Key Points
The use of barcoding to track lineages in 196 human CD34+ CB clones in serially sampled primary and secondary transplanted NSG mice is described.
Detection of early transient clones with later, more stable clones and definitive evidence of sustained self-renewal of multipotency is presented.
Abstract
Human cord blood (CB) offers an attractive source of cells for clinical transplants because of its rich content of cells with sustained repopulating ability in spite of an apparent deficiency of cells with rapid reconstituting ability. Nevertheless, the clonal dynamics of nonlimiting CB transplants remain poorly understood. To begin to address this question, we exposed CD34+ CB cells to a library of barcoded lentiviruses and used massively parallel sequencing to quantify the clonal distributions of lymphoid and myeloid cells subsequently detected in sequential marrow aspirates obtained from 2 primary NOD/SCID-IL2Rγ−/− mice, each transplanted with ∼105 of these cells, and for another 6 months in 2 secondary recipients. Of the 196 clones identified, 68 were detected at 4 weeks posttransplant and were often lympho-myeloid. The rest were detected later, after variable periods up to 13 months posttransplant, but with generally increasing stability throughout time, and they included clones in which different lineages were detected. However, definitive evidence of individual cells capable of generating T-, B-, and myeloid cells, for over a year, and self-renewal of this potential was also obtained. These findings highlight the caveats and utility of this model to analyze human hematopoietic stem cell control in vivo.
Introduction
Hematopoiesis is a complex, hierarchically ordered, multistep process that originates in cells with latent differentiation potentialities that can be maintained through many divisions.1 The features of this process have been inferred largely from studies of the in vitro or in vivo growth and differentiation properties of cells with distinct phenotypes. Additional contributions to our understanding of hematopoiesis have been obtained from studies of clones regenerated in vivo from mouse, monkey, or human cells, which were tracked in syngeneic, autologous, or xenogeneic recipients by limiting dilution analysis,2-4 by vector insert5-8 or barcoding strategies,9-12 or by using single-cell transplants.13-15 Interestingly, these analyses have shown evidence of extensive clonal heterogeneity in the rate of expansion, durability, and differentiation activity of individual hematopoietic cells with repopulating activity from these multiple species. However, more extensive investigation of these features in transplants of human hematopoietic cells remains a subject of active interest. The development of highly immunodeficient mice that live long enough to permit extensive periods of follow-up of engrafted human cells16,17 now offers an additional model to investigate these issues.
Improvements in the utility of clinical hematopoietic transplants require knowledge of how nonlimiting doses of transplanted cells will behave. Evidence of heterogeneity in the activity of serially transplantable clones generated in short-lived immunodeficient mice under nonlimiting transplantation conditions has been previously reported for human cord blood (CB) cells using viral integration site tracking.18-21 However, the sensitivity and resolution of these analyses have generally precluded investigation of the outputs of specific blood cell lineages within individual clones. The development of barcoded lentiviral libraries and their use in combination with massively parallel sequencing (MPS) and appropriate bioinformatic tools to infer cell numbers from barcode frequencies now allows these limitations to be circumvented (Nguyen et al, submitted 2013).22,23 Here we have exploited this technology to interrogate the patterns of clonal growth kinetics and differentiation detectable by serial sampling of bone marrow (BM) cells aspirated from primary and secondary NOD/SCID-IL2Rγ−/− (NSG) mice transplanted with nonlimiting transplants of purified human CD34+ CB cells during a combined period of 13 months.
Methods
Barcode library
The library of barcoded green fluorescence protein (GFP)-encoding lentiviruses used has been described in detail elsewhere (Nguyen et al, submitted 2013). Briefly, we constructed a plasmid library using forward and reverse oligonucleotide sequences (5′-TCGAGAAGTAANNATCNNGATSSAAANNGGTNNAACNNTGTAAAACGACGGCCAGTGAGC-3′ and 5′-CCGGGCTCACTGGCCGTCGTTTTACANNGTTNNACCNNTTTSSATCNNGATNNTTACTTC-3′) that were then annealed, purified, ligated into the MNDU3-PGK-GFP (MPG) vector24 (Figure 1A) and expanded in DH10B bacteria (Life Technologies). Deep sequencing of plasmids purified from the pooled amplified bacteria (MaxiPrep; Qiagen) showed these contain more than 2 × 105 unique barcodes. These plasmids were then used to produce a library of barcoded lentiviruses in a supernatant containing 109 infectious units/mL as titered on HeLa cells.
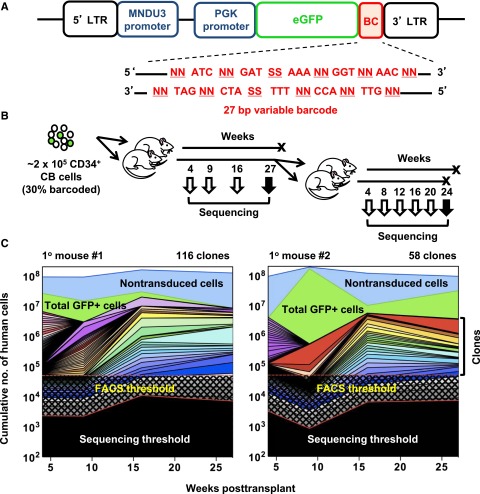
Figure 1.
