Background: Ferredoxin:NAD+-oxidoreductases (Rnf) found in many bacteria are novel ion-translocating electron transport chains.
Results: A Na+ requirement for the reaction and its reversible coupling to the transmembrane Na+ gradient are demonstrated.
Conclusion: Na+ is the coupling ion. Rnf not only generates a Na+ potential but also uses it to drive the reverse reaction.
Significance: Evidence for a function of Rnf in ferredoxin reduction is provided.
Keywords: Bioenergetics/Electron Transfer Complex, Electron Transfer, Energy Metabolism, Enzyme Kinetics, Membrane Energetics, Membrane Transport, Oxidation-Reduction, Physiology, Sodium Transport, Rnf
Abstract
The anaerobic acetogenic bacterium Acetobacterium woodii has a novel Na+-translocating electron transport chain that couples electron transfer from reduced ferredoxin to NAD+ with the generation of a primary electrochemical Na+ potential across its cytoplasmic membrane. In previous assays in which Ti3+ was used to reduce ferredoxin, Na+ transport was observed, but not a Na+ dependence of the electron transfer reaction. Here, we describe a new biological reduction system for ferredoxin in which ferredoxin is reduced with CO, catalyzed by the purified acetyl-CoA synthase/CO dehydrogenase from A. woodii. Using CO-reduced ferredoxin, NAD+ reduction was highly specific and strictly dependent on ferredoxin and occurred at a rate of 50 milliunits/mg of protein. Most important, this assay revealed for the first time a strict Na+ dependence of this electron transfer reaction. The Km was 0.2 mm. Na+ could be partly substituted by Li+. Na+ dependence was observed at neutral and acidic pH values, indicating the exclusive use of Na+ as a coupling ion. Electron transport from reduced ferredoxin to NAD+ was coupled to electrogenic Na+ transport, indicating the generation of Δμ̃Na+. Vice versa, endergonic ferredoxin reduction with NADH as reductant was possible, but only in the presence of Δμ̃Na+, and was accompanied by Na+ efflux out of the vesicles. This is consistent with the hypothesis that Rnf also catalyzes ferredoxin reduction at the expense of an electrochemical Na+ gradient. The physiological significance of this finding is discussed.
Introduction
Growth and acetate formation as carried out by the acetogenic bacterium Acetobacterium woodii are strictly sodium ion-dependent (1). Sodium ions are essential for ATP synthesis, rotation of the flagellar motor, and accumulation of nutrients that are taken up by secondary transport systems (2–5). Acetogenesis as carried out by resting cells is coupled to the generation of a transmembrane electrochemical Na+ gradient across the cytoplasmic membrane that is the driving force for the above-mentioned endergonic membrane processes. The nature of the enzyme(s) that generates Δμ̃Na+ has been obscure for decades, but we recently demonstrated a ferredoxin:NAD+ oxidoreductase activity at the cytoplasmic membrane of A. woodii (6). This enzyme is part of the electron transport chain leading from, for example, hydrogen to carbon dioxide or caffeate as electron acceptor. The enzyme was partially purified, and polypeptides in the preparation were identified to be encoded by rnf genes (7) that were hypothesized before in Rhodobacter capsulatus to encode a membrane-bound, ion-motive electron transport system (8). Rnf complexes are now widely believed to be of central importance for the bioenergetics of anaerobic bacteria (and some archaea), and very often, they are the only ion-motive electron transport chain present in these anaerobes (9). Inspection of the genome sequence of A. woodii did not reveal any candidate apart from the Rnf cluster that could potentially link acetogenesis or caffeate respiration to the generation of Δμ̃Na+ (10). Experiments using inverted membrane vesicles clearly revealed that the ferredoxin:NAD+ oxidoreductase is coupled to Na+ transport (11). However, the interpretation of these data as evidence for the use of Na+ as coupling ion was not unambiguous because a Na+ dependence of NAD+ reduction was not observed, and it could not be excluded that Na+ transport is only a side reaction of the enzyme. We have followed up this central question and present here an enzyme assay that enabled us to demonstrate a strict Na+ dependence of Rnf-catalyzed electron transport. Moreover, electron transport was coupled to the generation of Δμ̃Na+ that, vice versa, drove endergonic electron flow from NADH to ferredoxin.
EXPERIMENTAL PROCEDURES
Growth Conditions
For preparation of membranes and purification of acetyl-CoA synthase/CO dehydrogenase (Acs/CODH),2 A. woodii (DSM 1030) was grown at 30 °C on 20 mm fructose under anaerobic conditions as described previously (2). For preparation of vesicles, A. woodii was grown on 20 mm fructose as described (11) in 20-liter flasks (Glasgerätebau Ochs, Bovenden-Lenglern, Germany).
Preparation of Membranes
Membranes of fructose-grown cells were prepared as described previously (6). The cell-free extract was separated into cytoplasmic and membrane fractions by ultracentrifugation at 150,000 × g for 2 h at 4 °C. The resulting sediment was washed once with membrane buffer (50 mm Tris-HCl (pH 8.5) containing 20 mm MgSO4, 20% glycerol, 2 mm dithioerythritol (DTE) as a reducing agent, and 4 μm resazurin as a redox indicator).
Preparation of Vesicles
Vesicles were prepared as described (11) but with slight modifications. Protoplasts were harvested anaerobically by centrifugation at 6250 × g for 20 min at 4 °C and washed with vesicle buffer (50 mm Tris-HCl (pH 8) containing 25 mm MgSO4, 420 mm sucrose, 8 mm DTE, and 4 μm resazurin). After washing the protoplasts, they were resuspended in a total volume of 10 ml of vesicle buffer. These protoplasts were passed through a French pressure cell at 41 megapascals (MPa) and centrifuged three times at 4500 × g for 35 min at 4 °C. The resulting supernatant was centrifuged further by ultracentrifugation at 120,000 × g for 40 min at 4 °C. The pellet was washed with vesicle buffer and centrifuged again. The resulting pellet was resuspended in the same buffer in a volume of 5 ml.
