Background: The plant hormone jasmonoyl-isoleucine (JA-Ile) undergoes oxidative catabolism mediated by cytochrome P450 enzymes.
Results: Two amidohydrolases catalyze the cleavage of JA-Ile conjugates and generate 12OH-JA during Arabidopsis wound response.
Conclusion: IAR3 and ILL6 define an additional pathway for JA-Ile turnover and establish a biosynthetic route for 12OH-JA.
Significance: New enzymatic steps unravel the complexity in jasmonate metabolism.
Keywords: Arabidopsis, Enzymes, Hydrolases, Phytohormones, Plant Defense, Conjugate, Hormone Catabolism, Jasmonate Metabolism, Wound Response
Abstract
Jasmonates (JAs) are a class of signaling compounds that mediate complex developmental and adaptative responses in plants. JAs derive from jasmonic acid (JA) through various enzymatic modifications, including conjugation to amino acids or oxidation, yielding an array of derivatives. The main hormonal signal, jasmonoyl-l-isoleucine (JA-Ile), has been found recently to undergo catabolic inactivation by cytochrome P450-mediated oxidation. We characterize here two amidohydrolases, IAR3 and ILL6, that define a second pathway for JA-Ile turnover during the wound response in Arabidopsis leaves. Biochemical and genetic evidence indicates that these two enzymes cleave the JA-Ile signal, but act also on the 12OH-JA-Ile conjugate. We also show that unexpectedly, the abundant accumulation of tuberonic acid (12OH-JA) after wounding originates partly through a sequential pathway involving (i) conjugation of JA to Ile, (ii) oxidation of the JA-Ile conjugate, and (iii) cleavage under the action of the amidohydrolases. The coordinated actions of oxidative and hydrolytic branches in the jasmonate pathway highlight novel mechanisms of JA-Ile hormone turnover and redefine the dynamic metabolic grid of jasmonate conversion in the wound response.
Introduction
Recent years have seen significant progress in our understanding of how hormonal pathways orchestrate plant development and their adaptation to environmental cues. This is particularly the case for jasmonates (JAs),3 a term that refers to jasmonic acid (JA) and its derivatives, for which mechanistic insights into the biosynthesis, perception/action, and modification have been reported (1, 2).
JAs regulate important steps in plant development, but their roles are best documented as mediators of inducible responses to herbivorous insects and necrotrophic pathogens (3–5). These aggressors trigger the JA pathway, which in turn up-regulates overlapping yet distinct sets of genes that lead to the accumulation of a specific arsenal of chemical defenses and proteins (6, 7). Jasmonate metabolism is strongly stimulated by mechanical leaf wounding in Arabidopsis (8, 9) and in other plant species, and triggers many responses induced by leaf-eating insects (10).
The processes and enzymes leading to jasmonic acid (JA) biosynthesis are relatively well described (2), but the nature of the initial molecular event triggering the pathway is still elusive. In Arabidopsis, partially redundant lipases release the precursor fatty acid α-linolenic acid from plastidial membranes where it is converted to 12-oxo-phytodienoic acid through the sequential action of 13-lipoxygenases, the cytochrome P450 allene oxide synthase, and allene oxide cyclase. 12-Oxo-phytodienoic acid is then translocated to peroxisomes where it is reduced by 12-oxo-phytodienoic acid reductase 3 before three rounds of β-oxidation of the side chain. JA has been viewed initially as the active jasmonate in plants, due to its abundance and relative stability, and also because JA treatment recapitulates most known jasmonate responses.
JA is subjected to many enzymatic modifications that generate an array of derivatives, including oxidized and/or sulfonated forms or amino acid or sugar conjugates (2, 11). These individual jasmonates vary in abundance, biological activity, and distribution under various stimuli and in different tissues. For example, the JA metabolite 12OH-JA was first described in potato and Solanaceous species for its tuber-inducing activity, hence its trivial name tuberonic acid (TA) (12). More recently, TA and its sulfonated or glucosylated derivatives were found as commonly occurring metabolites of JA, particularly in the wound response (13, 14), but the mode of formation of TA and biological activity of its derivatives are still largely unknown. TA formation has, however, been proposed as a partial switch-off in JA signaling (14). However, TA could also be a metabolic intermediate, for example, TA-glucoside was described as a specific signal triggering leaf closing in the rain tree Samanea saman (15).
Among many existing JA modification routes, the JASMONATE RESISTANT 1 (JAR1)-catalyzed formation of (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile) is the critical signal activation step (16). JAR1 is a member of the GH3 family of acyl amidosynthases, whose other described members conjugate auxin to amino acids (17), indicating possible metabolic cross-talk between auxin and JA pathways. JA-Ile specifically promotes assembly of a co-receptor composed of the F-box protein CORONATINE INSENSITIVE 1 (COI1) and Jasmonate ZIM domain (JAZ) proteins. JAZ proteins constitute a family of transcriptional repressors that under low JA-Ile content prevent the transcription of target genes. JA-Ile-triggered co-receptor assembly results in JAZ ubiquitination by the SCFCOI1 E3 ligase and their subsequent proteolytic removal by the 26S proteasome, leading to the derepression of target genes, and the deployment of the JA response (18, 19).
Plants have also evolved mechanisms to reset JA signaling to pre-stimulation conditions. For example, the strong JAZ repressor gene co-induction with the JA metabolic pathway is believed to rapidly restore a repressed state on JA-responsive promoters (10). Another means to terminate signaling is to modify or eliminate the hormonal signal. We and others have recently described a cytochrome P450 (CYP)-based catabolic pathway that contributes to the turnover of JA-Ile. CYP94B3 and CYP94C1 oxidize JA-Ile at the C12 position, leading to sequential accumulation of the oxidized derivatives 12OH-JA-Ile and 12COOH-JA-Ile (20–22). This oxidative metabolism accounts largely for the transient nature of JA-Ile accumulation after wounding and, similarly to other plant hormones (23), corresponds to the attenuation of the hormonal signal, as plants with perturbed JA-Ile oxidizing activity have an altered JA response (20, 22).
Phytohormone conjugation to amino acids is generally associated with storage or inactivation purposes (24) like in the case of auxin, but the fascinating counter example of JA-Ile activity suggests that de-conjugation could be used to readily inactivate this hormone. Gene families encoding auxin amidohydrolases and their regulation by interacting microorganisms have been described in Brassica rapa (25) and Medicago truncatula (26). In Arabidopsis, they form a 7-gene family (IAR3, ILL1, ILL2, ILL3, ILL5, ILL6, and ILR1) with several members known to cleave auxin-amino acid conjugates (27). IAR3, encoding an indole acetic acid-alanine (IAA-Ala) hydrolase (28), was initially described as a wound- and jasmonate-induced gene named Jasmonate Responsive 3 (JR3) and used as a robust JA pathway marker (29). Recently, Woldemariam et al. (40) reported the characterization of the IAR3-related jasmonoyl-isoleucine hydrolase 1 (JIH1), an herbivore-induced amidohydrolase from the wild species Nicotiana attenuata, which contributes to JA-Ile signal attenuation by cleaving the hormone into JA and Ile. We describe here the characterization of IAR3 and ILL6, two of three amidohydrolase genes that are co-regulated with the JA pathway in Arabidopsis. IAR3 was previously characterized in the context of auxin conjugate hydrolysis (28), but no data were available for ILL6. Starting from a metabolic analysis of several JA pathway mutants, we hypothesized the action of amidohydrolases to explain the particular jasmonate profiles in these lines. Indeed, recombinant IAR3 and ILL6 were found to cleave JA-Ile in vitro, consistent with a role in hormonal turnover upon wounding. Interestingly, the enzymes also seem to act on the hydroxylated JA-Ile conjugate, with important consequences on the homeostasis of oxidized jasmonates. Particularly, we elucidate an unexpected mode of TA formation that, instead of direct hydroxylation of JA, proceeds through the cleavage of JAR1- and CYP94-generated 12OH-JA-Ile.
EXPERIMENTAL PROCEDURES
Plant Growth and Treatment
Arabidopsis thaliana genotypes used were all in the Col0 ecotype and grown under a 12/12-h photoperiod in a growth chamber. T-DNA insertion lines used were: coi1-1, jar1-1, the double mutant between cyp94c1-1 (SALK_55455) and cyp94b3-1 (CS302217) described in Ref. 20, iar3-5 (SALK_069047), iar3-6 (SALK_042101), ill6-1 (GK412E11), and ill6-2 (SALK_024894), all obtained from the Nottingham Arabidopsis Stock Center (NASC). For wounding experiments, between 4 and 6 fully expanded leaves of 6–7-week-old plants were wounded three times across the midvein with a hemostat. At increasing time points following mechanical damage, leaf samples were quickly harvested and flash-frozen in liquid nitrogen before storing at −80 °C until use.
RT-Quantitative PCR (qPCR) Gene Expression Assays
Total RNA was extracted from plant leaves with TRIzol reagent (Molecular Research Center). One μg of RNA was reverse transcribed using the ImProm-II reverse transcription system (Promega, Madison, WI). Real-time PCR was performed on 10 ng of cDNA as described in Ref. 30 using a LightCycler 480 II instrument (Roche). The housekeeping genes EXP (At4g26410) and GAPDH (At1g13440) were used as internal references. GAPDH was replaced by At4g26410 encoding an unknown protein (31) in the experiments with coi1 plants. Gene-specific primer sequences used for qPCR are listed in Table 1.
