Background: The identity of natural substrate proteins of small heat shock proteins is poorly understood.
Results: A total of 95 and 54 natural substrate proteins were identified for IbpB in living cells at 50 and 30 °C, respectively.
Conclusion: IbpB preferentially protects translation-related proteins and metabolic enzymes.
Significance: This study offers mechanistic insights into the chaperone functions of small heat shock proteins for cells against stresses.
Keywords: Heat Shock Protein, Molecular Chaperone, Protein Aggregation, Protein Folding, Ribosomes, Small Heat Shock Proteins, Translation, Metabolic Enzymes, Photocross-linking, Unnatural Amino Acid
Abstract
Small heat shock proteins (sHSPs), as ubiquitous molecular chaperones found in all forms of life, are known to be able to protect cells against stresses and suppress the aggregation of a variety of model substrate proteins under in vitro conditions. Nevertheless, it is poorly understood what natural substrate proteins are protected by sHSPs in living cells. Here, by using a genetically incorporated photo-cross-linker (p-benzoyl-l-phenylalanine), we identified a total of 95 and 54 natural substrate proteins of IbpB (an sHSP from Escherichia coli) in living cells with and without heat shock, respectively. Functional profiling of these proteins (110 in total) suggests that IbpB, although binding to a wide range of cellular proteins, has a remarkable substrate preference for translation-related proteins (e.g. ribosomal proteins and amino-acyl tRNA synthetases) and moderate preference for metabolic enzymes. Furthermore, these two classes of proteins were found to be more prone to aggregation and/or inactivation in cells lacking IbpB under stress conditions (e.g. heat shock). Together, our in vivo data offer novel insights into the chaperone function of IbpB, or sHSPs in general, and suggest that the preferential protection on the protein synthesis machine and metabolic enzymes may dominantly contribute to the well known protective effect of sHSPs on cell survival against stresses.
Introduction
Molecular chaperone proteins are essential for cells to maintain protein homeostasis by assisting the folding/assembly of other proteins as well as the degradation of their misfolded forms (1). Small heat shock proteins (sHSPs),4 a molecular chaperone family with a polypeptide size of 12–43 kDa, are ubiquitously found in prokaryotes, archaea, and eukaryotes (2–4), and their functions are linked to development, aging, and diseases in animals (5–8). The sHSPs are known to suppress protein aggregation and to hold the substrate proteins in a folding-competent state, which might be subsequently refolded with the assistance of other ATP-dependent molecular chaperones (9–16). Certain members of sHSPs were also reported to protect cell membranes from stress-induced damages (17–19). As such, the sHSPs are considered as the first line of defense against stress-induced cellular damages (20, 21) and, when overexpressed, were found to be able to increase cell tolerance against various stress treatments (8, 22–24). The primary structures of sHSPs are characterized by possessing a conserved α-crystallin domain of ∼100 amino acids, which is flanked by a highly variable N-terminal arm and a short flexible C-terminal extension (2). Under in vitro conditions, sHSPs are often found to assemble as large oligomers of 12–40 subunits, using dimers as the building block (20).
It was 2 decades ago when sHSPs were reported to exhibit chaperone-like activity (9, 16, 25). Since then, although the chaperone-like activities of sHSPs have been studied extensively (as reviewed in Refs. 20, 26 and references therein), most studies (including ours (27–31)) were performed under in vitro conditions and using a variety of model substrate proteins. Nevertheless, identification of their in vivo substrate proteins and characterization of their roles toward these natural substrates are of greater importance. In two attempts using conventional co-purification, a total of 13 proteins were identified to interact with cyanobacterium Synechocystis Hsp16.6 in heat-shocked cells (32), and around 100 proteins were identified to co-aggregate with the purified bacterium Deinococcus radiodurans Hsp20.2 in the heated cell extract (33). In addition, a few more proteins, including ubiquitin-conjugating enzyme, proteasomal subunit, F-box protein, myotonic dystrophy protein kinase, initiation factor 4G, actin, and intermediate filament proteins, have also been reported to interact with sHSPs in different systems (as reviewed in Ref. 32 and references therein).
We recently characterized the substrate-binding residues of IbpB (34), a representative sHSP from Escherichia coli that confers cell resistance against stresses (11–12, 16, 31–32, 36–40), by using in vivo site-specific photocross-linking as mediated by the genetically incorporated unnatural amino acid p-benzoyl-l-phenylalanine (Bpa) (35). As a continuous effort, we attempted to identify the substrate proteins that IbpB binds in living cells, as captured by such in vivo photocross-linking, which was successfully utilized in identifying unknown protein-protein interactions (36, 37). By contrast to conventional noncovalent co-purification (32, 33), the in vivo photocross-linking approach apparently has such advantages as covalently capturing those transiently or weakly interacting proteins, as well as facilitating the subsequent protein purification under denaturing conditions (e.g. in the presence of 8 m urea) that would efficiently remove proteins indirectly bound to the bait protein.
We here report the identification of a total of 110 substrate proteins of IbpB in living cells. Remarkably, we found that IbpB exhibits a high substrate preference for translation-related proteins and a moderate preference for metabolic enzymes. We further demonstrated that representative translation-related proteins and metabolic enzymes are more prone to aggregate in cells lacking IbpB under stress conditions. Together, our in vivo data offer critical insights into the chaperone function of sHSPs in cells.
EXPERIMENTAL PROCEDURES
Plasmid Construction, Protein Expression, and Purification
Coding sequences for EF-Tu and TnaA were amplified from E. coli genomic DNA and cloned into the pBAD plasmid at the sites of NcoI and HindIII, with a tag of six histidines at the C terminus. The plasmids for expressing Bpa variant proteins of IbpB with a tag of six histidines were constructed as we described previously (34). EF-Tu-His and TnaA-His recombinant proteins were expressed in wild-type BW25113 E. coli cells, purified using nickel-nitrilotriacetic acid affinity chromatography (GE Healthcare), and then subjected to polyclonal antibody production in mice.
Bpa-mediated in Vivo Photocross-linking, Purification of Photocross-linked Products, and LC-MS/MS Analysis
Bpa variant proteins of IbpB were expressed in ΔibpB BW25113 E. coli cells, and Bpa-mediated in vivo photocross-linking was performed as we described recently (34). Briefly, the cells (cultured at 30 °C or heat shocked at 50 °C for 30 min) were transferred to a glass plate of 20 cm in diameter and pre-cooled on ice before being subjected to UV irradiation at 365 nm for 5 min using a Hoefer UVC 500 cross-linker. The cells were maintained on ice (4 °C) during UV irradiation so as to eliminate the potential temperature effect on such a photocross-linking process and also to minimize the heating effect of the UV irradiation. The photocross-linked products of IbpB were purified by nickel-nitrilotriacetic acid affinity chromatography under denaturing conditions (i.e. in the presence of 8 m urea). The Phe-16 Bpa variant without in vivo photocross-linking was purified by the same procedure and taken as the control. The purified products were concentrated with Amicon Ultra-15 centrifugal filter units (Millipore) to a final volume of 500–1000 μl. Samples were then subjected to 10% Tricine SDS-PAGE and detected with Coomassie Blue staining. All the gel bands except IbpB monomers were cut for LC-MS/MS analysis (38, 39), similarly with we recently reported (40). Proteins were identified by running MASCOT against the entire NCBI database. The proteomic analysis of each gel region was performed in duplicate, and only those proteins that were found in both independent experimental runs were selected as identified hits. MASCOT score was calculated according to the earlier report (41). In our case, when it is larger than 35 (for Phe-16, 30 °C), 36 (for Phe-16, 50 °C), 33 (for Asn-25, 30 °C), 34 (for Ans-25, 50 °C), or 37 (for Tyr-45, 50 °C; Arg-67, 50 °C; and Ala-139, 50 °C), the MASCOT score indicates the identity or extensive homology between the experimental peptide set and the identified protein significantly (p < 0.05). The proteins identified in the sample of the purified Phe-16 Bpa variant protein without in vivo photocross-linking (Fig. 2, lane 2) were set as background to remove those proteins identified in the samples of purified in vivo photocross-linked products. Similarly, the protein aggregates purified from heat-shocked wild-type and ΔibpB cells were also subjected to LC-MS/MS analysis.