Purification of Acs/CODH
For purification of Acs/CODH, A. woodii (DSM 1030) was grown at 30 °C under anaerobic conditions in 20-liter flasks using 20 mm fructose to an A600 of ∼2.0 as described previously (2). The cytoplasmic fraction was prepared as described previously (12). The cytoplasmic fraction containing ∼1000 mg of protein was applied to Q-Sepharose high performance column (2.6 × 5 cm) equilibrated with buffer A (25 mm Tris-HCl, 20 mm MgSO4, 150 mm NaCl, and 20% glycerol (pH 7.5)). Protein was eluted with a 170-ml linear gradient of 150–400 mm NaCl in buffer A. Methyl viologen-dependent CO dehydrogenase activity eluted at ∼350 mm NaCl. Ammonium sulfate (0.8 m) was added to the pooled fractions, and these were loaded onto a phenyl-Sepharose high performance column (1.6 × 10 cm) equilibrated with buffer B (25 mm Tris-HCl, 20 mm MgSO4, 0.8 m (NH4)2SO4, and 20% glycerol (pH 7.5)). Protein was eluted with a 160-ml linear gradient from 0.8 to 0 m (NH4)2SO4 in buffer B. Acs/CODH activity eluted in a peak at ∼0.4 m (NH4)2SO4. Pooled fractions were concentrated by ultrafiltration in 50-kDa Vivaspin tubes (Sartorius Stedim Biotech GmbH, Göttingen, Germany), applied to a Sephacryl S-300 high resolution column (1.6 × 60 cm) equilibrated with buffer C (25 mm Tris-HCl, 20 mm MgSO4, and 20% glycerol (pH 7.5)), and eluted with buffer C at a flow rate of 0.8 ml/min. Acs/CODH activity eluted after 54 ml. The size of the protein was determined using a high molecular weight calibration kit (GE Healthcare) with conalbumin, aldolase, ferritin, and thyroglobulin as size standards run under identical conditions as Acs/CODH on a Sephacryl S-300 column. The enzyme was stable for weeks when stored at 4 °C.
Measurement of Rnf Activity
All measurements were performed at 30 °C in anaerobic cuvettes (Glasgerätebau Ochs) sealed by rubber stoppers with 1 ml of 20 mm Tris (sodium-free)-HCl (pH 7.7) containing 2 mm DTE and 2 μm resazurin in 3-fold distilled water. Measurement of CO-mediated ferredoxin:NAD+ oxidoreductase activity was performed at a pressure of 0.5 × 105 Pa of CO with 30 μg/ml Acs/CODH, 30 μm ferredoxin, and 150 μg/ml washed membranes. Ferredoxin was purified from Clostridium pasteurianum as described (13). The reaction was started by the addition of NAD+. Formation of NADH was measured photometrically at 340 nm (ϵNADH = 6.2 mm−1 cm−1). NaCl was added as indicated. For determination of electron transfer from reduced ferredoxin to NAD+, membrane vesicles were added at a concentration of 50 μg/ml. ETH2120, 3,3′,4′,5-tetrachlorosalicylanilide, valinomycin, and monensin were added at a concentration of 10 μm each. The potassium concentration inside the vesicles in the assay with valinomycin and potassium was 50 mm. Measurement of titanium citrate-mediated ferredoxin:NAD+ oxidoreductase activity was performed in a N2 atmosphere as described for the Acs/CODH-mediated assay, except that Acs/CODH was substituted with 5 mm titanium citrate, prepared according to Zehnder and Wuhrmann (14), and sodium salts were substituted with potassium salts. Electron transfer from reduced methyl viologen to NAD+ was measured at 340 nm as described for the CO-mediated ferredoxin:NAD+ oxidoreductase activity measurement, except that ferredoxin was substituted with 10 μm methyl viologen. Electron transfer from NADH to ferredoxin was measured in 100 mm Tris (pH 7.5), 5 mm MgCl2, 2 mm NaCl, 30 μm ferredoxin, 2 mm DTE, and 2 μm resazurin in a nitrogen atmosphere with 50 μg/ml washed membrane vesicles. ATP was added at a concentration of 2 mm. The reaction was started by the addition of 500 μm NADH. ETH2120 was used at a concentration of 10 μm.
Measurement of Acs/CODH Activity
Measurements were performed at 30 °C in anaerobic cuvettes filled with 1 ml of buffer containing 100 mm HEPES/NaOH (pH 7.0), 2 mm DTE, and 2 μm resazurin at a gas phase of 100% CO with an overpressure of 1.1 × 105 Pa. For CO-dependent reduction of methyl viologen (10 mm), activity was measured at 604 nm (ϵ = 13.9 mm−1 cm−1). Reduction of ferredoxin (20 μm) was assayed at 430 nm (ϵ = 13.1 mm−1 cm−1).
Measurement of Na+ Translocation
The experiments for Rnf-mediated Na+ import into the inverted membrane vesicles were performed under anaerobic conditions in 20 mm Tris (sodium-free)-HCl (pH 7.7) containing 2 mm DTE and 4 μm resazurin in 3-fold distilled water at 30 °C as described previously (2, 11). In 3.5-ml glass vials, 1 ml of buffer was supplemented with 30 μm ferredoxin and 3 mg/ml vesicles. The vials were pressurized with an atmosphere of CO or N2 (0.5 × 105 Pa). NaCl was added at the concentrations indicated; the contaminating amount of Na+ was 65 μm. Radioactive 22Na+ (carrier-free) was added to a final concentration of 0.5 μCi/ml. The sample was incubated for 20 min at 30 °C to ensure equilibration of 22Na+ before the reaction was started. Acs/CODH (30 μg/ml) or titanium citrate (5 mm) was added before the reaction start, which was done by the addition of NAD+ (concentrations as indicated). If CO-reduced methyl viologen was tested as electron donor, ferredoxin was substituted with 1 mm methyl viologen. The experiments for Rnf-mediated export of Na+ were performed as described above in buffer containing 100 mm Tris (pH 7.5), 5 mm MgCl2, 2 mm NaCl, 100 μm ferredoxin, 2 mm DTE, and 2 μm resazurin in a nitrogen atmosphere. ATP was added at a concentration of 2 mm. Sodium export was induced by the addition of 5 mm NADH.
