Background: Despite analyses of broadly neutralizing anti-HIV-1 antibodies directed against the gp41 MPER segment, there exists a paucity of structural information on MPER immunogenicity.
Results: Immunodominance of Trp-680 in the MPER arrayed on liposomes is modified by membrane anchoring.
Conclusion: Immunogenicity is manipulatable through subtle structural modification.
Significance: Learning about the structural basis of immunogenicity is critical for eliciting desired B cell antibody production through vaccination.
Keywords: Antibodies, HIV-1, Infectious Diseases, Protein Structure, Vaccine Development, Immunodominance, Immunogenicity
Abstract
Structural characterization of epitope-paratope pairs has contributed to the understanding of antigenicity. By contrast, few structural studies relate to immunogenicity, the process of antigen-induced immune responses in vivo. Using a lipid-arrayed membrane-proximal external region (MPER) of HIV-1 glycoprotein 41 as a model antigen, we investigated the influence of physicochemical properties on immunogenicity in relation to structural modifications of MPER/liposome vaccines. Anchoring the MPER to the membrane via an alkyl tail or transmembrane domain retained the MPER on liposomes in vivo, while preserving MPER secondary structure. However, structural modifications that affected MPER membrane orientation and antigenic residue accessibility strongly impacted induced antibody responses. The solvent-exposed MPER tryptophan residue (Trp-680) was immunodominant, focusing immune responses, despite sequence variability elsewhere. Nonetheless, immunogenicity could be readily manipulated using site-directed mutagenesis or structural constraints to modulate amino acid surface display. These studies provide fundamental insights for immunogen design aimed at targeting B cell antibody responses.
Introduction
Neutralizing antibodies bind to viral surface components involved in target cell attachment and fusion, interfering with virus entry (1). In the case of RNA viruses, antibodies are often highly strain-specific due to substantial sequence variability in the epitope regions. Unsurprisingly, broadly neutralizing antibodies (BNAb)6 are rarely elicited during the natural course of HIV-1 and influenza infections or by vaccination (2–5). Nonetheless, BNAbs against HIV-1 that neutralize multiple viral strains develop in about 20% of infected individuals, arising 2–4 years after initial HIV-1 infection (6, 7). Although these BNAbs are unusual, with long CDRH3 loops and/or extensive somatic hypermutation, their presence in infected individuals demonstrates that the immune system is capable of generating antibodies against conserved antigenic sites (8).
Understanding the structural and genetic heterogeneity of HIV-1 envelope protein antigens and the corresponding response pathways of B cell expansions is important for engineering vaccines capable of generating BNAbs (8–10). Most antibodies elicited by current vaccines or natural infection are non-neutralizing, stimulated by envelope proteins that are monomeric, denatured, degraded, or otherwise processed and non-native (11). Nonprotective antibodies are directed against epitopes whose ligation does not interfere with viral attachment or entry. Moreover, the poor immunogenic nature of conserved neutralizing determinants requires immune engineering to present or re-surface those epitopes to focus B cell responses away from immunodominant regions toward the conserved neutralizing determinants. Structural analysis of BNAb-antigen complexes allows for rational immunogen designs as a basis to achieve selective antibody responses directed to pre-determined functional epitopes (12–15).
Immunogenicity, the ability of antigen to elicit an immune response in vivo, involves cooperative biological interactions determined by the biophysical properties of the antigen and extrinsic host cellular factors, such as the frequency of epitope-specific germ line-encoded immunoglobulin variable regions in the B cell repertoire, tolerance mechanisms, affinity maturation, etc. (16–19). The immunogenicity of an epitope is dependent upon its context in a protein. Mutations in one segment can redirect B cell responses toward otherwise weakly immunogenic regions (20). Furthermore, the functionality of antibodies induced by West Nile virus is also strongly influenced by the quaternary structure of viral antigens, suggesting that the nature of the epitope in the native protein is key for interaction with B cell receptors (BCRs) (21). Although antigenicity has been previously predicted based on hydrophilicity, mobility of backbone atoms, accessibility, topology, and protein flexibility (see Refs. 22, 23 and references therein), the robustness of these correlations varies among proteins. Therefore, although the discipline of protein design is rapidly evolving, knowledge of the physicochemical and structural properties that determine antigen immunogenicity coupled with complementary experimental approaches needs to be obtained.
To date, no thorough examination has been performed at both molecular and immunological levels to assess quantitative and/or qualitative differences in immunogenicity. Here, we investigate the influence of antigenic structure on vaccine-induced antibody responses using the membrane-proximal external region (MPER) of the HIV-1 gp41 subunit as a model antigen. The MPER is a highly conserved tryptophan-rich hydrophobic segment (residues 662–683, according to HxB2 numbering) important for viral fusion (24). This region lies at the base of the gp41 ectodomain, immediately proximal to the transmembrane domain (TM). Unlike many BNAbs binding to discontinuous epitopes, the 2F5, 4E10, 10E8, and Z13el BNAbs recognize juxtaposed linear epitope residues in the MPER (25–28). The structure of the MPER, two helices connected to one another via a flexible hinge, is a unique feature formed in the membrane environment (29). In this study, we show how the membrane orientation of the MPER segment per se significantly impacts immunogenicity, including epitope immunodominance. Our observations have implications for therapeutic and preventive vaccine design.
EXPERIMENTAL PROCEDURES
Synthesis of MPER and Palmitoylated MPER Peptides
Various MPER sequences were synthesized according to standard procedures on an ABI 431 Peptide Synthesizer using Fmoc chemistry as follows: synthesis of HxB2 Npalm-MPER (ELDKWASLWNWFNITNWLWYIK), 089CON Npalm-MPER (ALDSWKNLWSWFSITNWLWYIK), and PB7 Npalm-MPER(ALDKWNSLWSWFDITKWLWYIK) from Tufts University (Boston, MA) and HxB2 MPER-Cpalm (ELDKWASLWNWFNITNWLWYIK), MPERkk-Cpalm (ELDKWASLWNWFNITNWLWYIKKK), and MPER-TM(ELDKWASLWNWFNITNWLWYIKIFIIIVGGLVGLRIVFAVLSIV) from Massachusetts Institute of Technology (Cambridge, MA). HPLC purification was performed on a reverse phase C18 or C5 column. Palmitic acid was conjugated to the N terminus of MPER (Npalm-MPER) overnight using the standard 2-(1H-benzotriazole-1-yl)1,1,3,3-tetramethyluronium activation.
To synthesize MPER coupled with palmitic acid at the C terminus (MPER-Cpalm), lysine pre-modified with palmitic acid at ϵ-amine (Fmoc-Lys(palmitoyl)-OH; Bachem, Torrance, CA) was used during peptide synthesis.
Preparation of MPER/Liposome Vaccine
To prepare standard liposomes, lipids in chloroform (typical lipid composition: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol)(DOPG)/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000])/monophosphoryl lipid A (MPLA)/MPER) or palmitoylated MPER = 72:18:10:0.4:0.5 molar ratio (lipids from Avanti Polar Lipids, Alabaster, AL, and MPLA from Sigma) were dispensed to glass vials, and the organic solvents were evaporated under vacuum overnight, resulting in dried thin lipid films. The lipid films were rehydrated with 1 mg/ml of LACK-1 (SPSLEHPIVVSGSWD) CD4 helper peptides at a lipid concentration of 20 mg/ml in PBS for 1 h with rigorous vortexing every 10 min, subjected to six freeze-thawing cycles (freezing in liquid nitrogen and thawing at 60 °C), and then sonicated (Misonix Microson XL probe tip sonicator, Farmingdale, NY) in alternating power cycles of 12 and 3 watts in 30-s intervals for 5 min on ice. In some experiments, MPER was loaded only on the external surface of liposomes by simply mixing MPER with liposomes pre-formed in the absence of MPER in a molar ratio of 1:100 = MPER/total lipid. In some experiments, the liposomes were extruded 11 times through polycarbonate membranes with 50-, 100-, 200-, or 400-nm pores. To synthesize liposomes encapsulating PADRE (AKXVAAWTLKAAA, where X is cyclohexylalanine) instead of LACK-1, PADRE was loaded in liposomes by the standard procedure of remote loading using ammonium sulfate gradient (30). The average size of standard liposomes used in immunization studies was 77 ± 12 nm with polydispersity index of 0.17 ± 0.093 as determined by dynamic light scattering using a 90Plus/ZetaPals particle size analyzer (Brookhaven Instruments).
