have been identified and annotated by Ensembl [74], as well as a further two non-coding transcripts, with differing nomenclature, suggested by Di Fonzo et al. [75]. The Ensembl annotated transcripts include three complete protein coding isoforms (figure 3b), and an additional seven transcripts, which are incomplete, non-coding or presumed to undergo nonsense-mediated decay. The three coding transcripts comprise of isoform 1, which is the most abundantly expressed form of the protein in most tissues and cultured cell lines, and two shorter transcripts, isoforms 2 and 3. Isoform 2 contains exons 3–9 of isoform 1, but has an alternative 5′ exon. This alternative exon, known as 1b, splices directly to exon 2b, skipping exon 2a, and produces a 443 amino acid protein with an entirely different N-terminal end, lacking the Ubl domain seen in isoform 1. This version of the protein is also detectable in mammalian tissues and cell lines. Although less well studied than isoform 1, isoform 2 interacts with some of the known Fbxo7 substrates [51,76,77]. Isoform 3 also contains exons 1b, 2a and 2b; however, protein translation starts from an alternate methionine distinct from that of isoform 2. Intriguingly, the initiating methionine of isoform 3 is located at amino acid 115 (according to isoform 1 numbering) and its coding is affected by a common single nucleotide polymorphism (SNP; rs11107; M115I) that changes the nucleotide sequence from ATG (methionine) to ATA (Isoleucine). This SNP, along with several others in FBXO7, is associated with changes to red blood cell volume in humans [78–80]. Although no evidence as yet has been published showing protein expression of isoform 3, this would suggest that people homozygous for the ATA allele would completely lack expression of this isoform.
6. Fbxo7 as an E3 ubiquitin ligase
Currently, the capacity of Fbxo7 to function as a canonical FBP has been described for three substrates: HURP, cIAP1 and TRAF2 (figure 3d) [70,76,81]. HURP is a cell cycle-regulated protein associated with the mitotic spindle, where it regulates chromosome congression [82,83]. HURP ubiquitination by SCFFbxo7 is preceded by its multisite phosphorylation by cyclin B/Cdk1 [70]. HURP was originally identified as a putative oncogene in hepatocellular carcinoma (HCC) and mechanistically has been shown to be a negative regulator of the tumour suppressor, p53 [84]. This raises the possibility that Fbxo7 could function as a tumour suppressor in HCC by negatively regulating HURP thus bolstering p53 activity, although this has not yet been investigated.
cIAP1 is a member of the IAP family, and it contains a C-terminal RING domain, enabling it to function as an E3 ubiquitin ligase [85]. It is this property of cIAP1 that allows it to inhibit apoptosis, both by targeting pro-apoptotic proteins, such as SMAC/DIABLO, for proteasomal degradation, and by stimulating anti-apoptotic NF-κB activity [85,86]. cIAP1 contributes to NF-κB signalling by forming part of the tumour necrosis factor-receptor signalling complex (TNF-RSC). Here, it interacts with TRAF2, another ubiquitin ligase and together they polyubiquitinate receptor interacting protein 1 (RIP) with Lys63-chains [86,87], producing a platform for the recruitment and activation of the inhibitor kappa B kinase (IKK) signalosome. IKK phosphorylates IκBα, stimulating its degradation and the concomitant release of NF-κB transcription factors into the nucleus. Using an siRNA screen targeting ubiquitin conjugating and de-conjugating enzymes, Kuiken et al. [81] recently identified Fbxo7 as an inhibitor of NF-κB activity, an effect that was mediated by the ubiquitination of cIAP1 and TRAF2. Thus, Fbxo7 has the potential to sever the link between the TNF-RSC core complex and the IKK signalosome, attenuating NF-κB activation. Dysregulation of NF-κB signalling has long been associated with cancer and oncogenesis and more recently has been linked to neurodegeneration and PD (reviewed in [88]). While it is tempting to speculate that the relationship between Fbxo7 and NF-κB signalling may be instrumental to its role in various pathologies, none of the currently known PD-associated Fbxo7 mutants (see §9) appear to affect NF-κB-dependent transcription ex vivo [81], and the ability of Fbxo7 to affect NF-κB signalling in a cancer context has yet to be probed.
A recent quantitative analysis of the Cul1 proteome revealed that SCFFbxo7 was the fifth most abundant SCF ligase in cultured cells [89], placing it in the ranks of FBPs like Skp2, which has at least 26 reported substrates [90], and β-Trcp1 (Fbxw1A), which has 10 verified substrates [91]. As evidenced by these three substrates, Fbxo7 has the potential to impact upon disease-associated signalling pathways, and given the association of Fbxo7 with human diseases, a more complete reckoning of Fbxo7 ubiquitination substrates is eagerly awaited. However, to truly comprehend Fbxo7 function and its role in disease, a full understanding of its alternative activities is also warranted.
7. Fbxo7 as a regulator of the cell cycle
The first SCF-independent function for Fbxo7 was uncovered when the protein was identified as interacting with an oncogenic viral cyclin [51]. Viral cyclins bypass normal cell cycle regulators and robustly activate G1 Cdks, promoting S phase entry [92]. Fbxo7 was found to associate specifically and directly via a bipartite interaction with Cdk6 (figure 3c). In this capacity, it functions as an assembly scaffold for the formation of cyclin D/Cdk6 complexes, rather than causing the ubiquitination of either subunit (figure 3d) [51]. As these G1/S regulators are themselves proto-oncogenes, as a direct positive regulator, it was thought that Fbxo7 could also be a putative oncogene. This was borne out in experiments showing that over-expression of Fbxo7 in mouse fibroblasts triggered changes associated with cellular transformation, including tumour formation in nude mice. In addition, Fbxo7 over-expression was observed in human tumour biopsies from lung squamous cell carcinoma and colon adenocarcinoma but was virtually absent from corresponding normal tissue, suggesting that Fbxo7 may be oncogenic in these tissue types [51].