Experimental design and analysis of clones detected. (A) Schematic outline of the MPG lentiviral vector that contained a 27-nucleotide non-coding DNA barcode sequence inserted downstream of the GFP reporter gene. The barcode sequence was designed with variable nucleotide doublets (NN) repeated 5 times, each separated by a constant nucleotide triplet sequence as previously described.10 (B) Schematic outline of the experimental design. CD34+ CB cells were transduced with the lentiviral barcode library for 6 hours in vitro, of which 105 cells (∼3.6 × 104 barcoded cells) were then intravenously transplanted into each of 2 sublethally irradiated NSG mice. BM aspirates were collected, total human CD45+ analyzed, and myeloid, B-, and T-cell subsets of GFP+ and GFP- cells were also separately isolated by FACS. After 27 weeks, all BM cells were harvested separately from both legs and pelvis of both primary mice. From one mouse, half the cells were sorted for clonal analysis and the other half were transplanted intravenously into 2 secondary mice. From the second primary mouse, all harvested BM cells were sorted and used for clonal analysis. (C) Analysis of clonal dynamics in the 2 transplanted primary mice. The upper blue line denotes the total number of human hematopoietic cells in the BM of each mouse (assumed to have a total cellularity of 2 × 108) and the upper green line denotes the total number of GFP+ cells in each mouse. Each color underneath denotes the total contribution of all of the lineages detected in each clone, as inferred from the BM sample analyzed. Black checked and shaded regions indicate the detection limit of the FACS (5 × 104 cells) and MPS (variable) methods, respectively.
CB cell preparation, transduction, and transplantation
We used an EasySep kit (STEMCELL Technologies, Vancouver, BC, Canada) to obtain a suspension of 2.4 × 105 cells containing 90% pure CD34+ cells from previously cryopreserved low-density (<1.077g/cm3) cells pooled from >100 anonymized normal CB samples obtained from women undergoing cesarian sections. After 16 hours of prestimulation in 100 µL of Stemspan medium (STEMCELL Technologies) containing 100 ng/mL FLT3-Ligand (FL; Immunex, Seattle, WA), 100 ng/mL Steel factor, 20 ng/mL granulocyte colony-stimulating factor (G-CSF; both from STEMCELL Technologies), 20 ng/mL interleukin-3 (IL-3; Novartis, Basel, Switzerland), and 20 ng/mL IL-6 (Cangene, Winnipeg, MA, Canada), the cells were washed and then incubated for another 6 hours in the same medium containing the library of barcoded GFP-encoding lentiviruses. The cells were then washed, one sixth (∼4 × 104) removed to measure the efficiency of gene transfer, and the remainder (∼2 × 105) immediately combined with 2 × 106 irradiated (15 Gy) human BM “carrier” cells and half of this combined suspension transplanted intravenously into each of 2 irradiated (315 cGy 137Cs γ-rays) 8 week-old NSG mice. Two secondary recipients were similarly transplanted, each with 25% of all the BM cells harvested from mouse #1 (∼7 × 106 cells/secondary mouse). The efficiency of gene transfer (∼30%) was obtained by flow cytometric assessment of the proportion of GFP+ cells in the aliquot that were removed after initial exposure to the virus and then incubated for 3 days. The Research Ethics Board and the Animal Care Committee of the University of British Columbia approved all studies.
Isolation of defined phenotypes of human cells from transplanted mice
BM cell samples were first incubated in 0.8% NH4Cl (STEMCELL Technologies) on ice for 10 minutes, and then suspended in PBS containing 10% human serum and an anti-mouse FcR antibody (2.4G2; STEMCELL Technologies) for another 10 minutes on ice before being stained with individual fluorochrome-labeled anti-human CD3 (SK7), CD15 (HI98), CD19 (4G7), CD45 (2D1), (all from BD Biosciences, San Jose, CA), CD33 (P67.6) (STEMCELL Technologies) and glycophorin A (GPA, 10F7MN; from P. Lansdorp, Terry Fox Laboratory, Vancouver, BC, Canada) antibodies for 30 minutes on ice. Cells were then washed and resuspended in Hanks’ buffered salt solution with 2% fetal calf serum and 0.2 µg/mL 4′,6-diamidino-2-phenylindole (DAPI). Specific phenotypes of viable (DAPI-) human cells were isolated using a FACSAria II cell sorter (BD Biosciences) using gates established to exclude all mouse BM cells based on simultaneously analyzed BM cells from nontransplanted NSG mice. Human phenotypes analyzed were defined as total hematopoietic (CD45+ and/or GPA+) cells, myeloid cells as CD15/33+ (granulocyte+macrophage, GM and occasional GPA+ erythroid) cells, B-lymphoid cells as CD19+ cells, and T-lymphoid cells as CD3+ cells. Samples were considered positive for a given phenotype when a minimum of 5 positive events was seen per 20 000 DAPI− cells analyzed.
MPS and barcode retrieval
Genomic DNA was extracted using PrepGEM DNA extraction kit (ZyGEM). A fault-tolerant sequence-based index was introduced during library construction to uniquely identify libraries pooled for sequencing in a single lane as described previously.25 Barcode amplicons were generated in an initial 35-cycle PCR reaction using sequence-specific primers with adaptors compatible with Illumina PE1 and PE2 primers (Illumina, Inc., Hayward, CA), and then indexed in a second 8 to 10 cycle PCR reaction. The individual amplicon libraries were then pooled at equimolar ratios, and sequenced by indexed paired-end sequencing on the Illumina HiSequation 2000 platform. A control phiX library was spiked into the library pool before sequencing (∼40% by mole) to improve cluster recognition. Barcode sequences were extracted from the resulting qseq files using custom scripts designed to identify the 2 flanking viral vector sequences and the 27 nucleotide barcode sequence allowing up to 3 mismatches and a minimum base quality of 20.
