Background: DNA-binding proteins help organize the nucleoid.
Results: The nucleic acid-binding properties of the nucleoid-associated protein YejK have been characterized.
Conclusion: YejK displays little preference for binding to any particular type of DNA but does affect the activity of DNA topoisomerases.
Significance: Understanding the binding characteristics of these types of DNA-binding proteins helps to understand how the nucleoid is organized.
Keywords: Chromosomes, Chromosomes/Non-histone Chromosomal Proteins, DNA-binding Protein, DNA Topoisomerase, DNA Topology
Abstract
Nucleoid-associated proteins play an important role in condensing chromosomal DNA and regulating gene expression. We report here the characterization of the nucleoid-associated protein YejK, which was detected in a yeast two-hybrid screen using the ParE subunit of topoisomerase IV as bait. The purified protein likely exists in a monomer-dimer equilibrium in solution and can form tetramers. Cross-linking of the protein bound to DNA suggests that the active form could be either a dimer or tetramer. YejK, which is present at about 24,000 copies of monomer per mid-log phase cell, binds double-stranded DNA with a site size of 12–14 base pairs/monomer, does not display a significant preference for either bent compared with straight DNA or supercoiled compared with relaxed DNA, and untwists DNA somewhat as it binds. YejK binds RNA, but not single-stranded DNA, with 65% of the avidity with which it binds DNA. However, cells deleted for yejK do not show defects in either RNA or protein synthesis. YejK interacts with all the subunits of both DNA gyrase and topoisomerase IV and has measurable effects on their activities. In the presence of YejK, relaxation of negatively supercoiled DNA by topoisomerase IV becomes distributive, whereas relaxation of positively supercoiled DNA is stimulated. Relaxation of negatively supercoiled DNA by DNA gyrase is inhibited, whereas the extent of supercoiling of relaxed DNA is limited. A YejK-GFP chimera is an effective marker for the nucleoid in live cell imaging.
Introduction
To fit within the nucleoid, the chromosomal DNA of Escherichia coli has to be compacted by at least 400-fold in a manner that does not compromise either DNA replication or transcription. This functional compaction is achieved by a combination of supercoiling of the DNA, macromolecular crowding, and the binding of (generally) small proteins that help shape and organize the DNA (1). These proteins are known collectively as nucleoid-associated proteins (NAPs)2 and can be divided into three groups: those that compact the DNA by either wrapping or bending it, or by bridging distal segments of the DNA (1, 2).
There are perhaps one dozen NAPs in E. coli that have been characterized to varying degrees (1, 2). Many of these proteins have effects on gene expression, which can be both repressive and stimulative, which can result from either global modulation of chromosomal DNA conformation or specific local effects on promoters. Global effects can come about because NAPs physically modify the path of the DNA by forming loops, bridging distant DNA segments, stabilizing bends in the DNA, or, in some instances, wrapping the DNA to generate supercoils. In addition, Fis, for example, can affect global supercoiling because it regulates the transcription of topoisomerase I and both subunits of DNA gyrase in a growth rate-dependent manner (3, 4). These two topoisomerases are responsible for homeostatically maintaining the optimal balance of chromosomal DNA supercoiling in E. coli (5). Fis can also act as a more typical transcriptional activator by making physical contact with RNA polymerase (6), and can repress and activate genes depending on the position of its binding site relative to the promoter (7, 8).
YejK has a predicted molecular mass of 37.8 kDa and was identified as a protein released from isolated spermidine nucleoids by digestion with DNase I (9). YejK was about one-third as abundant as RpoA in these preparations. The protein has an acidic pI and contains no known protein motifs. However, the protein is widely conserved across bacterial species.
We conducted a yeast two-hybrid analysis to find interacting partners of ParE and ParC, the two subunits of the cellular decatenase topoisomerase IV (Topo IV). Using ParE as bait, we isolated the intact YejK ORF. We report here the purification and characterization of this protein, its DNA binding preferences, its effect on the activities of Topo IV and DNA gyrase (with which it also interacts in a two-hybrid assay), and the effect of deleting the protein on cell growth. We also show that a YejK-GFP chimera is a non-toxic marker for chromosomal DNA in live cells.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Analysis
Construction of Yeast Two-hybrid Plasmids and Strains
Genomic DNA from E. coli strain W3110 was partially digested with FatI and DNA fragments 2.5–4 kbp in length were isolated by sedimentation through a neutral sucrose gradient. The resulting DNA fragments were ligated to NcoI-digested pACT2 (Clontech), generating gene fusions to the activator domain of the transcription factor Gal4. Plasmid DNA from ∼30,000 colonies was collected to create the plasmid library used in the yeast two-hybrid screen.
ORFs encoding the topoisomerase subunits ParE, ParC, GyrA, and GyrB were fused to the DNA-binding domain of LexA. For fusions to the C terminus of LexA, these ORFs were PCR amplified with primers including the indicated restriction sites and ligated to similarly digested pCA1 DNA (10): ParE and ParC, SalI and BglII; GyrA, SalI and BamHI; and GyrB, XhoI and BamHI. For fusions to the N terminus of LexA, these ORFs were PCR amplified with primers including the indicated restriction sites and ligated to similarly digested pCA11 DNA (10): ParE and ParC, NcoI and SpeI; GyrA and GyrB, PciI (an isoschizomer of NcoI) and SpeI.
Yeast Two-hybrid Screen
All yeast strains were grown in synthetic complete media. L40 (MATa, trp1, leu2, his3, LYS2::lexA-HIS3, URA3::lexA-lacZ) cells (Invitrogen) carrying the plasmid pCA1-parE were transformed with the pACT2-E. coli library by the lithium acetate method, as described by Clontech. To select for protein-protein interactions, strains were grown in minimal media lacking tryptophan, leucine, and histidine. Transformants were then grown in fresh minimal media to confirm the expression of histidine. Strains that also grew in the absence of His were then tested for expression of β-galactosidase.
To measure the production β-galactosidase, cells were grown in minimal media lacking tryptophan and leucine to an A600 of 0.5–1. Aliquots (0.5–1.5 ml) of cell cultures were used to assay β-galactosidase production (Miller units), as described by Clontech, using chlorophenol red-β-d-galactopyranoside (Calbiochem) as the substrate.
The pACT2 plasmids were recovered from the positive clones, transformed into the E. coli strain HB101, and grown in M9 minimal media lacking leucine. The plasmids were sequenced using as primers: 5′-GGCTTACCCATACGATGTTCCAG-3′ and 5′-GCACAGTTGAAGTGAACTTGCG-3′. Sequences were tested against the NCBI database using the blastn program. Plasmid clones with sequences in-frame with GAL4 were cotransformed with pCA1-parE into L40 to confirm the expression of His-3 and β-galactosidase.
All pCA1/pCA11-based plasmids and isolated positive pACT2 clones were tested for autoactivation of transcription. L40 cells carrying either the LexA plasmid or the Gal4 plasmid were maintained in minimal media lacking tryptophan or leucine, respectively, and assayed for His-3 and β-galactosidase expression as described above. Fusions that tested positive were not considered further. Positive Gal4 candidates that activated transcription only when co-expressed with LexA-ParE were also tested against fusions of LexA to ParC, GyrA, and GyrB.
Bacterial Strains, Plasmids, and DNA Substrates
All E. coli strains were cultured in either LB or in M9 minimal medium supplemented with 0.4% glucose and 0.2% casamino acids. The concentrations of antibiotics used in the media were as follows: ampicillin (Amp) (100 μg ml−1), kanamycin (Kan) (50 μg ml−1), and chloramphenicol (Cm) (35 μg ml−1). For cell viability analysis, aliquots of cultures were removed at the indicated time points, diluted into LB, and plated onto LB plates. Plates were incubated at 37 °C for 20 h and colony forming units were counted.