In a stringent test of its effects on proliferation, differentiation and transformation in primary cells, exogenous Fbxo7 expression was introduced into murine haematopoietic stem and progenitor cells (HSPCs) [93]. Haematopoietic cells were tested because of the selectivity of Fbxo7 for Cdk6, and because of the critical roles for Cdk6 in haematopoiesis: Cdk6 KO mice have thymic and splenic hypoplasia and reduced numbers of erythroid cells [94,95]. In this setting, increased Fbxo7 expression reduced both colony formation and proliferation of WT HSPCs along the granulocyte/macrophage lineages. However, in p53 null HSPCs, Fbxo7 expression enhanced proliferation in a growth factor-dependent manner and was also able to induce the formation of T-cell lymphomas when these cells were used to reconstitute irradiated mice [93]. Although this study did not test for Cdk6 dependence of the transformation, it nonetheless suggested that Fbxo7 has oncogenic capacity, which is held in check by p53.
Surprisingly, Fbxo7 has also been found to bind directly and to stabilize the levels of a second cell cycle regulator that acts at the G1/S transition, the Cdk inhibitor, p27. Cip/Kip inhibitors (p21, p27 and p57) usually function as inhibitors of Cdks, but they can operate as assembly and nuclear import factors for Cdk4 and 6. These competing functions of p27, inhibitor versus assembly factor, were originally postulated to be dependent on the stoichiometry of the Cip/Kip proteins relative to cyclin D/Cdk4/6 complexes [96]. However, more recent data would suggest that tyrosine phosphorylation of p27 converts it from a ‘bound inhibitor’ into a ‘bound non-inhibitory’ assembly factor [97,98]. The ability of Fbxo7 to act as a scaffold for the assembly of cyclin D/Cdk6/p27 complexes raises the possibility that it may facilitate p27 phosphorylation. It was an interaction with p27 that was thought to prevail and ultimately drive the phenotypic effects of Fbxo7 expression in a different haematopoietic cell type. In a separate study, it was shown that reducing Fbxo7 expression increased the proliferation rate in B cells by shortening the duration of the G1 phase of the cell cycle [99]. In this cell type, decreased Fbxo7 appeared to have no effect on the assembly or activity of cyclin D/Cdk6 complexes. Instead, a reduction in p27 levels was observed, along with enhanced Cdk2 activity. This study also demonstrated the ability of Fbxo7 to influence differentiation as well as cell cycle, two process that are thought to be closely coupled, but separable in lymphocytes [100–104]. Expressing Fbxo7 in Ba/F3 cells caused an apparent ‘de-differentiation’, as evidenced by a reversal in the expression of cell surface antigens from a more mature expression state to a less mature one. This effect was separable from the ability of Fbxo7 to regulate the cell cycle as over-expression of p27 alone, while altering cell cycle length, did not affect surface marker expression [99].
Together these studies illustrate how the activities of Fbxo7 are highly context-dependent: Fbxo7 can promote cell cycle entry by driving Cdk6 assembly or inhibit cell cycle progression by stabilizing p27. We speculate that this will be a consequence of the cell cycle regulatory circuitry in individual cell types or its stage of differentiation. As the outcome of Fbxo7 activity cannot be easily predicted by the mere abundance of its targets, we suggest that additional parameters, perhaps in the form of post-translational modifications of p27, Cdk6, Fbxo7 itself or other unknown factors, dictate the outcome of its participation at the G1/S boundary.
8. Fbxo7 as a regulator of proteasome activity
As with all FBPs, the link between Fbxo7 and the UPS could theoretically be limited to its ability to ubiquitinate proteins destined for proteasomal degradation. However, the presence of a Ubl domain in isoform 1 of Fbxo7, a motif commonly found in regulators of the proteasome, and its dimerization with PI31, a known proteasome regulator, suggest that Fbxo7 may itself act as a regulator of the proteasome (figure 3d). Nothing has been reported yet on the Ubl of Fbxo7 with regard to proteasome regulation; however, Fbxo7 binds with high affinity to PI31 [71], which was first characterized biochemically as a simple proteasome inhibitor [105]. PI31 binds the α-subunits of the 20S barrel via a conserved C-terminal HbYX motif, blocking access of substrates to the catalytic channel in vitro [72,106]. However, in intact cells, PI31 apparently does not inhibit proteasome activity but rather regulates maturation of the immunoproteasome, an inducible version of the proteasome with altered and enhanced proteolytic activity [73]. To date, the connection, if any, between PI31 and Fbxo7 in regulating mammalian proteasomal activity has not been published. This may be because in cultured cells, the localization of the two proteins is largely distinct. PI31 has been shown to localize almost entirely within the endoplasmic reticulum, while Fbxo7 is present throughout the cell, shuttling between the nucleus and cytoplasm in a cell cycle-dependent fashion [71,73,107]. At present, it is unclear what proportions of Fbxo7 and PI31 exist in heterodimeric complexes or if this changes in different cell types or culture conditions.
The clearest evidence for a relationship between PI31 and a partial Fbxo7 orthologue, Nutcracker, in the regulation of proteasomes comes from studies in Drosophila [52]. The effects of their loss are seen during ‘individualization’, the final stage of spermatogenesis, which is a process that uses components of the apoptotic system to purge organelles and excess cytoplasmic volume to produce mature sperm [108,109]. Bader et al. [52] found that DmPI31 stability was regulated by Nutcracker, and like their mammalian counterparts, the two proteins interact via their FP domains. This interaction inhibited DmPI31 cleavage, promoting proteasome activity and caspase activation. Intriguingly, the Nutcracker F-box domain was essential for DmPI31 stabilization, even though it did not mediated their interaction and DmPI31 was not found to be a substrate for Nutcracker-dependent ubiquitination.