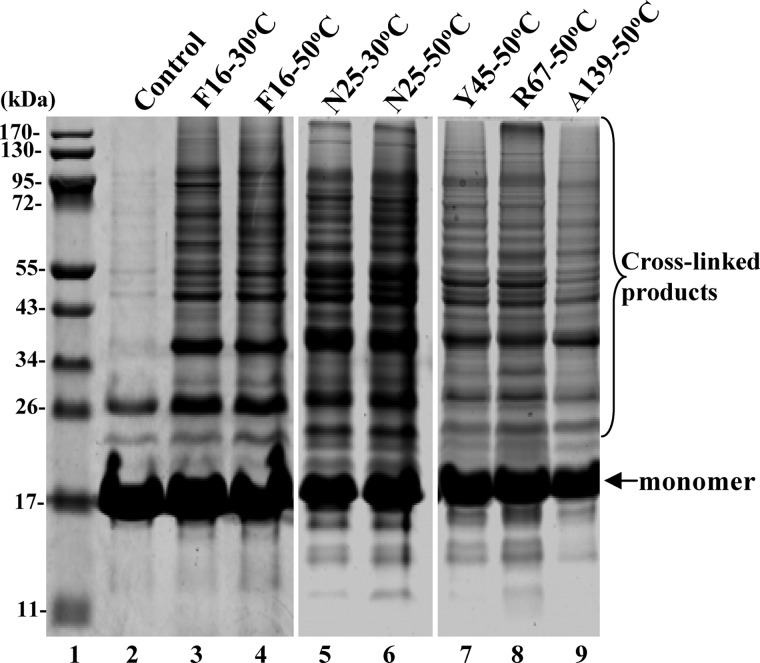
FIGURE 2.
Identification of IbpB-bound proteins in living cells. SDS-PAGE analysis of the purified in vivo photocross-linked products of the Phe-16 (F16) Bpa, Asn-25 (N25) Bpa, Tyr-45 (Y45) Bpa, Arg-67 (R67) Bpa, and Ala-139 (A139) Bpa variants of IbpB (as visualized by Coomassie Blue staining). The protein bands except for IbpB monomers were subjected to protein identification by LC-MS/MS, with the results shown in supplemental Tables S1–S3. The Phe-16 Bpa protein without in vivo photocross-linking was also purified in parallel (lane 2) and used as a negative control.
Isolation of Protein Aggregates from Cells after Heat Shock
Wild-type and ΔibpB BW25113 E. coli cells were grown at 30 °C to mid-exponential phase and then grown at 50 °C for 4 h before harvesting. Cells were washed and lysed by sonication. Unbroken cells and cell debris were removed by centrifugation at 5000 × g for 10 min. Cell extract was then centrifuged at 15,000 × g for 30 min to obtain the soluble fraction (supernatant) and the insoluble fraction (pellet of protein aggregates). All the fractions were analyzed by Tricine SDS-PAGE and immunoblotted using antibodies against the specified proteins. In particular, the protein aggregates were subjected to protein identification by LC-MS/MS.
Suppression of Protein Thermal Aggregation by IbpB in Vitro
The in vitro chaperone-like activity was assayed by measuring the capacity of purified His-tagged IbpB (0.5 mg/ml) to suppress the heat-induced aggregation of purified TnaA-His (0.5 mg/ml) in 50 mm sodium phosphate buffer (pH 7.2) at 50 °C. Aggregation was monitored on a UV-8500 spectrophotometer (Shanghai, TechCorp) at 360 nm. The temperature was controlled by connecting the cell surrounding the cuvette to a MultiTemp III water bath (Amersham Biosciences).
Catalase Activity Assay
Cells were washed three times with 50 mm sodium phosphate buffer (pH 7.4) and resuspended in the same buffer for subsequent sonication. Cell lysates were centrifuged at 15,000 × g for 10 min to obtain cell extract. The catalase activity was determined by adding 7 mm H2O2 to the cell extract and then immediately recording the absorbance at 240 nm on a UV-8500 spectrophotometer (Shanghai Tech Corp). Three independent experiments were performed.
Immunodetection of Protein-bound Carbonyl Groups
Cell lysates were subjected to Tricine SDS-PAGE, and the gel was incubated with 2,4-dinitrophenylhydrazine for 15 min. The 2,4-dinitrophenylhydrazine-derivatized proteins were immunodetected by anti-2,4-dinitrophenol antibodies (Sigma).
Bioinformatics Analysis
Protein function was classified according to an earlier report (42). The ratio of each functional protein class in the E. coli genome referred to the data deposited at NCBI (www.ncbi.nlm.nih.gov). The proteome of E. coli referred to the data deposited in the UnitProt database. At this end, 1648 proteins that are categorized according to their roles in a variety of biological processes, as well as nonredundant 51 ribosomal proteins, seven peptidyl-prolyl cis-trans isomerases, and 11 disulfide bond-catalyzing enzymes (a total of 1717 proteins), were taken together as the proteome of E. coli for calculating the basal ratio of each functional protein class (Table 2).
TABLE 2.
Functional classification of IbpB-interacting proteins
| Protein functiona | Total | 30 °C | 50 °C | 30–50 °C | Basal ratioa |
Aggregated proteinsb | |
|---|---|---|---|---|---|---|---|
| Proteome | Genome | ||||||
| Total | 113 (100%) | 55 (100%)c | 98 (100%) | 40 | 100.0% | 100.0% | 32 (100.0%) |
| Metabolism | 66 (58.4%)d | 36 (65.5%)e | 59 (60.2%)d | 29 | 46.9% | 42.9% | 18 (56.3%) |
| DNA replication and transcription, DNA/RNA modification | 8 (7%)f | 3 (5.5%)d | 6 (6.1%)f | 1 | 19.9% | 12.8% | 1 (3.1%)e |
| Translation | 19 (16.8%)f | 5 (9.1%) | 19 (19.4%)f | 5 | 5.7% | 4.1% | 7 (21.9%)e |
| Protein quality control | 3 (2.7%) | 1 (1.8%) | 3 (3.1%) | 1 | 3.8% | 3.2% | 2 (6.3%) |
| Other functions | 14 (12.4%)e | 8 (14.5%) | 10 (10.2%)e | 4 | 23.6% | 17.9% | 2 (6.3%) |
| Uncharacterized | 3 (2.7%) | 2 (3.6%) | 1 (1.0%) | 0 | NAg | 19.1% | 2 (6.3%) |
a Protein functional classification and basal level calculation were performed as described under “Experimental Procedures.”
b Protein species were counted based on the data displayed in supplemental Table S4.
c The value in parentheses is the percentage of each functional protein class.
d Significance level is indicated by p < 0.05 when compared with the basal ratio among the proteomes.
e Significance level is indicated by p < 0.01 when compared with the basal ratio among the proteomes.
f Significance level is indicated by p < 0.001 when compared with the basal ratio among the proteomes.
g NA means data were not available.