pET14b-yejK
The yejK ORF was PCR amplified with primers introducing NdeI and BglII sites to the 5′ and 3′ ends, respectively (YejKp14top and YejKp14bot, Table 1). The PCR product was digested with these enzymes and ligated into NdeI- and BamHI-digested pET14b (Novagen). The resulting protein expressed from this plasmid has a tag of Met-Gly-Ser2-His6-Ser2-Gly-Leu-Val-Pro-Arg-Gly-Ser-His added to the N terminus of YejK.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| gyrB0 | 5′-ATGTCGAATTCTTATGACTCCTCC-3′ |
| gyrB200rev | 5′-GTTATCGGCGTGAATGGTGACG-3′ |
| gyrB400rev | 5′-AACCAGCTCCAGTTTTTGCGA-3′ |
| gyrB800rev | 5′-GCAGTAGATGTTTTCCTGGAAGCC-3′ |
| 12bptop | 5′-GGCTTAAGCTGA-3′ |
| 12bpbot | 5′-TCAGCTTAAGCC-3′ |
| 18bptop | 5′-CATTGCAATCGCAGGTCA-3′ |
| 18bpbot | 5′-TGACCTGCGATTGCAATG-3′ |
| (bio)18bpbot | 5′-(bio)-TGACCTGCGATTGCAATG-3′ |
| 27bptop | 5′-CTCGAGGGGCAGGGCAGGGCAGGGTGCCG-3′ |
| 27bpbot | 5′-GCACCCTGCCCTGCCCTGCCCCTCGAGCG-3′ |
| 38bptop | 5′-ACGCTGTCTGCTAACATACTTCGTATTGAGGAGTCTAA-3′ |
| 38bpbot | 5′-TTAGACTCCTCAATACGAAGTATGTTAACAGACAGCGA-3′ |
| 38bptop-B | 5′-ACCCCCACACCCAAGATGCAACCGAAAGACCAGAAAAA-3′ |
| 38bpbot-B | 5′-TTTTTCTGGTCTTTCGGTTGCATCTTGGGTGTGGGGGT-3′ |
| 38bpRNA | 5′-ACCCCCACACCCAAGAUGCAACCGAAAGACCAGAAAAA-3′ |
| 62bptop | 5′-CATTGCAATCGCAGGTCATCGAGGACGCTGTCTGCTAACATACTTCGTATTGAGGAGTCTAA-3′ |
| 62bpbot | 5′-TTAGACTCCTCAATACGAAGTATGTTAGCAGACAGCGTCCTCGATGACCTGCGATTGCAATG-3′ |
| 42bpbenttop | 5′-GGCTGGGCAAAAAACGGGCAAAAAACGGCAAAAAACGGCTCC-3′ |
| 42bpbentbot | 5′-GGAGCCGTTTTTTGCCGTTTTTTGCCCGTTTTTTGCCCAGCC-3′ |
| 42bpstrttop | 5′-AACAGAGCAAAGGACGGCAATGAACGGCAAGGAACCGCTAAC-3′ |
| 42bpstrtbot | 5′-GTTAGCGGTTCCTTGCCGTTCATTGCCGTCCTTTGCTCTGTT-3′ |
| YejKp14btop | 5′-GCGGCATATGAGTCTGGATATCAACCAGA-3′ |
| YejKp14bbot | 5′-GTCGAGATCTTTAATTGCCGCCAGACGTC-3′ |
| YejK-top | 5′-GCGTTCACGCCGCATCTGGCATTCAGAACACAAAACCGAGTGTAGGCTGGAGCTGCTTCG-3′ |
| YejK-bot | 5′-CGGTGGTTGAATACCGCCCGGTCTTAAAGGAGAGTTTATCATGGGAATTAGCCATGGTCC-3′ |
| YejK-gfptop | 5′-CGCCGCCGAATTTGCGCGACCAATTGCAGCGCCGGACGTCTGGCGGCAATATGGCGAGCAAAGGTGAAGAG-3′ |
| YejK-gfpbot | 5′-GGCCAGATAAGGCGTTCACGCCGCATCTGGCATTCAGAACACAAAACCGATCCTCCTTAGTTCCTATTCC-3′ |
pGBM2-ecgfps
GFPuv (11) was modified for optimum codon expression in E. coli and the gene was straightened by reducing AT runs as described (12) to prevent inhibition of transcription by H-NS. The gene was synthesized by GeneScript and provided cloned in the vector pUC57. The gene was then transferred to pGBM2kan (13) using EcoRI and HindIII. Expression levels of genes modified with this GFP are identical to wild-type. Details will be provided elsewhere.
C600ΔyejK::CmR and MG1655ΔyejK::CmR
C600ΔyejK::Cmwas constructed using linear replacement as described (14). The resulting mutation is a complete replacement of yejK by cat flanked by FRT sites. The primers used to create the PCR product for the replacement were YejK-top and YejK-bot (Table 1). Loss of yejK was verified by PCR and DNA sequencing. The strain MG1655ΔyejK::CmR was created by bacteriophage P1 transduction of ΔyejK::Cm from C600ΔyejK::Cm.
C600yejK-ecgfps:kan and MG1655yejK-ecgfps:kan
C600yejK-ecgfps was constructed as described (14) using pGBM2-ecgfps as the PCR template and primers YejK-gfptop and YejK-gfpbot (Table 1). MG1655yejk-ecgfps was generated by bacteriophage P1 transduction of yejk-ecgfps:kan from C600yejk-ecgfps:kan.
Double-stranded, linear DNA substrates 200, 400, and 800 bp in size were generated by PCR amplification of the gyrB region of pCA1-gyrB using Ex-TaqHS DNA Polymerase (Takara) as instructed by the manufacturer. The following primer sets were used (Table 1): 200 bp, gyrB0 and gyrB200rev; 400 bp, gyrB0 and gyrB400rev; 800 bp, gyrB0 and gyrB800rev. PCR included [3H]TTP (PerkinElmer Life Sciences) and DNA yields were quantified by measuring incorporation of radioisotope into acid-insoluble product. The PCR products were treated with phenol:chloroform:isoamyl alcohol (25:24:1), precipitated with ethanol, and resuspended in TE (10 mm Tris-HCl (pH 7.5 at 25 °C), 1 mm EDTA).
For shorter duplex DNA, complementary oligonucleotides were annealed together as indicated (Table 1): 12bptop and 12bpbot; 18bptop and 18bpbot; 27bptop and 27bpbot; 38bptop and 38bpbot; 42bpbenttop and 42bpbentbot; and 42bpstrttop and 42bpstrtbot. Duplex oligonucleotides were dialyzed and stored in TE.
The 38-nt long RNA (38RNA, Table 1) used for binding assays was designed so that there was no secondary structure with a Tm >12 °C. Corresponding DNA oligonucleotides were 38bptop-B and 38bpbot-B (Table 2).
TABLE 2.
Two-hybrid interactions of DNA gyrase and Topo IV subunits with Gal4-YejK
β-Galactosidase activity of the indicated yeast two-hybrid combinations of full-length YejK and the subunits of either DNA gyrase or Topo IV.
| Binding fusion | Activity (Miller units)a |
|---|---|
| pACT2 | 1.1 ± 0.1 |
| LexA-GyrA | 1.8 ± 0.2 |
| GyrA-LexA | 29.1 ± 0.2 |
| LexA-GyrB | 95.3 ± 16.1 |
| GyrB-LexA | 2.7 ± 0.1 |
| LexA-ParC | 56.1 ± 13.9 |
| ParC-LexA | 1.2 ± 0.1 |
| LexA-ParE | 44.2 ± 0.8 |
| ParE-LexA | 20.3 ± 0.6 |
a Average of three determinations.
DNA and RNA substrates were end-labeled using bacteriophage T4 polynucleotide kinase and [γ-32P]ATP as suggested by the manufacturer (New England Biolabs). The labeled DNA was separated from unincorporated nucleotide by gel filtration through Sephadex G-50 in TE. The 800-bp substrate used in the Microccocal Nuclease protection assay (see below) was generated by PCR amplification as described above with [α-32P]dATP (PerkinElmer Life sciences) present in the reaction buffer.
Positively supercoiled ((+)ve sc) plasmid DNA was generated by treating negatively supercoiled ((−)ve sc) pBR322 DNA as described (15) with reverse gyrase (a gift of T. S. Hsieh, Duke University). (+)ve sc plasmid DNA was recovered from the reaction mixture using a Qiagen Qiaquick PCR Purification Kit as per the manufacturer's instructions.
Relaxed, circular plasmid DNA was generated by treating (−)ve sc pBR322 with Vaccinia virus topoisomerase I (a gift of Stewart Shuman, Memorial Sloan-Kettering Cancer Center). Multiple reaction mixtures (20 × 10 μl) containing (−)ve sc pBR322 (100 ng), 50 mm Tris-HCl (pH 7.5 at 4 °C), 20 mm NaCl, 5 mm DTT, 10 mm Mg(OAc)2, 2 mm ATP, 100 μg ml−1 BSA, and 1 mm EDTA were incubated at 37 °C for 20 min. Vaccinia topoisomerase I (4 ng) was added to each reaction mixture and the incubation continued for an additional 30 min. Proteinase K and SDS were then added to 100 μg ml−1 and 0.4%, respectively, and the incubation continued for an additional 30 min. The reaction mixtures were pooled and the DNA was recovered as above.
Purification of YejK
BL21/DE3(pLysS, pET14b-yejK) cells were grown in LB medium supplemented with Amp and Cm (20 liters of culture) at 37 °C to an A600 of 0.4 and induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. Cells were harvested by centrifugation, resuspended in 120 ml of Tris sucrose (50 mm Tris-HCl (pH 7.5 at 4 °C), 10% sucrose) and frozen at −80 °C. The cells were thawed and lysozyme, Tris-HCl (pH 8.8 at 25 °C), and NaCl were added to 200 μg ml−1, 50 mm, and 500 mm, respectively. The mixture was incubated on ice for 10 min, followed by treatment with 0.1% Brij and 10 units ml−1 benzonase (Sigma) for 2.5 h at 4 °C, with continuous mixing. The lysate (fraction 1) was cleared by centrifugation at 100,000 × g for 1 h at 4 °C, and applied to a 10-ml cobalt-charged Talon column (Clontech) equilibrated with Talon buffer (50 mm Tris-HCl (pH 7.5 at 4 °C), 10% glycerol, 500 mm NaCl). The column was washed with 1 column volume of Talon buffer, 1 volume of Talon buffer + 5 mm imidazole, and then eluted with a 10-column volume gradient of 5–350 mm imidazole in Talon buffer. Peak fractions (fractions 50–72 of 120), as determined by Bradford Assay and SDS-PAGE, were pooled (fraction 2, 123 mg), diluted in Buffer A (50 mm Tris-HCl (pH 7.5 at 4 °C), 10% glycerol) to an NaCl concentration of 50 mm, and applied to a 10-ml Q-Sepharose column (GE Healthcare) equilibrated with Buffer A + 50 mm NaCl. The column was washed with 1 column volume of Buffer A + 50 mm NaCl and eluted with a 10-column volume gradient of 50 mm to 1.5 m NaCl in Buffer A. The peak fractions (fractions 43–56 of 76) were pooled, protein was precipitated by the addition of (NH4)2SO4 to 65% saturation, recovered by centrifugation, and resuspended in Buffer A + 2 m NaCl (fraction 3, 65 mg). Half of fraction 3 was applied to a 125-ml Superdex 200 prep grade (GE Healthcare) column (1.6 × 58 cm) equilibrated and developed in Buffer A + 2 m NaCl. Two peaks were pooled separately as fraction 4B (3.7 mg), the peak that eluted first, and fraction 4A (3.5 mg), the second peak. Both pools were dialyzed against storage buffer (50 mm Tris-HCl (pH 7.5 at 4 °C), 150 mm NaCl, 38% glycerol, 5 mm DTT, 1 mm EDTA) and stored at −80 °C.