Nutcracker may also have an additional role to play in maintaining the delicate balance of caspase activity that is required during individualization. In a further parallel with mammalian Fbxo7 biology, Nutcracker also interacts with dBruce, a known regulator of spermatogenesis and a member of the IAP family of apoptosis inhibitors to which the Fbxo7 substrate cIAP1 belongs [108]. dBruce is a giant protein (approx. 500 kDa), with two principal anti-apoptotic activities: its N-terminal BIR domain enables it to function as a caspase inhibitor and its C-terminal UBC domain provides E2 ubiquitin conjugation activity, which has been shown to promote ubiquitination of the pro-apoptotic factor, Reaper [109,110]. Currently, the functional consequences of the Nutcracker : dBruce interaction are not clear. dBruce may indeed be a substrate of SCFNutcracker, but on the other hand it may simply act as an E2 enzyme for the complex in this setting, interacting directly with Nutcracker during ubiquitin transfer. Alternatively, Nutcracker could instead use its interactions with DmPI31 and dBruce, positive and negative regulators of caspases, respectively, to fine-tune their activity during spermatogenesis.
Most recently, Cho-Park & Steller [111] have shown that DmPI31 also controls the constitutive proteasome by regulating the attachment of the 19S regulatory particle to the 20S core. They showed that PI31 binding activity for the proteasome is ‘switchable’ and is controlled by ADP-ribosylation, promoting assembly of active 26S proteasomes. It has been postulated that this mechanism might tie proteasome activity to cellular metabolism as NAD+ is used for ADP-ribosylation of PI31 [111,112]. As a strong binding partner for DmPI31, it is possible that Nutcracker might impact on this aspect of PI31 regulation.
In Drosophila, dramatic phenotypes for DmPI31 and nutcracker loss were reported only in the testes. The requirement for PI31 and Fbxo7 in mammalian systems may also be tissue specific, necessitated in cells that stringently require high levels of proteasome activity. As mutations in FBXO7 have now been linked with PD, a disease in which UPS dysfunction and protein aggregation are potential contributors to its aetiology, it is tempting to speculate that Fbxo7 regulation of proteasome activity might also play a part in this disease. It is possible that in PD, as misfolded proteins accumulate and UPS stress mounts, the demand to ramp up proteasomal activity cannot be met by cells that have substandard Fbxo7 activity. This would hint at an underlying lack of fitness in the UPS. An alternative hypothesis is that the dimerization of PI31 with Fbxo7 might prevent or alter its SCF-dependent functions or its other atypical roles; thus PI31 might act as an inhibitor or refiner of Fbxo7-dependent substrate ubiquitination or its cell cycle regulatory roles.
9. Fbxo7 mutations cause Parkinson's disease
The identification of mutations within FBXO7 in patients presenting with an early-onset form of PD opened up new questions about the role of Fbxo7 in the preservation of neuronal health [75,113,114]. Whole-genome SNP arrays were instrumental in the discovery of the first disease-associated variant of the FBXO7 gene, revealing a homozygous mutation (R378G) in an Iranian family [114]. This was quickly followed by the identification of other mutations, including a homozygous truncating mutation (R498X) in an Italian family and compound heterozygous mutations consisting of a splice-site (IVS7 + 1G/T) and point mutation (T22M) in a Dutch family [75]. Another heterozygous mutation (R481C) was reported in an Italian family [115]. However, affected family members were also found to have a homozygous mutation within PARK9 (G877R), so it is unclear whether the mutation of a single FBXO7 allele contributed to the disease phenotype. What is striking about the mutations discovered to date is that they are distributed across the many functional domains of the protein (figure 3c), suggesting that a number of Fbxo7 functions are relevant to PD. Indeed, early characterization of the R378G mutant has shown that this mutation reduces Fbxo7 affinity for Skp1 [107]. In addition, as the R498X mutation removes 24 amino acids from the substrate-binding domain, these mutants hint that loss of Fbxo7's SCF-dependent E3 ligase activity will be an important aspect in the pathogenesis of the disease.
As a bona fide PD-associated gene, FBXO7 has been designated ‘PARK15’ and joins a small family of ‘PARK’ genes. These genes all have confirmed genetic association with PD and are classified as autosomal dominant or recessive. Dominant mutation of a single copy of PARK1, which encodes α-synuclein, can produce a gain-of-function, whereby the protein becomes prone to aggregation. α-synuclein is the main constituent of Lewy bodies, the intracellular proteinaceous plaques observed in the brains of idiopathic PD patients at autopsy. Lewy bodies are thought to develop from aggregated, ubiquitinated proteins, which accumulate at the microtubule-organizing centre (MTOC) of the cell in structures known as aggresomes [116,117]. Whether these deposits are part of a pathological process or a protective means to sequester cytotoxic oligomers of misfolded proteins remains controversial. In any case, they represent a residual signature for the inefficient functioning of the proteasome and/or autophagy pathways in the neurons of patients [118–120]. One study has been published suggesting that Fbxo7 may be involved in Lewy body formation [121]. Using immunohistochemistry, its expression was detected throughout ‘normal’ brain tissue, but was surprisingly more abundant in the neocortex, putamen and cerebellum than in the substantia nigra. Fbxo7 was also found in α-synuclein-positive Lewy bodies in idiopathic brain tissue [121]. This study lacked experiments that addressed the mechanism of Fbxo7 deposition in intracellular aggregates, so it is not known whether Fbxo7 is actively involved in the aggregation and disposal of misfolded proteins or whether Fbxo7 is merely a passenger that is itself prone to aggregation in stressed neurons. Answers to these questions await future molecular studies and analysis of PARK15 patient samples.