Determination of Na+ Concentration
The Na+ concentration in the buffer was determined with an Orion 84-11 ROSS sodium electrode (Thermo Electron Corp., Witchford, United Kingdom) as described (15).
Analytical Methods
Protein concentration was measured according to Bradford (16). Proteins were separated under denaturing conditions on 12% polyacrylamide gels (17) or under native conditions using clear native PAGE according to Wittig et al. (18) with a gradient of 5–11% polyacrylamide. Proteins were stained with Coomassie Brilliant Blue G-250.
RESULTS
Establishing a CO-dependent Enzyme Assay for Rnf
The ferredoxin:NAD+ oxidoreductase activity is difficult to assay because reduction of the electron donor ferredoxin (E0′ ∼ −500 mV) requires electrons of very low redox potential. In the assay that was used previously (6, 11), these were provided by Ti3+ (E0′ ∼ −480 mV) (15). However, Ti3+ is a very strong and unspecific reducing agent. For example, it is able to reduce NAD+ directly in a pH-dependent manner (19). This is the reason that the assay for ferredoxin:NAD+ oxidoreductase is usually done at pH 6. However, this assay did not reveal a Na+ dependence of ferredoxin-dependent NAD+ reduction.
An alternative biological reducing agent is carbon monoxide (E0′(CO/CO2) = −520 mV), which is oxidized to carbon dioxide by Acs/CODH from A. woodii. The electron acceptor in this reaction is ferredoxin (20). Acs/CODH was purified from a cell-free extract of A. woodii grown on fructose by chromatography on Q-Sepharose, phenyl-Sepharose, and Sephacryl S-300. This procedure led to an enrichment of 9-fold with a yield of 5%. The preparation contained primarily proteins of ∼63 and 78 kDa as derived from denaturing gel electrophoresis (Fig. 1A), which correspond in mass to the AcsA and AcsB subunits of Acs/CODH encoded by the genome of A. woodii. Separation by native gel electrophoresis revealed one major protein complex with an apparent mass of ∼350 kDa (Fig. 1B), which fitted well with the values obtained by calibrated gel filtration (Fig. 1C). The purified Acs/CODH catalyzed CO oxidation with methyl viologen (70 units/mg) or ferredoxin (11 units/mg) from C. pasteurianum as an electron acceptor.
FIGURE 1.
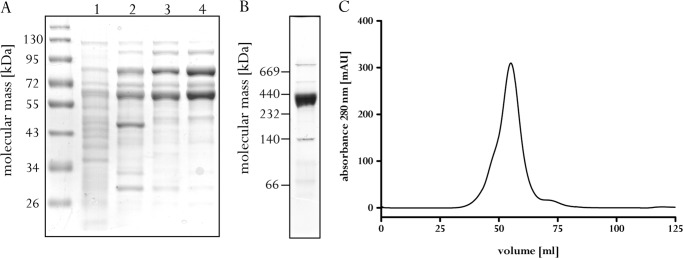
Monitoring the purification and subunit composition of Acs/CODH from A. woodii. A, protein samples (10 μg) of each purification step were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, cytoplasm; lane 2, sample after Q-Sepharose; lane 3, sample after phenyl-Sepharose; lane 4, sample after Sephacryl S-300. B, 15 μg of purified Acs/CODH was separated by clear native gel electrophoresis and stained with Coomassie Brilliant Blue. C, elution profile of Acs/CODH on Sephacryl S-300. mAU, milli-absorbance units.
Next, we tested whether the purified Acs/CODH was able to reduce NAD+ directly. As this was not the case (Fig. 2), we added washed membranes from A. woodii. This also did not result in NAD+ reduction, showing that the preparation did not contain ferredoxin and that there was no other entry port for electrons into the membrane. After the addition of ferredoxin, NAD+ was reduced at a rate of 50 milliunits/mg of protein. This activity was strictly dependent on the presence of ferredoxin, CO dehydrogenase, CO, and membranes.
FIGURE 2.
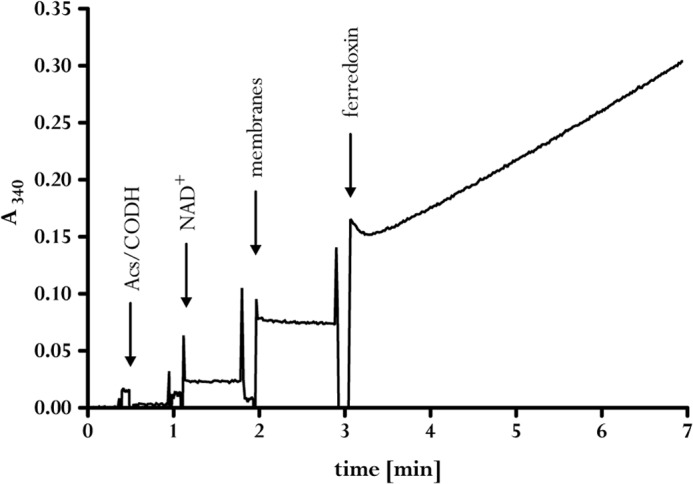
Membranes of A. woodii catalyze ferredoxin-dependent reduction of NAD+ with CO + Acs/CODH as reducing agent. Anaerobic cuvettes in an atmosphere of 0.5 × 105 Pa of CO contained 1 ml buffer containing 50 mm Tris-HCl, 20 mm NaCl (pH 7.7), 2 mm DTE, and 2 μm resazurin prepared in 3-fold distilled water. The addition of 30 μg of Acs/CODH, 4 mm NAD+, A. woodii membranes, and 30 μm ferredoxin led to an increase in absorption at 340 nm.