Analytical Gel Filtration
One milligram of fractions 4A and 4B were combined and the mixture was applied to a 25-ml Superose 12 FPLC gel filtration column (GE Healthcare) equilibrated and developed with Buffer A + 2 m NaCl, 1 mm DTT, and 200 mm imidazole. The elution profile was compared with that of the protein markers ribonuclease A (13.7 kDa), ovalbumin (43 kDa), aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa).
Protein-Protein Cross-linking
Reaction mixtures (10 μl) containing 5 μm YejK in 1× PBS and 1 mm EDTA and the indicated concentrations of Sulfo-SMPB were incubated at 23 °C for 80 min. The reaction was quenched by the addition of glycine to 50 mm and analyzed by SDS-PAGE.
To assay the cross-linking of YejK bound to a DNA substrate, a biotinylated duplex substrate (bio-18bp) was created by annealing the oligonucleotides 18bptop and (bio)18bpbot as indicated (Table 1). The biotinylated duplex (380 pmol) was bound to prewashed Dynabeads MyOne Streptavidin T1 beads (Invitrogen) (50 μl of packed beads) in a total volume of 600 μl, as per the manufacturer's instructions. The bio-18bp beads were washed as per the manufacturer's instructions and resuspended in 10 mm HEPES-KOH (pH 7.5). DNA binding reactions (30.8 μl) contained 50 mm HEPES-KOH (pH 7.5), 20 mm NaCl, 5 mm DTT, 1 mm EDTA, 1 mm ATP, 2 mm Mg(OAc)2, 5.3 μm YejK, and 1 μm bio-18bp beads were incubated at 37 °C with constant shaking at 850 rpm for 30 min. Either Sulfo-SMPB (in 10 mm HEPES-KOH (pH 7.5)) or 1 mm EDTA (control) was added to a final concentration of 460 μm in a total volume of 35 μl, and the reactions were incubated at 25 °C with constant shaking at 850 rpm for 80 min. The reaction was quenched by the addition of Tris-HCl to 13.9 mm and excess YejK was removed by applying the reaction to a magnet followed by washing with 10 mm HEPES-KOH (pH7.5), 1 mm EDTA. The washed beads were resuspended in loading dye (20 μl total), heated at 100 °C for 10 min, and reaction products were analyzed by SDS-PAGE. The gel was stained with SYPRO Ruby (Invitrogen) and imaged using a Kodak Image Station 4000R Pro and Carestream Molecular Imaging Software.
DNA Binding Assays
Linear Duplex Substrates
Reaction mixtures (15 μl) containing 50 mm Tris-HCl (pH 7.5 at 37 °C), 20 mm NaCl, 5 mm DTT, 4.75% glycerol, 1 mm EDTA, 2 mm Mg(OAc)2, 400 μg ml−1 of BSA, 1 mm ATP, the indicated DNA substrate (10 nm), and the indicated concentrations of either YejK (diluted in storage buffer) or storage buffer were incubated at 37 °C for 30 min and then loaded onto 6% polyacrylamide gels (80:1). The gels were electrophoresed at a constant current of 13 mA for 200–240 min at 4 °C, with constant recirculation of the buffer (6 mm Tris-HCl (pH 7.5 at 4 °C), 2 mm NaOAc, and 0.1 mm EDTA). Gels were dried, exposed to phosphorimager screens for quantification (using a Fujifilm FLA-7000 phosphorimager), and autoradiographed. Scans were analyzed with Fujifilm Image Gauge software. To calculate binding affinities, the amount of bound and free oligonucleotide was determined as a fraction of the total radioactivity present in the lane, and KD values were calculated using Scatchard analysis.
Plasmid DNA Substrates
Reaction mixtures (20 μl) containing Topo IV reaction buffer (see below) plasmid DNA (100 ng), and the indicated amounts of YejK were incubated for 20 min at 37 °C, loaded onto 1% agarose gels, and electrophoresed at 2 V/cm for 16 h at room temperature using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, and 1 mm EDTA (Tris acetate) as the running buffer. Gels were stained with ethidium bromide, imaged using a Kodak Image Station 4000R Pro, and the images analyzed using Carestream Molecular Imaging Software.
Microccocal Nuclease Protection Assay
Reaction mixtures (15 μl) containing 5′-32P-labeled 800-bp linear substrate (10 nm), 50 mm Tris-HCl (pH 7.5 at 37 °C), 1 mm CaCl2, 0.1 mm EDTA, 2 mm DTT, 50 μg ml−1 of BSA, 20 mm MgCl2, 4.75% glycerol, and 1 μm DNA gyrase, 4 μm YejK, or YejK storage buffer as indicated, were incubated for 20 min at 30 °C. CaCl2 and Microccocal Nuclease (Roche Applied Science) were added to 4 mm and 0.01–0.04 units μl−1, respectively, and the incubation continued for 10 min at 30 °C. The reactions were stopped by the addition of EDTA, Proteinase K, and SDS to final concentrations of 50 mm, 1 μg ml−1, and 0.2%, respectively, followed by incubation for 1 h at 30 °C. The reactions were loaded onto 8% polyacrylamide (80:1) gels and electrophoresed for 200 min at a constant current of 13 mA using 100 mm Tris base, 100 mm H3BO3, 4 mm EDTA (2× Tris borate, pH 8.3) as the buffer. The gel was dried and analyzed as described above.
Assay for Topological Alteration of DNA
Reaction mixtures (20 μl) in the linear duplex DNA binding assay reaction buffer containing 100 ng of either (−)ve sc pBR322 or relaxed pBR322 and the indicated concentrations of YejK were incubated for 20 min at 37 °C. Vaccinia topoisomerase I (4 ng) was then added and the incubation continued for 30 min at 37 °C. The reactions were stopped by the addition of SDS and Proteinase K to 0.4% and 100 μg ml−1, respectively, followed by incubation for 30 min at 37 °C. The DNA products were recovered by ethanol precipitation after extraction with phenol:chloroform, resuspended in 30 μl of TE, and electrophoresed through a 1% agarose gel containing 6.5 μg ml−1 of chloroquine phosphate for 16 h at a constant voltage of 2 V/cm using Tris acetate as the buffer (also containing chloroquine). The gel was stained and analyzed as described above.
DNA Relaxation and Supercoiling Assays
Supercoiled DNA relaxation by Topo IV. Master reaction mixtures (171 μl) contained 50 mm Tris-HCl (pH 7.5 at 37 °C), 20 mm NaCl, 5 mm DTT, 4.75% glycerol, 10 mm Mg(OAc)2, 2 mm ATP, 1 mm EDTA, 100 μg ml−1 of BSA, (−)ve sc pBR322 (900 ng), and either 8 μm YejK or an equivalent amount of YejK storage buffer. Aliquots (19 μl) were removed and placed in separate tubes to serve as negative controls without Topo IV and with YejK only. Topo IV was then added to 5 nm to the master reaction mixtures and all reactions were incubated at 37 °C. Aliquots (20 μl) were removed from the master reactions at the indicated time points and transferred to fresh tubes containing 2.5 μl of STOP Buffer (200 mm EDTA, 3 m NaCl). Terminated reactions were incubated at 37 °C for 5 min. SDS and Proteinase K were then added and the reactions processed and analyzed as described above for the superhelical DNA binding assays. The same protocol was followed for relaxation of (+)ve sc pBR322 by Topo IV, except that the Topo IV concentration was 50 pm.
Relaxation of (−)ve sc pBR322 by DNA gyrase was performed in a similar manner except that the reaction buffer contained 35 mm Tris-HCl (pH 7.5 at 37 °C), 24 mm KCl, 10 mm Mg(OAc)2, 4.75% glycerol, 5 mm DTT, 1 mm EDTA, and 20 nm DNA gyrase. Supercoiling of relaxed pBR322 by 1 nm DNA gyrase was performed in the same reaction buffer, including 2 mm ATP, and form I′ pBR322 was used instead of (−)ve sc pBR322. Reaction volumes were reduced 4-fold so that a total of 25 ng of DNA was electrophoresed. Gels were stained with SYBR Gold (Invitrogen), imaged with a Fujifilm FLA-5000 fluorimeter, and analyzed with Fujifilm Image Gauge software.