Preparation of Liposomes for EPR and SPR
Lipids were mixed in chloroform and dried as thin films under a nitrogen gas stream. To remove residual organic solvent, the lipid films were further dried by vacuum pump for ≈16 h. The lipids were resuspended in 20 mm HEPES and 150 mm KCl, pH 7.0, and subjected to 10–15 freeze-thaw cycles, followed by extrusion 15 times through two sheets of polycarbonate membrane with a pore size of 100 nm (Avanti Polar Lipids). Vesicles with virion membrane mimic were prepared at the molar ratio 9:18:20:9:45 of dioleoylphosphatidylcholine/sphingomyelin/dioleoylphosphatidylethanolamine/dioleoylphosphatidylglycerol/cholesterol (Avanti Polar Lipids). 1-Palmitoyl-2-oleoylphosphatidylcholine/1-palmitoyl-2-oleoylphosphatidylglycerol large unilamellar vesicles at a 4:1 molar ratio were used for EPR power saturation measurements. 1,2-Dioleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-snglycero-3-phospho-(1′-rac-glycerol) (DOPC/DOPG) large unilamellar vesicles at a 4:1 ratio were used for BIAcore analysis.
Electron Paramagnetic Resonance (EPR) Spectroscopy
The EPR experiments were carried out as described previously (31). To obtain EPR signals, peptides containing single cysteine substitutions were labeled with (1-oxyl-2,2,5,5,-tetramethylpyrroline-3-methyl)-methanethiosulfonate (Toronto Research Chemicals (Ontario, Canada)) as follows. 5 mg of synthetic peptides were dissolved in 150 μl of dimethyl sulfoxide (DMSO). Spin labeling was then performed by adding 3–5-fold excess of (1-oxyl-2,2,5,5,-tetramethylpyrroline-3-methyl)-methanethiosulfonate and incubating ∼16 h at room temperature. Peptides were purified and separated from free spin labels by reverse phase HPLC using a C4 or C5 column (Sigma). EPR spectra were recorded on a Bruker EMX spectrometer (Billerica, MA) at 2-milliwatt incident microwave power with a field modulation of 1.0–2.0 G at 100 kHz using a Bruker high sensitivity resonator. To measure immersion depths, solvent accessibility and power saturation measurements were performed on a loop-gap resonator (Molecular Specialties, Milwaukee, WI) with microwave power varied from 0.4 to 100 milliwatts. Samples were placed in gas-permeable TPX tubes (Molecular Specialties) and purged by either a stream of air or nitrogen gas. The immersion depth values were calculated by the ratio of the accessibility values of O2 to 50 mm nickel(II) ethylenediaminediacetic acid. Depth standard curves were determined using lipid vesicles containing trace amounts of spin-labeled lipids (1:500 by weight) as described (29).
Nuclear Magnetic Resonance (NMR)
NMR structural determination of HxB2, Con089A, and PB7 MPER peptides in dodecylphosphocholine micelles was carried out as described previously (29). Backbone amide and carbon chemical shift assignments for MPERTM peptide in lyso-myristoylphosphatidylglycerol micelles were completed using a 1 mm 15N-13C double-labeled sample with conventional NMR triple-resonance experiment data obtained on a Bruker (Billerica, MA) 750-MHz spectrometer. The preliminary MPERTM structural model was constructed using the software CYANA (32), based on three-dimensional 15N NOESY data and backbone dihedral angles predicted by software TALOS+ (33), and EPR membrane immersion depth restraints of the MPERTM and hypothetical immersion depths adopted from the N-terminal region of the HxB2 MPER peptide.
ELISA
A 96-well plate is coated with 50 μl of MPER peptide (2 μg/ml) or streptavidin (2 μg/ml) (Sigma S0677) in phosphate-buffered saline (PBS) overnight. The plates were washed with PBS containing 0.1% BSA three times and then blocked with 100 μl of 1% BSA in PBS/well at room temperature for 3 h. After incubation with 50 μl of biotinylated MPER peptides diluted in PBS, 1% BSA at 2 μg/ml for 2 h at room temperature, and an additional 2 h at 4 °C, pre- and post-immune sera serially diluted in PBS, 1% BSA were added to the plate for overnight incubation. The next day, the plates were washed three times with PBS, 0.1% BSA and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at 4 °C. These plates were then washed four times with PBS and developed with 50 μl of 3,3-tetramethylbenzidine substrate (SureBlue Reserve, Gaithersburg, MD)/well. The HRP reaction was stopped with 50 μl of 1 m HCl/well, and the well absorbance was determined at 490 nm.
Monoclonal Antibody Production
MPER-specific monoclonal antibodies were produced by fusion of murine myeloma cells (NS0 BCL2) with mesenteric lymph node cells from a mouse immunized with Npalm-7-MPER/liposome. Briefly, mice were immunized with Npalm-7-MPER/liposome three times every 3 weeks apart. 4 days after final immunization, mesenteric lymph nodes were isolated, and a single cell suspension was obtained. Electrofusion was carried out with the ratio of one mesenteric lymph node cell to two myeloma fusion partners, and the fused cell suspension was placed in culture at 1000 cells/well in 96-well plates (34). Twelve days after fusion, we harvested supernatants and screened for MPER reactivity by ELISA. The MPER-reactive wells were initially subcloned at 10 cells/well and followed by 3 cells/well and then 0.3 cells/well. The anti-MPER monoclonal antibodies were purified from supernatants of the subcloned hybridoma using protein G column.
Animals and Immunizations
BALB/c mice were purchased from Taconic and housed in the Vivarium at Dana-Farber Cancer Institute Animal Core Facility. Animals were studied under a protocol approved by the Animal Use and Care Committee of Dana Farber Cancer Institute. Mice (four or five animals per immunogen group) were immunized in the flanks of both thighs intradermally three times every 3–4 weeks with 50 μl/injection of MPER/liposome vaccine at the concentration of 20 mg/ml. Serum samples were collected 10 days after each immunization and stored at −20 °C until use.
Histology
To analyze the distribution of MPER and liposomes in draining lymph nodes, C57BL/6 mice were sacrificed 2 and 24 h after intradermal injection of rhodamine-labeled liposomes containing either FITC-labeled MPER peptide or FITC-labeled MPER peptide covalently attached to palmitic acid at the C terminus (50-μl injection at 20 mg/ml). Fluorophore-labeled liposomes were prepared from lipid films containing rhodamine-labeled 2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) and the standard lipid components in a 0.2:100 molar ratio, and rhodamine-liposome was mixed with FITC-labeled MPER peptides at 100:1 molar ratio. Inguinal lymph nodes were removed and embedded in TissueTek OCT compound (Fisher) by flash-freezing in 2-methylbutane chilled with liquid nitrogen. Sections (5 μm) were prepared using a cryostat and positively charged slides. After air-drying for 2 h, sections were fixed with 1:1 acetone/methanol for 10 min at −20 °C. Sections were blocked for 15 min with PBS containing 0.5% BSA, 5 μg/ml Fc block (BD Biosciences), and 5% rat serum. Sections were then labeled with biotin B220 and APC-conjugated TCRβ monoclonal antibodies (eBioscience). Biotinylated antibodies were detected with AlexaFluor 405-conjugated streptavidin (Invitrogen). Slides were then cover-slipped with Fluoromount-G (Southern Biotech) and left to dry overnight. Sections were imaged using a Leica SP5 confocal microscope (×10 objective, NA, 0.4; ×100 objective with oil, NA, 1.4) with LAS AF (version 2.6.0) software. Images were processed and merged in Adobe Photoshop.