Sodium Ion Dependence of Ferredoxin:NAD+ Oxidoreductase Activity
With Acs/CODH and CO as the reduction system for ferredoxin, we reinvestigated the effect of Na+ on ferredoxin-dependent NAD+ reduction. As shown in Fig. 3, there was only low activity in the absence of added NaCl; the Na+ concentration in this assay due to contamination was 65 μm. However, upon addition of NaCl, electron transfer and thus NAD+ reduction were restored in a Michaelis-Menten-type manner. Half-maximal activity was at 200 μm, and saturation was at 3 mm. Li+ could partly substitute for Na+, but K+ only to a small extent. Other sodium salts such as sodium glutamate and sodium bromide were as effective as NaCl, but not the corresponding potassium salts. These data demonstrate a strict Na+ dependence of ferredoxin:NAD+ oxidoreductase activity.
FIGURE 3.
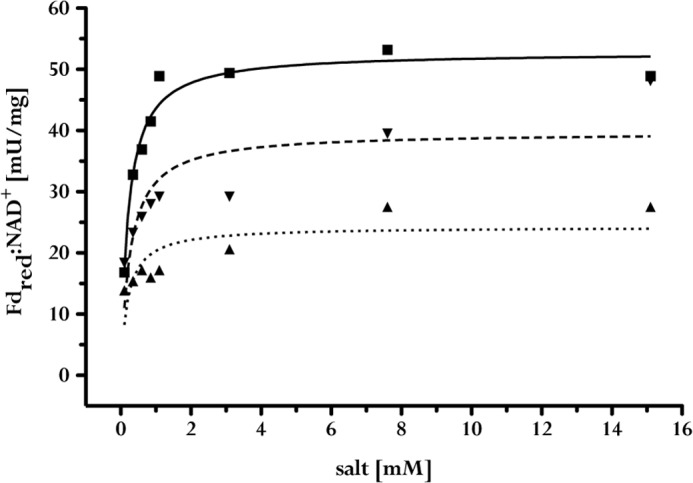
Na+ dependence of ferredoxin:NAD+ oxidoreductase activity at A. woodii membranes. Ferredoxin-dependent NADH formation at A. woodii membranes was monitored at 340 nm in 20 mm Tris-HCl (pH 7.7), 2 mm DTE, and 2 μm resazurin in 3-fold distilled water. Shown are the effects of NaCl (■), KCl (▴), and LiCl (▾) on NADH formation. The data shown are representative of the results of three separate experiments using independent membrane preparations. Curve fitting and determination of the kinetic parameters Km (Michaelis constant) and Vmax (maximal enzyme velocity) were performed using GraphPad Prism Version 4.03 and the Michaelis-Menten equation (Y = (Vmax × X)/(Km + X)). Fdred, reduced ferredoxin.
The ferredoxin:NAD+ oxidoreductase activity had a pH optimum at 7–8 and declined sharply below or above the optimum. When the enzyme was tested for its Na+ dependence at low pH (i.e. high proton concentrations), the results showed that the activity at low salt was similar at pH 6 and 7.7. The addition of NaCl led to the same Michaelis-Menten-type kinetics with a Km of 201 ± 30 μm and a Vmax of 50 milliunits/mg at pH 7.7 and 30 μm ferredoxin. At pH 6, the Km was 155 ± 39 μm, and the Vmax was 8 milliunits/mg at 30 μm ferredoxin. These analyses argue against a promiscuity of the enzyme toward Na+ and H+, as recently observed for other Na+-translocating enzymes such as ATP synthase (21) or the flagellar motor (22).
Different Electron Transfer Activities and Their Na+ Dependence
To dissect electron transfer and Na+ transport, we analyzed the effect of Na+ on the different activities catalyzed by Rnf. Again, Na+ stimulated NAD+ reduction by a factor of 5 (activity increased from ∼10 to 50 milliunits/mg after the addition of NaCl) when CO-reduced ferredoxin was used as reductant. When Ti3+ was added as reductant for ferredoxin, the NAD+ reduction rate was 65 milliunits/mg without salt, but the activity was stimulated by only a factor of 2 with 20 mm NaCl. There was even less stimulation when the reducing agent Ti3+ was used in the absence of ferredoxin (activity increased from 35 to 50 milliunits/mg). These data confirm our data from a previous study in which we could not unequivocally demonstrate a Na+ dependence of NAD+ reduction driven by Ti3+-reduced ferredoxin (6). In addition, they also demonstrate a ferredoxin-independent entry port(s) into the electron transport chain for electrons coming from Ti3+.
Reduced methyl viologen has a redox potential E0′ of −450 mV, so we tested whether methyl viologen reduced by Acs/CODH is used as an electron donor for NAD+ reduction. Indeed, NAD+ was reduced at a rate of 1 unit/mg, but this activity was independent of Na+ and also not coupled to Na+ transport.
