Background: Expanded CD3+CD4−CD8− T cells in SLE originate from CD8+ T cells.
Results: CREMα trans-represses CD8 gene transcription.
Conclusion: By trans-repressing CD8 expression, CREMα contributes to double-negative T cell expansion in SLE pathogenesis.
Significance: CREMα modulates T cell subset distribution and cytokine expression.
Keywords: Autoimmune Diseases, Cell Differentiation, Cellular Immune Response, Gene Regulation, T Cell, CD8, CREMα, SLE, Double-negative T Cells
Abstract
T cell receptor-αβ+ CD3+CD4−CD8− “double-negative” T cells are expanded in the peripheral blood of patients with systemic lupus erythematosus and autoimmune lymphoproliferative syndrome. In both disorders, double-negative T cells infiltrate tissues, induce immunoglobulin production, and secrete proinflammatory cytokines. Double-negative T cells derive from CD8+ T cells through down-regulation of CD8 surface co-receptors. However, the molecular mechanisms orchestrating this process remain unclear. Here, we demonstrate that the transcription factor cAMP-responsive element modulator α (CREMα), which is expressed at increased levels in T cells from systemic lupus erythematosus patients, contributes to transcriptional silencing of CD8A and CD8B. We provide the first evidence that CREMα trans-represses a regulatory element 5′ of the CD8B gene. Therefore, CREMα represents a promising candidate in the search for biomarkers and treatment options in diseases in which double-negative T cells contribute to the pathogenesis.
Introduction
Systemic lupus erythematosus (SLE)3 is an autoimmune/inflammatory disorder that can affect any organ of the human body. Tissue damage is caused by a number of mechanisms, including the deposition of autoantibodies and immune complexes, and organ infiltration by lymphocytes (1). In SLE, several molecular abnormalities contribute to the proinflammatory phenotype and tissue homing of B and T cells. T cell receptor (TCR)-αβ+ CD3+CD4−CD8− “double-negative” (DN) T cells are expanded in the peripheral blood and infiltrate tissues, where they induce immunoglobulin production and secrete proinflammatory cytokines (2–5). Recently, we demonstrated that human DN T cells derive from CD8+ T lymphocytes (3). Our studies suggested that following TCR stimulation, CD8+ T cells down-regulate the CD8 co-receptor on their surface and acquire a distinct DN T cell phenotype that includes increased production of proinflammatory IL-17A (3). However, the molecular mechanisms that contribute to the down-regulation of CD8 surface expression remain unclear.
The regulation of CD8 has been studied during the development of T lymphocytes in the thymus. Most thymus-derived TCR-αβ+ T cells express CD4 or CD8 co-receptors. Mature CD4+ or CD8+ T cells derive from common CD4+CD8+ “double-positive” progenitor cells, which derive from CD4−CD8− DN thymocytes (6, 7). Four clusters with increased DNase sensitivity have been identified within the murine CD8 locus, which are syntenic with six human clusters (8, 9). Transgenic reporter systems allowed the identification of several enhancer elements within the CD8 cluster (E8I–E8IV) (6, 8–19). This enhancer network is required for lineage-specific regulation of CD8α and CD8β during T cell development, and its elements undergo epigenetic remodeling during T cell development, either allowing or prohibiting the expression of CD8A and/or CD8B (9).
Here, we studied trans-regulatory mechanisms that contribute to the generation of peripheral DN T cells in health and disease. In addition, we studied Fas-deficient MRL/lpr mice that display an expanded DN T cell population (20, 21). Comparable to the lpr mutation in Fas-deficient MRL/lpr mice, a group of disorders, referred to as autoimmune lymphoproliferative syndrome, may result from mutations in the genes encoding Fas/CD95 (TNFRSF6) (22–26). In both Fas-deficient MRL/lpr mice and human autoimmune lymphoproliferative syndrome patients, the majority of DN T cells are believed to originate from CD8+ T cells through down-regulation of CD8 surface expression (21, 27). Autoimmune lymphoproliferative syndrome patients and MRL/lpr mice develop a lymphoproliferative disease with excessive autoantibody production (in humans, primarily anti-cardiolipin or direct Coombs antibodies; in MRL/lpr mice, mainly anti-dsDNA antibodies) and severe autoimmune phenomena and share features with SLE patients (e.g. cytopenias, glomerulonephritis, hepatosplenomegaly) (20, 23, 25, 26).
We present evidence that CD8+ T cells down-regulate CD8 surface expression in response to TCR stimulation through transcriptional silencing of the CD8A and CD8B genes. Transcriptional repression is mediated by the transcription factor cAMP-responsive element modulator α (CREMα), which is expressed at increased levels in T cells from SLE patients (28, 29). CREMα exerts direct trans-repressing effects on a region syntenic to the murine CD8b promoter (6, 30). Thus, CREMα represents the first described transcription factor to trans-repress CD8 gene expression.
EXPERIMENTAL PROCEDURES
Cell Culture
Peripheral blood mononuclear cells were enriched for T lymphocytes by negative selection (RosetteSep, STEMCELL Technologies). A second round of negative selection was used to isolate CD8+ T cells (Dynabeads, Invitrogen). CD8+ T lymphocytes were cultured at a concentration of 1 × 106 cells/ml in RPMI 1640 medium with 10% FCS in 12-well plates that had or had not been precoated with anti-CD3 and anti-CD28 antibodies (as indicated). Cells were collected after 120 h and harvested for quantitative RT-PCR, flow cytometry, or ChIP as indicated.