Thresholding and clone size calculations
For each of the 78 experimental datasets analyzed, the abundance of unique barcodes was enumerated along with those obtained from a set of 3 “spiked-in” reference controls of 20, 100, and 500 cells containing a known single barcode (supplemental Table 1A, available on the Blood Web site), as described (Nguyen et al, submitted 2013). The association curves for these spiked-in reference controls showed minimal variability between datasets (standard deviation <1 cell equivalent from calculated fraction-read abundance values, supplemental Table 1B-C). Raw barcode abundance values were transformed to absolute cell numbers using a polynomial regression fit to the reference control fractional read counts. The total number of cells of a particular phenotype within each clone in the entire mouse at a given time was then calculated from the total number of GFP+ cells of that phenotype detected in the BM, assuming a total of 2 × 108 BM cells per mouse. When the number of human cells fell below the frequency detectable by fluorescence-activated cell sorting (FACS) (0.025%), this latter lower limit value was used. Adding the total number of cells of all phenotypes analyzed yielded the total clone sizes. To calculate the clone-specific ratio of myeloid and lymphoid contributions to the total human myeloid and lymphoid cell outputs [GM/(B+T) ratio], separate clone-specific GM (FGM) and B (FB) + T (FT) contributions to the total GFP+ GM and GFP+B + GFP+T values in the same mouse were determined and then were expressed as a ratio (FGM)/(FB) + (FT).
Results
Experimental design and validation of the barcode monitoring strategy applied to CB transplants in NSG mice
Figure 1B shows the experimental design used in which 2 sublethally irradiated NSG mice were each injected intravenously with 105 CD34+ CB cells exposed to a highly diverse barcoded library of lentiviruses using a protocol designed to achieve a transduction efficiency (∼30%) that would be unlikely to yield more than a single barcode integrant per cell. BM aspirates containing ∼2 × 106 cells each were subsequently obtained for analysis 4, 9, and 16 weeks afterward. After 27 weeks, the 2 primary mice were sacrificed, and aliquots of BM cells from the pelvis and each hind leg were harvested and analyzed separately. In addition, 14.4 × 106 cells from primary mouse #1 (representing ∼25% of all the BM cells recovered) were transplanted into 2 secondary NSG mice (7.2 × 106 cells each), and these mice were then similarly followed for another 24 weeks. Each BM sample was stained with a panel of antibodies to allow multiple phenotypes of human hematopoietic cells to be quantified and the GFP+ subset of each isolated by FACS. Examples are shown in supplemental Figure 1, and all of the values obtained are shown in supplemental Table 2A-B. Genomic DNA was then extracted from each phenotypically defined isolate, and barcode contents were determined by MPS.
To quantify the clonal distributions of human myeloid and lymphoid cells within each sample, we first transformed the barcode frequency values obtained from the MPS data into cell numbers, based on the spiked-in reference control data included in each sample as described in Methods (see also supplemental Table 1A-C; supplemental Figure 2A). These standardized reference controls allowed technical biases to be normalized across all experimental samples. They also established confidence thresholds and allowed spurious barcodes to be identified based on their read abundance. The results predicted that clonal elements present at an absolute level of 100 cells or more would always be detected, but would be missed one third of the time if present at an absolute level of 20 cells. In the secondary recipients, the calculated likelihood of detecting a clonal contribution was 100% if 500 or more cells were present and 55% if 20 to 100 cells were present (supplemental Table 3A-B).
Conventional FACS analysis (which in these analyses had a lower limit of detection of 1 human cell in 2 × 104 viable cells analyzed) showed that human hematopoietic cells comprised from 46% to 80% and 47% to 95% of all the cells in the BM of the primary mouse #1 and #2, respectively, between 4 and 27 weeks after the initial transplant was performed (assuming the BM aspirates were representative of the total BM). These numbers indicate a high level of engraftment of both mice with CB cells for at least 7 months. The proportion of these that were GFP+ (ie, barcoded cells) generally mirrored the total human hematopoietic (CD45+) cell numbers in both primary mice throughout the same period, on average at levels of 20% to 30% (Figure 1C), similar to the gene transfer efficiency value of ∼30% obtained from the initial in vitro assessment of the bulk CD34+ cells. The numbers of human cells derived from the barcode-read abundance calculations and phenotypically by FACS were also highly correlated (Spearman r = 0.7; see supplemental Figure 2B). These findings reinforce the validity of the methodology used to derive clone size and composition values in the samples used to generate barcode-read numbers (complete data sets provided in supplemental Table 4). They also suggest that the data for the transduced cells are generally representative of the behavior of the nontransduced human cells present in the same mice and were not affected by any obvious insertional mutagenesis mechanism.
Clones produced in primary mice display multiple output patterns throughout a 7-month period
The barcode data for the 2 primary mice showed that they contained a large number of clones of human hematopoietic cells: 116 in mouse #1 and 58 in mouse #2 (Figure 1C; supplemental Figure 3A). Classification of these 174 clones according to the time at which each was first detected showed different kinetic patterns (Figure 2A). These included the rapid appearance of a large cohort of clones detectable at 4 weeks posttransplant but no longer at 9 weeks or subsequently (89% of the 76 clones detected at 4 weeks). A total of 58% of the 59 clones that were detected at 9 weeks posttransplant, were not evident in samples obtained after 16 weeks, and as many as a third of the clones that were not apparent in samples obtained before 16 weeks were no longer evident in the much larger samples of BM analyzed when the primary recipients were sacrificed at 27 weeks posttransplant. Conversely, most of the 50 clones that were detected at the end of the 7-month period of follow-up in the primary mice were usually not apparent until week 9 or even later (Figure 2A).