Statistics
Statistics were performed in Microsoft Excel software using the binominal distribution or U test. Significance levels (with p values being less than 0.05, 0.01, or 0.001) were indicated in Table 2 and supplemental Table S7.
RESULTS
IbpB Preferentially Functions under Heat Shock Conditions
Overexpression of IbpB was reported to confer E. coli cell resistance to oxidative, heat shock, and ethanol stresses (43). To find out the most physiologically relevant condition for identifying the in vivo substrate proteins of IbpB, we first examined the induction effect of these stresses on the protein level of endogenous IbpB in cells. Data presented in Fig. 1A clearly indicate that although the basal protein level of IbpB was marginally detectable in nonstressed cells, it was significantly increased after the cells were treated by H2O2, heat shock, or ethanol stresses. The induction effect of heat shock was apparently stronger than that of H2O2 and ethanol (Fig. 1A, lane 3 versus lanes 2 and 4), implicating that IbpB is more functionally relevant to the heat shock stress.
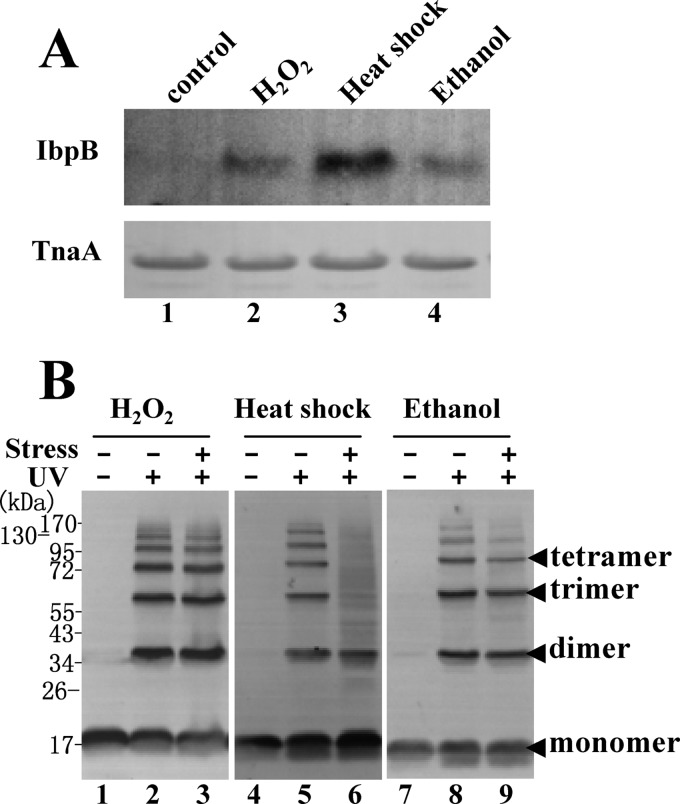
FIGURE 1.
IbpB preferentially functions under heat shock conditions. A, immunoblotting results of endogenous IbpB in wild-type cells cultured under the indicated stress conditions using the anti-IbpB polyclonal antibody. TnaA was used as a loading control. B, immunoblotting results of in vivo photocross-linked products of Tyr-45 Bpa expressed in ΔibpB cells using the anti-His tag antibody. Cells were grown at 30 °C and then subjected to stress treatment for half an hour by heating at 50 °C or by adding to a final concentration of 5 mm H2O2 or 10% ethanol before further analysis.
To further address this question, we took advantage of in vivo photocross-linking to characterize the interactions of IbpB with cellular proteins in living cells under different stress conditions. For this purpose, Tyr-45 Bpa was selected from the 71 Bpa variants of IbpB we generated earlier (34), as this variant was photocross-linked exclusively as IbpB homo-oligomers in nonstressed cells (Fig. 1B) (34). We observed that the interaction of the Tyr-45 Bpa variant with cellular proteins, as reflected by its photocross-linked products (exception of cross-linked homo-oligomers), was substantially enhanced under heat shock conditions (Fig. 1B, lane 6) but only slightly increased under H2O2- and ethanol-induced stress conditions (lanes 3 and 9). Similar results were obtained (supplemental Fig. S1A) when the in vivo photocross-linked products of another two Bpa variants (F4Bpa and M10Bpa) were analyzed. Together, these observations suggest that, although it also functions under H2O2- and ethanol-induced stress conditions, IbpB preferentially functions under the heat shock condition, in line with our earlier studies showing that the in vitro chaperone activity of IbpB was remarkably regulated by temperature (30, 31).
Identification of Proteins Bound to IbpB in Living Cells with or without Heat Shock as Captured by Site-specific Photocross-linking and Characterized by Mass Spectrometry
Our earlier study revealed at least 48 residues of IbpB participating in substrate binding (34). Furthermore, these residues are classified into three types as follows: type I generally being located in the N-terminal arm and capable of mediating substrate binding at both low and heat shock temperatures; type II being only activated at the heat shock temperature; and type III being involved in oligomerization at a low temperature (30 °C) but switching to substrate binding at heat shock temperatures (42–50 °C). Such information guided us here to rationally choose representative Bpa variants of IbpB to capture the substrate proteins in cells by in vivo photocross-linking.
Experimentally, five representative Bpa variants of IbpB out of the 48, with Bpa being individually incorporated at residues of type I (Phe-16 and Asn-25), type II (Ala-139) and type III (Tyr-45 and Arg-67) (34), were selected for capturing the natural substrate proteins of IbpB at both normal (30 °C) and heat shock (50 °C) temperatures. In addition, these five residues are positioned at the three characteristic domains of IbpB, with Phe-16 and Asn-25 being located at the N-terminal arm, Tyr-45 and Arg-67 at the α-crystallin domain, and Ala-139 at the C-terminal extension. Furthermore, the chaperone activities of these five Bpa variants were largely comparable with that of the wild-type IbpB protein (34), and their relatively high expression levels (34) also enabled us to obtain enough proteins for the subsequent protein identification experiment. Together, we consider that the total proteins identified from these variants somehow represent a full picture of IbpB-bound proteins, although such representations may not be exhaustive.
The bound proteins, being photocross-linked to IbpB Bpa variants (supplemental Fig. S1B), were purified by affinity chromatography under denaturing conditions (i.e. in the presence of 8 m urea, which would efficiently remove proteins indirectly bound to IbpB, as well as those proteins nonspecifically bound to the resin). After separation by SDS-PAGE (Fig. 2), the protein bands other than the monomers of IbpB were then collected and subjected to protein identification analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In our study, LC-MS/MS was performed repeatedly for each sample, and only those proteins with a MASCOT score (41) larger than the threshold value were considered as IbpB-bound proteins significantly (p < 0.05, for details, see under “Experimental Procedure”). Additionally, to remove the false-positive results, the Phe-16 Bpa variant without in vivo photocross-linking was purified by the same procedure (Fig. 2, lane 2) and subjected to protein identification analysis as a control.
All proteins identified with each Bpa variant are listed in supplemental Tables S1–S3 and summarized in Table 1. The photo-cross-linker (Bpa) appears to be different in the capacity of interaction with cellular proteins among these five variants (Table 1). Specifically, Bpa in Phe-16 exhibits the highest capacity, as the protein numbers identified with Phe-16 Bpa were up to 44 and 55 at 30 and 50 °C, respectively. In addition, 13 and 18 proteins were found to be specifically associated with Phe-16 Bpa but not with other Bpa variants at these two temperatures, respectively. These observations thus nicely support our earlier finding that the substrate-binding residues in IbpB were hierarchically activated (34).