SPR Measurements
BIAcore experiments were carried out with a BIAcore 3000 with the Pioneer L1 sensor chip at 25 °C. Data analyses were performed using BIAevaluation 3.1 software (BIAcore). The running buffer was 20 mm HEPES containing 0.15 m NaCl, pH 7.4 (HBS-N). The DOPC/DOPG liposome (30 μl, 150–250 μm) was applied to the sensor chip surface at a flow rate of 3 μl/min, and the liposomes were captured on the surface of the sensor chip and provided a supported lipid bilayer. To remove any multilamellar structures from the lipid surface, sodium hydroxide (20 μl, 25 mm) was injected at a flow rate of 100 μl/min, which resulted in a stable base line corresponding to the immobilized liposome bilayer membrane with response units of 4500–6000. For epitope mapping of polyclonal anti-MPER antibodies, polyclonal IgG was purified from immune sera using GammaBind Plus (GE Healthcare)-Sepharose column. Serial single alanine mutations were made to various parental MPER sequences used for immunization, respectively. MPER variant peptide solutions (0.5 μm) were prepared by dissolving in running buffer right before the injection, and the solution (60 μl) was injected over the lipid surface at a flow rate of 5 μl/min. Antibody solution (60–80 μg/ml) was passed over the peptide-liposome complex for 3 min at a flow rate of 10 μl/min. The immobilized liposomes were completely removed with an injection of 40 mm CHAPS (25 μl) at a flow rate of 5 μl/min, followed by a 10-μl injection of NaOH (50 mm)/isopropyl alcohol (6:4) at a 20 μl/min flow rate, and each peptide injection was performed on a freshly prepared liposome surface. The amount of each MPER variant peptide bound to DOPC/DOPG liposome was normalized to that of WT peptide. Relative percent binding activity of each purified polyclonal antibody for MPER alanine mutants in comparison with the WT peptide were measured by response units taken at the 3-min dissociation time point. A 3-min association and 3-min dissociation periods were monitored in each data collection sensorgram. More than three independent experiments were carried out to test relative binding reactivity of antibody with each experiment in different concentrations of peptides or antibody.
RESULTS
Configuration of the MPER Segment Embedded on Lipid-coated Vaccine Vehicles
As shown in Fig. 1A, the 22-amino acid MPER segment assumes a well defined helix-hinge-helix structure when embedded on a membrane surface (29). The lipid-immersed MPER structure presumably represents a configuration similar to the one found on HIV-1 viral particles. The BNAbs 4E10 and 2F5 each recognize several surface-exposed MPER residues and subsequently extract their buried core epitope residues from the membrane into their binding pockets (29, 31, 35). The extraction process is facilitated by BNAb interaction with the viral membrane via the tip of an elongated CDR-H3 loop in conjunction with the flexible hinge movement of the MPER segment itself. Given that the capacity of these BNAb to extract epitopic regions from a lipid membrane is crucial to their neutralizing activity, vaccine immunogens that select for antibody with this capacity to extract MPER epitopes are more likely to generate protective humoral immunity. Both antibody interactions with virion membranes and flexible hinge movement of the MPER are key extraction parameters. Because the helix-hinge-helix structure of the MPER is configured only in the membrane environment, stealth liposomes as well as lipid-coated poly(lactide-co-glycolide) nanoparticles (36) were employed as candidate vehicles to deliver the MPER peptide. As shown in Fig. 1B, a representative stealth liposome vaccine formulation used for immunogenicity studies consisted of the CD4 helper T cell epitope LACK(159–173) peptide from Leishmania major (37) encapsulated in the aqueous particle interior: 1 mol % MPLA, a TLR-4 agonist incorporated in the vesicle bilayer; and 10 mol % polyethylene glycol (PEG)-2000 lipid. The latter reduces nonspecific adsorption of the vesicles to the extracellular matrix, thereby efficiently accelerating the drainage of liposomes to the lymph node (38).
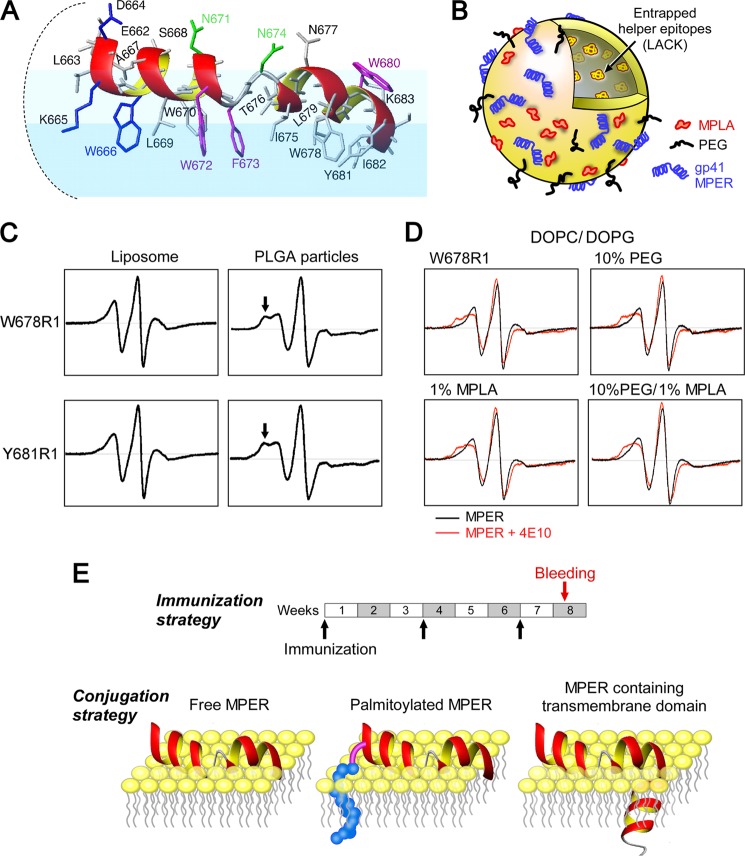
FIGURE 1.
Structural configuration of MPER segments in liposome vaccines. A, NMR structure of the HxB2 MPER in a virion mimic membrane surface. Residues essential for BNAb neutralization are color-coded as follows: blue for 2F5, green for Z13e1, and magenta for 4E10. B, schematic of a standard liposome vaccine formulation, including the various components used in immunization studies. C, comparison of EPR spectra of spin-labeled MPER Trp-678(R1) and Tyr-681(R1) peptides on liposomes versus poly(lactide-co-glycolide) (PLGA) particles. Arrows in the latter highlight immobilization. D, EPR spectra of a spin-labeled MPER Trp-678(R1) peptide bound to liposomes with varied PEG and MPLA compositions. Spectra were obtained in the absence (black) and presence (red) of 4E10. E, immunization schedule and various conjugation strategies employed for MPER display on the surface of the liposome vaccines. For clarity, only the outer leaflet of the membrane is shown.
First, we used site-directed spin labeling and EPR spectroscopy to monitor conformational changes in the MPER arrayed on the surface of vaccine vehicles. We investigated the mobility features of methanethiosulfonate spin probes covalently attached to individual cysteine residue (R1) replacing MPER residues. Immobilized components of the spectra were evident from the designated spectral peak of spin-labeled residues Trp-678(R1) and Tyr-681(R1) (arrow), respectively, in the poly(lactide-co-glycolide) particles preparation compared with that of the liposomes (Fig. 1C). Such immobility was clear in the spectra of Phe-673(R1), Ile-675(R1), and Asn-677(R1) as well (data not shown). This reduced mobility highlights an altered structural property of the MPER peptides when it is arrayed on the lipid-coated nanoparticles. Because conformational flexibility of MPER is important for 4E10 epitope extraction, the reduced mobility of core epitope residues (Phe-673 and Ile-675) when the MPER is placed on the surface of lipid-coated nanoparticles may hamper selection for 4E10-like BNAb responses. Therefore, liposomes were chosen as the delivery vehicle.