10. Fbxo7 as a regulator of mitophagy
FBXO7 is mutated in an autosomal recessive fashion, like several other PARK genes including Parkin (PARK2) and PTEN-induced kinase 1 (PINK1; PARK6), so it is thought that their normal functions are reduced or lost as a result of mutation. Parkin is a single-subunit RING-type E3 ubiquitin ligase that contributes to neuronal health by initiating the bulk-disposal of misfolded or aggregated proteins, and by regulating the mitochondrial quality control process, known as ‘mitophagy’ [122–125]. Mitophagy is a form of selective macroautophagy that enables the cell to constantly survey its mitochondrial network, identifying and removing damaged, depolarized mitochondria. This process runs continually, working alongside mitochondrial biogenesis to meet the energy requirements of the cell while minimizing its oxidative burden. Maintaining this balance is likely to be of particular importance for the long-lived dopaminergic neurons of the substantia nigra, which are lost during the progression of PD. These highly specialized neurons have massive, unmyelinated axonal arborizations that place extraordinary demands on their mitochondria [126,127]. The involvement of Parkin and PINK1 in the mitophagy pathway was uncovered in a series of genetic studies in Drosophila showing that loss of either gene resulted in identical mitochondrial defects in flight muscles and sperm. Their epistatic relationship was discovered when over-expression of Parkin was shown to rescue PINK1 null flies but not vice versa [128]. PINK1 acts as the sensor of mitochondrial membrane potential (Δψm), and under healthy conditions it is a highly labile protein, constitutively cleaved and shed from mitochondria before being degraded in the cytosol. However, when Δψm is lost, PINK1 accumulates and is integrated into the outer mitochondrial membrane. PINK1 recruits and activates cytosolic Parkin by phosphorylation [129,130], which directly or indirectly stimulates the Lys48 and Lys63 polyubiquitination of a number of proteins. This leads to the degradation of mitofusin (Mfn) 1/2, Drp1 and voltage-dependent anion channel 1 (VDAC1) [131–134], initiating the fragmentation and isolation of depolarized regions of the mitochondrial network. The coating of Lys63 polyubiquitin enables the LC3-adaptor protein, p62/SQSTM1, to bind these mitochondrial fragments, and traffic them to the nuclear periphery where they form aggresome-like structures and are eventually engulfed into autophagosomes.
Burchell et al. [77] have shown that Fbxo7 is also a component of this pathway (figure 3d), with Fbxo7 interacting directly with Parkin via its Ubl domain, helping to recruit it to mitochondria to initiate mitophagy. The T22M mutation in the Ubl of Fbxo7 ablates its interaction with Parkin, and thus prevents its recruitment to depolarized mitochondria. Fbxo7 was also required for the efficient ubiquitination of Mfn1 and the recruitment of p62. Importantly, the expression of human Fbxo7 rescues the phenotypes of parkin loss in a Drosophila model of neurodegeneration [77]. These data strongly suggest that the Parkin–Fbxo7 interaction is important for neuronal health.
In addition to interacting with Parkin, Fbxo7 also directly interacts with PINK1, via a domain that encompasses and extends beyond the Parkin binding site in the Fbxo7 Ubl domain (figure 3c). In vitro binding studies showed neither a cooperative nor a competitive interaction among the three proteins, suggesting that Fbxo7 may act as a scaffold to facilitate PINK1-mediated phosphorylation and activation of Parkin. Parkin is clearly downstream of PINK1 loss in the Drosophila model. However, the expression of human Fbxo7 did not rescue the mitochondrial defects of PINK1 null flies. Fbxo7 is not required for PINK1 accumulation at depolarized mitochondria, but these results might be explained by the requirement for PINK1 activity for rescue by Fbxo7 to take place.
At present, it is not known whether Fbxo7 solely acts as a recruitment factor for Parkin during mitophagy or if it has a more elaborate role to play. However, as Fbxo7 can compensate for all of the mitochondrial defects resulting from parkin loss in Drosophila, this would indicate that all the critical ubiquitination targets/functions for Parkin are fulfilled in the fly model. Do Fbxo7 and Parkin have overlapping substrate profiles in mammalian cells or does Fbxo7 have its own PD-relevant substrates? Certainly, the other pathogenic mutations within Fbxo7 suggest that its E3 activity is important for neuronal health even in the presence of Parkin, so discovering their identity is of great interest. Whichever substrate(s) are discovered to be the key targets, the finding that three PARK genes are involved in a common function to regulate mitochondrial homeostasis firmly focuses the PD field on the pathways regulating mitochondrial health for investigating the possibilities of targeted therapeutics and diagnostics.
11. Concluding remarks
With such a multifunctional protein that is relevant to human disease, there are a myriad of questions regarding the function, regulation and specificity of Fbxo7. For example, the sheer diversity of Fbxo7's canonical and non-canonical interactions raises questions of how Fbxo7 activity is dictated at a given moment. In other words, does Fbxo7 participate in all of these functions at all times, or do some activities pre-dominate over others? We note that many of Fbxo7's functions are restricted, spatially or temporally, at particular organelles, or in signalling hubs. For example, the assembly of active Cdk6 with D-type cyclins happens in a short window during G1 phase, and the need for boosting the proteasome activity may be a transient response to pathogens or other cellular stresses. In addition, although mitophagy in the Burchell et al. study used chemicals to induce the mass depolarization of the mitochondria, this is unlikely to occur so profoundly in cells. Instead, mitophagy more probably occurs at a low level and as a last resort to purge the network of irreparably damaged mitochondria. We speculate that Fbxo7 participates in mitophagy in other tissues as well as neurons, and it is also possible that in neurons, the atypical functions of Fbxo7, like proteasome regulation may also come into play. For example, where poor Fbxo7 function leads to mitochondrial dysfunction, this may contribute to the production of reactive oxygen species, which causes protein damage, misfolding and aggregation, and eventually overburdening of the UPS. Suboptimal Fbxo7 may contribute to the poor coping of proteasomes to this increased demand, directly or through its interactions with PI31. Additionally, Fbxo7's other atypical roles may also conceivably contribute to the development of PD, like in cell cycle regulation, where inappropriate cell cycle entry has been linked to the death of neurons, or in the NF-κB pathway, which has been linked to inflammation in the brain. Perhaps, it is due to the multifunctional character of PARK genes, like Fbxo7 and Parkin, that when they are defective, they disable numerous pathways, causing disease.