TABLE 1.
Primers used in this study
| Use | Gene/allele (locus) | Primer name | Sequence (5′ → 3′) |
|---|---|---|---|
| qPCR | CYP94B1 (At5g63450) | CYP94B1 qPCR F62 | caatgaggctttacccaccag |
| CYP94B1 qPCR R62 | aaatgtcgtcgtttgctgcat | ||
| CYP94B3 (At3g48520) | CYP94B3 qPCR F62 | tggcttacacgaaggcttgtc | |
| CYP94B3 qPCR R62 | agtcccacgaaactggaggat | ||
| CYP94C1 (At2g27690) | CYP94C1 qPCR F63 | ggcccggattacgaagagttt | |
| CYP94C1 qPCR R63 | ggccggaacttaccttcgtt | ||
| IAR3 (At1g51760) | IAR3 qPCR-F | gggctgatatggatgcacttg | |
| IAR3 qPCR-R | tgcatcttccctggaacctt | ||
| IAR3-69047-qFa | ttgcttcttgtacccctgct | ||
| IAR3-69047-qRa | tcggataagcaagtgaagga | ||
| ILL1 (At5g56650) | ILL1 qPCR-F | ttcaacaacatcgccaacatt | |
| ILL1 qPCR-R | caacgttcttcaacgctcct | ||
| ILL2 (At5g56660) | ILL2 qPCR-F | atgaacatcgccaccatttg | |
| ILL2 qPCR-R | ctcatcttctttgctccactcaa | ||
| ILL3 (At5g54140) | ILL3 qPCR-F | tgttgctctccgtgctgata | |
| ILL3 qPCR-R | tttgctcttatgatcccactcaa | ||
| ILL5 (At1g51780) | ILL5 qPCR-F | gtttcacgagaggcagatcct | |
| ILL5 qPCR-R | tcgggaatgacattaaaagca | ||
| ILL6 (At1g44350) | ILL6 qPCR-F | ggctgatatggacgcactacc | |
| ILL6 qPCR-F | tgcattttgcctgcaacttt | ||
| ILL6-GK412-Fa | gtgtcccatatccatccaacgg | ||
| ILL6-GK412-Ra | ggtccgacatacccatgatcc | ||
| ILL6–024894-Fa | cttggtgctgcccatattct | ||
| ILL6–024894-Ra | Tcatccaaagctccgtcttc | ||
| ILR1 (At3g02875) | ILR1 qPCR-F | Actggatccactcgaagctg | |
| ILR1 qPCR-R | Tgcggtattacgttttgagca | ||
| ST2a (At5g07010) | ST2a qPCR-F | Ggcttgcaactttcttagagctt | |
| ST2a qPCR-R | Cggcgatagccttcacaac | ||
| EXP (At4g26410) | EXP-qPCR-F | Gagctgaagtggcttcaatgac | |
| EXP-qPCR-R | Ggtccgacatacccatgatcc | ||
| GAPDH (At1g13440) | GAPDH-qPCR-F | Ttggtgacaacaggtcaagca | |
| GAPDH-qPCR-R | Aaacttgtcgctcaatgcaatc | ||
| At4g26410 | At4g26410-qPCR-F | Gagctgaagtggcttccatgac | |
| At4g26410-qPCR-R | Ggtccgacatacccatgatcc | ||
| IAR3 and ILL6 cloning in pENTR1a for recombinant protein expression | |||
| IAR3 | SalI-IAR3ΔN-F | Ctggtcgactcctctaatgggttatctcaaatac | |
| EcoRI-IAR3ΔN-R | Tgtgaattcaaagttcatcttttttgttactct | ||
| ILL6 | SalI-ILL6ΔN-F | Gtggtcgacaccaacttacctttctttgaagtg | |
| EcoRI-ILL6ΔN-R | Caagaattcttatgaatgtttatcatttaagtatctc | ||
| T-DNA genotyping | |||
| T-DNA | LBb1.3 (SALK) | Attttgccgatttcgaac | |
| o8409 (GABI-Kat) | Atattgaccatcatactcattgc | ||
| iar3-5 | SALK_069047-LP | Gttctccacgtgcgttatagc | |
| SALK_069047-RP | Aaaaagccacactgttccatg | ||
| iar3-6 | SALK_042101-LP | Gttctccacgtgcgttatagc | |
| SALK_042101-RP | Acaccagtaacagcaactggg | ||
| ill6-1 | GK412E11-LP | Gactatgcttcttggtgctgc | |
| GK412E11-RP | Cgcacctcttgaatacgtttc | ||
| ill6-2 | SALK_024894-LP | Gactatgcttcttggtgctgc | |
| SALK_024894-RP | Cgcacctcttgaatacgtttc |
a Primers used for iar3 and ill6 T-DNA lines.
Chemical Synthesis of Oxidized Jasmonates
JA-Ile was obtained according to the reported method (32). 12-OH-JA-Me-Ile was obtained as described in Refs. 21 and 33. The hydroxyl group and carboxyl function of 12OH-JA-Me-Ile were successively protected to provide 12-OAc-dioxaspiro-JA-Me-Ile. After saponification of the latter, the primary alcohol was oxidized using tetrapropylammonium perruthenate-N-methylmorpholine-N-oxide (34). Deprotection of the dioxolane function afforded 12COOH-JA-Me-Ile. Saponification of the carbomethoxy group was achieved using Me3SnOH (35) to yield the desired 12-COOH-JA-Ile. The complete procedures, the characterization of intermediates, and final product are given in supplemental Methods S1.
Recombinant IAR3 and ILL6 Production and Enzymatic Hydrolase Assay
For heterologous expression of IAR3 and ILL6, open reading frame sequences deleted of the 25 N-terminal signal peptide-encoding codons were amplified using Phusion Taq Polymerase (Thermo Scientific) prior to cloning in the pENTR1a (Invitrogen) plasmid in the DH5α Escherichia coli strain. Inserts with error-free sequences were recombined into the expression vector pHMGWA using GatewayTM technology (Invitrogen). Plasmids were further transformed into E. coli SoluBL21 strain (AMS Biotechnology, Abingdon, UK). Ice-cold bacterial pellets from isopropyl 1-thio-β-d-galactopyranoside-induced cultures were collected and resuspended in lysis buffer (50 mm Tris-HCl, pH 8, 300 mm NaCl, 5% glycerol, 1 mg/ml of lysozyme, 1 mm PMSF, 1 mm EDTA, 0,1% Tween 20) at an A600 of 20. Bacteria were lysed by sonication on ice for 2 min. Clarified protein lysates were filtered and loaded on a 1-ml HisTrap HP column (GE Healthcare Bio-Sciences) equilibrated in 50 mm Tris-HCl, pH 8, 300 mm NaCl, 5% glycerol installed on an Äkta Purifier10 Liquid chromatography system. The column was washed with equilibration buffer and bound protein was eluted with equilibration buffer complemented with 500 mm imidazole. Eluate was desalted on PD10 columns and the protein concentration was determined by polyacrylamide gel electrophoresis using a bovine serum albumin standard series.
Incubation conditions were based on those described for IAR3 activity with IAA-Ala (Sigma) (36). Incubations were performed in 200 μl containing 50 mm Tris-HCl, pH 8, 1 mm MnCl2, 1 mm dithiothreitol, and 50 μm JA-Ile or 12OH-JA-Ile or 12COOH-JA-Ile synthesized as described in the previous section as substrate. Control reactions were run with a similar amount of purified maltose-binding protein. Reactions were incubated at 30 °C and stopped by addition of 40 μl of 1 m HCl before extraction with 240 μl of ethyl acetate. After brief vortexing and centrifugation, the upper organic phase was dried under N2 flow, and reconstituted with 150 μl of MeOH before UPLC-MS analysis as described below.
Jasmonate Profiling
Jasmonates were identified and quantified in plant extracts and enzymatic incubations by ultra performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS), using MS transitions determined from pure standards (JA, 12OH-JA, JA-Ile, 12OH-JA-Ile, or 12COOH-JA-Ile). The relative quantification in samples was achieved by reporting MS peak areas relative to internal standard peak area and mass of biological material. Absolute quantifications of JA, 12OH-JA, JA-Ile, 12OH-JA-Ile, or 12COOH-JA-Ile were determined by comparison of a sample signal with dose-response curves established for each pure compound. For plant extraction, 5 volumes of ice-cold 90% methanol containing 9,10-dihydro-JA and 9,10-dihydro-JA-Ile as internal standards were added to one fresh weight (100–150 mg pre-weighed) of frozen leaf powder in a screw-capped tube containing glass beads. Material was ground twice for 30 s with a Precellys 24 tissue homogenizer (Bertin Technologies, Montigny-Le-Bretonneux, France). Homogenates were cleared by two successive centrifugations at 20,000 × g and supernatants were saved for UPLC-MS analysis. The conditions for analyzing jasmonates in enzymatic incubations or in plant extracts were described in Ref. 20. The multiple reaction monitoring transitions used for detection were (in negative mode): JA 209 > 59; 12OH-JA 225 > 59; 12OH-JA-Glc 387 > 89; 12-HSO4-JA 305 > 97; 12OH-JA-Ile 338 > 130; 12COOH-JA-Ile 352 > 130; IAA-Ala 247 > 130 and in positive mode: JA-Ile 324 > 151, IAA 176 > 130.