Human Subjects
All SLE patients included in our studies were diagnosed according to the American College of Rheumatology classification criteria and were recruited from the Division of Rheumatology at the Beth Israel Deaconess Medical Center; they gave written informed consent under Protocol 2006-P-0298. Healthy and age-, gender-, and ethnicity-matched individuals were chosen as controls. Peripheral venous blood was collected in heparin-lithium tubes, and total human T cells were purified as described before. Epidemiologic and clinical information is displayed in Table 1.
TABLE 1.
Demographic information on SLE patients
| Patient | Years of age | Gender | Ethnicity | SLEDAIa |
|---|---|---|---|---|
| 1 | 46 | F | C | 5 |
| 2 | 28 | F | C | 4 |
| 3 | 33 | F | AA | 4 |
| 4 | 40 | F | AA | 4 |
a SLEDAI, SLE disease activity index; F, female; C, Caucasian; AA, African American.
Mice
MRL/lpr mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed under specific pathogen-free conditions. Experimental procedures were approved by the Beth Israel Deaconess Medical Center Animal Care and Use Committee.
Flow Cytometry and Cell Sorting
Pacific Blue-conjugated anti-CD4, phycoerythrin-conjugated anti-CD8, and allophycocyanin/Cy7-conjugated anti-CD3 antibodies were purchased from BioLegend. Samples were acquired on an LSR II flow cytometer (BD Biosciences), and data were analyzed using FlowJo version 7.2.2 (TreeStar, Inc.). For the analysis of T lymphocyte populations, a first gate that included live cells was used. CD3+ T lymphocytes were then plotted in a CD4+ versus CD8+ dot plot that allowed the identification of discrete CD4+, CD8+, and DN T lymphocyte populations. For some experiments, stained cells were sorted in a FACSAria flow cytometer (BD Biosciences); post-sorting purity was >98%.
Semiquantitative RT-PCR
Total RNA from control and SLE T lymphocytes was isolated using an RNeasy mini kit (Qiagen). cDNA was generated using a first-strand cDNA synthesis kit (Invitrogen). For gene expression analyses, real-time PCR was performed using SYBR Green site-specific primers on the ABI StepOnePlus real-time PCR system. Results were normalized to 18 S.4
Gene Expression Plasmids
Expression plasmids for human CREMα have been described previously (31–33). Three million primary human CD8+ T lymphocytes were transfected with a total amount of 3 μg of the indicated expression plasmids using the Amaxa transfection system (Lonza). After 24 (RNA) or 120 h (flow cytometry), cells were harvested and assayed.
Generation of Luciferase Reporter Constructs
Reporter constructs of three conserved noncoding sequence (CNS) regions within the human CD8 cluster (CNS2 (271 bp), which is syntenic to the murine CD8b promoter, and CNS7 (352 bp) and CNS8 (209 bp), which map to previously reported enhancer regions (6)) were PCR-amplified and cloned into the luciferase vector pGL3-Basic (Promega) using primers with attached restriction sites for MluI and BglII. All plasmid DNAs were prepared using DNA purification kits (Qiagen) and sequence-verified (GENEWIZ, Cambridge, MA). Site-directed mutagenesis at two cAMP-responsive element (CRE) sites within the CNS2 reporter construct was performed using a DNA oligonucleotide harboring a mutated CRE and PfuTurbo® DNA polymerase (Stratagene) according to the manufacturer's instructions.
Luciferase Assays in Jurkat T Cells
One million Jurkat T cells were transfected with 500 ng of plasmid DNA using the Amaxa transfection system. Effector/reporter transfection experiments were performed at a molar ratio of 3:1. Each reporter experiment included 10 ng of Renilla luciferase construct as an internal control. Six hours after transfection, cells were collected and lysed, and luciferase activity was quantified using the Dual-Luciferase assay system (Promega) following the manufacturer's instructions. Luciferase experiments were repeated four times, and values in the bar graphs are given as means ± S.D.
ChIP Assays
Anti-CREMα polyclonal antibody detecting human CREMα has been described previously (6, 34). ChIP experiments were carried out with a ChIP kit (Upstate Biotechnology/Millipore) according to the manufacturer's instructions. For this assay, ChIP-grade Protein A/G Plus-agarose was used (Thermo Scientific). Briefly, 1–2 million cells were cross-linked with 1% formaldehyde, washed with cold phosphate-buffered saline, and lysed in buffer containing protease inhibitors (Roche Applied Science). Cell lysates were sonicated to shear DNA and sedimented, and diluted supernatants were immunoprecipitated with the indicated antibodies. A proportion (20%) of the diluted supernatants were kept as “input” (input represents PCR amplification of the total sample). The amount of immunoprecipitated DNA was subtracted from the amplified DNA that was bound by the nonspecific normal IgG and subsequently calculated relative to the respective input DNA.
Statistical Analysis
Paired two-tailed Student's t test was used for statistical analysis of all flow cytometry and transfection experiments as indicated. A p value of ≤0.05 was considered statistically significant. Results are indicated as means ± S.D. unless noted otherwise.