Comparison of Na+ Transport Coupled to Different Electron Transfer Activities
As observed before, electron transfer from Ti3+-reduced ferredoxin in one time-washed vesicles was coupled to Na+ transport into the lumen of the vesicles. However, when Ti3+ was used in the absence of added ferredoxin, the same Na+ transport was observed (Fig. 4A). This is in accordance with previous data showing that Na+ translocation with Ti3+ as reductant was ferredoxin-dependent only if the vesicles were washed three times (11). In contrast, when CO-reduced ferredoxin was used as reductant, there was a clear dependence of the rate and the final accumulation factor on the ferredoxin concentration, although the vesicles were washed only once (Fig. 4B). These data are in accord with the previous measurements and the hypothesis that there is a ferredoxin-independent entry port of electrons derived from Ti3+ into the Rnf complex.
FIGURE 4.
22Na+ transport depends on different ferredoxin-reducing systems. The assay contained membrane vesicles (3 mg/ml) in 20 mm Tris-HCl (pH 7.7), 2 mm DTE, 2 μm resazurin, and 4 mm NAD+. Ferredoxin (30 μm) was added (■ and ▾) or omitted (▴ and ♦). The reducing agent used was Ti3+ (A) or CO + Acs/CODH (B). The data shown are representative of three experiments using independent inverted membrane vesicle preparations.
The Magnitude of the Na+ Gradient Depends on the NAD+ Concentration
NAD+ is the electron acceptor of the electron transport chain. NAD+ reduction was dependent on the concentration of the electron acceptor, and the dependence followed Michaelis-Menten kinetics. The apparent Km for NAD+ at a ferredoxin concentration of 30 μm was 85 ± 10 μm. Interestingly, the magnitude of Na+ accumulation was dependent on the NAD+ concentration. After a 10-min incubation, it was 4-fold at 0.2 mm NAD+ but increased to 11-fold at 4 mm NAD+ (Fig. 5). This is consistent with the ΔG′ of the reaction being the limiting factor for Δμ̃Na+ generation. An increase in NAD+/NADH ratio shifts the redox potential of the acceptor couple to more positive values and increases ΔG.
FIGURE 5.
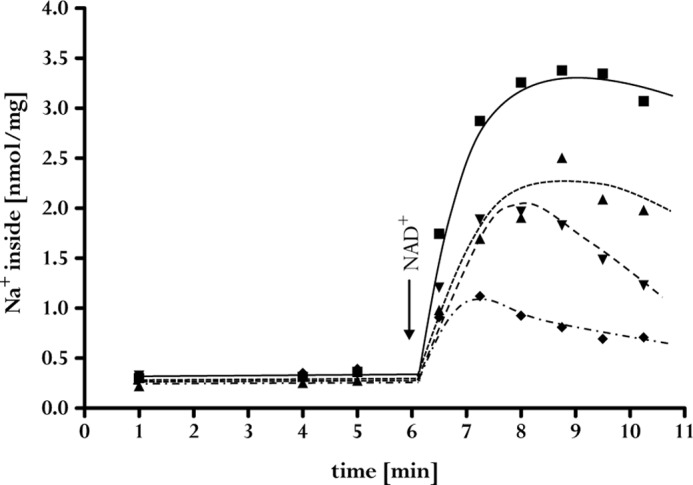
NAD+ dependence of Na+ transport. Membrane vesicles (3 mg/ml) were incubated in 20 mm Tris (pH 7.7), 2 mm DTE, 2 μm resazurin, 30 μg/ml Acs/CODH, and 30 μm ferredoxin in glass vials pressurized with 0.5 × 105 Pa of CO. The reaction was started by the addition of different NAD+ concentrations: 4 mm (■), 1.3 mm (▴), 0.6 mm (▾) or 0.2 mm (♦). The experiment was repeated three times using independent inverted membrane vesicle preparations.
Rnf-catalyzed Na+ Transport Is Electrogenic
After having established a sodium ion dependence of the enzymatic reaction and a dependence of the magnitude of the Na+ gradient on the electron acceptor concentration, we asked whether the Na+ transport is electrogenic. Reduced ferredoxin:NAD+ oxidoreductase activity at inverted membrane vesicles (100 ± 9%) was not stimulated by the protonophore 3,3′,4′,5-tetrachlorosalicylanilide but was stimulated by the sodium ionophore ETH2120 (155 ± 12%) and monensin (138 ± 15%), as well as by valinomycin in the presence of potassium (128 ± 20%). This is consistent with the generation of an electrical field during Na+ transport.
A Sodium Ion Gradient Drives NADH-dependent Ferredoxin Reduction
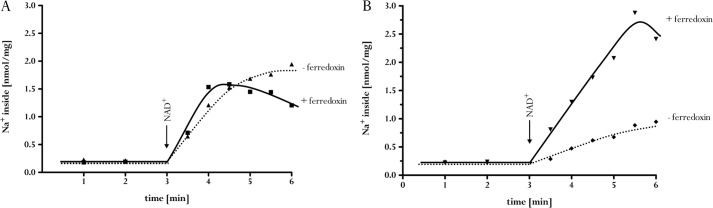
The experiments described so far are in full accord with the use of Na+ to couple the redox reaction to the electrical field across the membrane. Next, we determined whether the reaction is reversible. Therefore, a transmembrane Na+ gradient was generated by ATP hydrolysis, catalyzed by the Na+ F1F0-ATP synthase from A. woodii. ATP hydrolysis generated a much larger Na+ gradient than ferredoxin-dependent NAD+ reduction, as expected from the larger ΔG of the reaction. When NADH was added to the assay in the steady state of ATP hydrolysis, the Na+ content of the vesicles dropped from 36 to 6 nmol of Na+/mg (Fig. 6). This decrease was observed only in the presence of oxidized ferredoxin. The ferredoxin dependence indicated Na+ eflux from the inverted membrane vesicles coupled to ferredoxin reduction with NADH as reductant. This was indeed observed. When vesicles were incubated with ferredoxin and the membrane was energized by Na+ translocation coupled to ATP hydrolysis, ferredoxin was reduced (Fig. 7). Ferredoxin reduction was dependent on ATP and a transmembrane Na+ gradient, as evident from the observed inhibition of ferredoxin reduction by the sodium ionophore ETH2120.