TABLE 1.
Number of proteins identified with each Bpa variant
| Variant | Temperature | No. of proteinsa |
|
|---|---|---|---|
| Total | Unique | ||
| °C | |||
| Phe-16 | 30 | 44 | 13 |
| 50 | 55 | 18 | |
| Asn-25 | 30 | 15 | 2 |
| 50 | 24 | 7 | |
| Tyr-45 | 50 | 29 | 1 |
| Arg-67 | 50 | 31 | 6 |
| Ala-139 | 50 | 26 | 4 |
| Total | 30 | 55 | 15 |
| Total | 50 | 98 | 58 |
a Protein species identified with each variant were counted as total ones, and the protein species that are specifically identified with this variant but not with other variants were also counted as unique ones.
In sum, a total of 113 proteins were identified as IbpB-bound, by taking into account results of all the Bpa variants (Table 1). Specifically, 55 and 98 IbpB-bound proteins were identified at 30 and 50 °C, respectively, with 40 being common for both temperatures (Table 1). Among these IbpB-bound proteins, the three protein quality control factors, IbpA, ClpP, and Tig (part II of supplemental Table S3), are considered as functional partners, instead of substrate proteins of IbpB. As such, we identified a total of 110 natural substrate proteins of IbpB, with 54 at 30 °C and 95 at 50 °C.
Validation of the Interactions of IbpB with the Identified Proteins
We next validated the interaction of IbpB with these identified proteins using multiple approaches. First, we verified the presence of these proteins in the in vivo photocross-linked products by immunoblotting analysis. For this purpose, we chose EF-Tu and TnaA as representatives, with the former being identified, by mass spectrometry, only in the photocross-linked products formed at 50 °C (as protein 9 in supplemental Table S1), whereas the latter was identified at both 30 and 50 °C (as protein 27 in supplemental Table S2). Consistently, the cross-linked form of EF-Tu and IbpB (as indicated by asterisks in the upper part of Fig. 3A) was only detected in the in vivo photocross-linked products of Bpa variants of IbpB formed in cells cultured at 50 °C but not at 30 °C, whereas the cross-linked form of TnaA and IbpB was detected in cells cultured at both 30 and 50 °C (the lower part of Fig. 3A).
FIGURE 3.
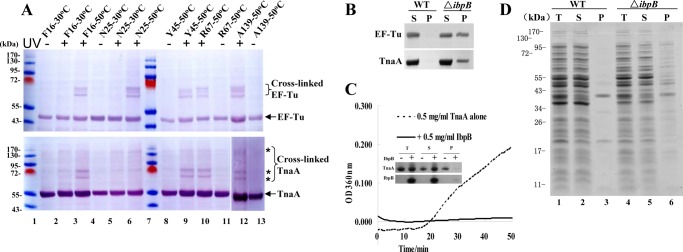
Validation of the interactions of representative IbpB-bound proteins with IbpB using multiple approaches. A, immunoblotting results of EF-Tu or TnaA in the in vivo photocross-linked products of the Bpa variants of IbpB (using polyclonal antibodies against EF-Tu or TnaA). B, immunoblotting results of EF-Tu and TnaA that were present in the protein aggregates isolated from wild-type and ΔibpB cells after being heat-shocked at 50 °C for 4 h. C, suppression of the in vitro thermal aggregation of purified TnaA at 50 °C by purified IbpB, as monitored by absorbance spectroscopic analysis at 360 nm. The inset shows the SDS-PAGE analysis results of proteins present in the soluble and pellet fractions after such thermal aggregation. D, SDS-PAGE analysis of the protein aggregates of the wild-type and ΔibpB cells, similarly to what described in B. Protein bands in the aggregates (lanes 3 and 6) were subjected to protein identification by LC-MS/MS, with the results shown in supplemental Table S4. B–D, T, total proteins; S, soluble proteins; P, protein aggregates (pellet).
We also performed genetic studies to find out whether these identified proteins are substrate proteins of IbpB. For this purpose, we analyzed the level of thermal aggregation of these proteins, also represented by EF-Tu and TnaA, in IbpB-deficient cells. Immunoblotting data presented in Fig. 3B demonstrate that the level of aggregation for both EF-Tu and TnaA was dramatically increased in the heat-shocked ΔibpB cells but was hardly detectable in wild-type cells. Furthermore, we found that the purified IbpB was able to efficiently suppress the thermal aggregation of the purified TnaA under in vitro conditions (Fig. 3C), a standard approach for assaying the chaperone activities of sHSPs in general (20).
Furthermore, we attempted to find out whether these IbpB-bound proteins as a whole, similar to EF-Tu and TnaA (Fig. 3B), are more prone to aggregation in IbpB-deficient cells under the heat shock condition. To this end, we did observe a significantly higher level of thermally aggregated proteins in ΔibpB cells than in the wild-type cells (Fig. 3D, lane 6 versus 3). This is in line with the earlier report that the deletion of ibpB and its homolog (ibpA) in E. coli cells resulted in an increase in protein aggregation (44). To reveal which proteins formed aggregates only in the ΔibpB cells but not in the wild-type cells, we then collected such aggregated proteins from both types of cells and subjected them to protein identification by using LC-MS/MS.
The results indicate that the identified IbpB-bound proteins as a whole are indeed more prone to aggregation in IbpB-deficient cells under the heat shock condition. In particular, among a total of 32 proteins identified as being specifically present in the aggregates of the ΔibpB cells, 21 were found as the IbpB-bound proteins from in vivo photocross-linking analysis (supplemental Table S4). As a validation, TnaA and EF-Tu that had been detected in the aggregates of the ΔibpB cells by immunoblotting analysis (Fig. 3B) were also found among the 32 proteins (supplemental Table S4). Together, all these observations indicate that IbpB is able to function as a chaperone in suppressing the thermal aggregation of its natural substrate proteins under both in vivo and in vitro conditions. The high deviation in the number of identified proteins from such aggregates and from photocross-linking might be due to the following reason. The aggregates would only be formed and isolatable for those proteins present at relatively high concentrations in the cells, although photocross-linking would be able to capture proteins with a far wider range of concentrations. Additionally, certain IbpB substrate proteins might also be protected by other molecular chaperones (e.g. IbpA, DnaK, and GroEL) and thus would not appear in the aggregates in the ΔibpB cells.
Last but not least, our protein identification data also find support from the earlier studies of others. For instance, a few of the proteins identified here (e.g. EF-Tu, GatY, LpdA, MreB, TnaA, and GadA/GadB) were found to be associated with IbpB via conventional noncovalent purifications (45, 46). In particular, both EF-Tu and TnaA were found to be pre-dominant IbpB-interacting proteins by using formaldehyde-medicated chemical cross-linking in our earlier study (34). Consistent with this, they were identified by photocross-linking with high MASCOT scores (for details see supplemental Tables S1 and S2), which, to certain degree (41), reflect the high amount of these two proteins in the purified photocross-linked products of IbpB. In addition, IbpA, which is known to interact and also functionally cooperate with IbpB (15, 45, 47), was also identified here (part II in supplemental Table S3). Furthermore, fructose-1,6-biphosphate aldolase (FbaB) and alcohol dehydrogenase (AdhE), both being protected by IbpA/IbpB in cells under heat shock conditions (44, 48), were identified here (supplemental Table S2).