RESULTS
A Subset of CD8+ T Cells Down-regulates CD8 in Response to Stimulation
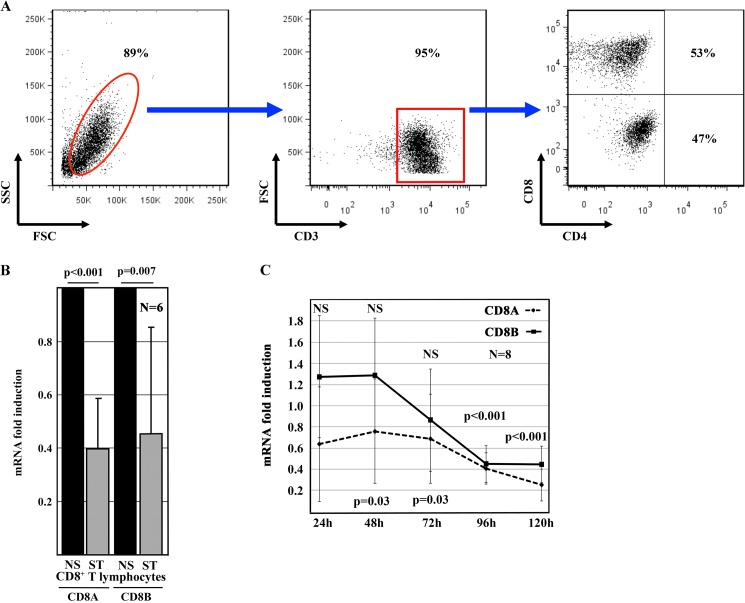
A significant fraction of CD8+ T cells down-regulates CD8 co-receptor surface expression in response to TCR activation with anti-CD3 and anti-CD28 antibodies (3). Although some CD8+ T cells underwent cell death in response to TCR stimulation in the absence of added cytokines, most cells survived (average of 94.6%, S.D. of 1.57) (Fig. 1A). After 120 h, CD8 co-receptor surface expression was down-regulated in 46.9% (S.D. of 12.9) of T cells (Fig. 1A and Table 2), allowing an enrichment of DN T cells (3). Reduced CD8 co-receptor expression was reflected by significant down-regulation of CD8A (p = 0.001) and CD8B (p = 0.007) mRNAs in response to stimulation, both of which followed individual kinetics (Fig. 1, B and C). Relative CD8 expression in unstimulated CD8+ T cells was assigned a value of 1. Changes in gene expression in response to TCR stimulation are displayed as an increase (>1) or decrease (<1) over CD8 expression in resting CD8+ T cells. During the initial 72 h, expression of the two CD8 isoforms was comparable in resting CD8+ T cells and in response to T cell activation. After 96 and 120 h, stimulated CD8+ T cells down-regulated CD8A and CD8B transcription (p < 0.001).
FIGURE 1.
Human CD8 protein and mRNA expression in response to TCR stimulation. A, primary human CD8+ T lymphocytes from healthy individuals were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 120 h. Most stimulated CD8 T cells were alive as shown by their forward (FSC) and side (SSC) scatter distribution (left panel) and expressed CD3 (middle panel). A large proportion of CD8+ T lymphocytes down-regulated CD8 and transformed into DN T cells (right panel). B, TCR stimulation with anti-CD3 and anti-CD28 antibodies for 120 h significantly decreased CD8A and CD8B mRNA expression in primary human CD8+ T lymphocytes from healthy individuals. Values are means ± S.D. NS, non-stimulated T cells; ST, TCR-stimulated T cells. C, kinetics of CD8A and CD8B mRNA expression in response to TCR stimulation with anti-CD3 and anti-CD28 antibodies over 120 h in primary human CD8+ T lymphocytes from healthy individuals. Values indicate -fold changes over the relative gene expression in unstimulated T cells, which were assigned a relative expression of 1. NS indicates time points at which TCR stimulation did not result in significant changes. p values indicate statistically significant changes in gene expression after 48 and 72 h (CD8A) or 96 and 120 h (CD8A and CD8B).
TABLE 2.
Effects of TCR stimulation on CD8+ T cells from healthy individuals
| Patient | Live cells | CD8+ T cells | DN T cells |
|---|---|---|---|
| % | |||
| 1 | 94.97 | 41.01 | 58.99 |
| 2 | 94.9 | 53.12 | 46.88 |
| 3 | 97.81 | 42.12 | 57.88 |
| 4 | 95.34 | 44.35 | 55.63 |
| 5 | 97.82 | 71.01 | 28.99 |
| 6 | 98.15 | 66.52 | 33.48 |
| Average | 96.49 | 53.02 | 46.97 |
| S.D. | 1.57 | 12.99 | 12.99 |
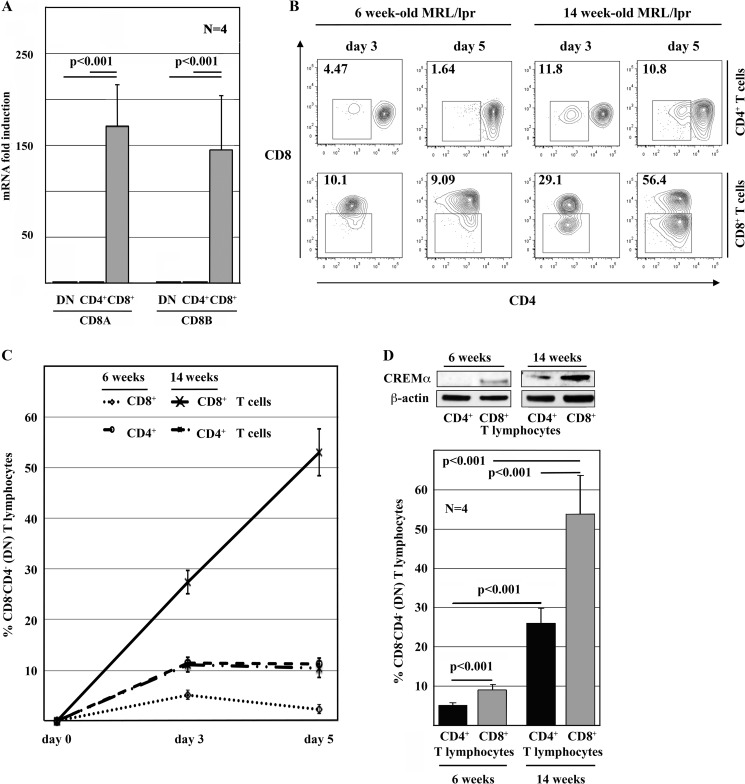
Fas-deficient MRL/lpr mice exhibit an enrichment of DN T cells and develop a lymphoproliferative disease with severe autoimmune phenomena mimicking SLE (20). In agreement with our observations in human T cells, in MRL/lpr T cells, the expression of CD8 was regulated at the transcriptional level (Fig. 2A). Recent studies suggest that the majority of DN T cells in MRL/lpr mice derive from CD8+ T cells (21). To confirm this observation, we stimulated sorted CD4+ and CD8+ T cells from asymptomatic MRL/lpr mice (6 weeks old) and from animals after disease onset (14 weeks old) with anti-CD3 and anti-CD28 antibodies (Fig. 2, B and C). TCR stimulation of cells from young animals resulted in lower numbers of DN T cells compared with T cells from diseased animals (p < 0.001). However, in both age groups, significantly more CD8+ T cells (6 weeks, 9.15%, S.D. of 1.32; 14 weeks, 53.7%, S.D. of 9.8) transformed into DN T cells compared with CD4+ T cells (6 weeks, 5.07%, S.D. of ±0.67; 14 weeks, 25.86%, S.D. of 3.99; p < 0.001).