Statistical Analysis
All statistical analysis were established by two-way ANOVA with Bonferroni post test using GraphPad Prism version 5.01.
RESULTS
Tuberonic Acid Abundance Is Not Correlated with JA Levels but with 12OH-JA-Ile Levels in JAR1- or CYP94B3/C1-deficient Plants Upon Wounding
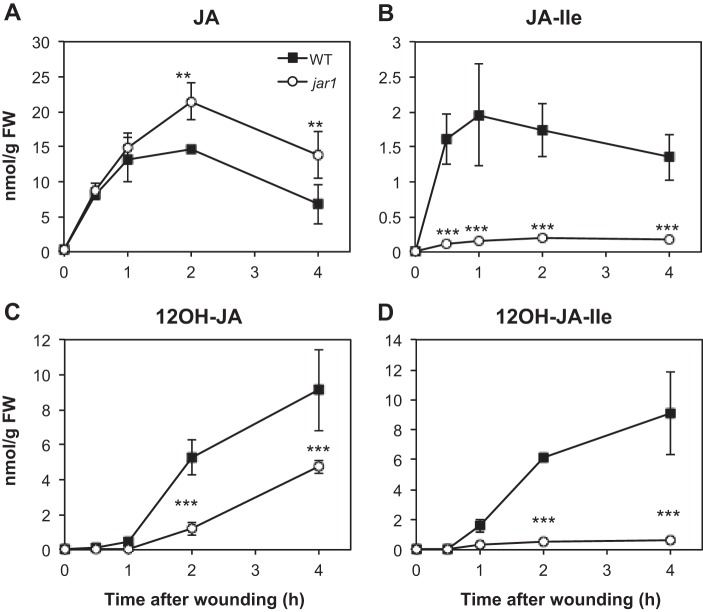
12OH-JA (tuberonic acid, TA) as well as its glucoside (12-O-Glc-JA, TA-Glc) and its sulfated derivative 12-HSO4-JA have been identified as common JA metabolites in several plant species (14). TA was further shown to accumulate in N. attenuata (37), tomato (14), and Arabidopsis (8) after leaf wounding. A kinetic analysis shows that TA is nearly undetectable in unstressed leaves and maximal levels are reached by 4 h after injury (Fig. 1C) in wild-type (WT) plants, which is later than those of JA that peak generally between 1 and 2 h (Fig. 1A). Despite the lack of data about TA biosynthesis, it is commonly thought that TA formation occurs through JA hydroxylation. In the course of the characterization of the impact of JA conjugation on the accumulation of oxidized JA-Ile derivatives (20), we analyzed the jasmonate profiles in jar1, a mutant impaired in JA-Ile formation. Levels of JA were increased (Fig. 1A) and those of JA-Ile depressed (Fig. 1B) in wounded jar1 compared with WT leaves, as expected for a block in JA-conjugating activity. Unexpectedly, despite JA overaccumulation, TA levels were reduced compared with WT (Fig. 1C). TA levels were severely reduced in jar1 at 2 h post-wounding and did not exceed 50% WT levels at 4 h, suggesting that excess JA in jar1 does not seem to function as a substrate for direct hydroxylation to form TA. Reduced TA levels were correlated with the strongly impaired accumulation of JA-Ile (Fig. 1B) and 12OH-JA-Ile (Fig. 1D). These observations suggest that the active hormone JA-Ile and/or the hydroxylated compound 12OH-JA-Ile are necessary for TA formation.
FIGURE 1.
Kinetic analysis of jasmonate accumulation in wild-type (WT) and in jar1 mutant plants upon leaf wounding. Leaves were harvested at increasing times after wounding and extracted for jasmonate determination by UPLC-MS/MS. A, JA; B, JA-Ile; C, 12OH-JA; D, 12OH-JA-Ile. Data are mean ± S.E. from three biological samples. Asterisks indicate significant differences between mutant and WT at p < 0.01 (**) or p < 0.001 (***) (two-way ANOVA). Similar results were obtained in two independent experiments.
To determine which of these compounds is required for full TA accumulation, we examined the levels of TA in the cyp94b3 and cyp94b3c1 oxidation mutants in which JA-Ile is durably overaccumulated (20) and 12OH-JA-Ile is strongly reduced compared with WT (Fig. 2C). We observed in both mutants a significantly reduced build-up of TA (Fig. 2B), demonstrating that prolonged JA-Ile accumulation is not sufficient to achieve high TA levels when 12OH-JA-Ile levels are decreased, and arguing that 12OH-JA-Ile is a precursor for TA formation. This latter hypothesis is also supported by 12OH-JA-Ile levels in cyp94b3c1, exceeding levels in cyp94b3 because of the impaired conversion to 12COOH-JA-Ile in the double mutant, which are correlated with commensurate effects on TA levels. Our previous in vitro data indicated that neither CYP94B3 nor CYP94C1 exhibited detectable JA-hydroxylase activity (20), therefore the reduced accumulation of TA in cyp94b3 and cyp94b3c1 mutants is unlikely due to a lack of CYP94-mediated JA hydroxylase activity. Similarly to jar1, JA is overaccumulated in cyp94b3c1 (Fig. 2A), yet TA levels are lower than in WT (Fig. 2B).
FIGURE 2.
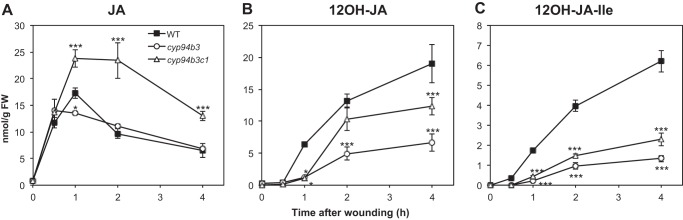
Kinetic analysis of hydroxylated jasmonate accumulation in wild-type (WT), cyp94b3, and cyp94b3c1 mutant plants upon leaf wounding. Leaves were harvested at increasing times after wounding and extracted for jasmonate determination by UPLC-MS/MS. A, JA; B, 12OH-JA; C, 12OH-JA-Ile. Data are mean ± S.E. from three biological samples. Asterisks indicate significant differences between mutant and WT at p < 0.05 (*) or p < 0.001 (***) (two-way ANOVA).
Together, these metabolic studies in different genetic backgrounds indicate that a large part of TA accumulation is dependent on JAR1 conjugation and on CYP94 conjugate oxidation activities, pointing to the hypothesis that 12OH-JA-Ile may be an important precursor for TA formation. This indirect evidence prompted us to investigate the missing step leading to the formation of TA, and namely to explore the possibility that it may proceed through cleavage of the 12OH-JA-Ile conjugate.
Several Amidohydrolase Genes Are Wound and COI1 Regulated in Arabidopsis
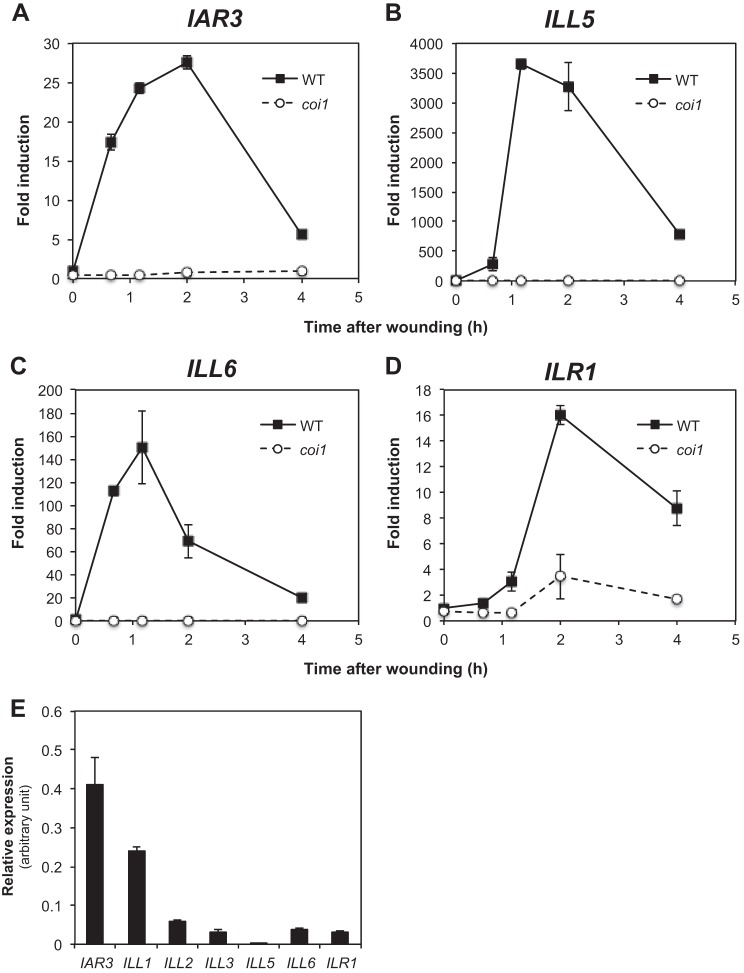
Woldemariam et al. (40) have recently described the herbivory-induced JIH amidohydrolase that cleaves JA-Ile to JA in N. attenuata. JIH defines an alternative JA-Ile hormone inactivation pathway that contributes to the attenuation of the JA-Ile burst. Arabidopsis possesses a family of 7 related amidohydrolases (36), with several members described as cleaving auxin conjugates (27, 28, 38). Recent large scale transcriptional analysis of public microarray data using co-regulation tools (39) revealed a strong coregulation of IAR3, ILL5, and ILL6 with the jasmonate metabolic and signaling pathways (data not shown), which includes the two CYP94 genes acting in JA-Ile catabolism (20–22). Experimental examination of the 7 gene expression profiles confirmed a rapid and transient wound induction of IAR3, ILL5, and ILL6 transcripts (Fig. 3, A–C). Kinetics were typical of early responsive genes, peaking by 1–2 h, and largely paralleled those of CYP94B1, CYP94B3, and CYP94C1 (20). The analysis also revealed the wound-responsive expression of ILR1, with a slight delay (Fig. 3D). Up-regulation of IAR3, ILL5, and ILL6 was essentially lost in the JA-insensitive mutant coi1 (Fig. 3, A–C), and strongly attenuated for ILR1. This suggests a functional link between these hydrolases and the JA pathway, and extends the previous report of COI1-dependent induction of IAR3 (29). Moderate (ILL1) or weak (ILL2 and ILL3) constitutive expression was evidenced for other genes, with ILL1 and ILL2 being down-regulated, and ILL3 unaffected by leaf wounding (data not shown). Among inducible genes, IAR3 had the highest basal expression level in leaves and ILL5 had the lowest (Fig. 3E), explaining the strong difference in induction factors between these genes in WT plants (Fig. 3). Furthermore, when induction factors are multiplied by basal expression, maximal expression levels appear about 20-fold lower for ILR1 than for IAR3, ILL5, or ILL6. These results prompted us to focus on the impact of the three wound-responsive, highly expressed amidohydrolase genes IAR3, ILL5, and ILL6 on jasmonate metabolism.