Fbxo7 is certainly an extraordinary case for the cell getting many disparate functions from a single protein and possibly for linking cellular pathways to each other. If the other 68 FBPs in the cell are so intricately engineered, there will be much to functionally dissect for the future. The challenge will be to understand the tremendous complexity and interplay of all the different types of activities of FBPs in the context of both development and disease, and tease out which aspects of their functions we can influence to benefit patients.
Funding statement
This work is funded by the BBSRC (BB/J007846/1).
References
- 1.Reed SI. 2003. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4, 855–864 (doi:10.1038/nrm1246) [DOI] [PubMed] [Google Scholar]
- 2.Ang XL, Harper JW. 2004. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci. STKE 2004, pe31 (doi:10.1126/stke.2422004pe31) [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (doi:10.1146/annurev.biochem.70.1.503) [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. 2004. Back to the future with ubiquitin. Cell 116, 181–190 (doi:10.1016/S0092-8674(03)01074-2) [DOI] [PubMed] [Google Scholar]
- 5.Dye BT, Schulman BA. 2007. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 (doi:10.1146/annurev.biophys.36.040306.132820) [DOI] [PubMed] [Google Scholar]
- 6.Skaar JR, Pagan JK, Pagano M. 2013. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (doi:10.1038/nrm3582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komander D. 2009. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953 (doi:10.1042/BST0370937) [DOI] [PubMed] [Google Scholar]
- 8.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (doi:10.1146/annurev.biochem.78.101807.093809) [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Seth AK, Aplin AE. 2006. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol. Cancer Res. 4, 695–707 (doi:10.1158/1541-7786.MCR-06-0182) [DOI] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (doi:10.1038/nrm1547) [DOI] [PubMed] [Google Scholar]
- 11.Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (doi:10.1038/nrm1471) [DOI] [PubMed] [Google Scholar]
- 12.Schulman BA, et al. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 (doi:10.1038/35042620) [DOI] [PubMed] [Google Scholar]
- 13.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274 (doi:10.1016/S0092-8674(00)80098-7) [DOI] [PubMed] [Google Scholar]
- 14.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9, 1177–1179 (doi:10.1016/S0960-9822(00)80020-2) [DOI] [PubMed] [Google Scholar]
- 15.Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. 1999. A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182 (doi:10.1016/S0960-9822(00)80021-4) [DOI] [PubMed] [Google Scholar]
- 16.Skaar JR, D'Angiolella V, Pagan JK, Pagano M. 2009. SnapShot: F box proteins II. Cell 137, 1358.e1–1358.e2 (doi:10.1016/j.cell.2009.05.040) [DOI] [PubMed] [Google Scholar]
- 17.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 (doi:10.1101/gad.1255304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu C, Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 (doi:10.1007/s13238-011-1018-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobe B, Kajava AV. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732 (doi:10.1016/S0959-440X(01)00266-4) [DOI] [PubMed] [Google Scholar]
- 20.Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. 2001. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol. 3, 321–324 (doi:10.1038/35060126) [DOI] [PubMed] [Google Scholar]
- 21.Tedesco D, Lukas J, Reed SI. 2002. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16, 2946–2957 (doi:10.1101/gad.1011202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan JM, Peter M. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl Acad. Sci. USA 96, 9124–9129 (doi:10.1073/pnas.96.16.9124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Howley PM. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2, 571–580 (doi:10.1016/S1097-2765(00)80156-2) [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Pan J, Wang X, Wu Y, Bauman WA, Cardozo CP. 2008. Expression of the muscle atrophy factor muscle atrophy F-box is suppressed by testosterone. Endocrinology 149, 5449–5460 (doi:10.1210/en.2008-0664) [DOI] [PubMed] [Google Scholar]
- 25.Santra MK, Wajapeyee N, Green MR. 2009. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 459, 722–725 (doi:10.1038/nature08011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Gazdoiu S, Pan ZQ, Fuchs SY. 2004. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J. Biol. Chem. 279, 11 074–11 080 (doi:10.1074/jbc.M312301200) [DOI] [PubMed] [Google Scholar]
- 27.Pierce NW, et al. 2013. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 (doi:10.1016/j.cell.2013.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA. 2013. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 4, 1642 (doi:10.1038/ncomms2636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (doi:10.1038/35107009) [DOI] [PubMed] [Google Scholar]
- 30.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (doi:10.1146/annurev-biochem-060310-170328) [DOI] [PubMed] [Google Scholar]
- 31.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (doi:10.1126/science.2538923) [DOI] [PubMed] [Google Scholar]
- 32.Kirkin V, McEwan DG, Novak I, Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 (doi:10.1016/j.molcel.2009.04.026) [DOI] [PubMed] [Google Scholar]
- 33.Al-Hakim A, Escribano-Diaz C, Landry MC, O'Donnell L, Panier S, Szilard RK, Durocher D. 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9, 1229–1240 (doi:10.1016/j.dnarep.2010.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A. 2009. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895 (doi:10.1038/emboj.2009.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petroski MD, Deshaies RJ. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 (doi:10.1016/j.cell.2005.09.033) [DOI] [PubMed] [Google Scholar]
- 36.Sadowski M, Suryadinata R, Lai X, Heierhorst J, Sarcevic B. 2010. Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol. Cell Biol. 30, 2316–2329 (doi:10.1128/MCB.01094-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao G, Fong A, Sun SC. 2004. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J. Biol. Chem. 279, 30 099–30 105 (doi:10.1074/jbc.M401428200) [DOI] [PubMed] [Google Scholar]