FIGURE 2.
CD8 expression in MRL/lpr mice is regulated at the transcriptional level. A, CD8a and CD8b mRNAs were differentially expressed in CD4+, CD8+, and DN T lymphocytes sorted from 14-week-old MRL/lpr mice. B and C, CD8+ T cells from MRL/lpr mice down-regulated CD8 surface receptors in response to TCR stimulation with anti-CD3 and anti-CD28 antibodies. This was more pronounced in 14-week-old symptomatic animals compared with younger animals (6 week old) before the onset of symptoms. Also, a subset of CD4+ T cells down-regulated CD4 in response to stimulation. However, the CD4-to-DN conversion rate was significantly lower than the CD8-to-DN conversion rate. Values are means ± S.D. D, CD4+ and CD8+ T cells from 6-week-old “healthy” MRL/lpr mice expressed reduced levels of CREMα compared with 14-week-old diseased animals. CD8+ T cells expressed more CREMα than CD4+ T cells from the same mouse, reflecting their capacity to transform into DN T lymphocytes in response to TCR stimulation.
We demonstrated previously that the transcription factor CREMα is expressed at increased levels in T cells from SLE patients (28, 29). After disease onset, MRL/lpr mice share key symptoms with SLE patients, including the enrichment of DN T cells and elevated T cell expression of CREMα. Thus, we asked whether the expression of CREMα in T cells from 6- and 14-week-old animals reflects the differential potential to down-regulate CD8. Indeed, ex vivo isolated CD4+ or CD8+ T cells from 14-week-old diseased animals exhibited increased CREMα expression compared with T cells from 6-week-old animals. Furthermore, CD4+ T cells expressed less CREMα compared with CD8+ T cells from the same mice (Fig. 2D).
CREMα Represses CD8 Gene Transcription
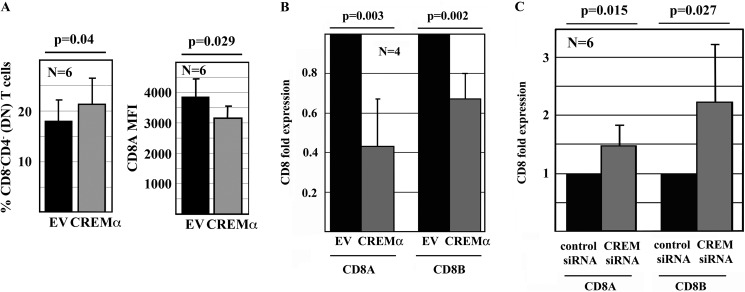
CREMα contributes to the dysregulation of cytokine expression and affects T cell subset distribution in SLE through transcriptional regulation of cytokines and the induction of epigenetic remodeling processes (28, 31–33, 35). Thus, we asked whether CREMα also trans-regulates CD8 expression. To this end, we overexpressed CREMα in human CD8+ T cells. After 120 h in culture, cells were collected and stained for CD3, CD4, and CD8 surface expression for flow cytometry (Fig. 3A). Forced expression of CREMα led to a significantly increased production of DN T cells (p = 0.04) (Fig. 3A, left panel). Furthermore, CD8+ T cells exhibited a significant decrease in the expression of the CD8 co-receptor (CD8 mean fluorescence intensity) in response to forced expression of CREMα (p = 0.029) (Fig. 3A, right panel). To determine whether CREMα regulates CD8 co-receptor expression at the transcriptional level, we assessed CD8A and CD8B mRNA expression in CD8+ T cells in response to forced expression of CREMα. Both CD8A (p = 0.003) and CD8B (p = 0.002) transcripts were significantly reduced in response to CREMα overexpression (Fig. 3B), suggesting trans-repressing effects on CD8A and CD8B. Because T cells constitutively express CREMα (28), we investigated the effects of CREM silencing by siRNAs. Indeed, CD8+ T cells exhibited increased CD8A (p = 0.015) and CD8B (p = 0.027) transcription in response to CREM knockdown (Fig. 3C).
FIGURE 3.
CREMα governs CD8 expression. A, CREMα enhanced the generation of DN T cells from primary human CD8+ T cells from healthy individuals (120 h). CD8− T cells were increased in response to CREMα compared with controls (empty vector (EV)). CREMα increased the relative numbers of DN T cells (left panel) by down-regulating CD8 surface expression on individual T cells (right panel). MFI, mean fluorescence intensity. B, CREMα negatively regulated CD8A and CD8B mRNA expression in primary human CD8+ T cells compared controls (24 h). C, CREM knockdown resulted in increased CD8A and CD8B expression in primary human CD8+ T cells compared with cells transfected with scrambled siRNA (24 h).