Measurement of Uridine and Methionine Incorporation in Vivo
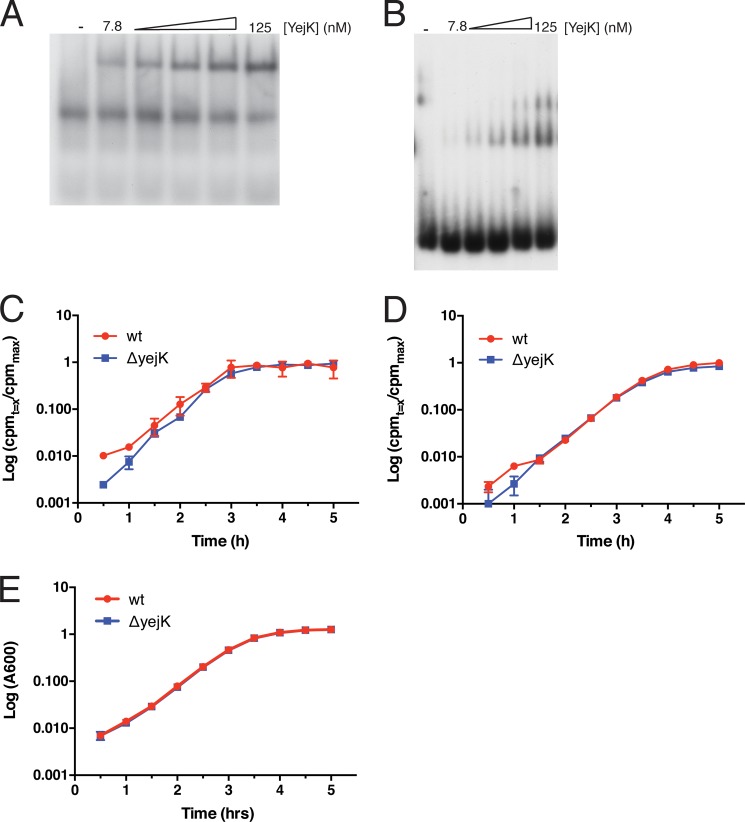
Stationary cultures of MG1655 and MG1655ΔyejK were diluted into LB medium containing either 2 μCi/ml of [5,6-3H]uridine (PerkinElmer Life Sciences) or 5 μCi/ml of l-[35S]methionine (PerkinElmer Life Sciences) to an A600 = 0.005 and grown at 37 °C. Aliquots (500 μl) of culture were removed at the indicated time points, chilled, made 5% in trichloroacetic acid, filtered through GF/C filters (Whatman), and the retained radioactivity measured by scintillation counting. Duplicates of each time point were averaged (cpmt = x), normalized to the average maximum radioactivity incorporated (cpmmax), and the values plotted against time.
Flow Cytometry
Stationary cultures were diluted into M9 minimal medium to an A600 = 0.005 and grown at 37 °C with aeration for 4 h to mid-log. Cultures were synchronized by the addition of dl-serine hydroxamate (Sigma) to 1 mg ml−1 and the incubation continued for 90 min (16). Cells were harvested by centrifugation at 16,000 × g for 5 min and resuspended in fresh medium containing 12 μg ml−1 of cephalexin (Sigma). The culture was aliquoted (1 ml samples, ∼3.5 × 107 cells) and growth was continued at 37 °C for 70 min. At the indicated times, rifampicin (Sigma) was added to an aliquot to 70 μg ml−1 (2.5 mg ml−1 stock in 10% DMSO) and the incubation continued with aeration for an additional 3 h. Cells were harvested by centrifugation at 16,000 × g for 1 min and resuspended in 100 μl of PBS. The resuspended cells were fixed overnight by the addition of 900 μl of cold 95% ethanol and stored at 4 °C. For staining with PicoGreen, ∼1.8 × 106 fixed cells were harvested at 16,000 × g for 1 min and resuspended in 400 μl of Staining buffer (10 mm Tris-HCl (pH 7.5 at 4 °C), 1 mm EDTA, 50 mm NaCl, 0.1% Triton X-100). PicoGreen (100 μl of 1:100 dilution in 25% DMSO) was added and the cells incubated for 2 h at room temperature (17). Stained cells were diluted with 1 ml of 1:1000 PicoGreen in Staining buffer, as described (16), and stored at 4 °C for 24–48 h prior to analysis with a BD Biosciences FACSCalibur flow cytometer using CellQuest Pro software. Data were analyzed using FlowJo software.
Fluorescence Microscopy
Stationary cultures of MG1655 and MG1655ΔyejK were diluted into fresh LB medium to an A600 = 0.0002 and grown at 37 °C. Cells were harvested at the indicated times by centrifugation at 6000 × g for 5 min, resuspended in 1× PBS at A600 = 0.4, and mixed with an equal volume of fixing solution (5% paraformaldehyde (EMS) and 0.06% glutaraldehyde (Sigma) in 1× PBS). DAPI (Sigma) and FM 4-64 FX (Invitrogen) were added to 1 μg ml−1. The cell suspension was layered onto slides coated with 0.1% poly-l-lysine (Sigma) and incubated at room temperature for 30 min in the dark. Slides were washed twice with 1× PBS and allowed to dry before a glycerol-based fluorescent mounting media was added. Slides were imaged using a Nikon Eclipse Ti microscope and analyzed using Nikon NIS Elements Imaging Software.
For the imaging of YejK-GFP. Stationary cultures of MG1655yejK-ecgfps:kan were diluted to A600 = 0.01 into M9 minimal medium supplemented with 0.4% glucose and 0.2% casamino acids and grown for 3 h at 37 °C. Cells were harvested, stained with DAPI, spread onto slides coated with 0.1% polylysine, and viewed immediately.
Abundance of YejK in Vivo
MG1655yejK-ecgfps:kan cells were grown in LB medium to mid-log phase and 2.03 × 109 cells (as determined by viable cell count) were harvested by centrifugation, resuspended in 500 μl of 50 mm Tris-HCl (pH 7.5 at 4°C), 10% sucrose, and frozen at −20 °C. The cells were thawed and lysozyme, Tris-HCl (pH 8.8 at 25°C), EDTA, and DTT were added to 200 μg ml−1, 50 mm, 2 mm, and 10 mm, respectively, in a total volume of 600 μl. The mixture was incubated on ice for 10 min, MgCl2 was added to 4 mm, followed by treatment with 250 units of benzonase (Sigma) and 0.1% Brij (Sigma) at 4°C for 1 h on a rotator. The lysate was sonicated using a microtip for 20 pulses at 50% duty cycle and cleared by centrifugation at 16,000 × g for 30 min. A total of 1.0 mg of protein was recovered in the cleared lysate.
The indicated amounts of lysate and recombinant GFP from Aequorea coerulescens (rACGFP1, Clontech) were electrophoresed through a 10–20% SDS-PAGE gradient gel at 12 V/cm. Proteins were transferred to a nitrocellulose membrane that was then blocked with 5% milk in phosphate-buffered saline containing 0.005% Tween 20 (PBST). The membrane was incubated with polyclonal α-GFP antibody (Invitrogen) at a dilution of 1:3,000 in PBST, 5% milk for 1 h at 25 °C and washed with PBST. The membrane was then incubated with goat anti-rabbit HRP conjugate (Bio-Rad), at a dilution of 1:10,000 in PBST, 5% milk and washed as above. The membrane was developed with ECL Plus (Pierce) prior to exposure to x-ray film. Films were scanned using an Epson Perfection V750 Pro scanner and the absorbance levels of the bands were quantified using Fujifilm Image Gauge software.
RESULTS AND DISCUSSION
Purification and Characterization of YejK
YejK was identified as an interacting partner of ParE, the ATPase subunit of Topo IV, in a yeast two-hybrid screen as described under “Experimental Procedures.” The cloned fragment of E. coli DNA contained nucleotides 2280685–2281969, bearing the intact yejK ORF and a truncated portion of rplY that was not expressed because it is transcribed in the opposite orientation to yejK. The Gal4-YejK fusion was tested for association with the full-length subunits of both of the E. coli type II topoisomerases, DNA gyrase and Topo IV, using the yeast two-hybrid assay. All four of the subunits were capable of eliciting significant β-galactosidase activity (Table 2), although only the ParE subunit of Topo IV was able to associate with Gal4-YejK as both an N-terminal and C-terminal fusion with the DNA-binding domain of LexA.
YejK had been identified previously, along with RdgC, as a protein released from isolated spermine nucleoids by treatment with DNase I (9). Little is known about this protein absent the initial publication. It has also been referred to as NdpA (18). Analysis against the protein databases did not uncover either any conserved domains or known motifs. Nevertheless, it is widely distributed in bacteria and the gene is present on some large plasmid DNAs as well (18). Because this was the only NAP identified in the two-hybrid screen, we characterized the protein and the effect on E. coli of deleting the gene.
YejK was expressed as an N-terminal fusion to a His6-thrombin cleavage site tag and purified by sequential chromatography on a Talon His tag affinity resin and Q-Sepharose, followed by gel filtration through Superdex 200. This last step yielded two peaks of YejK that were identical in composition by SDS-PAGE analysis (Fig. 1A). The smaller protein bands evident on the gel were identified using mass spectrometry as breakdown fragments of YejK (data not shown). Comparison to the elution of protein markers during gel filtration through a Superose 12 FPLC column in high salt showed that the two peaks were related in size by a factor of 2 with a derived molecular mass of 56 kDa for fraction 4A and 118 kDa for fraction 4B (Fig. 1, B and C). The calculated molecular mass of YejK with the tag from the cloning vector is about 40 kDa. These data suggest that fractions 4A and 4B represent a monomer-dimer equilibrium for a protein that is somewhat elongated.
FIGURE 1.
Characterization of purified YejK. A, YejK fractions 4A and 4B (2.5 μg) were analyzed by SDS-PAGE through a 12% gel. The gel was stained with Commasie Brilliant Blue and imaged. B, analytical gel filtration of an equal mixture of fractions 4A and 4B YejK as described under “Experimental Procedures.” C, analysis of the gel filtration experiment shown in panel B. The standard curve was generated by linear least squares regression analysis. D, SDS-PAGE analysis of YejK cross-linked using Sulfo-SMPB as described under “Experimental Procedures.” E, analysis of the cross-linking experiment shown in panel D. The standard curve was generating by fitting to a 4th order polynomial. The 25-kDa marker used in generating the standard curve is not shown on the SDS-PAGE gel image in panel D. F, SDS-PAGE analysis of cross-linking of YejK bound to DNA as described under “Experimental Procedures.” G, analysis of the cross-linking experiment shown in panel F. The standard curve was generated as for panel D.