- 38.Orian A, et al. 2000. SCF(β)(-TrCP) ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 19, 2580–2591 (doi:10.1093/emboj/19.11.2580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. 2001. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7, 627–637 (doi:10.1016/S1097-2765(01)00209-X) [DOI] [PubMed] [Google Scholar]
- 40.Chen BB, Coon TA, Glasser JR, Mallampalli RK. 2011. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol. Cell Biol. 31, 1905–1920 (doi:10.1128/MCB.00723-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar KG, et al. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-α receptor endocytosis. J. Cell Biol. 179, 935–950 (doi:10.1083/jcb.200706034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jonkers W, Rep M. 2009. Lessons from fungal F-box proteins. Eukaryot. Cell 8, 677–695 (doi:10.1128/EC.00386-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermand D. 2006. F-box proteins: more than baits for the SCF? Cell Div. 1, 30 (doi:10.1186/1747-1028-1-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Fiore B, Pines J. 2008. Defining the role of Emi1 in the DNA replication-segregation cycle. Chromosoma 117, 333–338 (doi:10.1007/s00412-008-0152-x) [DOI] [PubMed] [Google Scholar]
- 45.Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, Jackson PK. 2006. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 20, 2410–2420 (doi:10.1101/gad.1454006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smaldone S, Laub F, Else C, Dragomir C, Ramirez F. 2004. Identification of MoKA, a novel F-box protein that modulates Kruppel-like transcription factor 7 activity. Mol. Cell Biol. 24, 1058–1069 (doi:10.1128/MCB.24.3.1058-1069.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laub F, Aldabe R, Friedrich V, Jr, Ohnishi S, Yoshida T, Ramirez F. 2001. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 233, 305–318 (doi:10.1006/dbio.2001.0243) [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS. 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277, 24 530–24 537 (doi:10.1074/jbc.M201612200) [DOI] [PubMed] [Google Scholar]
- 49.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186, 655–663 (doi:10.1083/jcb.200812138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JH, Kim J, Kim DH, Ryu GH, Bae SH, Seo YS. 2004. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 32, 2287–2297 (doi:10.1093/nar/gkh534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E, Knowles P, McDonald N, Boshoff C. 2005. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 24, 3104–3116 (doi:10.1038/sj.emboj.7600775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. 2011. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145, 371–382 (doi:10.1016/j.cell.2011.03.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. 2001. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105, 645–655 (doi:10.1016/S0092-8674(01)00361-0) [DOI] [PubMed] [Google Scholar]
- 54.Tung JJ, Hansen DV, Ban KH, Loktev AV, Summers MK, Adler JR, III, Jackson PK. 2005. A role for the anaphase-promoting complex inhibitor Emi2/XErp1, a homolog of early mitotic inhibitor 1, in cytostatic factor arrest of Xenopus eggs. Proc. Natl Acad. Sci. USA 102, 4318–4323 (doi:10.1073/pnas.0501108102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong M, Barnes EA, Ollendorff V, Donoghue DJ. 2000. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 19, 1378–1388 (doi:10.1093/emboj/19.6.1378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell ID, Grancell AS, Sorger PK. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145, 933–950 (doi:10.1083/jcb.145.5.933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. 2007. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313 (doi:10.1038/nature06255) [DOI] [PubMed] [Google Scholar]
- 58.Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR. 2009. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc. Natl Acad. Sci. USA 106, 16 339–16 344 (doi:10.1073/pnas.0908560106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aso T, Haque D, Barstead RJ, Conaway RC, Conaway JW. 1996. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 15, 5557–5566 [PMC free article] [PubMed] [Google Scholar]
- 60.Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T. 2008. Mammalian elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 27, 3256–3266 (doi:10.1038/emboj.2008.249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. 2000. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127, 5265–5276 [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Nakatsukasa K, Kotera M, Kanada A, Nishimura T, Kishi T, Mimura S, Kamura T. 2011. Non-SCF-type F-box protein Roy1/Ymr258c interacts with a Rab5-like GTPase Ypt52 and inhibits Ypt52 function. Mol. Biol. Cell 22, 1575–1584 (doi:10.1091/mbc.E10-08-0716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell Biol. 21, 3105–3117 (doi:10.1128/MCB.21.9.3105-3117.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jourdain I, Spielewoy N, Thompson J, Dhut S, Yates JR, Toda T. 2009. Identification of a conserved F-box protein 6 interactor essential for endocytosis and cytokinesis in fission yeast. Biochem J. 420, 169–177 (doi:10.1042/BJ20081659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tafforeau L, Le Blastier S, Bamps S, Dewez M, Vandenhaute J, Hermand D. 2006. Repression of ergosterol level during oxidative stress by fission yeast F-box protein Pof14 independently of SCF. EMBO J. 25, 4547–4556 (doi:10.1038/sj.emboj.7601329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondo-Okamoto N, Ohkuni K, Kitagawa K, McCaffery JM, Shaw JM, Okamoto K. 2006. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol. Biol. Cell 17, 3756–3767 (doi:10.1091/mbc.E06-02-0145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durr M, Escobar-Henriques M, Merz S, Geimer S, Langer T, Westermann B. 2006. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell 17, 3745–3755 (doi:10.1091/mbc.E06-01-0053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duyvesteijn RG, van Wijk R, Boer Y, Rep M, Cornelissen BJ, Haring MA. 2005. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 57, 1051–1063 (doi:10.1111/j.1365-2958.2005.04751.x) [DOI] [PubMed] [Google Scholar]
- 69.Jonkers W, Rodrigues CD, Rep M. 2009. Impaired colonization and infection of tomato roots by the Deltafrp1 mutant of Fusarium oxysporum correlates with reduced CWDE gene expression. Mol. Plant Microbe Interact. 22, 507–518 (doi:10.1094/MPMI-22-5-0507) [DOI] [PubMed] [Google Scholar]