FIGURE 3.
Amidohydrolase gene expression in wounded WT and coi1 plants. Leaves were harvested at increasing times after wounding and submitted to RNA extraction. One μg of total RNA was reverse transcribed and expression of IAR3 (A), ILL5 (B), ILL6 (C), and ILR1 (D) was determined by real-time PCR using two reference genes. Expression is represented as fold-induction relative to the level at time 0 in WT that was set to 1 for each gene. E, relative expression of the 7 gene members of the Arabidopsis ILR1 family in unstressed leaves. Triplicate determinations ± S.E. are shown.
Amidohydrolase Activity of Recombinant IAR3 and ILL6 on JA-Ile and Oxidized Derivatives
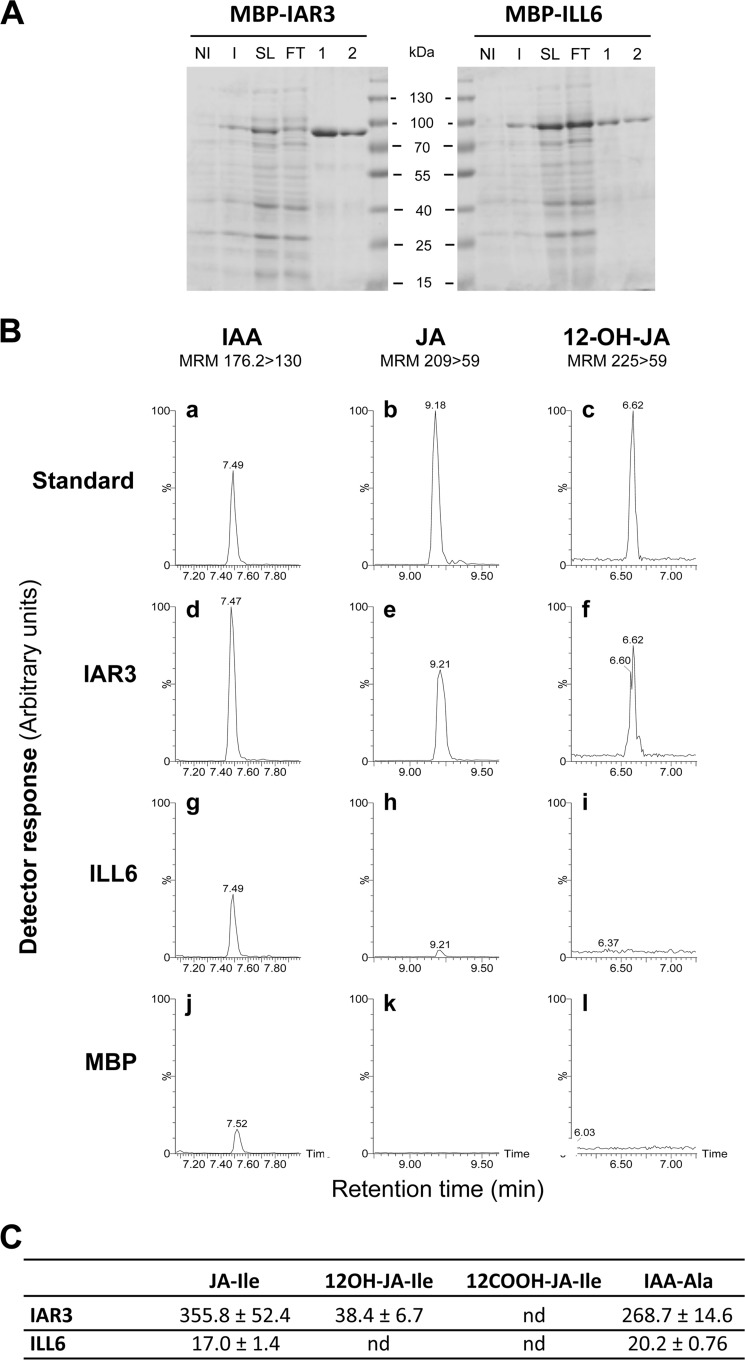
Recombinant IAR3, ILL1, ILR1, and ILL2 were described as cleaving several auxin-amino acid conjugates, with ILL2 displaying very high activity on IAA-alanine (28, 36). Accordingly, roots from several amidohydrolase mutants displayed altered growth responses to exogenously supplied conjugates (27, 36). In contrast, no conditions were found in which glutathione S-transferase-ILL6 fusions hydrolyze IAA-amino acids (36). Recently, the IAR3-related JIH protein, whose gene is wound- and jasmonate-induced in N. attenuata, was shown to cleave similarly JA-Ile and IAA-Ala in vitro (40). We expressed recombinant IAR3 and ILL6 devoid of their N-terminal signal peptides in E. coli as maltose-binding protein fusions before affinity purification through their His tag (Fig. 4A). ILL5 protein was not further studied here as its cloned cDNA was repeatedly found to bear a 7-nucleotide deletion introducing a frameshift and a premature stop codon compared with the TAIR gene model, comforting an earlier suspicion that ILL5 may be a pseudogene (28). Purified IAR3 and ILL6 were incubated in parallel with synthetic JA-Ile, 12OH-JA-Ile, 12COOH-JA-Ile, and IAA-Ala, and corresponding unconjugated compounds were searched for as a measurement of cleaving activity. Fig. 4B shows typical UPLC-MS/MS chromatograms obtained with either fusion protein or maltose-binding protein as a control. Both enzymes were found to release JA from JA-Ile (Fig. 4B, e and h) and IAA from IAA-Ala (Fig. 4B, d and g), recombinant ILL6 appearing less active than IAR3 on all compounds tested. When incubated with 12OH-JA-Ile, TA signal was detected only with IAR3 (Fig. 4B, f), whereas after incubation with 12COOH-JA-Ile, no cleavage activity was evidenced with either enzyme (data not shown). The different activities were evaluated quantitatively after establishing standard curves with pure compounds. Under the conditions used IAR3 was about 20-fold more active on JA-Ile than ILL6, and IAR3 was itself 9-fold more active on JA-Ile than on 12OH-JA-Ile (Fig. 4C). These results from in vitro experiments show that IAR3 and ILL6 release JA from JA-Ile and that at least IAR3 can also generate TA by cleaving 12OH-JA-Ile.
FIGURE 4.
Purification and enzymatic activity of recombinant IAR3 and ILL6 amidohydrolases. A, purification steps were analyzed by 12.5% SDS-PAGE. Total proteins from bacteria transformed with plasmids expressing fusion proteins were analyzed before (non-induced, NI) and after (induced, I) induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. Fusion proteins are visible after induction. His-tagged proteins present in soluble lysate (SL) were purified by affinity chromatography. FT, flow through. 1 and 2, elution fractions 1 and 2. Molecular mass markers are indicated. Proteins were visualized by Coomassie staining. B, 20 μg of either protein were incubated with the following amino acid conjugates as candidate substrates: IAA-Ala, JA-Ile, and 12OH-JA-Ile. LC chromatograms are shown where the cleavage products IAA (panels a, d, g, and j), JA (panels b, e, h, and k), or 12OH-JA (panels c, f, i, and l) were separated by UPLC-MS/MS and detected by their indicated multiple reaction monitoring transitions. C, specific activities (pmol/h/mg of protein) were determined by incubation of purified enzymes for 90 min with 100 μm substrates and quantification of cleavage products with authentic standards curves. Values represent mean ± S.E. (n = 3). nd, not detected.