To examine further the subunit structure, YejK was cross-linked in solution using varying concentrations of sulfosuccinimidyl 4-(p-maleimidophenyl) butyrate (Sulfo-SMPB) and the results analyzed by SDS-PAGE. Multiple higher molecular weight species were observed as the concentration of cross-linking agent was increased (Fig. 1D). The monomer band is converted to a species that migrates with a mobility slightly less than that of the uncross-linked monomer, suggesting formation of an internal cross-link that prevents the protein from being denatured completely. Comparison of the mobility of the species marked A, B, C, D, and E suggest a related family of bands with derived molecular masses of 39.8, 87.9, 168.8, 271.7, and 341.6 kDa, respectively (Fig. 1E). These data suggest that species A, B, and C represent monomer, dimer, and tetramer forms of YejK and that species D and E represent higher-order forms of the protein.
To obtain information as to the active DNA-binding form of the protein we cross-linked it when it was bound to DNA. Our DNA binding data described in the next section suggests that the DNA binding site size is 12–14 bp/monomer, thus a biotinylated 18-bp DNA was used to limit cross-linking of neighboring protomers on the DNA. YejK was bound to the 18-bp DNA substrate, which itself was bound to magnetic streptavidin beads, cross-linked as before, and YejK free in solution was removed by isolating and washing the beads. The cross-linked protein was then analyzed by SDS-PAGE (Fig. 1, F and G). The primary protein bands observed likely corresponded to dimer (B, 103.4 kDa) and tetramer (C, 163.6 kDa) forms of the protein. A third band was also observed (D′, 207 kDa). The higher molecular weight forms observed in Fig. 1D were not observed, although it is possible that forms D (Fig. 1D) and D′ (Fig. 1F) could be related by the formation of internal cross-links. Thus, although the primary form of the protein in solution is a monomer, at least the dimer and tetramer forms are capable of binding DNA. This observation does not rule out the possibility that higher molecular weight forms of the protein could bind longer DNA substrates.
Binding of YejK to DNA and RNA
Binding of YejK to DNA was tested in gel-shift assays using linear, double-stranded (ds) DNAs varying in size from 12 to 400 bp in length. Binding to a 12-bp substrate could be observed but was very weak, with a KD greater than 12 μm. As the DNA substrate increased in length, binding of YejK became more avid, with the KD decreasing to 58 nm for the 400-bp substrate (Fig. 2C). Binding to the 800-bp substrate improves marginally to 45 nm (data not shown), suggesting that the binding affinity of YejK to long chromosomal DNA is in the range of 50 nm monomer. Because it appears that higher order oligomeric forms of the protein bind DNA (Fig. 1F), the actual affinity of the protein for DNA in the cell is likely to be higher.
FIGURE 2.

Binding of YejK to linear duplex DNA substrates. A–C, gel shift analyses of the binding of YejK to the 27- (panel A), 38- (panel B), and 400-bp (panel C) DNA substrates as described under “Experimental Procedures.” D, YejK binding curves for various duplex DNA substrates. KD values for each substrate are: 12 bp, 12.2 μm; 18 bp, 5.9 μm; 27 bp, 218.3 nm; 38 bp, 224.4 nm; 200 bp, 70.1 nm; and 400 bp, 58.4 nm.
Multiple gel shifted bands could be observed with increasing concentrations of YejK (Figs. 2, A–C, and 3, A and B) with all the DNA substrates except the 12- and 18-bp substrates (which displayed only one gel shifted band, data not shown). These multiple gel shifts either represent addition of new protomeric binding units to the DNA or the aggregation of multiple YejK molecules onto one or a few nucleating protein molecules on the DNA. The former explanation is far more likely to be correct. If this is the case, one can estimate the length of DNA bound per molecular unit of YejK binding by counting the number of gel shifts as one approaches saturated binding on one of the smaller substrates. Two well defined gel shifts were observed for both the 27- and 38-bp substrates (Fig. 2, A and B), with a third shift evident using the 38-bp substrate at high concentrations of YejK. Binding to the 42-bp substrate (see below) gave three clear gel shifts. Therefore, the DNA binding site size per active binding unit of YejK is likely to be 12–14 bp. Because it appears that both dimers and tetramers of the protein can bind DNA and we cannot tell whether when any given form of the protein is bound to DNA all available binding sites are filled, we cannot give a more precise value. Given that binding to the 12-bp DNA substrate is very weak, we suspect that DNA in excess of the binding site size is required to stabilize the bound protein.
FIGURE 3.
YejK binds RNA. A and B, gel shift analysis of YejK binding to the 38-nt long RNA (panel A) and the equivalent sequence as duplex DNA (panel B). YejK bound to the RNA and DNA with KD values of 360 and 220 nm, respectively (average of three determinations). Our YejK preparations had a persistent RNase contamination, thus binding of YejK to RNA at concentrations greater than 125 nm was not reliable because of degradation of the substrate. C, deletion of yejK does not affect total RNA synthesis in vivo. Uptake of [3H]uridine and its incorporation into acid-insoluble product in wild-type and cells deleted for yejK was measured as described under “Experimental Procedures.” D, deletion of yejK does not affect total protein synthesis in vivo. Uptake of [35S]methionine and its incorporation into acid-insoluble product in wild-type and cells deleted for yejK was measured as described under “Experimental Procedures.” E, typical growth curves for the cells used.
Some NAPs, such as HU and H-NS, also display significant affinity for RNA (1, 2). We tested whether YejK bound RNA and single-stranded DNA using a 38-nt long oligonucleotide that was designed to be void of secondary structure (Table 1). YejK did not bind single-stranded DNA (data not shown) but did bind the 38-mer RNA with an affinity about 65% of what it displayed in binding the equivalent sequence as double-stranded DNA (Fig. 3, A and B). Binding to the RNA was clearly different from binding to the DNA because only one gel shift was observed, as opposed to the two gel shifts observed for the 38-bp DNA. The basis for this difference is not clear at this time. To assess whether deletion of yejK affected either RNA or protein synthesis in vivo, we measured uptake of [3H]uridine and [35S]methionine in wild-type cells and those deleted for yejK. No significant difference was observed (Fig. 3, C and D), indicating that the protein was unlikely to play a significant role in translational regulation.
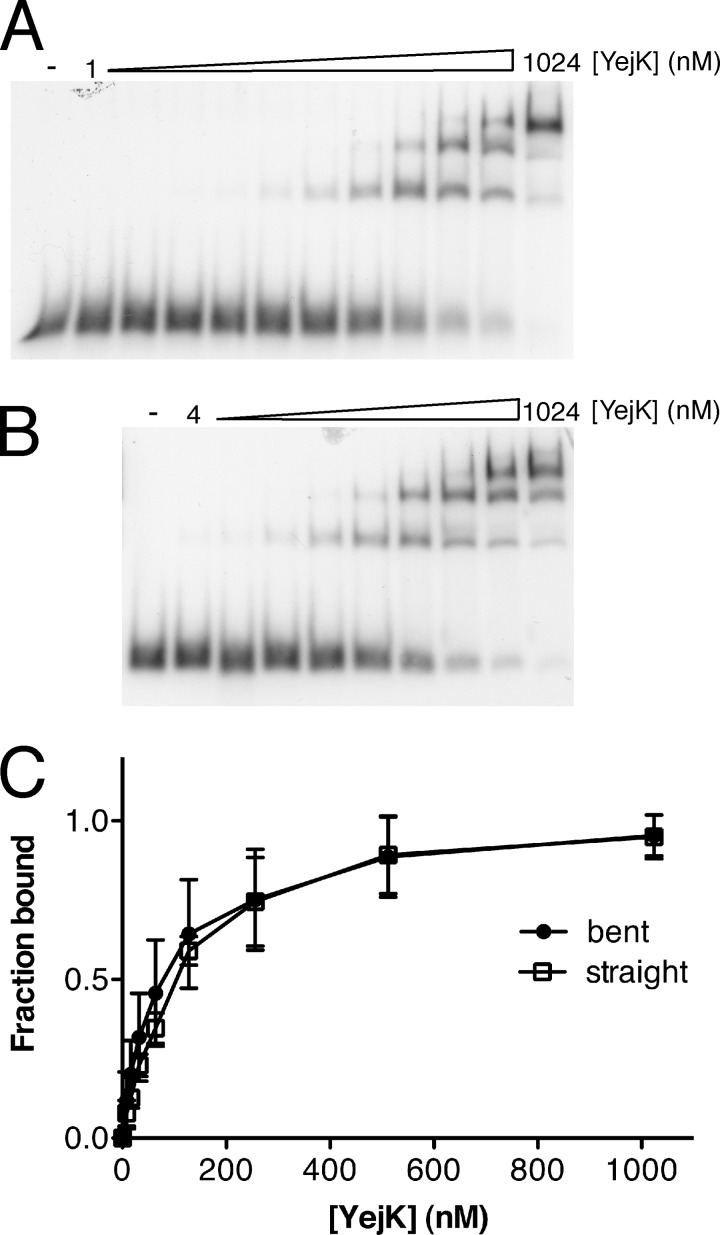
A number of NAPs, such as CpbA and Fis, display a preference for binding to curved DNA, whereas others, such as IHF, Fis, and HU, can bend DNA upon binding (2). The DNA used in the experiments displayed in Fig. 2 were derived from the sequence of gyrB, which does not possess any significant inherent curvature (as calculated using the Bend.It algorithim (19)). To test whether YejK had a preference for binding to curved DNA, we used a 42-bp substrate that had three phased A6 sequences and compared it to a control substrate of the same length and G + C content in which the phased A6 sequences had been scrambled (Fig. 4). The bent substrate had a predicted curvature of 18 degrees/10.5 bp, whereas the curvature of the sequence-scrambled substrate varied from 2.7–7 degrees/10.5 bp (19). YejK displayed little preference for either DNA (KD straight DNA, 88.0 nm; KD bent DNA, 104.9 nm).