- 70.Hsu JM, Lee YC, Yu CT, Huang CY. 2004. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J. Biol. Chem. 279, 32 592–32 602 (doi:10.1074/jbc.M404950200) [DOI] [PubMed] [Google Scholar]
- 71.Kirk R, Laman H, Knowles PP, Murray-Rust J, Lomonosov M, Meziane el K, McDonald NQ. 2008. Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J. Biol. Chem. 283, 22 325–22 335 (doi:10.1074/jbc.M709900200) [DOI] [PubMed] [Google Scholar]
- 72.Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ. 1999. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 457, 333–338 (doi:10.1016/S0014-5793(99)01072-8) [DOI] [PubMed] [Google Scholar]
- 73.Zaiss DM, Standera S, Kloetzel PM, Sijts AJ. 2002. PI31 is a modulator of proteasome formation and antigen processing. Proc. Natl Acad. Sci. USA 99, 14 344–14 349 (doi:10.1073/pnas.212257299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flicek P, et al. 2013. Ensembl 2013. Nucleic Acids Res. 41, D48–D55 (doi:10.1093/nar/gks1236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Fonzo A, et al. 2009. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 72, 240–245 (doi:10.1212/01.wnl.0000338144.10967.2b) [DOI] [PubMed] [Google Scholar]
- 76.Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. 2006. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem. Biophys. Res. Commun. 342, 1022–1026 (doi:10.1016/j.bbrc.2006.02.061) [DOI] [PubMed] [Google Scholar]
- 77.Burchell VS, et al. 2013. The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 16, 1257–1265 (doi:10.1038/nn.3489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Harst P, et al. 2012. Seventy-five genetic loci influencing the human red blood cell. Nature 492, 369–375 (doi:10.1038/nature11677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soranzo N, et al. 2009. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 41, 1182–1190 (doi:10.1038/ng.467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding K, et al. 2012. Genetic Loci implicated in erythroid differentiation and cell cycle regulation are associated with red blood cell traits. Mayo Clin. Proc. 87, 461–474 (doi:10.1016/j.mayocp.2012.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuiken HJ, Egan DA, Laman H, Bernards R, Beijersbergen RL, Dirac AM. 2012. Identification of F-box only protein 7 as a negative regulator of NF-κB signaling. J. Cell Mol. Med. 16, 2140–2149 (doi:10.1111/j.1582-4934.2012.01524.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye F, Tan L, Yang Q, Xia Y, Deng LW, Murata-Hori M, Liou YC. 2011. HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr. Biol. 21, 1584–1591 (doi:10.1016/j.cub.2011.08.024) [DOI] [PubMed] [Google Scholar]
- 83.Tsou AP, et al. 2003. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene 22, 298–307 (doi:10.1038/sj.onc.1206129) [DOI] [PubMed] [Google Scholar]
- 84.Kuo TC, Chang PY, Huang SF, Chou CK, Chao CC. 2012. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53. Biochem. Pharmacol. 83, 758–768 (doi:10.1016/j.bcp.2011.12.034) [DOI] [PubMed] [Google Scholar]
- 85.Hu S, Yang X. 2003. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 278, 10 055–10 060 (doi:10.1074/jbc.M207197200) [DOI] [PubMed] [Google Scholar]
- 86.Mahoney DJ, et al. 2008. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc. Natl Acad. Sci. USA 105, 11 778–11 783 (doi:10.1073/pnas.0711122105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertrand MJ, et al. 2008. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 (doi:10.1016/j.molcel.2008.05.014) [DOI] [PubMed] [Google Scholar]
- 88.Flood PM, Qian L, Peterson LJ, Zhang F, Shi JS, Gao HM, Hong JS. 2011. Transcriptional Factor NF-κB as a Target for Therapy in Parkinson's Disease. Parkinsons Dis. 2011, 216298 (doi:10.4061/2011/216298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. 2011. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell Proteomics 10, M110 006460 (doi:10.1074/mcp.M110.006460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frescas D, Pagano M. 2008. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 (doi:10.1038/nrc2396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fuchs SY, Spiegelman VS, Kumar KG. 2004. The many faces of β-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene 23, 2028–2036 (doi:10.1038/sj.onc.1207389) [DOI] [PubMed] [Google Scholar]
- 92.Laman H, Mann DJ, Jones NC. 2000. Viral-encoded cyclins. Curr. Opin. Genet. Dev. 10, 70–74 (doi:10.1016/S0959-437X(99)00045-3) [DOI] [PubMed] [Google Scholar]
- 93.Lomonosov M, Meziane el K, Ye H, Nelson DE, Randle SJ, Laman H. 2011. Expression of Fbxo7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. PLoS ONE 6, e21165 (doi:10.1371/journal.pone.0021165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu MG, et al. 2009. A requirement for cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res. 69, 810–818 (doi:10.1158/0008-5472.CAN-08-2473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, Dubus P, Barbacid M. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118, 493–504 (doi:10.1016/j.cell.2004.08.002) [DOI] [PubMed] [Google Scholar]
- 96.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. 1999. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18, 1571–1583 (doi:10.1093/emboj/18.6.1571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.James MK, Ray A, Leznova D, Blain SW. 2008. Differential modification of p27Kip1 controls its cyclin D-cdk4 inhibitory activity. Mol. Cell Biol. 28, 498–510 (doi:10.1128/MCB.02171-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blain SW. 2008. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle 7, 892–898 (doi:10.4161/cc.7.7.5637) [DOI] [PubMed] [Google Scholar]
- 99.Meziane el K, Randle SJ, Nelson DE, Lomonosov M, Laman H. 2011. Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. J. Cell Sci. 124, 2175–2186 (doi:10.1242/jcs.080465) [DOI] [PubMed] [Google Scholar]
- 100.Carroll M, Zhu Y, D'Andrea AD. 1995. Erythropoietin-induced cellular differentiation requires prolongation of the G1 phase of the cell cycle. Proc. Natl Acad. Sci. USA 92, 2869–2873 (doi:10.1073/pnas.92.7.2869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson P, Chung S, Benchimol S. 1993. Growth suppression of Friend virus-transformed erythroleukemia cells by p53 protein is accompanied by hemoglobin production and is sensitive to erythropoietin. Mol. Cell Biol. 13, 1456–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kato JY, Sherr CJ. 1993. Inhibition of granulocyte differentiation by G1 cyclins D2 and D3 but not D1. Proc. Natl Acad. Sci. USA 90, 11 513–11 517 (doi:10.1073/pnas.90.24.11513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lea NC, Orr SJ, Stoeber K, Williams GH, Lam EW, Ibrahim MA, Mufti GJ, Thomas NS. 2003. Commitment point during G0→G1 that controls entry into the cell cycle. Mol. Cell Biol. 23, 2351–2361 (doi:10.1128/MCB.23.7.2351-2361.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rush JS, Hasbold J, Hodgkin PD. 2002. Cross-linking surface Ig delays CD40 ligand- and IL-4-induced B cell Ig class switching and reveals evidence for independent regulation of B cell proliferation and differentiation. J. Immunol. 168, 2676–2682 [DOI] [PubMed] [Google Scholar]