Bioinformatic Analysis of the CD8A/CD8B Gene Cluster
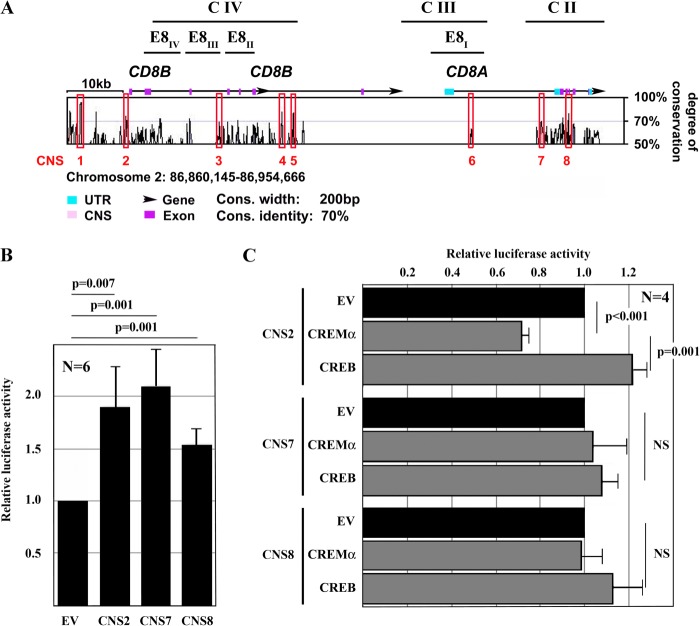
The human CD8 gene cluster on chromosome 2q12 harbors both the CD8A and CD8B genes, which encode the CD8α and CD8β proteins, respectively. Applying bioinformatic approaches, we aligned the murine CD8a and CD8b genes with the human CD8A and CD8B genes (VISTA Genome Browser) and defined CNS regions (Fig. 4A, pink), exons (magenta), and UTRs (turquoise). Eight CNS sites were defined that display sequence homologies of >70% over at least 200 bp between species, and they were used as targets in the search for CREM-responsive regulatory elements.
FIGURE 4.
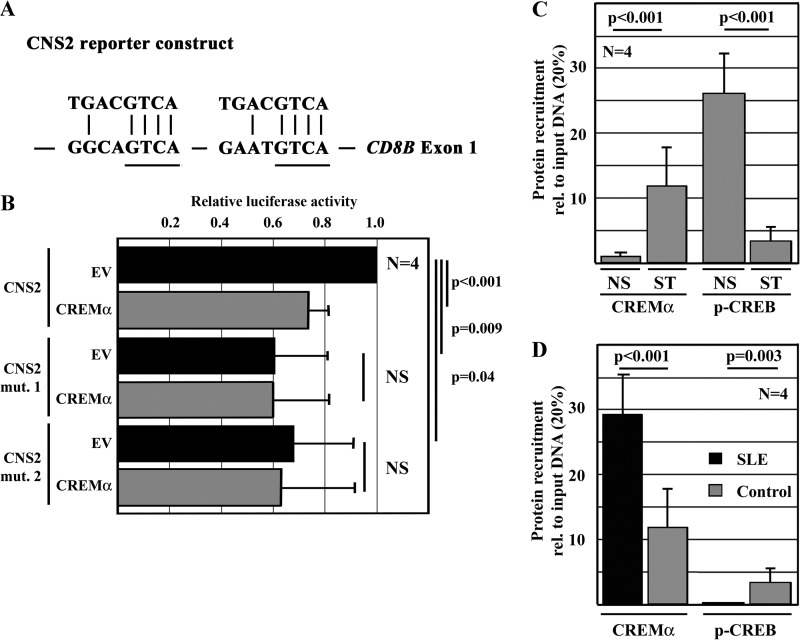
trans-Regulation of the CD8 cluster. A, to investigate trans- and cis-regulatory elements across the human and murine CD8 locus, we defined regions of interest based on bioinformatic approaches. We aligned the mouse and human CD8 genes (VISTA Genome Browser) and searched for CNS sites. CNS sites were defined as regions of >200 bp with >70% homology between human and mouse. Eight regions of interest (CNS1–CNS8) were defined based on the degree of sequence conservation and the presence of reported regulatory regions. C II–C IV are previously reported DNase hypersensitivity clusters with regulatory capacities; E8I–E8IV are previously defined enhancer elements (6). B, reporter constructs of CNS regions within the human CD8 cluster were generated and transfected into Jurkat T cells. CNS2 is syntenic to the murine CD8b promoter, and CNS7 and CNS8 are within previously reported regulatory elements. All constructs exhibited enhanced activity over an empty pGL3 plasmid (empty vector (EV)). C, CREMα reduced the activity of CNS2, whereas CREB enhanced promoter activity. Both CREMα and CREB did not show significant effects on either CNS7 or CNS8.