Next, to examine the impact of PEG and MPLA surface modifications of liposomes on MPER mobility and 4E10-induced conformational changes, a representative spin-labeled MPER peptide (Trp-678(R1)) was adsorbed to liposomes (1:100 ratio, mol/mol), containing 10% PEG-2000 and/or 1% MPLA on the liposome surface. Comparable EPR spectra in the absence and presence of 4E10 were observed for all liposomes (Fig. 1D), indicating that the MPER configuration and its antigenic features in lipids were not affected by PEG and MPLA. The immunization schedule and peptide modifications, including palmitoylation and TM segment addition used to anchor the MPER to the liposome surfaces, are shown in Fig. 1E.
Enhanced Immunogenicity through Covalent MPER Attachment to Lipid
Given that multiple MPER hydrophobic residues were shown to be embedded into the acyl chain of lipids in our structural studies (Fig. 1A), initial pilot immunizations were carried out with “free” MPER adsorbed onto the surface of liposome vaccines (1:100 ratio, mol/mol) in the absence of peptide lipophilic modifications. Note that when noncovalent binding of MPER 678R1 peptide to liposomes (100 nm) at various peptide to lipid molar ratios was titrated, maximal density of peptides on the liposome surface was achieved at a peptide to lipid ratio of 1:25 (data not shown) based on the measurements of peak intensity of Trp-678(R1) spectra by EPR. Immune sera were collected from BALB/c mice immunized intradermally with the MPER/liposome preparation following the second boost. MPER antibody titers were measured by ELISA against MPER peptide either directly coated onto plastic, or alternatively, N-terminally biotinylated MPER peptide was bound to streptavidin-coated plates. ELISA titrations revealed that MPER immune sera preferentially recognized directly coated MPER peptide (MPER plate) compared with biotin-MPER bound to streptavidin (biotin-MPER plate) (Fig. 2). Of note, 4E10 is sensitive to MPER configuration, being significantly more reactive with the biotin-MPER plate, whereas 2F5 reactivity is comparable for both types of antigen. The reduced 4E10 binding suggests that the random hydrophobic interaction between MPER and the plate alters MPER structure and/or occludes 4E10 binding to hydrophobic core epitope residues. The results indicated that a majority of antibodies in immune sera recognize undesirable MPER configurations, distinct from that detected by 4E10.
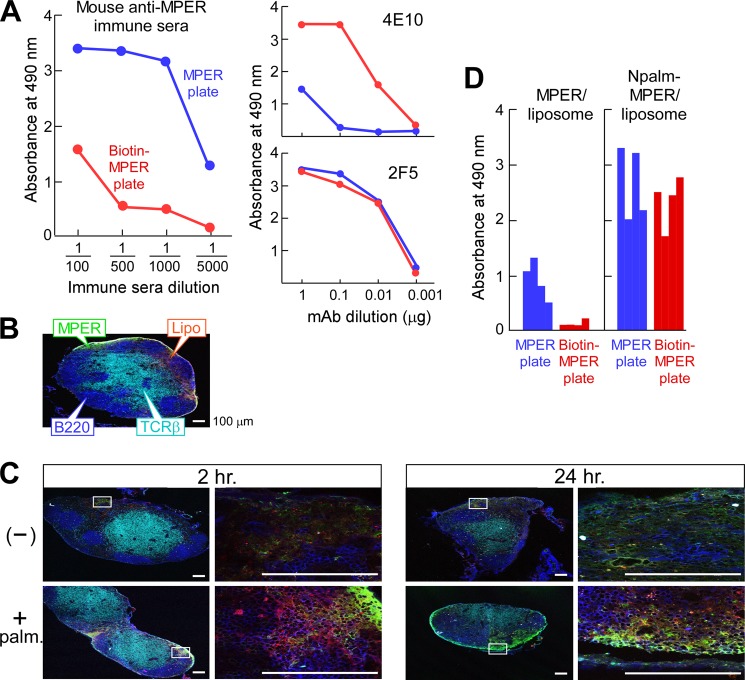
FIGURE 2.
Enhanced immunogenicity of the MPER through covalent attachment to lipid. A, serum IgG responses of a representative BALB/c mouse immunized with noncovalently attached MPER/liposome. Anti-MPER-specific IgG in the sera was determined using ELISA plates either directly coated with MPER (blue) or via biotin-MPER peptide bound to a streptavidin-coated plate (red). mAbs 4E10 and 2F5 were separately assessed as positive controls. B, localization of the palmitoylated MPER/liposome in inguinal LN 2 h post-injection. C57BL/6 mice were injected with FITC-labeled palmitoylated MPER/rhodamine-labeled liposomes. T and B cell areas were identified by anti-TCRβ (H57 mAb) (cyan) and anti-B220 (blue), respectively. C, co-localization of MPER and liposome. The association of FITC-MPER with rhodamine-liposome (top row, −) or of palmitoylated FITC-MPER with rhodamine-liposome (bottom row, + palm) in inguinal LN was monitored by confocal microscopy at 2 and 24 h after injection. Scale bars, 100 μm. D, enhanced serum IgG responses specific to MPER via immunization with palmitoylated MPER/liposome compared with free MPER/liposome. The anti-MPER specific IgG in a 1:5000 dilution of the sera from BALB/c mice (n = 4) immunized with MPER/liposome or Npalm-MPER/liposome was determined by ELISA using a MPER plate (blue) or biotin-MPER plate (red).
To improve MPER presentation on the liposome surface, the MPER was anchored by palmitic acid. The impact of this modification in the vaccine was then tested to assess the association between liposome and MPER in vivo. Fluorescent liposomes containing rhodamine-labeled lipids mixed with FITC-labeled free MPER or palmitoylated MPER were injected intradermally over the thigh; post-immunization, we monitored MPER association with the injected liposomes in the draining inguinal lymph node (LN) by microscopic examination of cryosections. T and B cell zones were visualized with fluorochrome-labeled antibodies specific for TCRβ (H57 mAb) and B220, respectively (Fig. 2B). Histological analyses of liposomes adsorbed with free MPER revealed diffuse FITC (MPER) and rhodamine (liposome) staining in the subcapsular region by 2 h post-injection (Fig. 2C). Although the FITC signal at this time was largely confined to the areas directly beneath the subcapsular sinus, the rhodamine signal was dispersed more deeply into the LN, suggesting dissociation of MPER and liposome. After the injection of palmitoylated MPER/liposomes, the FITC signal was more intense at 2 h (Fig. 2, B and C) and was also evident at 24 h, suggesting that the FITC and rhodamine signals are more closely associated with palmitoylated MPER than the noncovalently adsorbed MPER. Nonetheless, even with palmitoylated MPER liposome, some areas within the LN show distinct FITC or rhodamine fluorescence, indicating dissociation.
These results demonstrate that the adsorbed free MPER is readily dissociated from liposomes during their passage into regional LNs compared with MPER adducted with a lipid anchor. This dissociation of the MPER from liposomes may correlate with weaker antibody responses and the generation of disfavored forms of MPER antibodies, i.e. those that cannot extract MPER determinants buried in lipid membranes. Consistent with this notion, immunizations with Npalm-MPER yielded significantly higher titers of MPER antibodies than did adsorbed MPER (Fig. 2D). In addition, MPER antibodies elicited by Npalm-MPER reacted significantly better with biotin-MPER on streptavidin plates than those elicited by absorbed MPER. This change in specificity suggests that stability of the helix-hinge-helix MPER structure is conferred by lipophilic anchoring of the immunogen to the liposome in vivo, facilitating extraction of BNAb epitope residues by flexible hinge movement.