FIGURE 6.
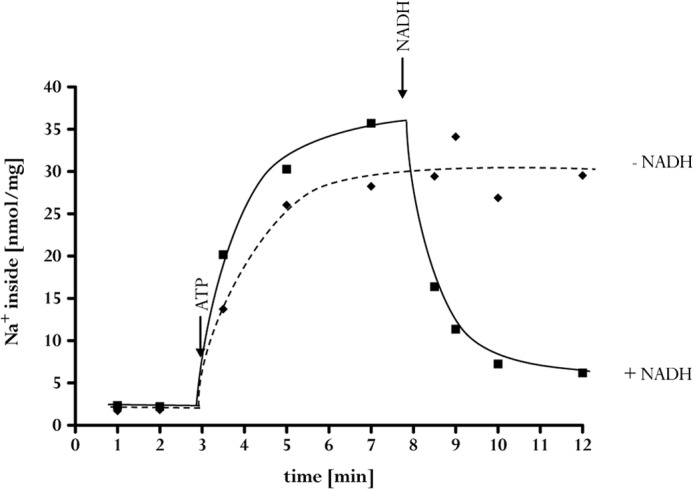
NADH-dependent 22Na+ export from inverted membrane vesicles. For sodium transport, membrane vesicles (3 mg) were incubated in 1 ml of 100 mm Tris (pH 7.5), 5 mm MgCl2, 2 mm NaCl, 100 μm ferredoxin, 2 mm DTE, and 2 μm resazurin in a nitrogen atmosphere. The reaction was started by the addition of 2 mm ATP. In the assay with NADH (■), the electron carrier was added at a concentration of 5 mm. In the assay without NADH (♦), water was added in an equal volume. The data shown are representative of three experiments using independent inverted membrane vesicle preparations.
FIGURE 7.
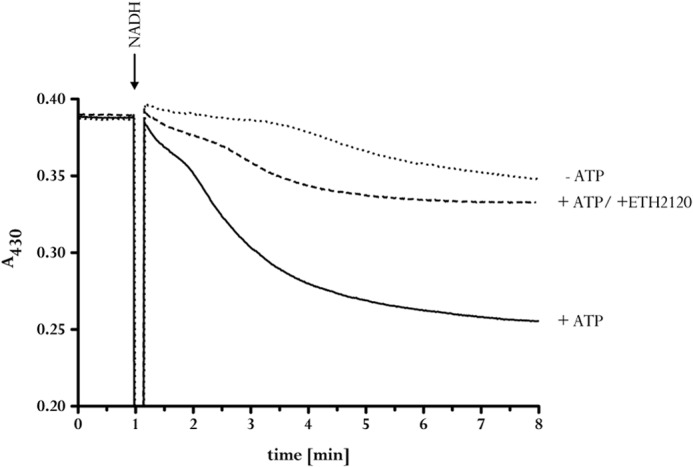
ATP- and NADH-dependent ferredoxin reduction at inverted membrane vesicles. For measurement of ferredoxin reduction at 430 nm, membrane vesicles (50 μg) were incubated in 1 ml of 100 mm Tris (pH 7.5), 5 mm MgCl2, 2 mm NaCl, 30 μm ferredoxin, 2 mm DTE, and 2 μm resazurin in a nitrogen atmosphere. The reaction was started by the addition of 2 mm ATP. ETH2120 was added at a concentration of 10 μm. The data shown are representative of three experiments using independent inverted membrane vesicle preparations.
DISCUSSION
A. woodii is one of the rare organisms that rely only on a sodium ion potential for cellular bioenergetics: its F1F0-ATP synthase is strictly sodium ion-dependent (3, 23–25), its flagellar motor is powered by a sodium-motive force (4), and it grows in the absence of a transmembrane electrochemical proton gradient, indicating that the essential secondary transporters are also Na+-coupled. A search for the enzyme that couples metabolism to the generation of the electrochemical sodium ion potential revealed that none of the enzymes that catalyze carbon flow in the Wood-Ljungdahl pathway are membrane-bound, excluding their role as Na+ pumps. This was recently corroborated by analyzing the proteins predicted from the genome sequence (10). Instead, we found a membrane-bound electron transfer chain that transfers electrons from reduced ferredoxin to NAD+ (6). Ferredoxin has a low redox potential (E0′ ∼ −500 mV), and its reduction with H2 (E0′ = −414 mm) as reductant is catalyzed by a soluble iron-only hydrogenase that uses the exergonic electron transfer from H2 to NAD+ (E0′ = −320 mm) to drive the endergonic ferredoxin reduction by a flavin-based electron bifurcation (12). NADH produced by the hydrogenase and the ferredoxin:NAD+ oxidoreductase is then used as reductant for reduction of carbon dioxide to acetate. A preparation of the partially purified ferredoxin:NAD+ oxidoreductase contained polypeptides encoded by the rnf genes of A. woodii (7). The Rnf complex is widespread in bacteria, and it was almost common sense that it uses exergonic electron transfer from ferredoxin to NAD+ to drive export of ions from the cytoplasm to the medium, thus establishing a primary electrochemical ion gradient across the bacterial cytoplasmic membrane (9). Indeed, we have demonstrated that ferredoxin-dependent NAD+ reduction is accompanied by primary Na+ transport in inside-out vesicles from A. woodii (11). This is clear evidence for Rnf-catalyzed Na+ transport under physiological conditions, but the interpretation was hampered by the fact that we could not observe a Na+ dependence of the electron transfer reaction. This is in contrast to other Na+-translocating enzymes such as decarboxylases (26, 27), methyltransferases (28, 29), and ATP synthases (25, 30, 31) or flagellar motors (4, 32), in which the enzymatic activity/rotation was clearly Na+-dependent.