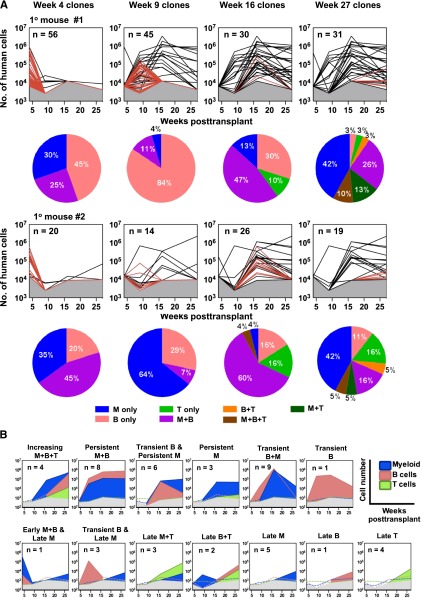
Figure 2.
Kinetics of clone appearance, size, persistence, and lineage content. (A) Lines depict changes throughout time in the total size of each barcoded (GFP+) clone in primary mice, extrapolated from the BM sample and distinguishing those only seen at the time indicated in the panel (shown as red lines) vs those also detected at other times (shown as black lines). Gray shaded region denotes the limit of detection by MPS. Each pie chart shows the relative distributions of different types of clones present at different times (of the line plot, above) according to the diversity of lineages present at that time. (B) Different patterns of lineage content of clones present at 27 weeks posttransplant. 13 clonal patterns were defined based on the changes (absolute increases or decreases) detected in the lineage content of each clone through time as illustrated by the representative plots shown. Colored dotted lines denote the absence of specific cell lineages at the indicated detection limit.
Examination of the clonal distribution of the human GM, B-, and T-lineages present in each sample analyzed showed that 29% (20/68) of the clones evident only in the 4-week BM aspirates contained exclusively GM (CD15/33+) cells (Figure 2A; supplemental Figure 3A). The remaining 48 of these early “transient” clones contained only B-lineage (CD19+) cells, or both GM and B cells. T cells (CD3+) were detected in 19 of the 174 clones identified in the primary mice, and only in clones that were first detected 16 weeks posttransplant (Figure 2A). Interestingly, 11 of these T-cell–containing clones contained at least one other lineage and continued to be detectable at 27 weeks posttransplant, although the additional lineages seen in them were variable (one with B cells only, 3 with myeloid cells only, and 7 with all 3 lineages at some point, although not necessarily simultaneously). The more durable clones showed no consistent pattern of mature cell content, although a sustained, high level content of lymphoid and myeloid cells (increasing M+B+T cells and/or persistent M+B cells) in them was frequently evident (Figure 2B; supplemental Figure 3B). However, 11 of the clones that were first seen in the harvested week-27 BM cells were still relatively small at that time (<6 × 104 cells/clone), with only myeloid (5/11) or only lymphoid cells (6/11) detected in them (supplemental Figure 3B).
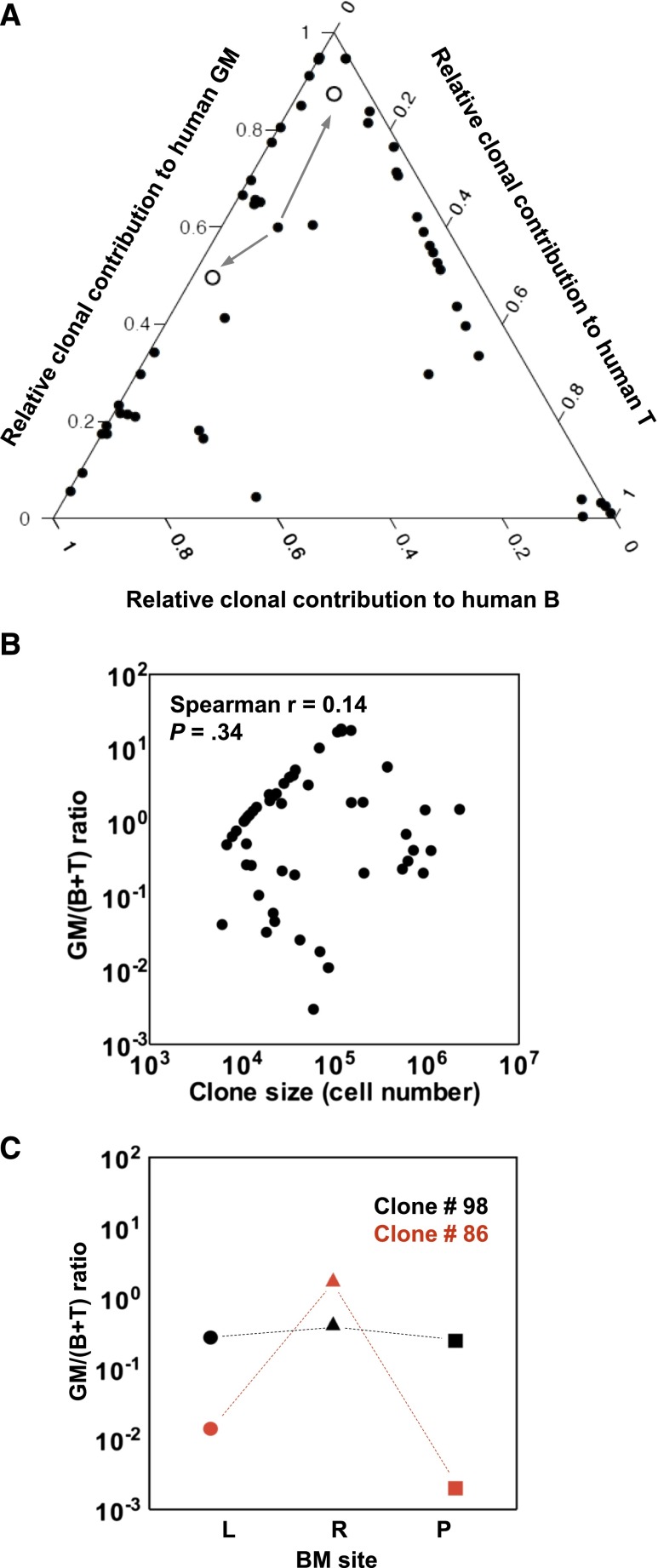
Previous studies of the clonal outputs of syngeneic transplants of mouse hematopoietic cells have shown that clones that can be serially propagated retain robust myeloid cell production but may vary markedly in their lymphopoietic activity.9,15,26,27 It was therefore of interest to determine whether the long-term clones generated from human CB cells (ie, the 50 evident at 27 weeks posttransplant) might show a similar segregated pattern of mature cell output, using the same definitions applied to the mouse clones. This involved first determining the ratio of the myeloid (GM) and lymphoid (B+T) cell contributions of each