Optimization of Palmitic Acid Adduction, Liposome Formulation, and Antigenicity
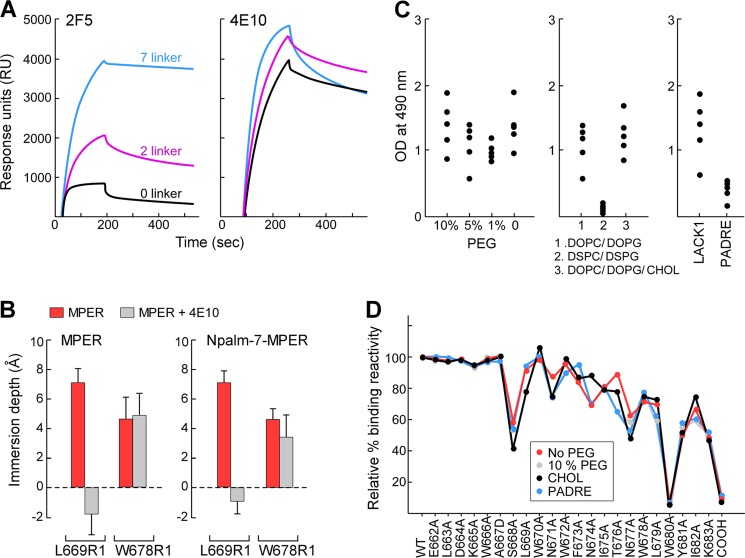
Because acyl modification of the MPER improved the desirable antibody response to the Npalm-MPER/liposome immunogen, we examined the effects of immunogen modifications on antigenicity and immunogenicity in greater detail. Three different amino acid spacer sequences were introduced between the lipid anchor and the MPER peptide. Their influence on antigenicity as well as MPER structural configuration on liposomes was determined by BNAb binding measurements and by EPR. Although 4E10 binding was not affected by lipid attachment to the MPER in DOPC/DOPG membranes, as shown by SPR, 2F5 reactivity was significantly dependent upon spacer length. Maximal 2F5 binding was achieved when a 7-residue glycine/serine spacer was inserted in comparison with placement of a short 2-residue spacer or spacer elimination (0 residue) (Fig. 3A). Given that the N-terminal region of the MPER is tilted ∼15° upward relative to the membrane surface (29) and 2F5 binding requires extraction of epitope residues out of the membrane (35), the result suggests that the structural constraint imposed by lipid attachment on the mobility of the MPER N-terminal region is alleviated by the 7-residue linker.
FIGURE 3.
Effect of biophysical properties of Npalm-7-MPER/liposome on immunogenicity. A, antigenicity of Npalm-MPER antigens with various linker residues. The binding reactivity of 2F5 and 4E10 was measured for Npalm-MPER peptides in DOPC/DOPG membrane by Biacore. B, comparison of membrane immersion depth values of MPER and Npalm-7-MPER peptides in the presence and absence of 4E10. Depth values between −2 to 0 Å and larger than 0 Å correspond to the lipid headgroup and acyl chain region, respectively. C, effect of PEG density (left), lipid compositions (middle), and helper peptides (right) on MPER-specific IgG responses. BALB/c mice (n = 5) were immunized three times with various MPER/liposome vaccines, and the anti-MPER-specific IgGs in a 1:5000 dilution of the sera were determined in an ELISA on biotin-MPER plate. D, immunogenicity of C variants by epitope map analysis relative to the standard liposome vaccine containing 10% PEG. The x axis labels WT and 23 variant peptides tested. The single mutation converts each residue to alanine, except the 667 mutated to aspartic acid and the C-terminal amide to carboxylate (COOH).
EPR analysis was performed to assess changes in MPER orientation that were induced by the 7-residue linker and lipid modification in Npalm-MPER. To this end, two residues deeply buried in the acyl chain region of the lipid bilayer in membrane-adsorbed MPER, Leu-669 and Trp-678, were chosen as reference residues (29). The membrane immersion depths of spin-labeled Leu-669(R1) and Trp-678(R1) in Npalm-MPER were comparable with those of Leu-669(R1) and Trp-678(R1) in the free MPER segment adsorbed on the surface of the membrane (Fig. 3B). Consistent with 4E10-induced depth change of Leu-669(R1) previously observed, Leu-669(R1) was lifted out of the membrane and exposed to the aqueous phase upon 4E10 binding to Npalm-MPER (Fig. 3B). Trp-678(R1) in Npalm-MPER was slightly raised in the acyl chain region compared with that in MPER upon 4E10 binding. Comparable EPR spectral mobility of Npalm-MPER and MPER residues in the presence and absence of antibody is also indicative of similar conformation (data not shown), suggesting that MPER orientation is preserved on the surface of Npalm-7-MPER (7 spacer)/liposome.
Using Npalm-7-MPER peptide as a model antigen, we examined the impact on immunogenicity of varying biophysical properties of liposomes such as lipid composition, fluidity, and surface PEG density. MPER-specific IgG responses were significantly affected by lipid composition. The MPER in DSPC/DSPG was poorly immunogenic compared with that in DOPC/DOPG or DOPC/DOPG/cholesterol. However, no significant differences in the magnitude of MPER-specific immune responses were observed in the sera of mice immunized with 10% versus 0% PEG-liposome formulations. In addition, the I-Ad binding LACK1 peptide provided better CD4 T cell help in BALB/c mice than the universal T cell epitope PADRE (Fig. 3C). Notwithstanding, single residue MPER alanine-scanning mutagenesis coupled with Biacore analysis using purified polyclonal IgG from each immune sera showed that antibodies with similar distribution of antigenic specificities were engendered, regardless of changes in biophysical properties of the vaccine formulations (Fig. 3D).
Antibody Responses Directed to the C-terminal Region of the MPER
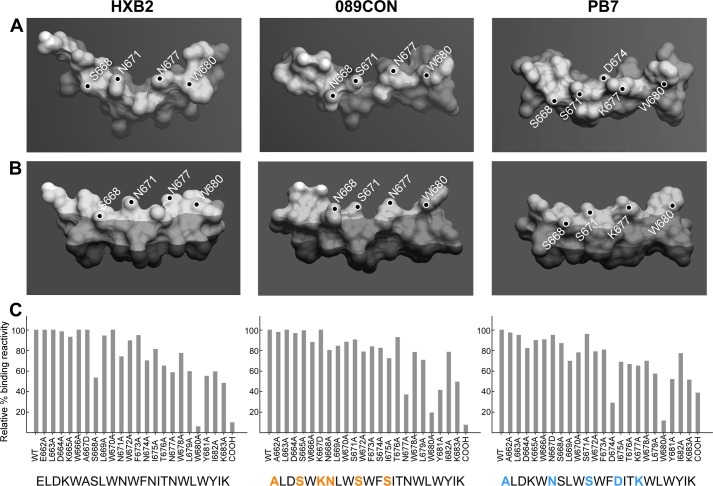
We selected three MPER sequences from two viral clades (B and C) to determine how sequence differences resulting in alterations of overall backbone structure and/or local side chain features would impact immune responses. The structures of HxB2, 089CON, and PB7 MPER segments were modeled on the membrane based on NMR structural determinations through NOE distance constraints combined with EPR membrane immersion depths of each residue in three distinct MPER segments, respectively.7 The mean positions of all three peptides are located in the membrane headgroup region near the aliphatic region interface. Fig. 4A provides the view looking down on the membrane surface from above indicating that all three MPER peptides adopted a segment helix-hinge-helix conformation with varying degrees of an L-shaped bend due to intrinsic flexibility in the central hinge. Fig. 4B shows the side view of each MPER as suspended in the lipid bilayer where the darkest shade represents the membrane aliphatic region; the boundary between medium and light shade marks the phosphates in the headgroup region, and the occasional bright spots indicate complete exposure outside of the membrane. Although there are subtle differences in side chain orientation and membrane immersion depth among surface-exposed and membrane-buried residues, the overall MPER structures from HxB2, 089CON, and PB7 strains were not affected by their sequence.