- 105.Chu-Ping M, Slaughter CA, DeMartino GN. 1992. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochim. Biophys. Acta 1119, 303–311 (doi:10.1016/0167-4838(92)90218-3) [DOI] [PubMed] [Google Scholar]
- 106.McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, DeMartino GN. 2000. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 275, 18 557–18 565 (doi:10.1074/jbc.M001697200) [DOI] [PubMed] [Google Scholar]
- 107.Nelson DE, Laman H. 2011. A Competitive binding mechanism between Skp1 and exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J. Biol. Chem. 286, 19 804–19 815 (doi:10.1074/jbc.M111.220079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bader M, Arama E, Steller H. 2010. A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development 137, 1679–1688 (doi:10.1242/dev.050088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arama E, Agapite J, Steller H. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697 (doi:10.1016/S1534-5807(03)00120-5) [DOI] [PubMed] [Google Scholar]
- 110.Domingues C, Ryoo HD. 2012. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 19, 470–477 (doi:10.1038/cdd.2011.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cho-Park PF, Steller H. 2013. Proteasome regulation by ADP-ribosylation. Cell 153, 614–627 (doi:10.1016/j.cell.2013.03.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (doi:10.1038/nrm3376) [DOI] [PubMed] [Google Scholar]
- 113.Paisan-Ruiz C, et al. 2010. Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov. Disord. 25, 1791–1800 (doi:10.1002/mds.23221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shojaee S, Sina F, Banihosseini SS, Kazemi MH, Kalhor R, Shahidi GA, Fakhrai-Rad H, Ronaghi M, Elahi E. 2008. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am. J. Hum. Genet. 82, 1375–1384 (doi:10.1016/j.ajhg.2008.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santoro L, et al. 2011. Novel ATP13A2 (PARK9) homozygous mutation in a family with marked phenotype variability. Neurogenetics 12, 33–39 (doi:10.1007/s10048-010-0259-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kopito RR. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (doi:10.1016/S0962-8924(00)01852-3) [DOI] [PubMed] [Google Scholar]
- 117.Garcia-Mata R, Gao YS, Sztul E. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3, 388–396 (doi:10.1034/j.1600-0854.2002.30602.x) [DOI] [PubMed] [Google Scholar]
- 118.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr 2000. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl Acad. Sci. USA 97, 571–576 (doi:10.1073/pnas.97.2.571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kalia LV, Kalia SK, McLean PJ, Lozano AM, Lang AE. 2013. α-Synuclein oligomers and clinical implications for Parkinson disease. Ann. Neurol. 73, 155–169 (doi:10.1002/ana.23746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wan OW, Chung KK. 2012. The role of alpha-synuclein oligomerization and aggregation in cellular and animal models of Parkinson's disease. PLoS ONE 7, e38545 (doi:10.1371/journal.pone.0038545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao T, Severijnen LA, van der Weiden M, Zheng PP, Oostra BA, Hukema RK, Willemsen R, Kros JM, Bonifati V. 2013. FBXO7 immunoreactivity in alpha-synuclein-containing inclusions in Parkinson disease and multiple system atrophy. J. Neuropathol. Exp. Neurol. 72, 482–488 (doi:10.1097/NEN.0b013e318293c586) [DOI] [PubMed] [Google Scholar]
- 122.Perier C, Vila M. 2012. Mitochondrial biology and Parkinson's disease. Cold Spring Harb. Perspect. Med. 2, a009332 (doi:10.1101/cshperspect.a009332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cook C, Petrucelli L. 2009. A critical evaluation of the ubiquitin-proteasome system in Parkinson's disease. Biochim. Biophys. Acta 1792, 664–675 (doi:10.1016/j.bbadis.2009.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schneider L, Zhang J. 2010. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson's disease. Mol. Neurodegener. 5, 14 (doi:10.1186/1750-1326-5-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Keane PC, Kurzawa M, Blain PG, Morris CM. 2011. Mitochondrial dysfunction in Parkinson's disease. Parkinsons Dis. 2011, 716871 (doi:10.4061/2011/716871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, Kaneko T. 2009. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci. 29, 444–453 (doi:10.1523/JNEUROSCI.4029-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pissadaki EK, Bolam JP. 2013. The energy cost of action potential propagation in dopamine neurons: clues to susceptibility in Parkinson's disease. Front. Comput. Neurosci. 7, 13 (doi:10.3389/fncom.2013.00013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Park J, et al. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (doi:10.1038/nature04788) [DOI] [PubMed] [Google Scholar]
- 129.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. 2008. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 377, 975–980 (doi:10.1016/j.bbrc.2008.10.104) [DOI] [PubMed] [Google Scholar]
- 130.Sha D, Chin LS, Li L. 2010. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-κB signaling. Hum. Mol. Genet. 19, 352–363 (doi:10.1093/hmg/ddp501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Glauser L, Sonnay S, Stafa K, Moore DJ. 2011. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 (doi:10.1111/j.1471-4159.2011.07318.x) [DOI] [PubMed] [Google Scholar]
- 132.Ziviani E, Tao RN, Whitworth AJ. 2010. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA 107, 5018–5023 (doi:10.1073/pnas.0913485107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 (doi:10.1083/jcb.201007013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. 2010. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE 5, e10054 (doi:10.1371/journal.pone.0010054) [DOI] [PMC free article] [PubMed] [Google Scholar]