clone detected at 27 weeks to the total concomitant outputs of human myeloid and lymphoid cells in the same mouse at the same time. We then displayed these relative contributions to the total human myeloid and lymphoid lineages in each mouse as a ternary plot (Figure 3A). The derived GM/(B+T) ratios varied over a wide range (>6000-fold), similar to the spread of values seen in mice,9,15 and were also unrelated to the size of the clone (Figure 3B). However, it is interesting to note that a majority (52%) of the human CB cell-derived clones present 27 weeks posttransplant showed a high GM/(B+T) ratio (>1), which is highly predictive of durable self-renewal potential in the mouse.26
Figure 3.
Clonal contributions to the total human GFP+ GM, B- and T-cell compartments in all clones present at 27 weeks posttransplant. (A) Ternary plot showing the relative contribution to the GM, B-, and T-cell lineages of each clone present at 27 weeks posttransplant in the 2 primary mice (calculated as described in the text). Clone #110 persisted until weeks 20 and 24 posttransplant in the 2 secondary mice. Its contributions to the lineages assessed in both are indicated (arrows from clone #110 to the open circles, one for each of the secondary recipient mice). (B) Lack of correlation between GM/(B+T) value and clone size. (C) Examples of clones with similar and highly dissimilar GM/(B+T) ratio in different BM sites. The GM/(B+T) ratios of clone #98 (in black) and clone #86 (in red) in the BM of left leg (L, circles), right leg (R, triangles) and pelvis (P, squares) are shown.
In conclusion, a large number of human barcoded clones could be detected in serial BM aspirates obtained from NSG mice transplanted with sufficient numbers of CD34+ CB cells to produce high levels of repopulation for 7 months (∼5% to 10% of the mouse BM). Overall, the results showed a pattern of appearance and longevity expected from previous analyses of transplants of purified subsets of CB cells.1 Analysis of the lineage content of each clone sampled provided formal evidence of a few that produced GM, B-, and T cells, although the apparent representation of these lineages was diverse between clones and within clones throughout time.
Spatial heterogeneity in the lineage content of clones present 7 months posttransplant
To determine the extent of heterogeneity in the distribution of long-lived clones in different BM sites, we examined the clonal representation of human myeloid, B- and T cells in the pelvis and each of the hind legs of both primary mice at the time of their sacrifice 27 weeks posttransplant. One third of the 50 clones detected were present in all 3 BM sites examined (supplemental Figure 4A), although the lineage representation in the different BM sites was variable (Figure 3C; supplemental Figure 4B). The remaining clones were less homogeneously distributed, particularly those that were relatively small in size (<5.5 × 105 cells/clone; see supplemental Figure 4C). These findings suggest constrained mobility of the progeny of CD34+ CB cells when these are transplanted at nonlimiting conditions.