IbpB Exhibits Remarkable Substrate Preference to Translation-related Proteins That Are Prone to Aggregation in IbpB-deficient Cells under the Heat Shock Condition
A literature search indicates that these IbpB-bound proteins, with a total number of 113, function in a variety of essential cellular processes (Table 2), including metabolism, DNA replication, recombination and repair, gene transcription, translation, protein quality control, and so on. This is in line with the earlier report on Hsp16.6 (32).
The most novel observation is that, among the 95 substrate proteins bound to IbpB at 50 °C, 19 are translation-related proteins (Table 2; for details see supplemental Table S1), including ribosomal proteins, translation factors like EF-Tu, and many amino-acyl tRNA synthetases. The occurrence of such translation-related proteins is 19.6%, more than three times higher than their basal ratio being 5.7% in the proteome or 4.1% in the genome of the E. coli cells (p < 0.001; Table 2). This result strongly suggests that IbpB preferentially protects translation-related proteins under the heat shock condition. In support of this, among the 32 thermally aggregated proteins in ΔibpB cells, 7 or 21.9% are translation-related (Table 2), being almost three times higher than their basal level in the proteome (p < 0.01), and 5 of the 7 aggregated translation-related proteins were already identified as IbpB-bound proteins by the in vivo photocross-linking (supplemental Table S5). Taken together, our observations suggest that, on the one hand, that the translation-related proteins are prone to aggregation under the heat shock condition and, on the other hand, that IbpB is able to protect them under such conditions in living cells.
IbpB Exhibits Moderate Substrate Preference to Metabolic Enzymes That Are Prone to Inactivation in IbpB-deficient Cells under Heat Shock and Oxidative Conditions
Another notable point from functional profiling of IbpB-bound proteins is that the occurrence of metabolic enzymes among them is as high as 65.5% at 30 °C and 60.2% at 50 °C, significantly higher than their occurrence among the E. coli proteomes (46.9%) (p < 0.01 and p < 0.05, respectively; Table 2). These metabolic enzymes (listed in supplemental Table S2) include those involved in carbohydrate metabolism (e.g. AceB, GltA, and AcnB), amino acid metabolism (e.g. PutA, AspC, and TnaA), lipid metabolism (e.g. FabB and GlpK), oxidative respiration (e.g. NuoC and CyoA), and ATP synthesis (e.g. AtpA and AtpD).
We next examined whether IbpB is able to protect stress-induced enzymatic inactivation in cells. In retrospect, fructose-1,6-biphosphate aldolase and alcohol dehydrogenase, two enzymes here identified as IbpB-bound proteins (supplemental Table S2), had been reported to be protected from thermal inactivation in cells by IbpB (44, 48). Here, we examined the effect of ibpB deletion on the enzymatic activity of two E. coli catalases (KatE and KatG) that were both identified as IbpB-bound proteins (supplemental Table S2). Data presented in Fig. 4A revealed a significant decrease in the cellular catalase activity of ΔibpB cells when treated by heat shock or H2O2, but not in wild-type cells. In particular, when a combination stress of heat shock and H2O2 was applied, the catalase activity of ΔibpB was hardly detectable but was maintained at a significant level in wild-type cells (Fig. 4A). In addition, the loss of the catalase activity of ΔibpB cells could be rescued by a complementary expression of wild-type or Bpa variants of IbpB (data not shown). Because we also identified KatG as an aggregated protein in heat-shocked ΔibpB cells (supplemental Table S4), the loss of catalase activity in ΔibpB cells should most likely result from the aggregation of the functional enzyme, which is presumed to be protected by IbpB in wild-type cells.
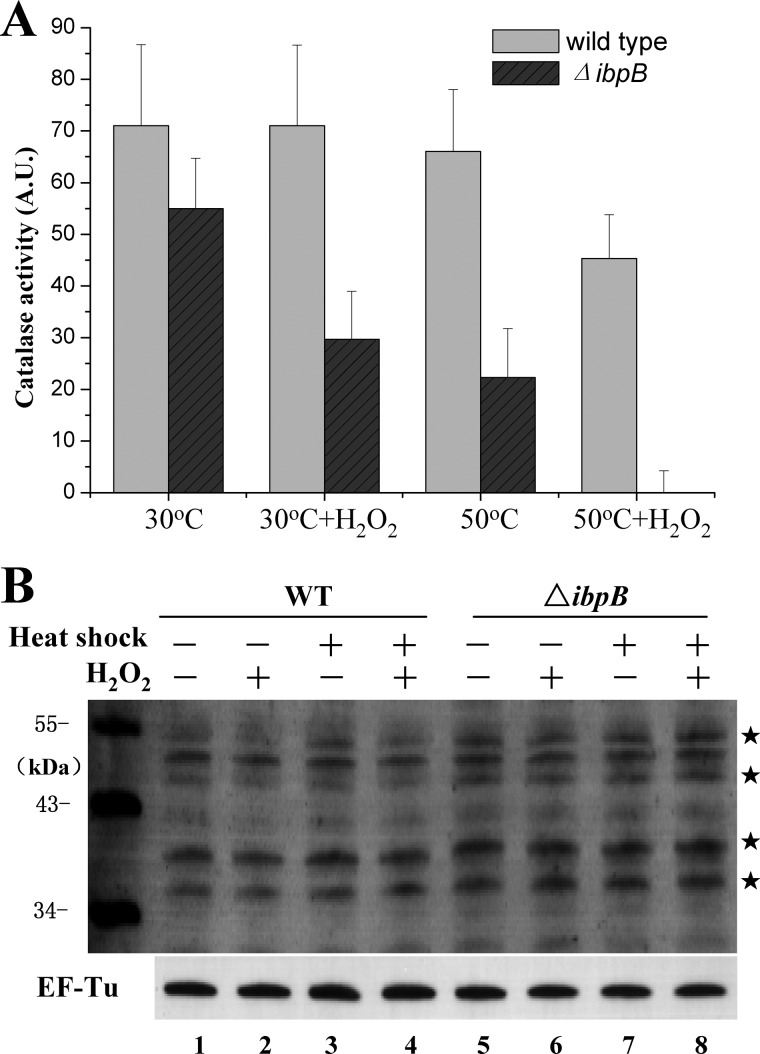
FIGURE 4.
IbpB protects representative metabolic enzymes from stress-induced inactivation. A, relative levels of catalase activity of the wild-type and ΔibpB cells (from three independent assays). Cells were heat-shocked at 50 °C for 0.5 h and/or treated with 5 mm H2O2 for 0.5 h, sonicated, and centrifuged. Cell extracts were then subjected to catalase activity assay as described under “Experimental Procedures.” B, immunoblotting detection of carbonylated proteins in the wild-type and ΔibpB cells after heat shock at 50 °C for 0.5 h and/or treatment with 5 mm H2O2 for 0.5 h. Protein bands with a higher level of carbonylation signals in ΔibpB cells than that in wild-type cells are indicated by asterisks. EF-Tu was immunoblotted as a loading control.
Such a potential protection effect of IbpB on the enzymatic activity of catalases prompted us to further determine whether IbpB would influence protein oxidation in living cells, given that H2O2, as a main cause of intracellular protein oxidation (49), is decomposed by catalases. To this end, the level of protein carbonylation, a major form of protein oxidation (50), was found to be significantly higher in ΔibpB cells than in wild-type cells (as represented by the protein bands labeled with asterisks in Fig. 4B), suggesting that certain E. coli proteins became more vulnerable to oxidation in the absence of IbpB.