Amidohydrolase Mutants Display Elevated Oxidized JA-Ile Conjugates and Reduced TA Levels
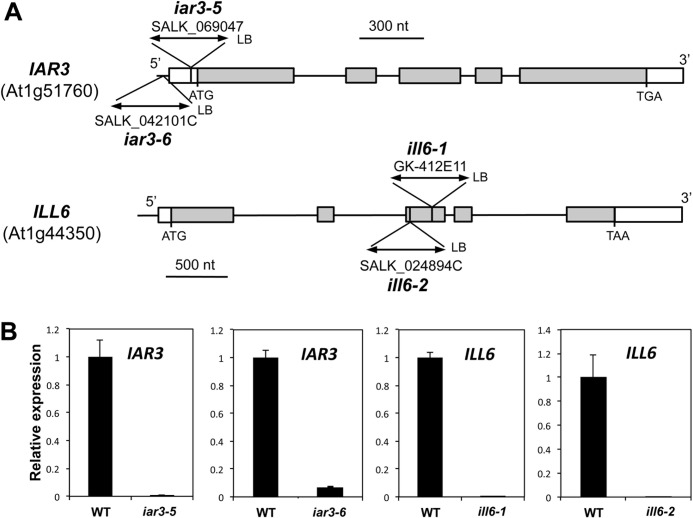
To obtain direct genetic evidence for amidohydrolase involvement in jasmonate metabolism during the wound response, we used T-DNA insertion mutant lines that are impaired in the expression of IAR3 and ILL6. The EMS alleles iar3-1 through iar3-4 were previously described (28), therefore we named the T-DNA alleles used here iar3-5 and iar3-6 (Fig. 5A). No ill6 alleles were previously reported, and we introduce in this study ill6-1 and ill6-2. All alleles were found to have undetectable transcript levels, except iar3-6, which had an insertion in the promoter region and appeared as a strong knock-down allele (Fig. 5B). The wound-induced content in five JAs of iar3-5, ill6-1, and iar3-6, ill6-2 plants was determined relative to WT in two independent series of kinetic experiments. As shown in Fig. 6A, JA-Ile was found similar to WT levels, except in iar3-5 that showed increased levels at early time points, a result that is consistent with the respective in vitro JA-Ile cleaving activities found for IAR3 and ILL6. Mutations had little impact on JA levels that fluctuated near WT levels in the different alleles (Fig. 6B). We next examined the profiles of oxidized jasmonates. 12OH-JA-Ile was significantly hyperaccumulated in ill6 and in iar3 mutants (Fig. 6C), and this increase was accompanied by a corresponding reduction in 12OH-JA (TA) levels in all examined alleles (Fig. 6D). By 4 h post-wounding, amidohydrolase mutants accumulated about 50–60% WT TA levels. Finally, we quantified 12COOH-JA-Ile levels in planta, the second oxidation product, generated mainly by CYP94C1 (20). This required its prior chemical synthesis that is extensively described under supplemental Methods S1 as an original method. We determined that 12COOH-JA-Ile accumulated abundantly to about 10–25 nmol/g of fresh weight by 4 h post-wounding (Fig. 6E), making it a highly abundant jasmonate in wounded Arabidopsis leaves. Its levels were further enhanced in amidohydrolase mutants relative to WT, particularly in iar3-5 and ill6-1. The overall differences in levels of metabolites, especially of Ile conjugates, in the two independent experiments (Fig. 6, left versus right panels) may illustrate adaptation of jasmonate dynamics to different growth periods or slight changes in experimental setup. The milder chemotypes recorded for all compounds in iar3-6 are consistent with this line being a weaker allele than iar3-5 (Fig. 5B). The simplest interpretation of these genetic data are that similarly to JIH in N. attenuata (40), IAR3 and ILL6 readily hydrolyze JA-Ile in vivo and the two Arabidopsis enzymes likely also act on 12OH-JA-Ile. Consequently, they establish that a significant part of the TA accumulated in response to wounding is generated via the cleavage of 12OH-JA-Ile.
FIGURE 5.
Molecular characterization of iar3 and ill6 insertion alleles. A, positions of T-DNA insertions in the different mutant lines are shown, and for each allele the position of the left border (LB) is indicated. Boxes denote exons and thin vertical lines denote introns or promoter regions. White boxes depict untranslated regions and gray boxes coding regions. B, RT-qPCR analysis of gene expression in WT and mutant leaves. Leaves were harvested 1 h after wounding and abundance of transcript regions spanning the insertion site was monitored. Expression in mutant lines was calculated relative to WT levels set to 1. Expression was normalized using two reference genes.
FIGURE 6.
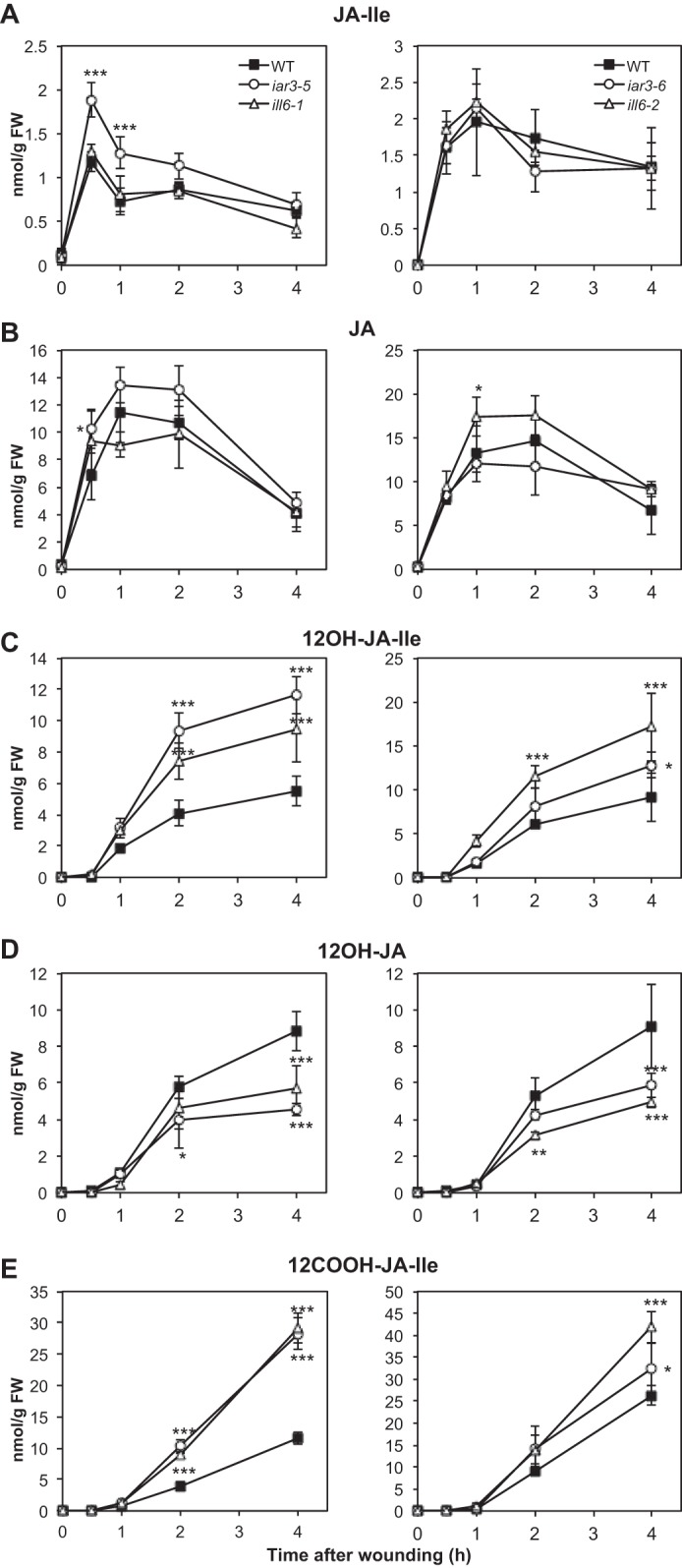
Kinetic analysis of jasmonate accumulation in wild-type (WT) and in iar3 and ill6 mutant plants upon leaf wounding. iar3-5 with ill6-1 and iar3-6 with ill6-2 lines were analyzed along with WT plants in independent experiments. Leaves were harvested at increasing times after wounding and extracted for jasmonate determination by UPLC-MS/MS. A, JA-Ile; B, JA; C, 12OH-JA-Ile; D, 12OH-JA; E, 12COOH-JA-Ile. Data are mean ± S.E. from three biological samples. Asterisks indicate significant differences between mutant and WT at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***) (two-way ANOVA).