The ferredoxin:NAD+ oxidoreductase is usually measured using ferredoxin that has been reduced by Ti3+ (6, 14, 19, 33). This assay has a number of inherent problems such as a low concentration of “electrons,” and the assay does not reliably require ferredoxin, indicating that Ti3+-derived electrons enter the enzyme at different sites and direct non-enzymatic electron flow from Ti3+ to NAD+ at pH values above 7. Furthermore, the assay is hardly reproducible, probably due to the high instability of Ti3+. These data indicate a weak coupling and/or uncoupling of electron transport from Na+ transport in the Ti3+-based assay and that Na+ is not rate-limiting. Using CO as reductant and the Acs/CODH from A. woodii as catalyst for ferredoxin reduction overcomes these problems. CO can be provided in large excess over NAD+, and electron transfer to NAD+ is strictly dependent on ferredoxin; thus, there is only one entry point for electrons into the Rnf complex. Using this assay, we observed a ferredoxin-dependent, highly reproducible NAD+ reduction; enhanced Na+ transport, which also leads to higher accumulation factors; and most importantly, a clear Na+ dependence of the ferredoxin:NAD+ oxidoreductase activity. These data are in line with the hypothesis that the ferredoxin:NAD+ oxidoreductase from A. woodii is a Na+ pump. Evidence for additional H+ transport was not found, which is in agreement with previous data that demonstrate a tight coupling between acetogenesis and ATP synthesis via a sodium ion potential (1). Δμ̃H+-driven ATP synthesis was excluded in that study.
The Rnf complex of A. woodii has six subunits (Fig. 8). RnfB has a predicted polyferredoxin and is hypothesized to be the ferredoxin-binding site and RnfC to be the NAD+-binding site. RnfG has only one transmembrane helix, which leaves RnfD (with its covalently bound flavin), RnfE, and RnfA as likely candidates for the Na+-translocating subunit(s). It is interesting to note that Ti3+ can reduce NAD+, but this activity is hardly Na+-dependent, indicating more than one entry point for Ti3+-based electrons. This would then also argue for a entry site that is “after” the Na+ translocation site. The highest enzymatic activity was found for methyl viologen:NAD+ oxidoreductase activity, but this activity was Na+-independent and also not accompanied by Na+ transport. These data are consistent with the hypothesis that these activities are mediated by subcomplexes not harboring the Na+ translocation site. Possible candidates would be RnfC, RnfG, and RnfB.
FIGURE 8.
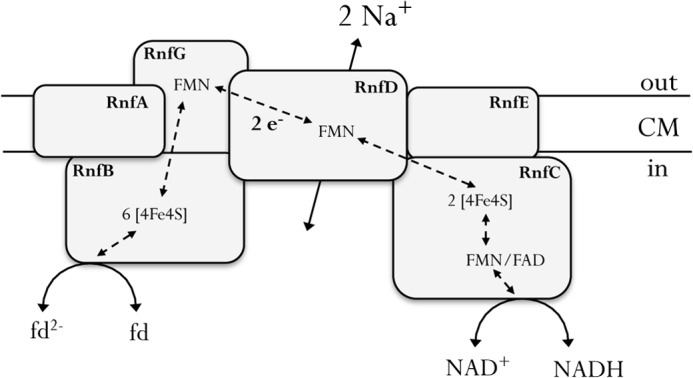
Model of the Rnf complex of A. woodii and its coupling to the membrane potential. The Rnf complex couples the flow of electrons from reduced ferredoxin (fd) to NAD+ with the formation of a sodium gradient across the cytoplasmic membrane (CM). This reaction is reversible, and endergonic ferredoxin reduction is driven at the expose of Δμ̃Na+.
As demonstrated here, Rnf activity at inverted membrane vesicles is stimulated by sodium ionophores. However, stimulation is weak, indicating a weak coupling. The redox potential difference between reduced ferredoxin and NAD+ is small, leading to a ΔG0′ of the reaction of only −34.7 kJ/mol. During the course of the reaction, NAD+ is reduced, and the energetics becomes even worse through an increase in the NADH concentration of the reaction. Increasing the NAD+ concentration shifts the ΔG′ of the reaction to more negative values, thus leading to higher Na+ gradients. However, they are still much smaller compared with ATP hydrolysis as driving force for Na+ export (ΔG′ ≈ −60 kJ/mol). Moreover, the activity of the Rnf complex is in the milliunits/mg range, whereas ATP hydrolysis is in the units/mg range. Both low activities and small ΔG values may explain why we were not able to detect the generation of ΔΨ or ATP synthesis so far. However, we were able to reverse the reaction: a Na+ gradient established by ATP hydrolysis drove ferredoxin reduction. This not only demonstrates the reversibility of the reaction in vitro but also supports the idea that the first postulated function of the Rnf complex, a reverse electron transport system that uses an ion-motive force to drive reverse endergonic transport (8), may be correct. Indeed, metabolism of low potential substrates such as lactate (E0′ = −190 mV) and ethanol (E0′ = −190 mV) would require an Rnf complex to drive ferredoxin reduction. This is currently under investigation.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
- Acs/CODH
- acetyl-CoA synthase/CO dehydrogenase
- DTE
- dithioerythritol
- Pa
- pascals.