FIGURE 4.
Stereochemistry and immunogenicity of MPER segments from three different viral strains. A, top, and B, side views of HxB2 (left), 089CON (middle), and PB7 (right) MPER peptides shown as molecular surface representations embedded on viral membrane surfaces. Dark-shaded region indicates acyl chain region of the lipid bilayer, boundary between light- and medium-shaded region marks the position of phosphate in the lipid headgroup area, and the small, unshaded parts indicate complete solvent exposure. Key solvent-exposed residues are labeled. C, fine epitope specificities of MPER responses elicited by three different MPER/liposome vaccines. Single alanine-substituted peptides of each position of the various Npalm-MPER immunogens were tested for their reactivity with the purified polyclonal IgG antibody elicited against the corresponding immunogen as measured by SPR. Sequence variations in 089CON and PB7 MPER compared with HxB2 MPER are color-coded in orange (089CON) and blue (PB7). Data are representative of three independent measurements varying peptide or antibody amounts.
Given these similar structural features, we determined how MPER sequence differences impact immunogenicity by immunizing mice with three different MPER/liposome vaccines using Npalm-7-MPER and the same liposome formulation. Subsequently, purified serum IgG samples were tested by Biacore for binding reactivity to each MPER-alanine mutation relative to that of WT MPER arrayed on the surface of DOPC/DOPG liposomes. Although the Trp-680 residue and the amide adduct at the C terminus of the MPER are essential for anti-HxB2 MPER (6 and 10% wild type) and anti-089CON MPER antibody binding (20 and 8% wild type), anti-PB7 MPER antibody was virtually abolished by W680A mutation (8% wild type) but only diminished by removal of the amide adduct (39% wild type) (Fig. 4C). In addition, PB7 MPER binding was significantly reduced by D674A mutation (28% wild type), whereas the binding of anti-HxB2 MPER antibody was affected by mutations at the surface-exposed S668 (53%) and anti-089CON MPER antibody by Asn-677 (37%). Despite isolated differences, the epitope mapping analysis indicates immunodominance focused toward the C-terminal end of the MPER, most notably on Trp-680, a bulky hydrophobic residue that is fully solvent-exposed (Fig. 4, A and B). Epitope mapping analysis of three anti-HxB2 MPER mAbs generated is consistent with the majority of polyclonal IgG in the HxB2 MPER immune sera (data not shown). In TZM/bl pseudovirus assays, no neutralization was detected against the tier 1 virus SF162.LS using purified polyclonal antibodies tested at the concentration of 100 μg/ml, at least in part due to response directed against the synthetic amide adduct.
Immunogenicity Modulated by MPER Membrane Orientation
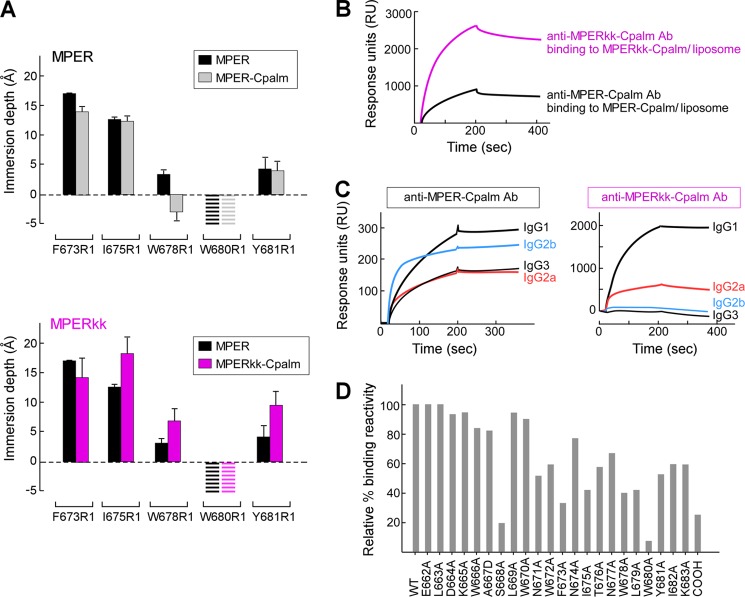
To determine whether the lack of B cell recognition of N-terminal residues in Npalm-7-MPER resulted from covalent attachment of palmitic acid, this linkage was moved to the C terminus. Specifically, the MPER was anchored via palmitic acid linkage to lysine at the C terminus directly (MPER-Cpalm) or to the second lysine of a two-residue lysine spacer at the C terminus (MPERkk-Cpalm). Although binding of 2F5 and 4E10 to MPER-Cpalm and MPERkk-Cpalm as measured by Biacore are similar, the impact of C-terminal modification on the MPER orientation relative to membrane was distinct. The membrane depths of the acyl chain-immersed reference residues Phe-673(R1) and the surface-exposed Trp-680(R1) in MPER-Cpalm are comparable with those in MPER. Trp-678(R1) translocates into the headgroup region in MPER-Cpalm (Fig. 5A, upper panel). In contrast, the changes in immersion depths of Ile-675(R1), Trp-678(R1), and Tyr-681(R1) in MPERkk-Cpalm indicate that the MPER C-helix (residues 673–682) becomes more immersed into the membrane acyl chain in MPERkk-Cpalm compared with that of the MPER (Fig. 5A, lower panel).
FIGURE 5.
Orientations of MPER-Cpalm versus MPERkk-Cpalm segments relative to membrane and their corresponding immunogenicity. A, EPR membrane immersion depths of MPER, MPER-Cpalm, and MPERkk-Cpalm in liposomes. The striped bars indicate complete exposure to aqueous phase (depth < −5 Å). B, binding of anti-MPER-Cpalm and anti-MPERkk-Cpalm antibodies to MPER-Cpalm or MPERkk-Cpalm/liposome (DOPC/DOPG) by Biacore. Each purified polyclonal antibody at 50 μg/ml from immune sera pooled from five immunized mice was injected over the L1 chip-bound peptide-liposome complex. C, analysis of anti-MPER-Cpalm and MPERkk-Cpalm IgG subclasses as measured by Biacore. Isotype-specific monoclonal anti-mouse IgG (10 μg/ml each) was passed over the surface of anti-MPER-Cpalm- or MPERkk-Cpalm-specific antibody bound to MPER-Cpalm/liposome or MPERkk-Cpalm/liposome arrayed on the surface of L1 chip, respectively. D, epitope mapping of anti-MPERkk-Cpalm polyclonal IgG antibody. Three independent immunizations of MPERkk-Cpalm/liposomes were carried out, with data representative of anti-MPERkk-Cpalm-specific antibody analysis from pooled serum (n = 5). The purified polyclonal IgG antibody was tested for binding affinity by Biacore.
To determine further whether the altered membrane orientation of the MPER through C-terminal conjugation alters the immunodominance involving the C-terminal region observed using Npalm-7-MPER immunogens (Fig. 4C), we immunized BALB/c mice three times with either MPER-Cpalm/liposome or MPERkk-Cpalm/liposomes and purified IgG from immune sera. Qualitative binding of the purified polyclonal antibody to each immunogen peptide was determined by Biacore. MPER-Cpalm was only weakly immunogenic compared with that of MPERkk-Cpalm, despite comparable levels of peptide incorporation into the liposomes, implying that the antigen configuration influences immunogenicity (Fig. 5B). Note that T cell-dependent isotype switching to various IgG subclasses resulting from these two immunogens was also different between the two immunogen peptides (Fig. 5C).