CREMα trans-Represses CNS2 Activity
A comparative analysis of the murine and human CD8 genes yielded multiple putative CREs that could potentially recruit CREMα. Thus, we focused our search for CREs within the identified CNS regions (Fig. 4, A and B). We generated luciferase constructs of CNS2 (syntenic to the murine CD8b promoter) (30), CNS7, and CNS8, all of which exhibited promoter/enhancer activities (Fig. 4B): CNS2 by a 1.9-fold increase (S.D. of 0.34; p = 0.007), CNS7 by a 2.1-fold increase (S.D. of 0.35; p = 0.001), and CNS8 by a 1.5-fold increase (S.D. of 0.15; p = 0.001). To investigate whether CREMα accounts for CD8 repression, we performed luciferase assays using the CNS2, CNS7, and CNS8 reporter constructs in the presence and absence of CREMα (Fig. 4C). Forced expression of CREMα did not have a significant effect on the activities of CNS7 or CNS8. However, CREMα significantly decreased the promoter activity of CNS2 (by >20%), whereas forced expression of the CREMα counteractor CRE-binding protein (CREB), another member of this transcription factor superfamily, increased its activity.
CNS2, which is syntenic to the murine CD8b promoter, harbors two CRE sites (Fig. 5A). Deletion of either of the CRE sites within the 271-bp CNS2 reporter construct (Fig. 5A) reduced the promoter activity of CNS2 by 35–40% and abolished additional effects by CREMα (Fig. 5B). This suggests trans-activating effects of the transcription factor CREB on the CNS2 enhancer that are dependent on the two intact CRE half-sites.
FIGURE 5.
CREMα represses the activity of CNS2 through binding to CRE motifs. A, CNS2, which is trans-regulated by CREB and CREMα, harbors two CREs that bind CREB or CREMα. Displayed are the consensus sequence of the palindromic CRE site (upper) and two putative CRE sites within CNS2 (lower). B, deletion of either of the two CRE sites within CNS2 resulted in reduced promoter activity and abrogated CREMα effects after transfection into Jurkat T cells. C, CREMα and phospho-CREB (p-CREB) competed for the recruitment to CNS2 as assessed by ChIP. CD8+ T cells from healthy donors exhibited mostly phospho-CREB and almost no CREMα recruitment to CNS2 (bars 1 and 3). In response to TCR stimulation with anti-CD3 and anti-CD28 antibodies, phospho-CREB was partially replaced with CREMα (bars 2 and 4). D, in stimulated CD8+ T cells from SLE patients, CREMα recruitment to CNS2 was even more pronounced (bars 1 and 2). Phospho-CREB was almost completely replaced with CREMα in SLE T cells. NS, non-stimulated cells; ST, T cells after TCR stimulation with anti-CD3 and anti-CD28 antibodies for 120 h; rel., relative; EV, empty vector.
To investigate whether CREMα trans-represses the promoter activity of CNS2 by replacing CREB from these binding sites, we performed ChIP experiments in CD8+ T cells from SLE patients and healthy controls (Fig. 5C). In both CD8+ T cells from SLE patients and healthy controls, CREMα was recruited to CNS2 in response to T cell activation with anti-CD3 and anti-CD28 antibodies where it replaces CREB. Notably, SLE T cells showed enriched (28, 29) CREMα recruitment to the CNS2 region, whereas CREB was less recruited to this site (Fig. 5D).
DISCUSSION
Both the origin and function of DN T cells have been the focus of research for several years. As a result from Vα and Vβ gene studies, phenotypic differences between immature DN thymocytes and also within mature DN T cells have been recognized (36–40). In the peripheral blood of SLE patients, DN T cells are expanded and derive from CD8+ T cells that undergo phenotypic transformation (2–5), involving the down-regulation of CD8 and the acquisition of a distinct effector phenotype (2, 3). Because DN T cells infiltrate tissues, they are believed to be of pathophysiological relevance in SLE (2, 3). We demonstrated previously that a considerably large cohort of CD8+ T cells down-regulates CD8 in vitro as a response to TCR stimulation (3). However, the molecular mechanisms directing the transformation of CD8+ into DN T cells remained elusive. In this study, we addressed regulatory mechanisms governing CD8 expression contributing to the generation of DN T cells.
Studies focusing on the development of T cells in the thymus have documented that the two CD8 genes, CD8A and CD8B, are regulated at the transcriptional level (7, 6). The CD8 genes are controlled by a number of cis-regulatory regions that respond to trans-activation. Hitherto, no trans-repressing factors have been identified (6–18, 30, 41–44). We have demonstrated that CREMα, which is expressed at increased levels in T cells from SLE patients and in activated T cells, controls CD8A and CD8B expression. Applying luciferase constructs, we showed that transcription factors CREB and CREMα trans-regulate a region syntenic to the murine CD8b promoter (CNS2) in a diametric fashion. Upon TCR activation, CREMα replaces the trans-activator CREB, suggesting that the balance between CREMα and CREB controls the activity of CNS2. This is in line with our observation that forced CREMα expression in CD8+ T cells suppresses CD8A and CD8B transcription. Increased CREMα expression in SLE T cells may therefore centrally contribute to the enhanced generation of DN T cells through trans-repression. Thus, we have provided the first evidence for the presence of trans-suppressors of CD8. However, effects of forced CREMα expression on the generation of DN T cells are limited, indicating that additional mechanisms, such as additional transcription factors, may be involved in the expansion of DN T cells in SLE patients.
Taken together, our data reveal a novel role of CREMα in the regulation of CD8. During the induction of DN T cells through stimulation of CD8+ T cells, CREMα contributes to the down-regulation of CD8 through trans-repression of CNS2, making CREMα the first transcription factor reported to trans-repress the CD8 genes. Because CREMα is increased in T cells from SLE patients and MRL/lpr mice, these mechanisms could be central in the generation of DN T cells in SLE and other autoimmune diseases with increased numbers of DN T cells. This makes CREMα a promising molecule in the search for disease biomarkers and therapeutic targets.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI42269, R01 AI49954, and R01 AI85567 (to G. C. T.).
Primer sequences for quantitative RT-PCR, plasmid generation, methylated DNA immunoprecipitation, and ChIP-PCR will be provided upon request.