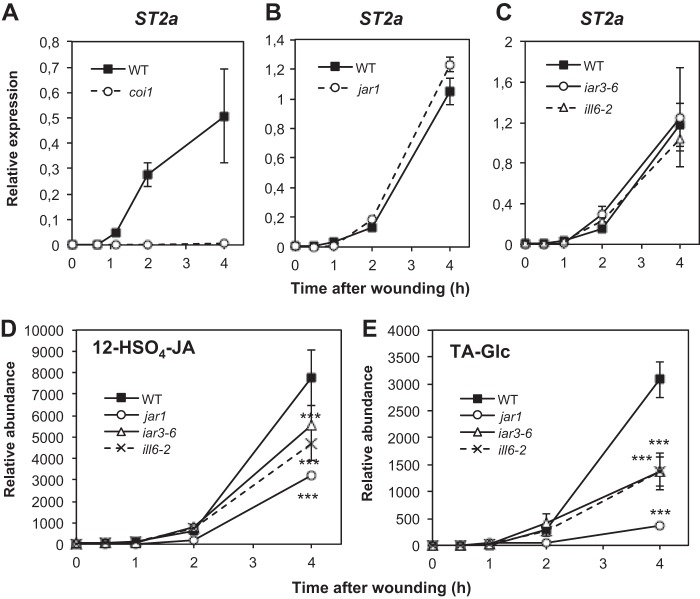
TA, one of the products of amidohydrolase activity, is also known to undergo further metabolism via two competing routes (2), one leading to the TA glucoside (TA-Glc), the other yielding the sulfonated derivative 12-HSO4-JA (13, 14). We therefore investigated if differential TA conversion could result in variable TA levels in the different mutant backgrounds. Transcripts of ST2a, the gene encoding the TA-sulfotransferase (13), were strongly up-regulated in wounded leaves in a coi1-dependent manner (Fig. 7A), whereas this induction was similar to WT in jar1 (Fig. 7B) or in iar3-6 or ill6-2 mutants (Fig. 7C). Therefore, consumption of TA substrate by ST2a is not predicted to be affected differentially in these latter lines. Direct measurement of 12-HSO4-JA established the rather late wound accumulation of this compound, and its relative abundance was reduced by about 60% in jar1 and 40% in amidohydrolase mutants by 4 h (Fig. 7D). No TA-specific glucosyltransferase is characterized in Arabidopsis, but such an activity has been described in rice (41). We found that TA-Glc accumulation was strongly impaired in jar1 and significantly reduced in iar3-6 or ill6-2 mutants (Fig. 7E). Collectively, these results suggest that the two known TA conversion pathways are not enhanced in iar3-6 or ill6-2 and that reduced TA accumulation is mostly due to impaired hydrolytic activity. In addition, they provide evidence that 12-HSO4-JA and TA-Glc partly derive from TA generated through the deconjugation pathway uncovered in this work.
FIGURE 7.
Analysis of tuberonic acid metabolization pathways in jar1, coi1, and amidohydrolase mutants upon wounding. Leaves were harvested at increasing times after wounding and submitted to RNA extraction. Expression of ST2a was examined by real-time PCR in coi1 (A), jar1 (B), and iar3-6 and ill6-2 (C) mutants relative to WT and represented as ΔΔCt values. Two reference genes were used for normalization. The TA derivatives 12-HSO4-JA (D) and tuberonic acid glucoside (TA-Glc, E) levels were determined in WT, jar1, iar3-6, and ill6-2 plants. Triplicate determinations ± S.E. are shown for all analyses. Asterisks indicate significant differences between mutant and WT at p < 0.001 (two-way ANOVA).
JA-Ile Turnover Is Tightly Controlled by Compensating Oxidative and Hydrolytic Catabolic Pathways
The observation that JA-Ile levels are only moderately or not altered by mutations in amidohydrolase genes (Fig. 6A), is in contrast to the hyperaccumulation of its two oxidized derivatives (Fig. 6, C and E) and may be indicative of enhanced hormone turnover in these lines. To examine this possibility, we monitored by RT-qPCR the wound-induced expression of genes encoding known JA-Ile-metabolizing enzymes, either from the CYP94 family or the respective non-mutated amidohydrolase gene. Fig. 8A shows that in either ill6 allele, IAR3 transcript levels significantly exceeded those in WT at 1 h post-wounding. In contrast, ILL6 expression was only marginally affected by IAR3 deficiency (Fig. 8B). In addition, CYP94B3 transcripts, encoding the primary enzyme for oxidative JA-Ile inactivation (20–22) were found hyperaccumulated in both ill6 alleles, but not in iar3 alleles (Fig. 8D). CYP94B1, that also catalyzes JA-Ile hydroxylation in vitro,4 showed more variable transcript levels between mutants (Fig. 8C). CYP94C1 catalyzes the second JA-Ile oxidation step (20), and was hyperinduced in all four alleles (Fig. 8E), consistent with a pronounced 12COOH-JA-Ile overaccumulation in these mutants (Fig. 6E). This expression survey points to the existence of compensation mechanisms (mainly when ILL6 is impaired) where the reduced JA-Ile-cleaving activity results in a stronger or earlier up-regulation of the other genes encoding JA-Ile-consuming enzymes to prevent an excessive accumulation of the hormone. Conversely, the weaker reciprocal compensation in iar3-5 correlated with higher impacts of IAR3 deficiency on JA-Ile levels (Fig. 6A). We finally checked if cyp94 mutants exhibited a similar compensation by the amidohydrolase pathway. The expression patterns of IAR3 and ILL6 in cyp94b3 and cyp94b3c1 were similar to WT (data not shown), in good agreement with the highly persistent JA-Ile accumulation that was described previously in these lines (20, 22).
FIGURE 8.
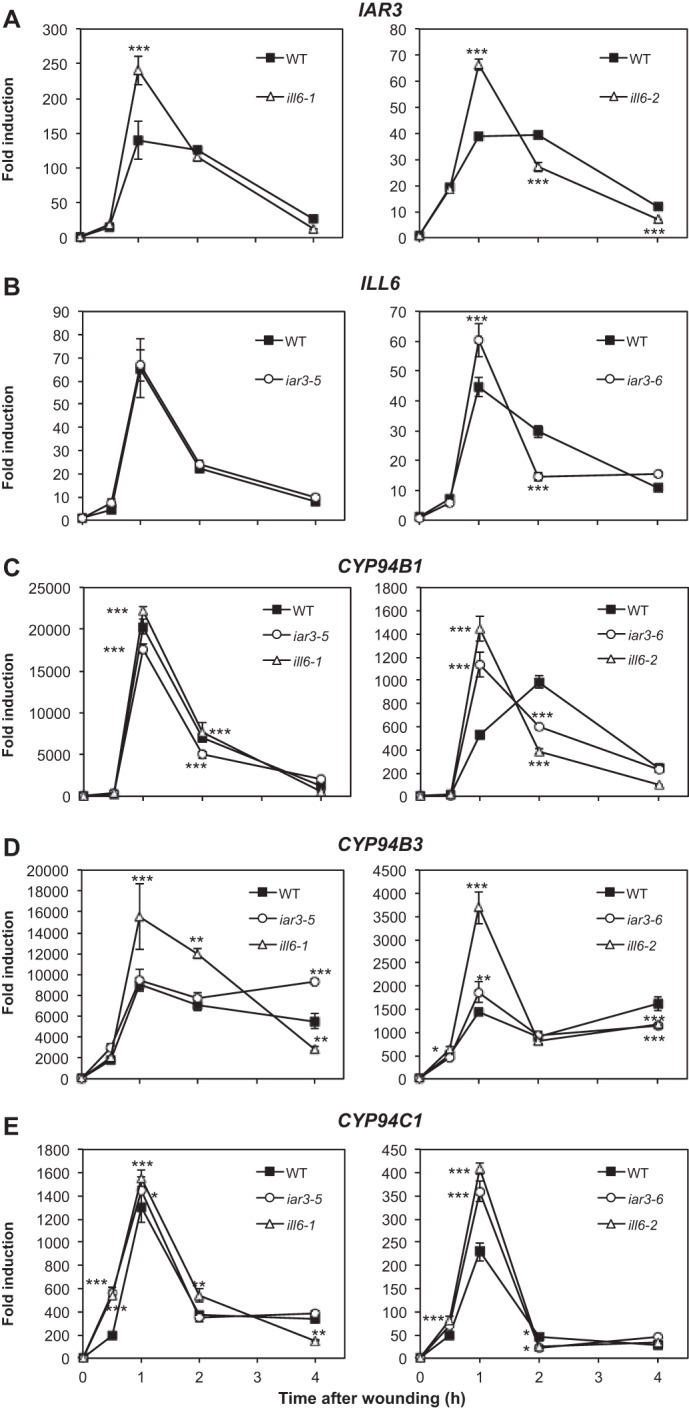
Impact of amidohydrolase mutations on expression of genes encoding JA-Ile metabolizing enzymes. Total RNA was extracted from wounded plants analyzed kinetically for jasmonate content in Fig. 6 and reverse-transcribed. Gene expression time courses were established for the following genes by real-time PCR: A, IAR3; B, ILL6; C, CYP94B1; D, CYP94B3; and E, CYP94C1. Expression is represented as fold-induction relative to level at time 0 in WT plants, which was set to 1 for each gene. Differences between the left and right panel fold-inductions are due to differences in basal expression levels between independent batches of plants. Two reference genes were used for normalization. Triplicate determinations ± S.E. are shown. Asterisks indicate significant differences between mutant and WT at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***) (two-way ANOVA).
DISCUSSION
Plant hormone inactivation and turnover generally proceeds through two major pathways. The first route leads to sugar or amino acid conjugates and has been mostly studied for auxin (42), but was also reported for other hormones. The second pathway operates through oxidations typically catalyzed by CYP enzymes belonging to distinct subfamilies (23). Amide conjugates are synthesized by the so-called GH3 family and in the case of auxin their formation is believed to ensure removal or storage of excessive free active hormone (43). This conjugation is reversible as several members of the ILR1-like family of amidohydrolases release free auxin from such compounds (36).