DISCUSSION
Here, we utilized the in vivo photocross-linking approach to characterize the substrate proteins of sHSPs in living cells, and we identified over 100 natural substrate proteins for IbpB. Importantly, we show that IbpB, although able to protect a wide range of cellular proteins as commonly believed for sHSPs (32), remarkably shows a significant substrate preference to translation-related proteins and metabolic enzymes. These findings offer novel insights into the chaperone function and mechanism of sHSPs.
A comparison of the protein abundance histograms of the E. coli proteome and the IbpB substrate proteins (supplemental Fig. S2, A and B) indicates that the latter are found to be relatively abundant. However, our conclusion that IbpB has a substrate preference is validated by the following three facts. First, among the eight most abundant proteins present in the E. coli proteome, only TufB, TufA, and MDH were identified as IbpB substrate proteins in our study (TufB and TufA, being different at only one residue, are usually termed together as EF-Tu), although the others (i.e. RplL, CspC, GapA, Eno, and GroEL) were not identified as IbpB substrates. Second, some IbpB substrate proteins (e.g. FdnG, GatC, and PolB) were found to be expressed at relatively low levels, only accounting for around one ten-thousandth of EF-Tu level (for details, see supplemental Table S6). Third, and more convincingly, EF-Tu, as the most abundant protein among the E. coli proteomes, was found to be an IbpB substrate protein only at 50 °C but not at 30 °C (supplemental Table S1 and Fig. 3A) although present at the same level at both temperatures (Figs. 3A and 4B).
Apparently, protein abundance is not the determining factor for a protein to be bound with IbpB. Rather, the principal factor determining such interaction is most likely the aggregation tendency of the protein during its folding/unfolding, which has been well recorded earlier using other sHSPs (51–56).
Functional Diversity of Substrate Proteins of sHSPs
In retrospect, sHSPs were reported to nearly interact with all types of aggregation-prone model substrate proteins under in vitro conditions (9, 16, 25, 56). These substrate proteins are structurally and functionally diversified and include but are not limited to metabolic enzymes (e.g. α-glucosidase, phosphoglucose isomerase, glutathione S-transferase, enolase, aldolase, lactate dehydrogenase, citrate synthase, carbonic anhydrase, malate dehydrogenase, alcohol dehydrogenase, xylose reductase, and β-galactosidase), cytoskeleton proteins (e.g. β-actin and β-tubulin), and other functions (e.g. elastase, luciferase, lysozyme, Ataxin-3, Sup35, α-synuclein, α-lactalbumin, Abrin, β-crystallin, γ-crystallin, and titin), and even include peptides (insulin B chain and melittin). More importantly, identification of substrate proteins of a single sHSP (bacterial Hsp16.6 or Hsp20.2) by conventional co-purification indicated that the sHSP was able to protect a wide range of cellular proteins (32, 33).
The broad substrate spectrum of bacterial sHSPs has been unequivocally demonstrated by our study reported here, as IbpB-bound proteins were found to participate in metabolism, DNA replication, recombination and repair, gene transcription, protein synthesis, transportation, cell shape, and division (supplemental Tables S1–S3). Nevertheless, the substrate functional diversity of eukaryotic sHSPs is largely unknown. Given that the genomes of eukaryotes usually encode a larger number of sHSPs than those of prokaryotes (4) and that the eukaryotic sHSPs may have different subcellular compartments (57, 58) and cell/tissue specificity (59), it is speculated that the function spectrum of substrate proteins of eukaryotic sHSPs would not be so broad as that of prokaryotic sHSPs.
Preferential Protection on Translation-related Proteins and Metabolic Enzymes May Dominantly Contribute to the Protective Effect of sHSPs on Cells against Stresses
Despite the functional diversity of its substrate, our data also reveal that IbpB has a remarkable substrate preference to translation-related proteins, which appear more prone to aggregation in cells lacking IbpB under the heat shock conditions. We noticed that translation-related proteins were even more substantially enriched in the substrate proteins of Hsp20.2 that were identified from the thermally treated cell extract (33). In light of these observations, it is conceivable that preferential protection on the protein synthesis machine apparently represents one of the primary functions of sHSPs in cells under heat shock conditions. It follows that such a protective effect on the protein synthesis machines would substantially contribute to the commonly observed increase of thermotolerance of E. coli cells elicited by the overexpression of sHSPs (e.g. IbpB) (43, 60–62), although protection on other cellular components, including cell membranes (17–19), may also be involved.
We also detected a significant substrate preference of IbpB to metabolic enzymes (Table 2), and the representative ones were found by us (Fig. 4A) and others (44, 48) to be more vulnerable to stress-induced inactivation in cells lacking IbpB. Whereas preferential protection of each metabolic enzyme by IbpB may have specific biological effects, as indicated by the decrease of catalase activity and the related increase of protein carbonylation in ΔibpB cells under stress conditions (Fig. 4), one primary benefit from the protection of IbpB on metabolic enzymes as a whole may be linked to protein synthesis, a cellular process whose functional integrity definitely requires both abundant amino acids and ATP. In support of this, IbpB-bound metabolic enzymes include many proteins involved in amino acid metabolism and energy production/conversion (supplemental Table S2). In this respect, preferential protection of IbpB on metabolic enzymes is partially intrinsically linked to its preferential protection on the translation-related proteins.
In addition, functional profiling of the substrates identified for each Bpa variant of IbpB (supplemental Table S7) reveals that metabolic enzymes seem to be preferentially bound to residues located in the N-terminal region (Phe-16 and Asn-25), whereas translation-related proteins are preferentially bound to residues located in both the N-terminal region (Phe-16) and α-crystallin domain (Tyr-45 and Arg-67). These observations suggest that each functional class of substrate proteins might be preferentially bound to certain positions in IbpB.
Other Notable Observations ClpP
Given that molecular chaperones are believed to function as a network in cells (1), it is of interest to notice that ClpP, a protease associated with Hsp100, was identified here as an IbpB-interacting protein (part II in supplemental Table S3). Given that ClpB, the chaperone subunit of Hsp100, was found to be functionally related to IbpB (11, 15, 47, 63), identification of ClpP as an IbpB-interacting protein strongly implicates that Hsp100 would not only help release the IbpB-bound substrate proteins (via the ClpB subunit) but also further degrade them (via the ClpP subunit), which is worth further examination.
DnaK
Surprisingly, DnaK, which has been reported to assist the refolding/reactivation of the substrate proteins of IbpB (10–11, 15, 47), was not identified here as an IbpB-interacting protein. One likely explanation for this observation is that DnaK functionally cooperates with IbpB by interacting with the substrate protein but not directly with IbpB. Alternatively, it might result from the nonexhaustive nature of our photocross-linking experiments, i.e. residues other than these five Bpa incorporations may interact with DnaK.
Secretory Proteins
Additionally, seven proteins in the outer membrane (LamB, OmpC, OmpF, Lpp, OsmE, Pal, and YbaY) and two in the periplasm (FdnG and MdoG) were also identified as IbpB-bound proteins. We noticed that the cytoplasmic chaperone GroEL was also reported to interact with some proteins localized in the outer membrane and periplasm (64). Whether these secretory nascent polypeptides bound by cytoplasmic chaperones (IbpB or GroEL) are processed for the downstream biogenesis (e.g. the Sec system-assisted translocation across the inner membrane (65)) or are destined for degradation by cytoplasmic proteases (e.g. ClpP) merits further explorations. In addition, three integral membrane proteins (CcmH, CyoA, and GatC) and one membrane-anchored protein (MetQ) were identified, implicating that the observed protective effect of sHSPs on cell membrane (18) may partially result from their chaperone functions on those membrane proteins.