- SLE
- systemic lupus erythematosus
- TCR
- T cell receptor
- DN
- double-negative
- CREMα
- cAMP-responsive element modulator α
- CNS
- conserved noncoding sequence
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein.
REFERENCES
- 1. Tsokos G. C. (2011) Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 2. Crispín J. C., Oukka M., Bayliss G., Cohen R. A., Van Beek C. A., Stillman I. E., Kyttaris V. C., Juang Y. T., Tsokos G. C. (2008) Expanded double-negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 181, 8761–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crispín J. C., Tsokos G. C. (2009) Human TCR-αβ+ CD4−CD8− T cells can derive from CD8+ T cells and display an inflammatory effector phenotype. J. Immunol. 183, 4675–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shivakumar S., Tsokos G. C., Datta S. K. (1989) T cell receptor α/β expressing double-negative (CD4−/CD8−) and CD4+ T helper cells in humans augment the production of pathogenic anti-DNA autoantibodies associated with lupus nephritis. J. Immunol. 143, 103–112 [PubMed] [Google Scholar]
- 5. Anand A., Dean G. S., Quereshi K., Isenberg D. A., Lydyard P. M. (2002) Characterization of CD3+CD4−CD8− (double-negative) T cells in patients with systemic lupus erythematosus: activation markers. Lupus 11, 493–500 [DOI] [PubMed] [Google Scholar]
- 6. Kioussis D., Ellmeier W. (2002) Chromatin and CD4, CD8a and CD8b gene expression during thymic differentiation. Nat. Rev. Immunol. 2, 909–919 [DOI] [PubMed] [Google Scholar]
- 7. Kappes D. J., He X., He X. (2005) CD4-CD8 lineage commitment: an inside view. Nat. Immunol. 6, 761–766 [DOI] [PubMed] [Google Scholar]
- 8. Hostert A., Tolaini M., Festenstein R., McNeill L., Malissen B., Williams O., Zamoyska R., Kioussis D. (1997) A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J. Immunol. 158, 4270–4281 [PubMed] [Google Scholar]
- 9. Kieffer L. J., Yan L., Hanke J. H., Kavathas P. B. (1997) Appropriate developmental expression of human CD8β in transgenic mice. J. Immunol. 159, 4907–4912 [PubMed] [Google Scholar]
- 10. Ellmeier W., Sunshine M. J., Losos K., Hatam F., Littman D. R. (1997) An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 [DOI] [PubMed] [Google Scholar]
- 11. Ellmeier W., Sunshine M. J., Losos K., Littman D. R. (1998) Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity 9, 485–496 [DOI] [PubMed] [Google Scholar]
- 12. Ellmeier W., Sunshine M. J., Maschek R., Littman D. R. (2002) Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity 16, 623–634 [DOI] [PubMed] [Google Scholar]
- 13. Garefalaki A., Coles M., Hirschberg S., Mavria G., Norton T., Hostert A., Kioussis D. (2002) Variegated expression of CD8α resulting from in situ deletion of regulatory sequences. Immunity 16, 635–647 [DOI] [PubMed] [Google Scholar]
- 14. Hostert A., Tolaini M., Roderick K., Harker N., Norton T., Kioussis D. (1997) A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity 7, 525–536 [DOI] [PubMed] [Google Scholar]
- 15. Hostert A., Garefalaki A., Mavria G., Tolaini M., Roderick K., Norton T., Mee P. J., Tybulewicz V. L., Coles M., Kioussis D. (1998) Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity 9, 497–508 [DOI] [PubMed] [Google Scholar]
- 16. Kieffer L. J., Bennett J. A., Cunningham A. C., Gladue R. P., McNeish J., Kavathas P. B., Hanke J. H. (1996) Human CD8α expression in NK cells but not cytotoxic T cells of transgenic mice. Int. Immunol. 8, 1617–1626 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X. L., Seong R., Piracha R., Larijani M., Heeney M., Parnes J. R., Chamberlain J. W. (1998) Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8α at double- and single-positive stages of thymic development. J. Immunol. 161, 2254–2266 [PubMed] [Google Scholar]
- 18. Zhang X. L., Zhao S., Borenstein S. H., Liu Y., Jayabalasingham B., Chamberlain J. W. (2001) CD8 expression up to the double-positive CD3Low/Intermediate stage of thymic differentiation is sufficient for development of peripheral functional cytotoxic T lymphocytes. J. Exp. Med. 194, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feik N., Bilic I., Tinhofer J., Unger B., Littman D. R., Ellmeier W. (2005) Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J. Immunol. 174, 1513–1524 [DOI] [PubMed] [Google Scholar]
- 20. Cohen P. L., Eisenberg R. A. (1991) Lpr and gld: single gene models of systemic autoimmunity and lymphoproliferative disease. Annu. Rev. Immunol. 9, 243–269 [DOI] [PubMed] [Google Scholar]
- 21. Gonzalez-Quintial R., Lawson B. R., Scatizzi J. C., Craft J., Kono D. H., Baccala R., Theofilopoulos A. N. (2011) Systemic autoimmunity and lymphoproliferation are associated with excess IL-7 and inhibited by IL-7Rα blockade. PLoS ONE 6, e27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holzelova E., Vonarbourg C., Stolzenberg M. C., Arkwright P. D., Selz F., Prieur A. M., Blanche S., Bartunkova J., Vilmer E., Fischer A., Le Deist F., Rieux-Laucat F. (2004) Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 351, 1409–1418 [DOI] [PubMed] [Google Scholar]
- 23. Notarangelo L. D. (2009) Primary immunodeficiencies (PIDs) presenting with cytopenias. Hematology Am. Soc. Hematol. Educ. Program 2009, 139–143 [DOI] [PubMed] [Google Scholar]