The case of jasmonate is a remarkable exception to this scheme as conjugation of JA to Ile by JAR1 (GH3.11) is the critical hormone activation step. We and others have characterized recently a CYP94-based JA-Ile catabolic pathway that generates hydroxylated and carboxylated JA-Ile derivatives and contributes largely to the rapid hormone clearance after leaf wounding (20–22). Unconjugated jasmonates also occur as oxidized derivatives, namely tuberonic acid (2, 13, 14, 41). Despite its abundance in several plant species, the question of the mode of TA formation has remained unsolved, as no enzyme for direct hydroxylation of JA could be characterized. We have provided in this work biochemical and genetic evidence that JA-Ile catabolism and TA formation are linked through the action of two amidohydrolases. Although the family of ILR1-like (ILL) amidohydrolases was discovered in the context of auxin metabolism (42, 44), a closer examination revealed that IAR3, ILL5, and ILL6 genes are strongly co-regulated with the JA metabolic and signaling pathway. Consistently, we demonstrated their rapid and COI1-dependent up-regulation upon wounding, with kinetics similar to the oxidative branch of JA-Ile catabolism represented by the CYP94B3 and CYP94C1 genes (20). ILR1, despite a lower expression level, also displays COI-dependent wound induction, and must be analyzed in the context of jasmonate metabolism. Based on their transcriptional behavior and abundant expression, IAR3 and ILL6 were the prime candidates for a role in jasmonate metabolism, whereas ILL5 is a non-coding sequence, and ILL1, ILL2, and ILL3 transcripts do not correlate with jasmonate conjugate abundance. Similarly to the recently described related JIH enzyme in N. attenuata (40), recombinant IAR3 and ILL6 hydrolyzed almost equally JA-Ile and IAA-Ala, and IAR3 was also found to act on the oxidized derivative 12OH-JA-Ile. IAR3 was previously shown to cleave abscisic acid amino acid conjugates in vitro (45) and appears to be a versatile enzyme, but actual catalysis may be determined in vivo by substrate abundance. In vitro activity of recombinant ILL6 was much lower than IAR3 on either JA-Ile or IAA-Ala and no activity could be detected on 12OH-JA-Ile under our assay conditions. Previous attempts to produce active AtILL6 (36) or the Brassica homolog BrILL6 (25) in bacteria have failed, one possible reason being the presence of potential transmembrane domain(s). These limitations leave open the possibility that the in vitro enzymatic data presented here have under evaluated the catalytic capacities of AtILL6.
Genetic analysis has provided novel insights into the impact of IAR3 and ILL6 in metabolism of jasmonate conjugates in Arabidopsis. Although JA-Ile appeared as the best in vitro substrate tested, IAR3 or ILL6 deficiency had little apparent impact on the accumulation of the hormone, in contrast to JIH-silenced N. attenuata plants (40). This observation points to JA-Ile as a central component in the JA pathway, not only by its hormonal activity, but also as a metabolic hub with links to other conjugated and non-conjugated jasmonates. We established that the increased JA-Ile accumulation expected from reduced amidohydrolase activity is likely counterbalanced by a stronger induction of other genes encoding JA-Ile-consuming enzymes, such as IAR3 and CYP94B3 and CYP94C1 transcripts in ill6 alleles. In contrast, in the iar3-5 allele, such a transcriptional compensation was not evidenced, and accordingly, this line accumulated more JA-Ile than WT. Conversely, it is worth noting that the higher and persistent JA-Ile hyperaccumulation in cyp94b3 and cyp94b3c1 mutants blocked in oxidative JA-Ile catabolism (20) was not associated with hyperinduction of amidohydrolase genes. When manipulating Arabidopsis plants genetically to increase or stabilize JA-Ile levels, selective compensation can occur by enhanced stimulation of other enzymes catabolizing JA-Ile. This flexible response illustrates the metabolic plasticity of the JA pathway to maintain proper regulation of active hormone levels. JA-Ile dynamics are under high flux through both oxidation and cleavage pathways that prevent excessively high levels to build up. Consequently, if metabolic control of JA signaling is determined solely by JA-Ile levels, one may anticipate little impact of amidohydrolase deficiency on physiological JA responses. Also, the exact extent of functional redundancy between IAR3 and ILL6, as well as the role of ILR1, needs to be investigated by combining mutations.
A close examination of metabolic profiles in different JA pathway mutants led us to the first description of a metabolic route leading to the abundant jasmonate TA. Several lines of genetic evidence indicate that TA accumulation is achieved by a route that is dependent on JA conjugation, oxidation, and cleavage. First, the jar1 mutation lowers by more than half the TA content compared with WT. The remaining TA accumulating in jar1 may arise through a JAR1-independent route, for example, by cleaving other oxidized JA-amino acid conjugates generated by distinct GH3 enzymes, but such conjugate pools seem quantitatively minor in wounded Arabidopsis leaves (46) to account for the significant levels of TA in jar1. Alternatively, the jar1-independent TA pool may reflect the existence of a direct JA-hydroxylation route by still unknown hydroxylase(s). Under our conditions, none of the 6 recombinant Arabidopsis CYP94 proteins showed JA-hydroxylase activity.4 Second, mutants in cyp94 genes that govern JA-Ile oxidation displayed reduced TA levels despite of abundant JA supply similarly to jar1, questioning the importance of direct hydroxylation for TA formation. The higher abundance of TA in cyp94b3c1 than in cyp94c1 is well correlated with more 12OH-JA-Ile being available in the double mutant because of impaired 12COOH-JA-Ile formation (Fig. 2). Third, depleting either IAR3 or ILL6 amidohydrolase resulted in hyperaccumulation of 12OH-JA-Ile with a concomitant reduction in TA levels (Fig. 6), pointing to a direct substrate-product link between these two JAs. The analysis of two known TA modification routes confirmed that TA itself is a metabolic intermediate. Abundance of both 12-HSO4-JA and TA-Glc was correlated with TA levels in the different genetic backgrounds. This characteristic makes them direct TA derivatives and extends the number of JAs whose formation requires a conjugation-oxidation-deconjugation cascade.
The impact of amidohydrolase inactivation was much stronger on the two oxidized conjugates than on JA-Ile. As discussed above, an excess of uncleaved JA-Ile or 12OH-JA-Ile can alternatively be cleared by enhanced CYP94-mediated oxidation that shifts the pool of jasmonates toward more oxidized conjugates. The fate of the long-lived 12COOH-JA-Ile is the less well known. We have described its first chemical synthesis that allowed its quantification, and revealed high levels in plant extracts. No cleavage of 12COOH-JA-Ile has been evidenced. Precise analysis of potential in vitro and in planta cleavage of this compound awaits the availability of synthetic 12COOH-JA. In any case, as the most downstream JA-Ile catabolite, largely enhanced 12COOH-JA-Ile levels in iar3 and ill6 mutants result from the cumulative effects of uncleaved JA-Ile and 12OH-JA-Ile that are available for oxidation.
Collectively, the mutant analysis along with partial in vitro enzymatic data establish an original and indirect metabolic route to TA formation that requires the sequential conjugation, oxidation, and cleavage of JA. It also reveals that part of the wound-induced TA accumulates in Arabidopsis as a by-product of JA-Ile turnover rather than a JA shunt upstream of hormone activation. Our study proposes a novel function for IAR3 in jasmonate metabolism and describes the first functional data for ILL6, both enzymes reversing the action of JAR1. Whereas previous IAR3 impact on metabolism was evidenced after supplying exogenous auxin conjugates (28, 36) to Arabidopsis seedlings, the data presented here show the impact of its activity on endogenously generated substrates.
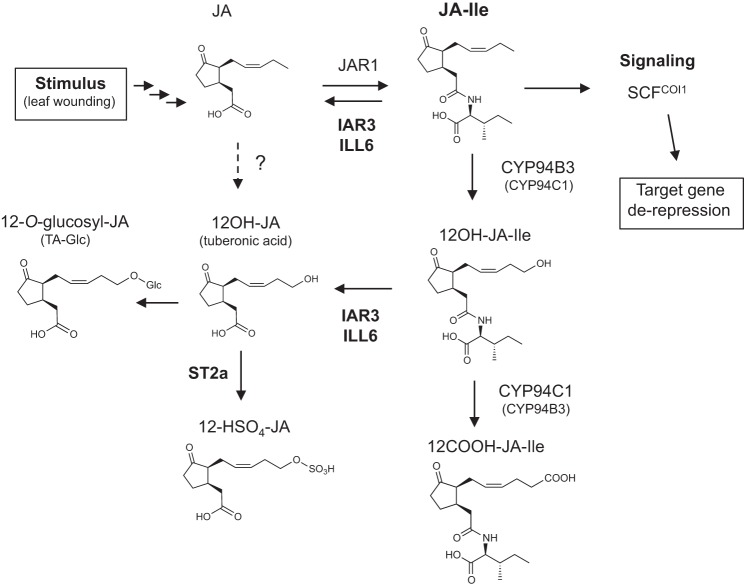
In summary, the transient nature of the JA-Ile burst triggered by wounding is shaped by the rapid, simultaneous and COI1-dependent up-regulation of two hormone inactivation pathways (Fig. 9): the first route is contributed by the CYP94-based oxidation to 12OH-JA-Ile and 12COOH-JA-Ile (20–22); the second pathway examined in this study is represented by the IAR3 and ILL6 amidohydrolases that proceed to the cleavage of JA-Ile and the 12OH-JA-Ile oxidized conjugate. The fact that JA pathway activation triggers both conjugation and deconjugation steps to form and eliminate the active hormone is intriguing. It is not known if JA issued from deconjugation is recycled for further JA-Ile signaling or if it engages into an independent metabolic route. The combined action of both oxidative and hydrolytic pathways identifies an additional metabolic function by producing a significant part of wound-induced TA and TA derivatives. The availability of these novel genetic backgrounds impaired in jasmonate metabolism will allow deeper exploration of the complex regulation of JA-Ile hormone homeostasis.