FIGURE 4.
YejK shows little preference for binding to either bent or straight DNA. A and B, gel shift analyses of the binding of YejK to the straight (panel A) and bent (panel B) DNA substrates as described under “Experimental Procedures.” C, binding curves for the experiments shown in panels A and B.
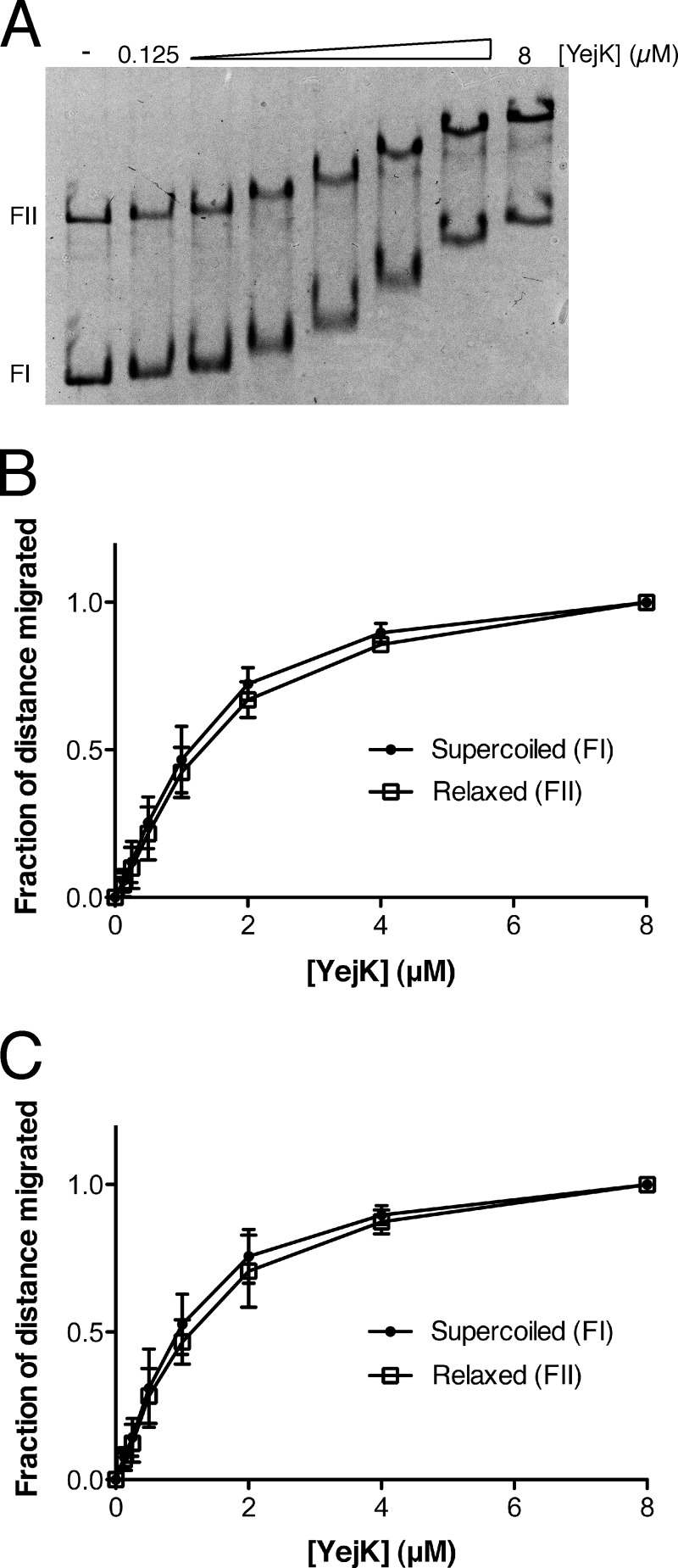
We also examined the binding of YejK to another version of curved DNA, (+)ve sc and (−)ve sc plasmid DNA (Fig. 5). In the gel mobility shift assay used, as the mass of YejK bound to the 4.5-kbp plasmid increases, the mobility of the protein-DNA complex will decrease in a manner that is directly proportional to the amount of protein bound. Therefore, the difference in mobility of the bound and free forms of DNA can be used as a relative approximation of the amount of YejK bound. Using this measure as a comparison, YejK displayed little preference for binding to either the (−)ve sc form of pBR322 (apparent KD, 1.0 μm) or the (+)ve sc form (apparent KD, 1.2 μm), binding each DNA form roughly the same as it bound the relaxed, form II (nicked) DNA present in the DNA preparations (apparent KD for the form II DNA present in the (−)ve sc DNA preparation, 1.2 μm; apparent KD for the form II DNA present in the (+)ve sc DNA preparation, 1.0 μm). It is important to note that because it requires considerable mass of YejK to gel shift the plasmid DNA, one cannot compare the apparent affinities derived from this experiment to those calculated from the data in Fig. 2.
FIGURE 5.
Binding of YejK to superhelical DNA. A, an example of gel shift analysis of YejK binding to (−)ve sc DNA. FI, supercoiled form I DNA; FII, relaxed (nicked) form II DNA. B, binding curves for YejK binding to (−)ve sc and the form II DNA present in the DNA preparation. C, binding curves of YejK binding to (+)ve sc DNA and the form II DNA present in the DNA preparation.
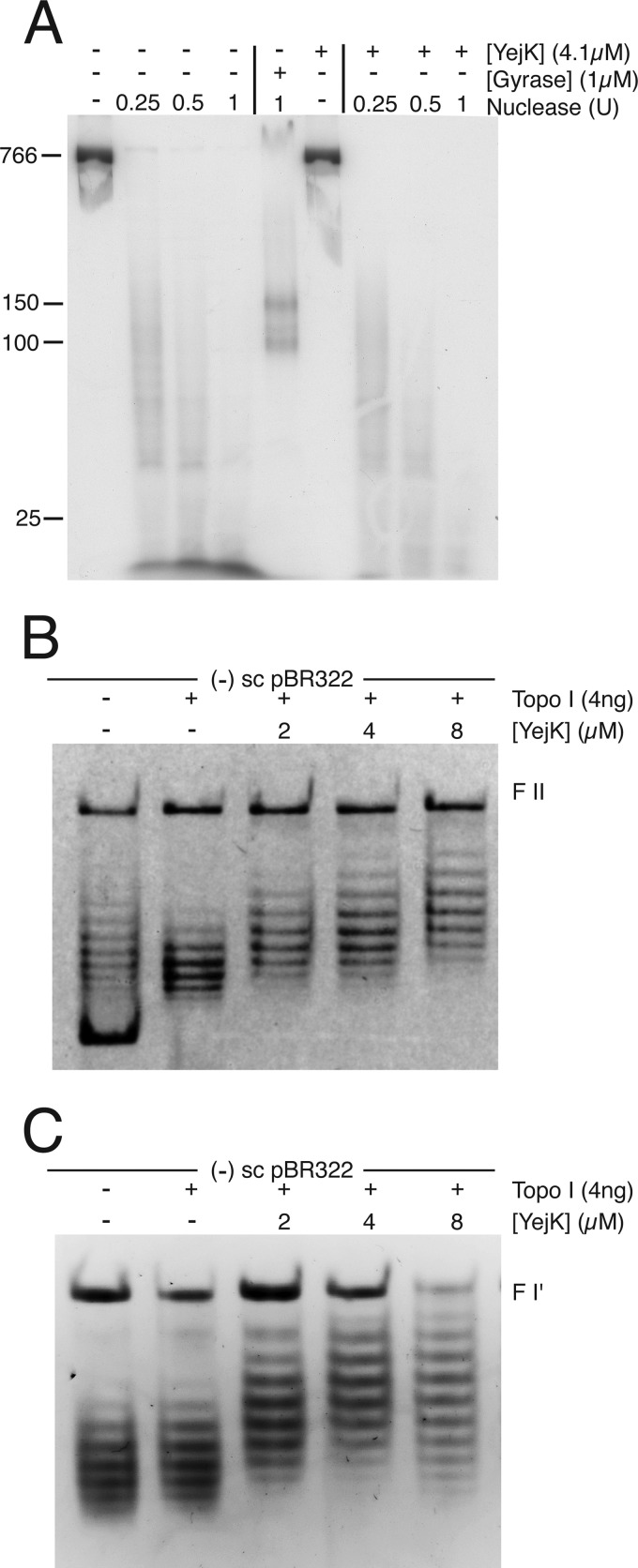
Two NAPs from Gram-negative bacteria may wrap DNA about themselves. The crystal structure of HU bound to DNA suggests this possibility, whereas LRP can wrap DNA at specific locations (20, 21). To assess whether YejK wrapped DNA in a consistent, global manner, we asked whether YejK could protect fragments of dsDNA from digestion with microccocal nuclease (Fig. 6). YejK was bound to an 800-bp dsDNA that had been uniformly labeled with 32P by including [α-32P]dATP in the PCR, the DNA was then digested with varying amounts of microccocal nuclease, and the products analyzed by electrophoresis through an 8% polyacrylamide gel. As a positive control we used DNA gyrase, which is known to wrap about 150 bp of DNA (22). Two bands in the range of 100–150 bp in size were evident after nuclease digestion when gyrase was bound to the DNA (Fig. 6A). There were no similar products evident for YejK, which, if it bound DNA as a dimer, would be expected to produce fragments in the range of 25–30 bp. Of course, this assay would not detect YejK wrapping of DNA at a specific sequence.