Clones perpetuated in secondary mice also show varied growth and differentiation patterns
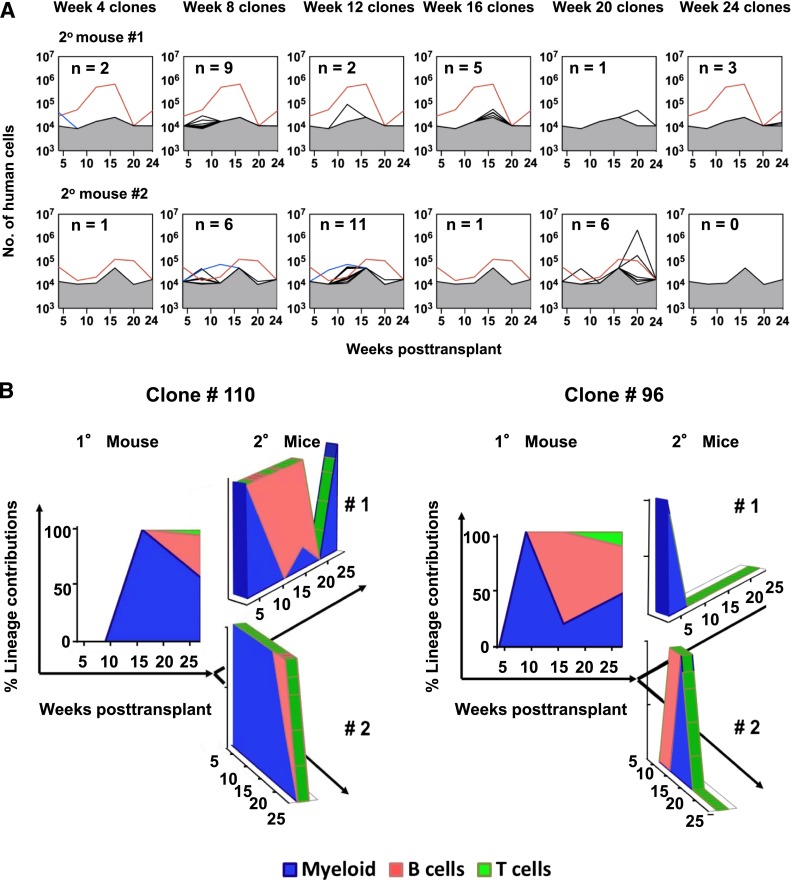
We next examined the number, time of appearance, size, lineage content, and origin of clones detected in serial BM aspirates obtained from secondary recipients of BM cells harvested from one of the primary mice (Figures 1B and 4A). 19% of the clones apparent in the primary mouse when it was sacrificed 7 months posttransplant gave rise to detectable clones in secondary mice, and the lineages detected in these were again highly variable over time (Table 1). The 2 most robust clones (#96 and #110) showed lympho-myeloid differentiation activity in both the primary and the secondary mice (Figure 4B). Clone #96 appeared early in the primary mouse (week 9), but persisted only to week 4 and 12 in the 2 secondary recipient mice, respectively. In contrast, clone #110 appeared later (week 16) in the primary mouse and persisted to week 24 in both secondary recipients (Figure 4A-B), demonstrating a sustained robust output of multiple types of blood cells for at least 51 weeks. In another 5 clones, B cells were exclusively and transiently evident in the samples obtained at week 8 in the secondary mice, including one (#94) in which only B cells had been transiently detected at week 9 in the primary mouse. Detectable T-cell production in the primary host did not appear to be associated with secondary repopulating activity (supplemental Table 5).
Figure 4.
Changes throughout time in the lineage content of clones regenerated in secondary mice. (A) Changes through time in the total size of each barcoded (GFP+) clone detected in the 2 secondary mice. Clone #110 (red line) and clone #96 (blue line) are shown separately. Gray shaded region denotes the limit of detection by MPS. (B) Relative contributions of GM, B-, and T-cells to clones #110 and #96 over time in the secondary mice as compared with the primary mouse in which they were first seen.
Table 1.
Mature cell output (×103) of clones serially repopulating 1 primary and 2 secondary recipients
| Week | Clone 110 | Clone 96 | Clone 65 | Clone 99 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | B | T | M | B | T | M | B | T | M | B | T | ||
| Primary recipient | 4 | — | — | — | — | — | — | — | — | — | — | — | — |
| 9 | — | — | — | 23 | — | — | 32 | 121 | — | — | — | — | |
| 16 | 78 | — | — | 198 | 792 | — | 122 | 618 | — | 242 | 335 | — | |
| 27 | 511 | 364 | 13 | 318 | 239 | 87 | 113 | 759 | — | 117 | — | — | |
| Secondary recipients |
4 | 18/42 | −/− | −/− | 31/- | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 8 | 23/5 | 23/- | −/− | 0.3/- | -/19 | −/− | −/− | 2/- | −/− | −/− | 12/- | −/− | |
| 12 | -/10 | 481/- | −/− | -/62 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| 16 | 123/68 | 517/- | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| 20 | -/36 | -/50 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| 24 |
39/- |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
−/− |
|
|
Week |
Clone 94 |
Clone 98 |
Clone 113 |
||||||||||
|
M |
B |
T |
M |
B |
T |
M |
B |
T |
|||||
| Primary recipient | 4 | — | — | — | — | — | — | — | — | — | |||
| 9 | — | 1 | — | — | — | — | — | — | — | ||||
| 16 | — | — | — | 173 | 1372 | — | — | — | — | ||||
| 27 | — | — | — | 236 | 788 | — | — | 20 | 38 | ||||
| Secondary recipients | 4 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |||
| 8 | −/− | 1/- | −/− | −/− | 5/4 | −/− | −/− | 2/6 | −/− | ||||
| 12 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | ||||
| 16 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | ||||
| 20 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | ||||
| 24 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | ||||
B, B cells; M, myeloid; T, T cells; —, no detectable output.
A total of 22 small clones (<8 × 104 cells, S1-S22) were detected for the first time and only at a single time in secondary mice. In most of these, only a single lineage was evident (supplemental Table 6), although 3 (#S2, #S3, and #S11) were evident in both of the secondary recipients, and, for 5, the identifying barcode sequence was detected in the primary recipient but was below the reliable detection threshold (supplemental Table 7). None of the human cells produced in the secondary mice included a T-cell component (supplemental Table 6).
The results obtained from the secondary mice thus establish the ability of multipotent CB cells with long-term repopulating activity to execute self-renewal divisions in NSG mice. They also suggest a very delayed onset of production of differentiated cells from some transplanted CB cells.