The resulting IgG responses were further characterized as to their fine epitope specificities. Much broader epitope specificity involving residues in the central hinge region of MPER was generated against HxB2 MPERkk-Cpalm compared with the Npalm-MPER antigens (Fig. 5D). For example, Asn-671, Trp-672, Phe-673, and Ile-675 in HxB2 MPERkk-Cpalm contribute more to anti-MPER antibody binding (52, 59, 33, and 42% of wild type levels) compared with those residues in HxB2 Npalm-7-MPER (74, 90, 95, and 81% of wild type levels) (Fig. 4C). Nevertheless, lack of antibody reactivity to the 2F5-like region (residue 662–666) and preferential recognition of Trp-680 and the C-terminal NH2 adduct remain features independent of MPER lipid anchor position.
Trp-680 as an Immunogenicity Hot Spot
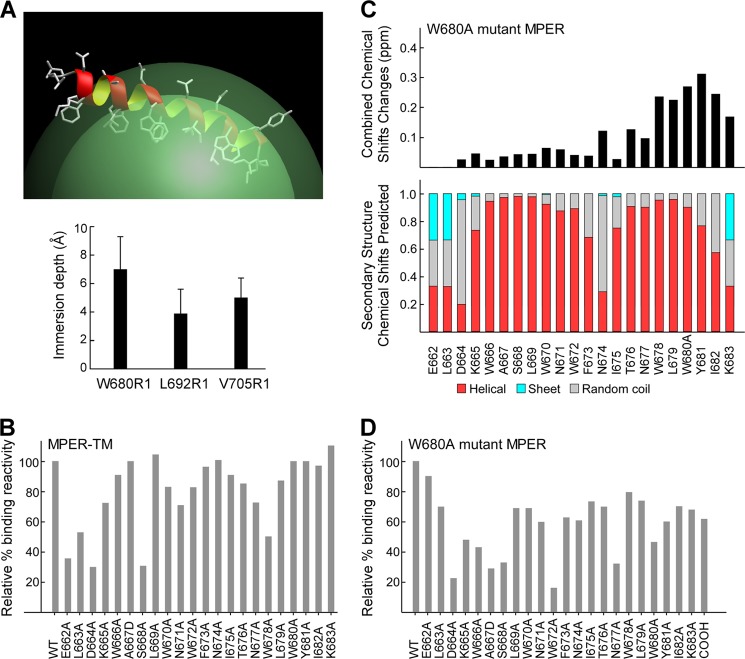
To better mimic the covalent attachment of MPER to a virion membrane, we synthesized an MPER peptide containing the contiguous transmembrane domain (MPER-TM) from Glu-662 to Val-705. Fig. 6A (upper panel) shows an MPER-TM model based upon EPR immersion depths (Fig. 6, lower panel) and NMR secondary structure analysis. Because the C-terminal MPER residues, including Trp-680, are buried in the acyl chain region in the context of MPER-TM/liposomes, we examined the effects of epitope accessibility on immunogenicity by vaccinating mice with the MPER-TM containing liposomes. In contrast to palmitoylated MPER immunogen, the immune response against MPER-TM was more directed to the N-helix region of the MPER (Fig. 6B). Residues critical for antibody binding include Glu-662, Asp-664, and Ser-668 (36, 30, and 31% of wild type) and Leu-663 with moderate reduction of binding (53% wild type) as shown by alanine mutagenesis. The lack of antibody recognition from Trp-680 to Tyr-682 correlates well with the membrane immersion depths of those residues by EPR (Fig. 6A, lower panel).
FIGURE 6.
Shifted immunogenicity of MPER-TM and W680A mutant MPER segments as a result of the loss of the accessible Trp-680 antibody-binding “hot spot.” A, MPER segment of the MPER-TM peptide is shown in ribbon representation on a micelle surface (top) to illustrate NMR secondary structural analysis with the corresponding EPR membrane immersion depth measurements of spin-labeled residues (bottom). B, representative epitope specificity of anti-MPER-TM polyclonal IgG antibody shown as binding of alanine mutants relative to the wild type sequence. C, combined 15N/1H amide chemical shift changes of the W680A mutant MPER, indicating local perturbations near the C terminus (top) with similar overall secondary structures (bottom) relative to the wild type MPER peptide. D, epitope mapping of W680A mutant MPER specific antibody. The purified polyclonal IgG antibody was tested for binding affinity by Biacore. Data are representative of three independent immunizations of a group of five mice each with W680A mutant MPER/liposomes.
To address whether the differences in epitope specificity between palmitoylated MPER and MPER-TM immunogens were attributable to the immersion depth of the hydrophobic Trp-680, an additional experiment was performed. The Npalm-7-MPER antigen with a W680A mutation was synthesized and used to immunize mice on liposomes to evaluate its epitope specificity. NMR analysis showed local chemical shift changes in the C-terminal region without any perturbation in the N-terminal helix and the central hinge region (Fig. 6C, upper panel). The overall secondary structure and orientation of the MPER is preserved in the W680A MPER peptide with Ala-680 exposed on the membrane surface (Fig. 6C, lower panel). Strikingly, many residues important for serum IgG binding were located in the N-helix of W680A mutant MPER (Fig. 6D). These include Asp-664, Ala-667, Ser-668, and Trp-672 (23, 29, 33, and 16% of wild type) followed by Lys-665 and Trp-666 (48 and 43% of wild type). Although the C-helix of the W680A mutant MPER remained immunogenic, the contributions to antibody binding from residues Ala-680 and the NH2 adduct at the C terminus were substantially diminished (48 and 62% of wild type compared with 6 and 10% of wild type in Npalm-7-MPER). Collectively, these data suggest that W680A shifts immune focus toward the N-terminal region of MPER. In line with results from the MPER-TM immunogen, our observations suggest that exposure of Trp-680 in the MPER is a dominant key residue modulating a common spectrum of epitope specificities for MPER/liposome formulations.
DISCUSSION
We have characterized the structural configuration of MPER peptides after integration into liposome vaccines and determined key parameters influencing their immunogenicity. The liposome vaccines used in this study were identical in terms of lipid composition, peptide display density, the ratio of lipid to peptide, T cell helper epitopes, and adjuvant content. However, stereochemical properties either intrinsic to the individual MPER peptide sequences or arising as a consequence of conjugation were specifically varied. Membrane anchoring via acyl modifications of the MPER antigen was necessary to limit dissociation from the liposome vehicle and, consequently, to maintain helix-hinge-helix peptide configuration during transport to the regional LN. While covalent linkage enhanced conformation-specific MPER antibody responses (Fig. 2D), the conjugation per se affected mobility of adjacent amino acid residues and/or accessibility of potential antigenic residues in the MPER/liposome formulation without altering secondary structure (Figs. 3A, 5, and 6). The reduced binding of 2F5 to Npalm-MPER with 0–2-amino acid spacers (Fig. 3A) suggests that constrained mobility in the N-helix of Npalm-MPER may also reduce its fitness to elicit 2F5-like antibodies. Constrained movement of adjacent local residues imposed by short spacers also produced membrane depth changes in specific residues and in the MPER orientation, resulting in significant differences in the magnitude of the anti-MPER-specific antibody responses as well as its IgG subclass distribution. Nonetheless, Npalm- and Cpalm-MPER immunogens elicited a majority of antibody directed to the C-terminal MPER region, irrespective of stereochemical and biophysical properties of various MPER/liposomes, T cell helper epitopes, and adjuvants (Figs. 3D, 4 and 5D and data not shown).
Although immune tolerance toward the 2F5 epitope sequence in the MPER has been proposed (19, 39), the intrinsic chemical nature of the N-helix per se may not account for the poor immunogenicity in this region; indeed, antibodies directed to the MPER N-helix have been elicited by a variety immunogens (40, 41). Earlier studies revealed a correlation between local mobility of short protein segments and their antigenicity (42, 43). However, limited flexibility imposed by N-terminal palmitoylation is not the key impediment, because a similar lack of 2F5-like elicitation is observed with HxB2 MPERkk-Cpalm immunization.