- 24. Oliveira J. B., Bleesing J. J., Dianzani U., Fleisher T. A., Jaffe E. S., Lenardo M. J., Rieux-Laucat F., Siegel R. M., Su H. C., Teachey D. T., Rao V. K. (2010) Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): report from the 2009 NIH International Workshop. Blood 116, e35–e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teachey D. T., Seif A. E., Grupp S. A. (2010) Advances in the management and understanding of autoimmune lymphoproliferative syndrome (ALPS). Br. J. Haematol. 148, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sneller M. C., Dale J. K., Straus S. E. (2003) Autoimmune lymphoproliferative syndrome. Curr. Opin. Rheumatol. 15, 417–421 [DOI] [PubMed] [Google Scholar]
- 27. Bristeau-Leprince A., Mateo V., Lim A., Magerus-Chatinet A., Solary E., Fischer A., Rieux-Laucat F., Gougeon M. L. (2008) Human TCR-α/β+ CD4−CD8− double-negative T cells in patients with autoimmune lymphoproliferative syndrome express restricted Vβ TCR diversity and are clonally related to CD8+ T cells. J. Immunol. 181, 440–448 [DOI] [PubMed] [Google Scholar]
- 28. Rauen T., Hedrich C. M., Tenbrock K., Tsokos G. C. (2013) cAMP responsive element modulator: a critical regulator of cytokine production. Trends Mol. Med. 19, 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Solomou E. E., Juang Y. T., Gourley M. F., Kammer G. M., Tsokos G. C. (2001) Molecular basis of deficient IL-2 production in T cells from patients with systemic lupus erythematosus. J. Immunol. 166, 4216–4222 [DOI] [PubMed] [Google Scholar]
- 30. Kawachi Y., Otsuka F., Nakauchi H. (1996) Characterization of the mouse CD8β chain-encoding gene promoter region. Immunogenetics 44, 358–365 [DOI] [PubMed] [Google Scholar]
- 31. Hedrich C. M., Rauen T., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM) α protein signaling mediates epigenetic remodeling of the human interleukin-2 gene: implications in systemic lupus erythematosus. J. Biol. Chem. 286, 43429–43436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hedrich C. M., Crispin J. C., Rauen T., Ioannidis C., Apostolidis S. A., Lo M. S., Kyttaris V. C., Tsokos G. C. (2012) cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. U.S.A. 109, 16606–16611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rauen T., Hedrich C. M., Juang Y. T., Tenbrock K., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM) α protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J. Biol. Chem. 286, 43437–43446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Juang Y. T., Wang Y., Solomou E. E., Li Y., Mawrin C., Tenbrock K., Kyttaris V. C., Tsokos G. C. (2005) Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J. Clin. Invest. 115, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hedrich C. M., Rauen T., Kis-Toth K., Kyttaris V. C., Tsokos G. C. (2012) cAMP-responsive element modulator α (CREMα) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). J. Biol. Chem. 287, 4715–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bender A., Kabelitz D. (1990) CD4−CD8− human T cells: phenotypic heterogeneity and activation requirements of freshly isolated “double-negative” T cells. Cell. Immunol. 128, 542–554 [DOI] [PubMed] [Google Scholar]
- 37. Fischer K., Voelkl S., Heymann J., Przybylski G. K., Mondal K., Laumer M., Kunz-Schughart L., Schmidt C. A., Andreesen R., Mackensen A. (2005) Isolation and characterization of human antigen-specific TCR-αβ+ CD4−CD8− double-negative regulatory T cells. Blood 105, 2828–2835 [DOI] [PubMed] [Google Scholar]
- 38. Hamad A. R., Mohamood A. S., Trujillo C. J., Huang C. T., Yuan E., Schneck J. P. (2003) B220+ double-negative T cells suppress polyclonal T cell activation by a Fas-independent mechanism that involves inhibition of IL-2 production. J. Immunol. 171, 2421–2426 [DOI] [PubMed] [Google Scholar]
- 39. Treiner E., Lantz O. (2006) CD1d- and MR1-restricted invariant T cells: of mice and men. Curr. Opin. Immunol. 18, 519–526 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z. X., Yang L., Young K. J., DuTemple B., Zhang L. (2000) Identification of a previously unknown antigen-specific regulatory T cell and its mechanism of suppression. Nat. Med. 6, 782–789 [DOI] [PubMed] [Google Scholar]
- 41. Gorman S. D., Sun Y. H., Zamoyska R., Parnes J. R. (1988) Molecular linkage of the Ly-3 and Ly-2 genes. Requirement of Ly-2 for Ly-3 surface expression. J. Immunol. 140, 3646–3653 [PubMed] [Google Scholar]
- 42. Jarry A., Cerf-Bensussan N., Brousse N., Selz F., Guy-Grand D. (1990) Subsets of CD3+ (T cell receptor α/β or γ/δ) and CD3− lymphocytes isolated from normal human gut epithelium display phenotypical features different from their counterparts in peripheral blood. Eur. J. Immunol. 20, 1097–1103 [DOI] [PubMed] [Google Scholar]
- 43. Lefrancois L. (1991) Phenotypic complexity of intraepithelial lymphocytes of the small intestine. J. Immunol. 147, 1746–1751 [PubMed] [Google Scholar]
- 44. Vremec D., Zorbas M., Scollay R., Saunders D. J., Ardavin C. F., Wu L., Shortman K. (1992) The surface phenotype of dendritic cells purified from mouse thymus and spleen: investigation of the CD8 expression by a subpopulation of dendritic cells. J. Exp. Med. 176, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]