FIGURE 6.
YejK binding untwists DNA. A, YejK does not wrap DNA as it binds. The [32P]-labeled 800-bp linear DNA substrate was treated with MDNase either in the absence (left-hand lanes) or presence (right-hand lanes) of 4 μm YejK or in the absence and presence of 1 μm DNA gyrase (center lanes) as described under “Experimental Procedures” and the products analyzed by electrophoresis through an 8% polyacrylamide gel. B and C, YejK untwists DNA as it binds. The indicated amounts of YejK were bound to either (−)ve sc (panel B) or relaxed (form I′) (panel C) pBR322 DNA and the protein-DNA complex was then treated with vacinnia virus DNA topoisomerase I. After treatment with SDS and proteinase K, the DNA products were analyzed by electrophoresis through 1% agarose gels containing chloroquine phosphate in both the gel and the electrophoresis buffer. FII, form II DNA.
A more sensitive test for the alteration of DNA topology by bound proteins is to bind the protein to a covalently closed, circular plasmid DNA and then treat with an excess of a topoisomerase that can remove both (+)ve and (−)ve supercoils. Following removal of all bound protein, the residual, altered topology is reported as residual supercoils. DNA topology can be altered in this assay either by affecting the writhe of the DNA because of DNA wrapping, or affecting the twist of the DNA because of either an over- or unwinding. We performed this experiment using YejK, vacinnia virus topoisomerase I, and either (−)ve sc (Fig. 6B) or relaxed form I′ (Fig. 6C) pBR322 DNA. The DNA products were analyzed by electrophoresis through agarose gels in the presence of saturating amounts of chloroquine phosphate. Under these conditions, all DNA topoisomers become positively supercoiled. With both DNA substrates, shifts of ΔLk ≈ −4 were observed at saturating concentrations of YejK. It is very unlikely that this represents the sequestration of four negative supercoils because of wrapping of DNA by YejK. We found no evidence of protection of DNA from nuclease digestion (Fig. 6A), and there are many more that four binding units of YejK present on the DNA at this concentration. We therefore interpret this finding as indicating that YejK untwists DNA as it binds. If we assume that the binding site size for YejK is 12 bp, then the observed ΔLk implies an untwisting of the DNA by about 4 degrees per YejK binding unit.
Effect of YejK on the Type II Topoisomerases
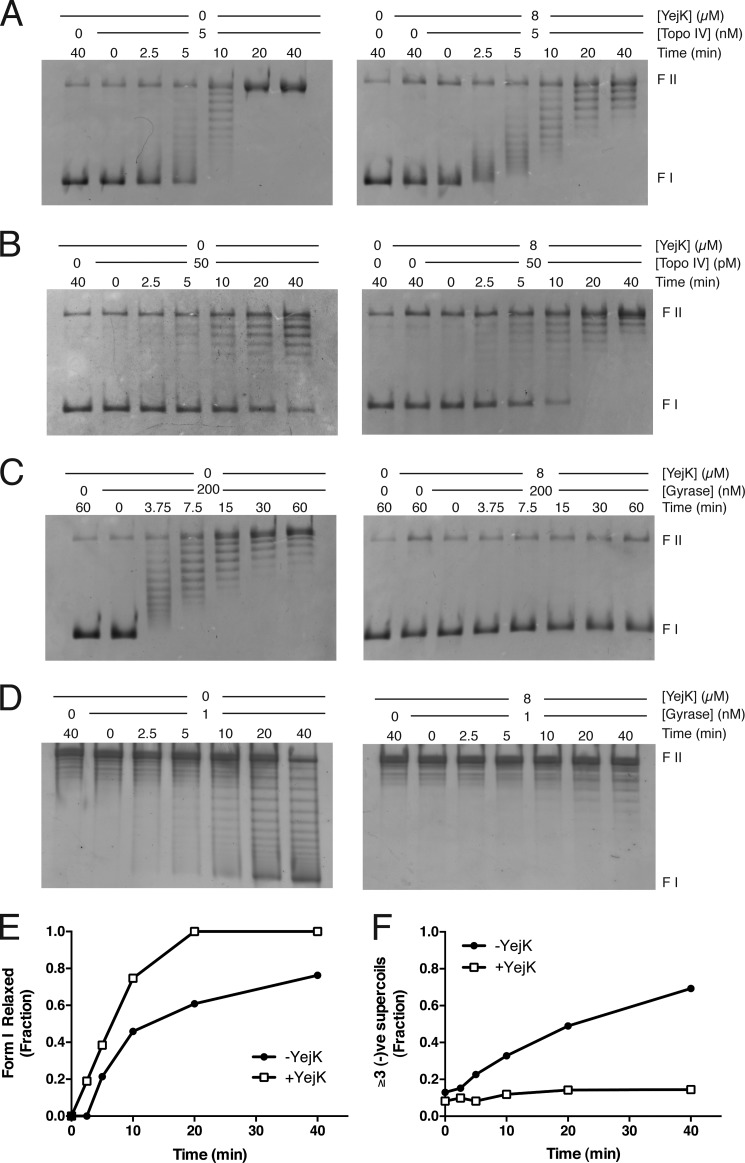
Because YejK was identified in a two-hybrid screen using the ParE subunit of Topo IV as bait, and could interact with all the subunits of Topo IV and DNA gyrase in this manner, we examined the effect of binding YejK to DNA on the action of these type II topoisomerases (Fig. 7).
FIGURE 7.
Effect of YejK binding to DNA on the activity of the type II topoisomerases. A, relaxation of (−)ve sc DNA by Topo IV. B, relaxation of (+)ve sc DNA by Topo IV. C, relaxation of (−)ve sc DNA by DNA gyrase. D, supercoiling of form I′ DNA by DNA gyrase. E, YejK binding to DNA stimulates relaxation of (+)ve sc DNA by Topo IV. Quantification of the extent of form I DNA relaxation in the experiment shown in panel B. The fraction of form I DNA present in the input DNA preparation (65% of the total DNA) was set at 100% and the loss of the form I band was quantified. F, YejK binding to DNA inhibits DNA supercoiling by DNA gyrase. Quantification of the extent of generation of form I DNA in the experiment shown in panel D. The fraction of DNA with three or more supercoils was quantified. Three supercoils was taken as the starting point because this allowed a clear distinction between the supercoiled and relaxed DNA on the gel.
YejK had a subtle, but obvious, effect on Topo IV-catalyzed relaxation of (−)ve sc DNA (Fig. 7A). Although there was no overt stimulation or inhibition, the apparent nature of the reaction changed from processive to distributive. This is most clearly evident by comparing the 2.5–10-min time points in each experiment. In the absence of YejK, starting material (i.e. sc form I DNA) is evident at the same time as fully relaxed DNA, as would be expected for a processive reaction where Topo IV binds the substrate and relaxes it to completion. In contrast, in the presence of YejK, relaxation of all DNA molecules occurred roughly equally and at the same time, generating a pattern where a tight distribution of relaxed DNA could be observed becoming progressively more relaxed as the reaction proceeded. This switch in mode may be because of a lowered affinity of Topo IV for the YejK-DNA complex compared with DNA alone. On the other hand, YejK stimulated Topo IV-catalyzed relaxation of (+)ve sc DNA slightly (Fig. 7, B and E) but no switch was observed in the nature of the reaction; it was processive either with or without YejK bound to the DNA.
The effect of YejK on DNA gyrase-catalyzed reactions was, for the most part, inhibitory. Relaxation of (−)ve sc DNA was completely inhibited (Fig. 7C), whereas the rate of supercoiling of form I′ DNA was reduced (Fig. 7, D and F). This pronounced difference in effect was interesting. Gyrase could clearly access the YejK-bound DNA for the supercoiling reaction without much problem, the concentration of gyrase in the supercoiling reaction is 1/200 of that in the relaxation reaction. Instead, it may be that YejK has some influence on how DNA can access gyrase. The relaxation reaction is thought to be the reverse of the supercoiling reaction, with the T segment entering the enzyme through the protein exit gate and exiting through the ATP-dependent clamp (23). Furthermore, the relaxation reaction is unlikely to require DNA wrapping by the enzyme. The differential inhibition observed suggests that gyrase with DNA wrapped about itself is stable on the YejK-DNA complex, whereas any other mode of DNA binding is unstable.
The YejK-induced switch in mode of DNA relaxation by Topo IV points out how little we know about the interaction of DNA enzymes with the real substrate in the cell, a protein-DNA complex, rather than naked DNA. We had previously shown that another NAP, HU, stimulated DNA gyrase-catalyzed decatenation of multiply linked DNA dimers (24). However, the vast majority of the biochemical analyses of the action of DNA enzymes are conducted in the absence of proteins that act to compact and shape the nucleoid. Whether the effect observed here on Topo IV can be extended to its action in the cell is difficult to estimate. Here we used a DNA substrate that was saturated with one particular NAP, YejK, whereas in the cell the DNA is bound by many different NAPs and we know little about how they are distributed.