Last but not least, we analyzed the properties of these IbpB substrate proteins in terms of the content of hydrophobic residues, charged residues, and secondary structural elements by referring to the E. coli proteome. The histogram data (data not shown) reveal that IbpB substrate proteins are more concentrated in regions with a high percentage of negatively or positively charged residues and also a high percentage of coil or β-sheet structures. However, they are concentrated in regions of the middle percentage of hydrophobic residues or α-helix structures (data not shown). All these observations indicate that IbpB may selectively bind substrates of certain structural features, the biological significance of which merits further investigations.
Acknowledgments
We thank the Nara Institute of Science and Technology (Ikoma, Nara, Japan) for the BW25113 ΔibpB E. coli strain and Prof. Peter Schultz for the aminoacyl-tRNA synthetase and tRNA expression vectors.
This work was supported by National Natural Science Foundation of China Research Grants 31170738 (to Z. C.) and 31100559 and 31270804 (to X. F.) and National Basic Research Program of China 973 Program Grant 2012CB917300 (to X. F. and Z. C.).

This article contains supplemental Tables S1–S7, Figs. S1 and S2, and additional references.
- sHSP
- small heat shock protein
- Bpa
- p-benzoyl-l-phenylalanine
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 2. de Jong W. W., Leunissen J. A., Voorter C. E. (1993) Evolution of the α-crystallin/small heat-shock protein family. Mol. Biol. Evol. 10, 103–126 [DOI] [PubMed] [Google Scholar]
- 3. Lindquist S., Craig E. A. (1988) The heat-shock proteins. Annu. Rev. Genet. 22, 631–677 [DOI] [PubMed] [Google Scholar]
- 4. Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 5. Laskowska E., Matuszewska E., Kuczyńska-Wiśnik D. (2010) Small heat shock proteins and protein-misfolding diseases. Curr. Pharm. Biotechnol. 11, 146–157 [DOI] [PubMed] [Google Scholar]
- 6. Clark J. I., Muchowski P. J. (2000) Small heat-shock proteins and their potential role in human disease. Curr. Opin. Struct. Biol. 10, 52–59 [DOI] [PubMed] [Google Scholar]
- 7. Morrow G., Battistini S., Zhang P., Tanguay R. M. (2004) Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J. Biol. Chem. 279, 43382–43385 [DOI] [PubMed] [Google Scholar]
- 8. Richards E. H., Hickey E., Weber L., Master J. R. (1996) Effect of overexpression of the small heat shock protein HSP27 on the heat and drug sensitivities of human testis tumor cells. Cancer Res. 56, 2446–2451 [PubMed] [Google Scholar]
- 9. Horwitz J. (1992) α-Crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. U.S.A. 89, 10449–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veinger L., Diamant S., Buchner J., Goloubinoff P. (1998) The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 273, 11032–11037 [DOI] [PubMed] [Google Scholar]
- 11. Mogk A., Deuerling E., Vorderwülbecke S., Vierling E., Bukau B. (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triad in reversing protein aggregation. Mol. Microbiol. 50, 585–595 [DOI] [PubMed] [Google Scholar]
- 12. Lee G. J., Roseman A. M., Saibil H. R., Vierling E. (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cashikar A. G., Duennwald M., Lindquist S. L. (2005) A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 280, 23869–23875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ehrnsperger M., Gräber S., Gaestel M., Buchner J. (1997) Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matuszewska M., Kuczyńska-Wiśnik D., Laskowska E., Liberek K. (2005) The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J. Biol. Chem. 280, 12292–12298 [DOI] [PubMed] [Google Scholar]
- 16. Jakob U., Gaestel M., Engel K., Buchner J. (1993) Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268, 1517–1520 [PubMed] [Google Scholar]
- 17. Welker S., Rudolph B., Frenzel E., Hagn F., Liebisch G., Schmitz G., Scheuring J., Kerth A., Blume A., Weinkauf S., Haslbeck M., Kessler H., Buchner J. (2010) Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 39, 507–520 [DOI] [PubMed] [Google Scholar]
- 18. Török Z., Goloubinoff P., Horváth I., Tsvetkova N. M., Glatz A., Balogh G., Varvasovszki V., Los D. A., Vierling E., Crowe J. H., Vigh L. (2001) Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc. Natl. Acad. Sci. U.S.A. 98, 3098–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamoto H., Vígh L. (2007) The small heat shock proteins and their clients. Cell. Mol. Life Sci. 64, 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basha E., O'Neill H., Vierling E. (2012) Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jaya N., Garcia V., Vierling E. (2009) Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. U.S.A. 106, 15604–15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arata S., Hamaguchi S., Nose K. (1995) Effects of the overexpression of the small heat shock protein, HSP27, on the sensitivity of human fibroblast cells exposed to oxidative stress. J. Cell. Physiol. 163, 458–465 [DOI] [PubMed] [Google Scholar]
- 23. Balogi Z., Cheregi O., Giese K. C., Juhász K., Vierling E., Vass I., Vígh L., Horváth I. (2008) A mutant small heat shock protein with increased thylakoid association provides an elevated resistance against UV-B damage in synechocystis 6803. J. Biol. Chem. 283, 22983–22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salinthone S., Ba M., Hanson L., Martin J. L., Halayko A. J., Gerthoffer W. T. (2007) Overexpression of human Hsp27 inhibits serum-induced proliferation in airway smooth muscle myocytes and confers resistance to hydrogen peroxide cytotoxicity. Am. J. Physiol. Lung Cell Mol Physiol 293, L1194–L1207 [DOI] [PubMed] [Google Scholar]
- 25. Merck K. B., Groenen P. J., Voorter C. E., de Haard-Hoekman W. A., Horwitz J., Bloemendal H., de Jong W. W. (1993) Structural and functional similarities of bovine α-crystallin and mouse small heat-shock protein. A family of chaperones. J. Biol. Chem. 268, 1046–1052 [PubMed] [Google Scholar]
- 26. Narberhaus F. (2002) α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66, 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang Z., Primm T. P., Jakana J., Lee I. H., Serysheva I., Chiu W., Gilbert H. F., Quiocho F. A. (1996) Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J. Biol. Chem. 271, 7218–7223 [PubMed] [Google Scholar]
- 28. Gu L., Abulimiti A., Li W., Chang Z. (2002) Monodisperse Hsp16.3 nonamer exhibits dynamic dissociation and reassociation, with the nonamer dissociation prerequisite for chaperone-like activity. J. Mol. Biol. 319, 517–526 [DOI] [PubMed] [Google Scholar]
- 29. Fu X., Zhang H., Zhang X., Cao Y., Jiao W., Liu C., Song Y., Abulimiti A., Chang Z. (2005) A dual role for the N-terminal region of Mycobacterium tuberculosis Hsp16.3 in self-oligomerization and binding denaturing substrate proteins. J. Biol. Chem. 280, 6337–6348 [DOI] [PubMed] [Google Scholar]
- 30. Jiao W., Qian M., Li P., Zhao L., Chang Z. (2005) The essential role of the flexible termini in the temperature-responsiveness of the oligomeric state and chaperone-like activity for the polydisperse small heat shock protein IbpB from Escherichia coli. J. Mol. Biol. 347, 871–884 [DOI] [PubMed] [Google Scholar]