REFERENCES
- 1. Heise R., Müller V., Gottschalk G. (1989) Sodium dependence of acetate formation by the acetogenic bacterium Acetobacterium woodii. J. Bacteriol. 171, 5473–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heise R., Müller V., Gottschalk G. (1992) Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur. J. Biochem. 206, 553–557 [DOI] [PubMed] [Google Scholar]
- 3. Fritz M., Müller V. (2007) An intermediate step in the evolution of ATPases–the F1FO-ATPase from Acetobacterium woodii contains F-type and V-type rotor subunits and is capable of ATP synthesis. FEBS J. 274, 3421–3428 [DOI] [PubMed] [Google Scholar]
- 4. Müller V., Bowien S. (1995) Differential effects of sodium ions on motility in the homoacetogenic bacteria Acetobacterium woodii and Sporomusa sphaeroides. Arch. Microbiol. 164, 363–369 [Google Scholar]
- 5. Müller V. (2003) Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69, 6345–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imkamp F., Biegel E., Jayamani E., Buckel W., Müller V. (2007) Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: indications for a Rnf-type NADH dehydrogenase as coupling site. J. Bacteriol. 189, 8145–8153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biegel E., Schmidt S., Müller V. (2009) Genetic, immunological and biochemical evidence of a Rnf complex in the acetogen Acetobacterium woodii. Environ. Microbiol. 11, 1438–1443 [DOI] [PubMed] [Google Scholar]
- 8. Schmehl M., Jahn A., Meyer zu Vilsendorf A., Hennecke S., Masepohl B., Schuppler M., Marxer M., Oelze J., Klipp W. (1993) Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241, 602–615 [DOI] [PubMed] [Google Scholar]
- 9. Biegel E., Schmidt S., González J. M., Müller V. (2011) Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68, 613–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poehlein A., Schmidt S., Kaster A.-K., Goenrich M., Vollmers J., Thürmer A., Bertsch J., Schuchmann K., Voigt B., Hecker M., Daniel R., Thauer R. K., Gottschalk G., Müller V. (2012) An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS ONE 7, e33439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biegel E., Müller V. (2010) Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 107, 18138–18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuchmann K., Müller V. (2012) A bacterial electron bifurcating hydrogenase. J. Biol. Chem. 287, 31165–31171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schönheit P., Wäscher C., Thauer R. K. (1978) A rapid procedure for the purification of ferredoxin from Clostridia using polyethylenimine. FEBS Lett. 89, 219–222 [DOI] [PubMed] [Google Scholar]
- 14. Zehnder A. J., Wuhrmann K. (1976) Titanium(III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194, 1165–1166 [DOI] [PubMed] [Google Scholar]
- 15. Pisa K. Y., Huber H., Thomm M., Müller V. (2007) A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274, 3928–3938 [DOI] [PubMed] [Google Scholar]
- 16. Bradford M. M. (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 17. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 18. Wittig I., Carrozzo R., Santorelli F. M., Schägger H. (2007) Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis 28, 3811–3820 [DOI] [PubMed] [Google Scholar]
- 19. Jayamani E. (2008) A Unique Way of Energy Conservation in Glutamate Fermenting Clostridia. Ph.D. thesis, Philipps University Marburg, Marburg, Germany [Google Scholar]
- 20. Ragsdale S. W., Ljungdahl L. G., DerVartanian D. V. (1983) Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J. Bacteriol. 155, 1224–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schlegel K., Leone V., Faraldo-Gómez J. D., Müller V. (2012) Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U.S.A. 109, 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terahara N., Krulwich T. A., Ito M. (2008) Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. U.S.A. 105, 14359–14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Müller V., Aufurth S., Rahlfs S. (2001) The Na+ cycle in Acetobacterium woodii: identification and characterization of a Na+-translocating F1FO-ATPase with a mixed oligomer of 8 and 16 kDa proteolipids. Biochim. Biophys. Acta 1505, 108–120 [DOI] [PubMed] [Google Scholar]
- 24. Schmidt S., Biegel E., Müller V. (2009) The ins and outs of Na+ bioenergetics in Acetobacterium woodii. Biochim. Biophys. Acta 1787, 691–696 [DOI] [PubMed] [Google Scholar]
- 25. Reidlinger J., Müller V. (1994) Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1FO-type enzyme. Eur. J. Biochem. 223, 275–283 [DOI] [PubMed] [Google Scholar]
- 26. Dimroth P. (1982) The generation of an electrochemical gradient of sodium ions upon decarboxylation of oxaloacetate by the membrane-bound and Na+-activated oxaloacetate decarboxylase from Klebsiella aerogenes. Eur. J. Biochem. 121, 443–449 [DOI] [PubMed] [Google Scholar]
- 27. Buckel W., Semmler R. (1982) A biotin-dependent sodium pump: glutaconyl-CoA decarboxylase from Acidaminococcus fermentans. FEBS Lett. 148, 35–38 [DOI] [PubMed] [Google Scholar]
- 28. Becher B., Müller V., Gottschalk G. (1992) N5-Methyl-tetrahydromethanopterin:coenzyme M methyltransferase of Methanosarcina strain Gö1 is an Na+-translocating membrane protein. J. Bacteriol. 174, 7656–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gärtner P., Weiss D. S., Harms U., Thauer R. K. (1994) N5-Methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum–catalytic mechanism and sodium ion dependence. Eur. J. Biochem. 226, 465–472 [DOI] [PubMed] [Google Scholar]
- 30. Takase K., Kakinuma S., Yamato I., Konishi K., Igarashi K., Kakinuma Y. (1994) Sequencing and characterization of the ntp gene cluster for vacuolar-type Na+-translocating ATPase of Enterococcus hirae. J. Biol. Chem. 269, 11037–11044 [PubMed] [Google Scholar]
- 31. Laubinger W., Dimroth P. (1988) Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry 27, 7531–7537 [DOI] [PubMed] [Google Scholar]
- 32. Imae Y., Atsumi T. (1989) Na+-driven flagellar motors. J. Bioenerg. Biomembr. 21, 705–716 [DOI] [PubMed] [Google Scholar]
- 33. Buckel W., Thauer R. K. (2013) Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 [DOI] [PubMed] [Google Scholar]