Mutants of yejK Have an Elongated C Period
In a genome-wide, ORF knock-out survey, deletion of yejK was indicated to have little effect on growth (25). We constructed a yejK deletion using the linear replacement technique (14). The resulting strains, either C600ΔyejK::Cm or MG1655ΔyejK::Cm, have a complete replacement of yejK with cat flanked by FRT sites. The yejK mutant strain grew in LB with the same doubling time as the wild-type and displayed the same distribution of cell sizes (Fig. 8, A–D). To examine DNA replication, the wild-type and mutant strains were grown in minimal medium containing casamino acids and glucose and synchronized using dl-serine hydroxamate as described by Ferullo et al. (16). Cells were released from synchrony by resuspending in fresh medium containing cephalexin to prevent cell division. Aliquots of the cell suspension were removed with time, treated with rifampicin for 3 h to allow replication run out, and then analyzed by flow cytometry (Fig. 8, E and F). Based on when a change in the initial distribution of cells could be detected, resumption of replication appeared to be delayed slightly in the yejK mutant strain compared with wild-type and the C period (determined here as the time required for all 1N cells in the culture to be converted to 2N cells) was elongated by 18% (64 versus 56 min). This effect, whereas minor, is consistent with an ancillary role for YejK in maintaining DNA structure in the nucleoid.
FIGURE 8.
Characterization of a mutant strain deleted of yejK. A and B, MG1655 (panel A) and MG1655ΔyejK (panel B) cells were grown in LB medium at 37 °C for 2 h, fixed, stained with DAPI (a DNA stain, blue channel) and FM 4-64 FX (a live membrane stain, red channel), and imaged by fluorescence microscopy. C, growth curves of MG1655 and MG1655ΔyejK cells grown in LB medium at 37 °C. D, distribution of cell lengths for the cultures imaged in panels A and B. E and F, flow cytometric analyses of DNA replication in MG1655 (panel E) and MG1655ΔyejK (panel F). Cells growing in minimal medium were synchronized using serine hydroxamate, released by resuspension in fresh medium containing cephalexin, and grown at 37 °C. Aliquots were removed every 5 min and treated with rifampicin for 3 h at 37 °C. The distribution of chromosomal content was then determined by flow cytometry as described under “Experimental Procedures.”
YejK-GFP Is an Effective Marker for the Nucleoid
We constructed an allelic replacement of yejK with yejK-gfp. The GFP-tagged YejK did not have any effect on cell growth (Fig. 9E), and the pattern of GFP fluorescence was identical to that of the DAPI stain (Fig. 9, A–D). Thus, at this level of analysis, YejK is likely bound uniformly to the chromosomal DNA, similar to NAPs such as HU. The non-toxicity of the YejK-GFP construct should make it a useful marker for the nucleoid in live cell imaging.
FIGURE 9.

YejK-GFP is an effective marker for the nucleoid. A–D, MG1655yejK-ecgfps:kan cells were grown in minimal medium supplemented with 0.4% glucose and 0.2% casamino acids, stained with DAPI, spread onto polylysine-coated slides, and imaged by fluorescence microscopy. A, overlay of panels B (DIC), C (DAPI, a DNA stain, purple channel), and D (GFP, green channel). E, expression of YejK-GFP does not effect cell growth. Growth curves in LB medium at 37 °C of MG1655 and MG1655yejK-ecgfps:kan.
YejK Expression Level in Vivo
In the original report (9), YejK was released from spermine nucleoids treated with DNase I. Its abundance was estimated to be one-third of that of the large subunit of RNA polymerase, suggesting that there are about 600 copies in the cell. We used the strain containing the YejK-GFP chimera to make a more accurate determination of its abundance. The amount of YejK-GFP in a lysate prepared from mid-log phase MG1655yejK-ecgfps:kan cells grown in LB medium was compared with a reference standard of recombinant GFP from A. coerulescens (rACGFP1) by quantitative Western blotting using polyclonal anti-GFP antibodies (Fig. 10). We used a straightened version of the GFP gene to avoid silencing of the chimeric yejK-gfp gene by binding of H-NS to the naturally curved GFP gene (12) (see “Experimental Procedures”). The results indicated that there were about 24,000 copies of the YejK monomer per cell (the rACGFP1 is a monomer). Given that the DNA-binding form of the protein is likely either a dimer or tetramer, this puts the active form of the protein between 6,000 and 12,000 copies per cell. This level of abundance is in the same range of that of StpA, H-NS, IHF, and CbpB, but lower than that of HU and Fis (26). YejK levels were reduced by about one-third in cells grown in minimal medium containing glucose (determined by calculating the mean flourescence present in 200 cells) compared with those grown in LB medium.
FIGURE 10.

Abundance of YejK in vivo. A, an example of Western blots of a lysate prepared from mid-log phase MG1655yejK-ecgfps:kan cells and recombinant ACGFP1 as a standard. B and C, quantification (average of three experiments) of the blots shown in panel D. The amount of YejK per cell was calculated as described under “Experimental Procedures.”
This work was supported, in whole or in part, by National Institutes of Health Grant GM34558 (to K. J. M.).
- NAP
- nucleoid-associated protein
- bp
- base pair(s)
- (+)sc
- positively supercoiled
- (−)sc
- negatively supercoiled
- Topo IV
- topoisomerase IV
- nt
- nucleotide.
REFERENCES
- 1. Luijsterburg M. S., White M. F., van Driel R., Dame R. T. (2008) The major architects of chromatin. Architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43, 393–418 [DOI] [PubMed] [Google Scholar]
- 2. Dillon S. C., Dorman C. J. (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 [DOI] [PubMed] [Google Scholar]
- 3. Keane O. M., Dorman C. J. (2003) The gyr genes of Salmonella enterica serovar Typhimurium are repressed by the factor for inversion stimulation, Fis. Mol. Genet. Genomics 270, 56–65 [DOI] [PubMed] [Google Scholar]
- 4. Weinstein-Fischer D., Altuvia S. (2007) Differential regulation of Escherichia coli topoisomerase I by Fis. Mol. Microbiol. 63, 1131–1144 [DOI] [PubMed] [Google Scholar]
- 5. Menzel R., Gellert M. (1983) Regulation of the genes for E. coli DNA gyrase. Homeostatic control of DNA supercoiling. Cell 34, 105–113 [DOI] [PubMed] [Google Scholar]
- 6. McLeod S. M., Aiyar S. E., Gourse R. L., Johnson R. C. (2002) The C-terminal domains of the RNA polymerase α subunits. Contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 316, 517–529 [DOI] [PubMed] [Google Scholar]
- 7. Grainger D. C., Goldberg M. D., Lee D. J., Busby S. J. (2008) Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68, 1366–1377 [DOI] [PubMed] [Google Scholar]
- 8. Schneider R., Travers A., Kutateladze T., Muskhelishvili G. (1999) A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34, 953–964 [DOI] [PubMed] [Google Scholar]
- 9. Murphy L. D., Rosner J. L., Zimmerman S. B., Esposito D. (1999) Identification of two new proteins in spermidine nucleoids isolated from Escherichia coli. J. Bacteriol. 181, 3842–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arora C., Kee K., Maleki S., Keeney S. (2004) Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13, 549–559 [DOI] [PubMed] [Google Scholar]
- 11. Crameri A., Whitehorn E. A., Tate E., Stemmer W. P. (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14, 315–319 [DOI] [PubMed] [Google Scholar]
- 12. Corcoran C. P., Cameron A. D., Dorman C. J. (2010) H-NS silences gfp, the green fluorescent protein gene. gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J. Bacteriol. 192, 4790–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Espeli O., Nurse P., Levine C., Lee C., Marians K. J. (2003) SetB: an integral membrane protein that affects chromosome segregation in Escherichia coli. Mol. Microbiol. 50, 495–509 [DOI] [PubMed] [Google Scholar]
- 14. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McClendon A. K., Rodriguez A. C., Osheroff N. (2005) Human topoisomerase IIα rapidly relaxes positively supercoiled DNA. Implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345 [DOI] [PubMed] [Google Scholar]
- 16. Ferullo D. J., Cooper D. L., Moore H. R., Lovett S. T. (2009) Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods 48, 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marie D., Vaulot D., Partensky F. (1996) Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl. Environ. Microbiol. 62, 1649–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeda T., Yun C. S., Shintani M., Yamane H., Nojiri H. (2011) Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int. J. Evol. Biol. 10.4061/2011/685015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vlahovicek K., Kaján L., Pongor S. (2003) DNA analysis servers. Plot.it, bend.it, model.it and IS. Nucleic Acids Res. 31, 3686–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo F., Adhya S. (2007) Spiral structure of Escherichia coli HUαβ provides foundation for DNA supercoiling. Proc. Natl. Acad. Sci. U.S.A. 104, 4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q., Calvo J. M. (1993) Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 12, 2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klevan L., Wang J. C. (1980) Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an α2β2 protein core. Biochemistry 19, 5229–5234 [DOI] [PubMed] [Google Scholar]
- 23. Williams N. L., Howells A. J., Maxwell A. (2001) Locking the ATP-operated clamp of DNA gyrase. Probing the mechanism of strand passage. J. Mol. Biol. 306, 969–984 [DOI] [PubMed] [Google Scholar]
- 24. Marians K. J. (1987) DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J. Biol. Chem. 262, 10362–10368 [PubMed] [Google Scholar]
- 25. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants. The Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A. (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]