- 31. Jiao W., Hong W., Li P., Sun S., Ma J., Qian M., Hu M., Chang Z. (2008) The dramatically increased chaperone activity of small heat-shock protein IbpB is retained for an extended period of time after the stress condition is removed. Biochem. J. 410, 63–70 [DOI] [PubMed] [Google Scholar]
- 32. Basha E., Lee G. J., Breci L. A., Hausrath A. C., Buan N. R., Giese K. C., Vierling E. (2004) The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J. Biol. Chem. 279, 7566–7575 [DOI] [PubMed] [Google Scholar]
- 33. Bepperling A., Alte F., Kriehuber T., Braun N., Weinkauf S., Groll M., Haslbeck M., Buchner J. (2012) Alternative bacterial two-component small heat shock protein systems. Proc. Natl. Acad. Sci. U.S.A. 109, 20407–20412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu X., Shi X., Yin L., Liu J., Joo K., Lee J., Chang Z. (2013) Small heat shock protein IbpB acts as a robust chaperone in living cells by hierarchically activating its multitype substrate-binding residues. J. Biol. Chem. 288, 11897–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chin J. W., Martin A. B., King D. S., Wang L., Schultz P. G. (2002) Addition of a photocross-linking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farrell I. S., Toroney R., Hazen J. L., Mehl R. A., Chin J. W. (2005) Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods 2, 377–384 [DOI] [PubMed] [Google Scholar]
- 37. Mohibullah N., Hahn S. (2008) Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22, 2994–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leng F. (2012) Opportunity and challenge: 10 years of proteomics in China. Sci. China Life Sci. 55, 837–839 [DOI] [PubMed] [Google Scholar]
- 39. Hu Y. (2012) Progress in protein structure and function studies in China during 2010–2011. Sci. China Life Sci. 55, 927–930 [DOI] [PubMed] [Google Scholar]
- 40. Zhang M., Lin S., Song X., Liu J., Fu Y., Ge X., Fu X., Chang Z., Chen P. R. (2011) A genetically incorporated cross-linker reveals chaperone cooperation in acid resistance. Nat. Chem. Biol. 7, 671–677 [DOI] [PubMed] [Google Scholar]
- 41. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 42. Tatusov R. L., Koonin E. V., Lipman D. J. (1997) A genomic perspective on protein families. Science 278, 631–637 [DOI] [PubMed] [Google Scholar]
- 43. Kitagawa M., Matsumura Y., Tsuchido T. (2000) Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 184, 165–171 [DOI] [PubMed] [Google Scholar]
- 44. Kuczyńska-Wiśnik D., Kedzierska S., Matuszewska E., Lund P., Taylor A., Lipińska B., Laskowska E. (2002) The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 45. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 46. Rinas U., Hoffmann F., Betiku E., Estapé D., Marten S. (2007) Inclusion body anatomy and functioning of chaperone-mediated in vivo inclusion body disassembly during high-level recombinant protein production in Escherichia coli. J. Biotechnol. 127, 244–257 [DOI] [PubMed] [Google Scholar]
- 47. Ratajczak E., Zietkiewicz S., Liberek K. (2009) Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J. Mol. Biol. 386, 178–189 [DOI] [PubMed] [Google Scholar]
- 48. Matuszewska E., Kwiatkowska J., Ratajczak E., Kuczyńska-Wiśnik D., Laskowska E. (2009) Role of Escherichia coli heat shock proteins IbpA and IbpB in protection of alcohol dehydrogenase AdhE against heat inactivation in the presence of oxygen. Acta Biochim. Pol. 56, 55–61 [PubMed] [Google Scholar]
- 49. Matuszewska E., Kwiatkowska J., Kuczynska-Wisnik D., Laskowska E. (2008) Escherichia coli heat-shock proteins IbpA/B are involved in resistance to oxidative stress induced by copper. Microbiology 154, 1739–1747 [DOI] [PubMed] [Google Scholar]
- 50. Møller I. M., Rogowska-Wrzesinska A., Rao R. S. (2011) Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J. Proteomics 74, 2228–2242 [DOI] [PubMed] [Google Scholar]
- 51. Carver J. A., Lindner R. A., Lyon C., Canet D., Hernandez H., Dobson C. M., Redfield C. (2002) The interaction of the molecular chaperone α-crystallin with unfolding α-lactalbumin: a structural and kinetic spectroscopic study. J. Mol. Biol. 318, 815–827 [DOI] [PubMed] [Google Scholar]
- 52. Carver J. A., Guerreiro N., Nicholls K. A., Truscott R. J. (1995) On the interaction of α-crystallin with unfolded proteins. Biochim. Biophys. Acta 1252, 251–260 [DOI] [PubMed] [Google Scholar]
- 53. Treweek T. M., Lindner R. A., Mariani M., Carver J. A. (2000) The small heat-shock chaperone protein, α-crystallin, does not recognize stable molten globule states of cytosolic proteins. Biochim. Biophys. Acta 1481, 175–188 [DOI] [PubMed] [Google Scholar]
- 54. Farahbakhsh Z. T., Huang Q. L., Ding L. L., Altenbach C., Steinhoff H. J., Horwitz J., Hubbell W. L. (1995) Interaction of α-crystallin with spin-labeled peptides. Biochemistry 34, 509–516 [DOI] [PubMed] [Google Scholar]
- 55. Lindner R. A., Kapur A., Carver J. A. (1997) The interaction of the molecular chaperone, α-crystallin, with molten globule states of bovine α-lactalbumin. J. Biol. Chem. 272, 27722–27729 [DOI] [PubMed] [Google Scholar]
- 56. Das K. P., Petrash J. M., Surewicz W. K. (1996) Conformational properties of substrate proteins bound to a molecular chaperone α-crystallin. J. Biol. Chem. 271, 10449–10452 [DOI] [PubMed] [Google Scholar]
- 57. Waters E. R., Nguyen S. L., Eskandar R., Behan J., Sanders-Reed Z. (2008) The recent evolution of a pseudogene: diversity and divergence of a mitochondria-localized small heat shock protein in Arabidopsis thaliana. Genome 51, 177–186 [DOI] [PubMed] [Google Scholar]
- 58. Waters E. R., Vierling E. (1999) Chloroplast small heat shock proteins: evidence for atypical evolution of an organelle-localized protein. Proc. Natl. Acad. Sci. U.S.A. 96, 14394–14399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morrow G., Tanguay R. M. (2012) Small heat shock protein expression and functions during development. Int. J. Biochem. Cell Biol. 44, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 60. Yeh C. H., Chang P. F., Yeh K. W., Lin W. C., Chen Y. M., Lin C. Y. (1997) Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc. Natl. Acad. Sci. U.S.A. 94, 10967–10972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Muchowski P. J., Clark J. I. (1998) ATP-enhanced molecular chaperone functions of the small heat shock protein human α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 95, 1004–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Soto A., Allona I., Collada C., Guevara M. A., Casado R., Rodriguez-Cerezo E., Aragoncillo C., Gomez L. (1999) Heterologous expression of a plant small heat-shock protein enhances Escherichia coli viability under heat and cold stress. Plant Physiol. 120, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mogk A., Schlieker C., Friedrich K. L., Schönfeld H. J., Vierling E., Bukau B. (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042 [DOI] [PubMed] [Google Scholar]
- 64. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 65. Lycklama A Nijeholt J. A., Driessen A. J. (2012) The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]