FIGURE 9.
Proposed model for interconversion routes between JA and its oxidized and/or Ile-conjugated derivatives in Arabidopsis. Upon stimulation following leaf wounding, JA biosynthesis is strongly activated, including further metabolism to an array of derivatives. Part of JA is conjugated to isoleucine, forming JA-Ile that is transiently accumulated as the active hormonal signal. JA-Ile is rapidly inactivated/eliminated by two simultaneously up-regulated COI1-dependent enzymatic pathways, one that is CYP94-based and leads to oxidized derivatives, and the second by conjugate cleavage under the action of the amidohydrolases IAR3 and ILL6. The compounds are shown in the demonstrated (up to JA-Ile) or supposed (3,7)-cis stereochemistry occurring in planta.
Acknowledgments
We are grateful to Dr. Rozenn Ménard for critical reading of the manuscript and Dr. Nicolas Baumberger for help in recombinant protein production. We thank the gardener team for producing the numerous plants used in this study, and the Nottingham Arabidopsis Stock Center for providing seeds of T-DNA insertion lines. The UPLC-MS/MS instrument was co-financed by the Centre National de la Recherche Scientifique, the Université de Strasbourg, the Région Alsace, the Institut National de la Recherche Agronomique, and the Tepral Company.
Addendum
During the review of this paper, a paper was published (47) that partially describes ill6 insertion mutants. The ill6-1 line used in Ref. 47 corresponds to the ill6-2 allele analyzed in the present study.
This work was supported in part by Agence Nationale de la Recherche Contract ANR-12-BSV8-005 (to Y. A).

This article contains supplemental Methods S1.
E. Widemann and F. Pinot, unpublished data.
- JA
- jasmonates or jasmonic acid
- TA
- tuberonic acid
- JA-Ile
- (+)-7-iso-jasmonoyl-l-isoleucine
- JAR1
- JASMONATE RESISTANT 1
- COI1
- CORONATINE INSENSITIVE 1
- JAZ
- Jasmonate ZIM domain
- IAA-Ala
- indole acetic acid-alanine
- UPLC
- ultra performance liquid chromatography
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR.
REFERENCES
- 1. Browse J. (2009) Jasmonate passes muster. A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 [DOI] [PubMed] [Google Scholar]
- 2. Wasternack C., Hause B. (2013) Jasmonates. Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 111, 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erb M., Meldau S., Howe G. A. (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koo A. J., Howe G. A. (2009) The wound hormone jasmonate. Phytochemistry 70, 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhage A., Vlaardingerbroek I., Raaymakers C., Van Dam N. M., Dicke M., Van Wees S. C., Pieterse C. M. (2011) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mithöfer A., Boland W. (2012) Plant defense against herbivores. Chemical aspects. Annu. Rev. Plant Biol. 63, 431–450 [DOI] [PubMed] [Google Scholar]
- 7. Wu J., Baldwin I. T. (2010) New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24 [DOI] [PubMed] [Google Scholar]
- 8. Glauser G., Grata E., Dubugnon L., Rudaz S., Farmer E. E., Wolfender J. L. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 283, 16400–16407 [DOI] [PubMed] [Google Scholar]
- 9. Glauser G., Wolfender J. L. (2013) A non-targeted approach for extended liquid chromatography-mass spectrometry profiling of free and esterified jasmonates after wounding. Methods Mol. Biol. 1011, 123–134 [DOI] [PubMed] [Google Scholar]
- 10. Chung H. S., Koo A. J., Gao X., Jayanty S., Thines B., Jones A. D., Howe G. A. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koo A. J., Howe G. A. (2012) Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshihara T., Omer E.-L. A., Koshino H., Sakamura S., Kikuta Y., Koda Y. (1989) Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.). Agric. Biol. Chem. 53, 2835–2837 [Google Scholar]
- 13. Gidda S. K., Miersch O., Levitin A., Schmidt J., Wasternack C., Varin L. (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900 [DOI] [PubMed] [Google Scholar]
- 14. Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127 [DOI] [PubMed] [Google Scholar]
- 15. Nakamura Y., Mithöfer A., Kombrink E., Boland W., Hamamoto S., Uozumi N., Tohma K., Ueda M. (2011) 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol. 155, 1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Staswick P. E., Tiryaki I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fonseca S., Chico J. M., Solano R. (2009) The jasmonate pathway. The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547 [DOI] [PubMed] [Google Scholar]
- 19. Katsir L., Chung H. S., Koo A. J., Howe G. A. (2008) Jasmonate signaling. A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., Pinot F. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287, 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell. Physiol. 52, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 22. Koo A. J., Cooke T. F., Howe G. A. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizutani M., Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315 [DOI] [PubMed] [Google Scholar]
- 24. Piotrowska A., Bajguz A. (2011) Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72, 2097–2112 [DOI] [PubMed] [Google Scholar]
- 25. Schuller A., Ludwig-Müller J. (2006) A family of auxin conjugate hydrolases from Brassica rapa. Characterization and expression during clubroot disease. New Phytol 171, 145–157 [DOI] [PubMed] [Google Scholar]
- 26. Campanella J. J., Smith S. M., Leibu D., Wexler S., Ludwig-Müller J. (2008) The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 27, 26–38 [Google Scholar]
- 27. Rampey R. A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., Bartel B. (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davies R. T., Goetz D. H., Lasswell J., Anderson M. N., Bartel B. (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Titarenko E., Rojo E., León J., Sánchez-Serrano J. J. (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115, 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berr A., McCallum E. J., Alioua A., Heintz D., Heitz T., Shen W. H. (2010) Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Birkenbihl R. P., Diezel C., Somssich I. E. (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kramell T., Schmidt J., Schneider G., Sembdner G., Schreiber K. (1988) Synthesis of n-(jasmonoyl)amino acid conjugates. Tetrahedron 44, 5791–5807 [Google Scholar]
- 33. Matsuura H., Ohmori F., Kobayashi M., Sakurai A., Yoshihara T. (2000) Qualitative and quantitative analysis of endogenous jasmonoids in potato plant (Solanum tuberosum L.). Biosci. Biotechnol. Biochem. 64, 2380–2387 [DOI] [PubMed] [Google Scholar]
- 34. Ogawa N., Kobayashi Y. (2012) Synthesis of the amino acid conjugates of epi-jasmonic acid. Amino Acids 42, 1955–1966 [DOI] [PubMed] [Google Scholar]
- 35. Nicolaou K. C., Estrada A. A., Zak M., Lee S. H., Safina B. S. (2005) A mild and selective method for the hydrolysis of esters with trimethyltin hydroxide. Angew. Chem. Int. Ed. Engl. 44, 1378–1382 [DOI] [PubMed] [Google Scholar]
- 36. LeClere S., Tellez R., Rampey R. A., Matsuda S. P., Bartel B. (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 [DOI] [PubMed] [Google Scholar]
- 37. VanDoorn A., Bonaventure G., Schmidt D. D., Baldwin I. T. (2011) Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum. COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 190, 640–652 [DOI] [PubMed] [Google Scholar]
- 38. Campanella J. J., Larko D., Smalley J. (2003) A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp. Funct. Genomics 4, 584–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toufighi K., Brady S. M., Austin R., Ly E., Provart N. J. (2005) The Botany Array Resource. e-Northerns, expression angling, and promoter analyses. Plant J. 43, 153–163 [DOI] [PubMed] [Google Scholar]
- 40. Woldemariam M. G., Onkokesung N., Baldwin I. T., Galis I. (2012) Jasmonoyl-l-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-l-isoleucine levels and attenuates plant defenses against herbivores. Plant J. 72, 758–767 [DOI] [PubMed] [Google Scholar]
- 41. Seto Y., Hamada S., Matsuura H., Matsushige M., Satou C., Takahashi K., Masuta C., Ito H., Matsui H., Nabeta K. (2009) Purification and cDNA cloning of a wound inducible glucosyltransferase active toward 12-hydroxy jasmonic acid. Phytochemistry 70, 370–379 [DOI] [PubMed] [Google Scholar]
- 42. Ludwig-Müller J. (2011) Auxin conjugates. Their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 [DOI] [PubMed] [Google Scholar]
- 43. Staswick P. (2009) Plant hormone conjugation. A signal decision. Plant Signal. Behav. 4, 757–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bartel B., Fink G. R. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748 [DOI] [PubMed] [Google Scholar]
- 45. Todoroki Y., Narita K., Muramatsu T., Shimomura H., Ohnishi T., Mizutani M., Ueno K., Hirai N. (2011) Synthesis and biological activity of amino acid conjugates of abscisic acid. Bioorg. Med. Chem. 19, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 46. Koo A. J., Gao X., Jones A. D., Howe G. A. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986 [DOI] [PubMed] [Google Scholar]
- 47. Bhosale R., Jewell J. B., Hollunder J., Koo A. J., Vuylsteke M., Michoel T., Hilson P., Goossens A., Howe G. A., Browse J., Maere S. (2013) Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell. 10.1105/tpc.113.112268 [DOI] [PMC free article] [PubMed] [Google Scholar]