Discussion
This study provides a first detailed description of the in vivo growth and differentiation dynamics of clones generated by transplants of nonlimiting numbers of human CD34+ CB cells evident from serial BM aspirates obtained during a period of 13 months in primary and secondary NSG mice. In the 2 primary mice that were each transplanted with ∼3.6 × 104 barcoded cells (plus ∼6.4 × 104 nontransduced cells), 174 clones surpassed our detection threshold in the BM aspirate or final BM harvest assessed. This number indicates a minimal frequency of repopulating cells within the CD34+ cell compartment of ∼0.2%. Of the 174 clones, 50 were still large enough to be detected at 7 months posttransplant. This frequency translates to ∼0.05% to 0.1% of the CD34+ CB population, a value that is remarkably consistent with previously reported frequencies of total CB repopulating cells derived from limiting dilution transplant data.28 However, it is quite possible that the number of clones detected in our study represents an underestimate given that, before sacrifice, all clone numbers were derived from the small fraction (0.01% to 1%) of the total BM obtained in serial femoral aspirates. The heterogeneity in lineage representation of many clones between different BM sites as late as 27 weeks posttransplant also suggests that primitive human hematopoietic cells recirculate throughout the host less frequently than anticipated from experiments in mouse parabionts.29 This finding has important implications for tracking studies that typically rely on sampling a single BM site.
Importantly, 2 clones from one of the primary mice (#1) showed robust self-renewing, multilineage differentiation activity in secondary recipients. Thus, the frequency of CD34+ CB cells capable of sustained self-maintenance of multipotency must be at least 0.02% (2 out of 3.6 × 104 barcoded cells transplanted into the first primary mouse), and is likely higher because of the multiple detection caveats inherent in deriving this estimate. In addition to sampling limitations, differences in the supportive properties of the microenvironment within the mice used as recipients, would affect the extent to which different lineage progenitors are stimulated to produce mature progeny, and hence, influence the timed appearance, size, and composition of individual clones. This is well exemplified by the relatively exaggerated output of maturing human B-lineage cells relative to GM cells, and the almost complete lack of human erythropoiesis in this xenograft model.
It was thus reassuring to obtain results that are consistent with previous evidence that human myeloid and B-lymphoid cell production in transplanted recipients is initially derived from cells with transient repopulating activity and later (after 16 weeks) from cells that establish more durable clones.6,8,17,21,30-35 These findings also mirror the prolonged periods (≥16 weeks) required to distinguish outputs of mature blood cells from biologically distinct subsets of mouse11,26,36 and monkey cells7 with durable vs extensive, but limited, self-renewal ability.26,36 They are also consistent with the demonstrated association of rapid but short-term repopulating activity of human CD34+ hematopoietic cells that are CD38+30,34 and lack aldehyde dehydrogenase activity.35 However, our barcoding data detected a greater lineage output diversity in the early transient clones than has been previously documented. Thus, more extensive clonal tracking strategies using the approach described here should be valuable to delineate molecularly the process of lineage restriction in primitive human hematopoietic cells.
An interesting finding was the high number of small clones that were detected late in both primary and secondary mice including some in which only a single lineage of mature cells was detected. Assuming such clonal growth behavior is not entirely a result of sampling variations, it could suggest that the cells from which these clones arose, or their initial progeny, remained largely quiescent, died, or, in some cases, executed exclusively self-renewal divisions in the primary mice before generating an expanded, albeit limited, population of differentiated cells in the secondary mice. Alternatively, such a result could reflect the operation of mechanisms that limit the activation of repopulating cells when nonlimiting numbers of cells are present in the same environment, or it could reflect inadequacies of the mouse environment to elicit the potential of transplanted human cells. Further studies will be useful to discriminate among these possibilities.
This study also provides the first definitive evidence of human CD3+ T-cell generation from CD34+ CB cells with multilineage differentiation potential. Consistent with a recent study of bulk transplants of ALDH+ CB cells,33 clones containing T cells were not detected within the first month posttransplant. We also noted a progressive time-dependent decline in human lymphoid cell output without a concomitant decrease in myeloid cell production both within clones and in clones that appeared at later times (supplemental Figure 4C) as noted in long-term studies of nontransduced cells.37
The extensive and complex patterns of clonal activity evident from an examination of 2 xeno-transplanted NSG mice attests to the feasibility and power of the barcoding methodology to identify many clones generated in the BM of mice transplanted with nonlimiting numbers of human hematopoietic cells. Although this transplant model has many caveats, the high resolution and sensitivity of clone size detection afforded by MPS of barcoded cells should enable this strategy to be applied usefully for interrogating the effects of many types of manipulations of the human cells with in vivo repopulating activity.
Supplementary Material
Acknowledgments
The authors thank the staff of the British Columbia Women’s Hospital and Health Centre and the staff of the British Columbia Cancer Agency Stem Cell Assay Laboratory for assistance in procuring, processing, and cryopreserving CB samples, they thank G. Edin and M. Hale for excellent technical assistance, and they also thank other members of the Eaves’ laboratory for helpful discussions.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), the Canadian Stem Cell Network, a Terry Fox Foundation program project grant (TFF-122869), Health Canada and the Public Health Agency of Canada. A.M.S.C. held a Croucher Foundation Fellowship from Hong Kong. L.V.N. and D.J.H.F.K. held CIHR Vanier Canada Studentships. P.B. held a Kay Kendall Leukemia Fund Intermediate Fellowship from the United Kingdom. P.H.M. held a CIHR Transplantation Training Program Scholarship and a CIHR Doctoral Award. K.D. held a Co-op Studentship award from the Canadian Stem Cell Network.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.M.S.C., P.B., P.H.M., and C.J.E. designed the experiments; A.M.S.C., L.V.N., P.H.M., P.B., and K.D. performed the experiments; L.V.N., A.C., D.J.H.F.K., and M.H. performed the bioinformatics analysis; A.M.S.C., L.V.N., P.B., M.H., and C.J.E. wrote the manuscript; and all authors read and agreed with the content of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Connie J. Eaves, Terry Fox Laboratory, BC Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: ceaves@bccrc.ca.
References
- 1.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci USA. 1997;94(10):5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conneally E, Cashman J, Petzer A, Eaves C. Expansion in vitro of transplantable human cord blood stem cells demonstrated using a quantitative assay of their lympho-myeloid repopulating activity in nonobese diabetic-scid/scid mice. Proc Natl Acad Sci USA. 1997;94(18):9836–9841. doi: 10.1073/pnas.94.18.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller-Sieburg CE, Sieburg HB. Clonal diversity of the stem cell compartment. Curr Opin Hematol. 2006;13(4):243–248. doi: 10.1097/01.moh.0000231421.00407.65. [DOI] [PubMed] [Google Scholar]
- 5.Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4(2):220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt M, Zickler P, Hoffmann G, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100(8):2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 7.Kiem HP, Sellers S, Thomasson B, et al. Long-term clinical and molecular follow-up of large animals receiving retrovirally transduced stem and progenitor cells: no progression to clonal hematopoiesis or leukemia. Mol Ther. 2004;9(3):389–395. doi: 10.1016/j.ymthe.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Glimm H, Schmidt M, Fischer M, et al. Efficient marking of human cells with rapid but transient repopulating activity in autografted recipients. Blood. 2005;106(3):893–898. doi: 10.1182/blood-2004-07-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29(10):928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerrits A, Dykstra B, Kalmykowa OJ, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115(13):2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 11.Verovskaya E, Broekhuis MJ, Zwart E, Ritsema M, van Os R, de Haan G, Bystrykh LV. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013;122(4):523–532. doi: 10.1182/blood-2013-01-481135. [DOI] [PubMed] [Google Scholar]
- 12.Grosselin J, Sii-Felice K, Payen E, Chretien S, Roux DT, Leboulch P. Arrayed lentiviral barcoding for quantification analysis of hematopoietic dynamics. Stem Cells. 2013 doi: 10.1002/stem.1383. [DOI] [PubMed] [Google Scholar]
- 13.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 14.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 15.Benz C, Copley MR, Kent DG, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12(11):786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 18.Björgvinsdóttir H, Bryder D, Sitnicka E, et al. Efficient oncoretroviral transduction of extended long-term culture-initiating cells and NOD/SCID repopulating cells: enhanced reconstitution with gene-marked cells through an ex vivo expansion approach. Hum Gene Ther. 2002;13(9):1061–1073. doi: 10.1089/104303402753812467. [DOI] [PubMed] [Google Scholar]
- 19.Leurs C, Jansen M, Pollok KE, et al. Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34+ cord blood cells. Hum Gene Ther. 2003;14(6):509–519. doi: 10.1089/104303403764539305. [DOI] [PubMed] [Google Scholar]
- 20.Ando K, Yahata T, Sato T, et al. Direct evidence for ex vivo expansion of human hematopoietic stem cells. Blood. 2006;107(8):3371–3377. doi: 10.1182/blood-2005-08-3108. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7(11):1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- 22.Schepers K, Swart E, van Heijst JW, et al. Dissecting T cell lineage relationships by cellular barcoding. J Exp Med. 2008;205(10):2309–2318. doi: 10.1084/jem.20072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bystrykh LV, Verovskaya E, Zwart E, Broekhuis M, de Haan G. Counting stem cells: methodological constraints. Nat Methods. 2012;9(6):567–574. doi: 10.1038/nmeth.2043. [DOI] [PubMed] [Google Scholar]
- 24.Logan AC, Nightingale SJ, Haas DL, Cho GJ, Pepper KA, Kohn DB. Factors influencing the titer and infectivity of lentiviral vectors. Hum Gene Ther. 2004;15(10):976–988. doi: 10.1089/hum.2004.15.976. [DOI] [PubMed] [Google Scholar]
- 25.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dykstra B, Kent D, Bowie M, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111(12):5553–5561. doi: 10.1182/blood-2007-11-123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294(5548):1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 30.Glimm H, Eisterer W, Lee K, et al. Previously undetected human hematopoietic cell populations with short-term repopulating activity selectively engraft NOD/SCID-beta2 microglobulin-null mice. J Clin Invest. 2001;107(2):199–206. doi: 10.1172/JCI11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazurier F, Doedens M, Gan OI, Dick JE. Rapid myeloerythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nat Med. 2003;9(7):959–963. doi: 10.1038/nm886. [DOI] [PubMed] [Google Scholar]
- 32.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1(6):635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Chen BJ, Deoliveira D, Sempowski GD, Chao NJ, Storms RW. Progenitor cell dose determines the pace and completeness of engraftment in a xenograft model for cord blood transplantation. Blood. 2010;116(25):5518–5527. doi: 10.1182/blood-2009-12-260810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanjani ED, Flake AW, Rice H, Hedrick M, Tavassoli M. Long-term repopulating ability of xenogeneic transplanted human fetal liver hematopoietic stem cells in sheep. J Clin Invest. 1994;93(3):1051–1055. doi: 10.1172/JCI117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung AM, Leung D, Rostamirad S, et al. Distinct but phenotypically heterogeneous human cell populations produce rapid recovery of platelets and neutrophils after transplantation. Blood. 2012;119(15):3431–3439. doi: 10.1182/blood-2011-12-398024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benveniste P, Frelin C, Janmohamed S, et al. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 2010;6(1):48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Sloma I, Beer PA, Saw KM, et al. Genotypic and functional diversity of phenotypically defined primitive hematopoietic cells in patients with chronic myeloid leukemia. Exp Hematol. 2013 doi: 10.1016/j.exphem.2013.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.