Previous studies observed that hot spot residues critical for binding specificity and antibody-antigen complexes most commonly include tryptophan, arginine, and tyrosine residues (23, 44). Charged and polar residues are also found enriched in the continuous epitopes (45). Our immunization study revealed an immunogenicity hot spot in membrane-bound MPER antigen. The surface-exposed bulky hydrophobic Trp-680 and CONH2 at the C terminus of the MPER are hot spot residues for anti-Npalm-MPER and anti-MPERkk-Cpalm antibodies. Although tyrosine, isoleucine, and lysine residues adjoining Trp-680 are juxtaposed at the C-terminal end of the MPER segment, the Trp-680 is uniquely surface-exposed relative to those other acyl chain buried residues. We anticipate that Trp-680 assumes a significant role in the initial interaction with antibody combining sites to create a binding strength sufficient for B cell selection. The influence of a terminal CONH2 on the antigenic characteristics of synthetic peptides has been described previously in other systems (46). Whether the major contribution of the terminal CONH2 to antibody binding herein is a consequence of its juxtaposition to Trp-680 and the neighboring YIK segment remains to be determined.
Our results show that either the W680A MPER mutation or an engineered covalent TM attachment to the MPER peptide shifted the antibody recognition from the C terminus toward the N-terminal end and central region of the MPER. Characterization of the MPER-TM by NMR and EPR showed that the immersion depth and structure of the N-helix was the same as in the MPER alone. In contrast, the C-helix had become less structured near the central hinge region with more residues in the C-helix buried in the membrane acyl chain region. The membrane immersion depth of the Val-705(R1) TM residue indicates it is shallower in the acyl chain region than expected within the gp41 TM on the virion. The buried Trp-680 configuration resulting from TM attachment to MPER argues that future modification of the MPER-TM configuration is required for liposome vaccination, based on 4E10 and 10E8 binding to virion (26). However, in the context of MPER-TM/liposome, the inaccessibility of the BCR to Trp-680 in MPER-TM was associated with the lack of immune response to C-helix as a consequence of shifting antigen specificity toward the surface-exposed N-helix residues. Our results highlight the exposure of Trp-680 as a key intrinsic factor influencing C-terminal region immunodominance.
Early humoral responses are heterogeneous in character and often of low affinity (47–49). As the response progresses, antibodies become higher affinity and manifest reduced diversity, sharpening immunological focus (50–52). Previous studies showed that immunodominance is positively correlated with antibody affinity in the primary response and differential binding kinetics (53). Whether the observed antibody C-helix selection specificity arising from the secondary responses to MPER/liposome is a consequence of strong B cell binding to the region involving Trp-680, pre-empting expansion of other B cells directed to neighboring or overlapping epitopes in the Npalm-MPER peptide requires future studies. In addition, which features of the W680A Npalm-MPER immunogen foster B cell selection with broader antigen specificities is unknown, given equivalent stereochemical properties of W680A mutant Npalm-MPER and Npalm-MPER except at position 680.
The immune response focuses on one or a few immunodominant structural features of a protein, influencing initial BCR selection and subsequent B cell fate. At a molecular level, immunodominance is highly context-dependent; a B cell epitope may be either immunodominant or nonimmunodominant depending on its relationship to other B cell epitopes in a given protein. Our finding that exposure of Trp-680 in the MPER can modulate immunodominance of the anti-MPER response has broad significance for HIV-1 immunity, given the genetic divergence of HIV-1 strains and quasispecies evolving in each individual.
The differences in isotype subclass distribution and immunogenicity observed in the MPER-Cpalm compared with MPERkk-Cpalm vaccines, however, cannot be accounted for by Trp-680 exposure, structural, and/or sequence differences. Additional regulatory mechanisms that direct selection of fine epitope specificity must be invoked. These findings, in conjunction with the W680A results, emphasize the interplay between the intrinsic nature of antigen and extrinsic cellular factors, including the naive repertoire, in impacting immunization outcome. Indeed, a previous study suggests that the differences in on-rates for engagement of B cell antigen receptors caused by minor antigen sequence changes outside the core epitope determinant directly influence the ability of antigen-primed B cells to recruit T cell help (54). In this regard, Lys-683 is involved in binding for the anti-MPERkk-Cpalm and Npalm-MPER antibodies. The loss of its binding contribution due to direct conjugation of palmitic acid to Lys-683 in the MPER-Cpalm immunogen may weaken the affinity of B cells for the C-terminal region. In addition, the limited flexibility and subtle changes in the MPER C-helix orientation in the MPER-Cpalm/liposome immunogen may produce an unfavorable conformational fit between paratope and epitope, translating into reduced B cell activation and resulting in alternative cellular fates responsible for low antibody production. However, interaction of the BCR with Lys-683 and additional spacer Lys residues in MPERkk-Cpalm/liposome immunogens may compensate for the energetically unfavorable binding of BCR to the C-terminal region with a strong isotype-switched response.
The discordance between antigenicity of anti-MPER antibodies elicited through vaccination and those immunoprotective monoclonal BNAbs suggest that MPER immunogens may improperly mimic the three-dimensional structure of the targeted region in the native protein (Ref. 55 and references therein). However, as shown here, the chemical modifications of antigens impact antigenic determinants without inducing structural alterations in the segment, thereby creating additional challenges as well as facilitating tools for MPER immunogen design. Similar caution applies to delivery systems enforcing the chemical or physical association of synthetic peptide segments or soluble proteins and for alum or other vaccine vehicles chosen to enhance antibody responses. Future rational vaccine design requires knowledge of immunogen structure in the vaccine and the rules of immunodominance, coupled with the ability to manipulate the latter to focus responses on those affording protection.
Acknowledgments
We thank Dongmei Liao for monoclonal antibody production and Lu Yu for EPR experiments.
This work was supported by National Institutes of Health Grants AI 84785 (to E. L. R. and G. W.), AI 91693 (to E. L. R., G. W., G. H. K., and D. I.), AI 81579 and HHSN272201000053C from NIAID and a Collaboration for AIDS Vaccine Discovery grant from the Bill and Melinda Gates Foundation (to E. L. R., G. W. and D. I.) and a Global Health grant from the Bill and Melinda Gates Foundation (to G. H. K.).
Sun, Z. Y., Cheng, Y., Kim, M., Song, L., Choi, J., Kudahl, U. J., Brusic, V., Chowdhury, B., Yu, L., Seaman, M. S., Bellot, G., Shih, W. M., Wagner, G., and Reinherz, E. L. (2013) Disruption of helix-capping residues 671 and 674 reveals a role in HIV-1 entry for a specialized hinge segment of the membrane proximal external region of gp41. J. Mol. Biol., in press.
- BNAb
- broadly neutralizing antibody
- MPER
- membrane proximal external region
- gp41
- glycoprotein 41
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPG
- 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol)
- MPLA
- monophosphoryl lipid A
- SPR
- surface plasmon resonance
- Npalm
- palmitic acid conjugated to the N terminus of MPER
- TM
- transmembrane domain
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- Cpalm
- palmitic acid at the C terminus
- LN
- lymph node
- BCR
- B cell receptor
- TCRβ
- T cell receptor β
- PEG
- polyethylene glycol
- MPERTM
- membrane-proximal external region containing the transmembrane segment.
REFERENCES
- 1. Burton D. R., Poignard P., Stanfield R. L., Wilson I. A. (2012) Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ekiert D. C., Bhabha G., Elsliger M. A., Friesen R. H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I. A. (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kashyap A. K., Steel J., Oner A. F., Dillon M. A., Swale R. E., Wall K. M., Perry K. J., Faynboym A., Ilhan M., Horowitz M., Horowitz L., Palese P., Bhatt R. R., Lerner R. A. (2008) Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. U.S.A. 105, 5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y., Migueles S. A., Welcher B., Svehla K., Phogat A., Louder M. K., Wu X., Shaw G. M., Connors M., Wyatt R. T., Mascola J. R. (2007) Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sather D. N., Armann J., Ching L. K., Mavrantoni A., Sellhorn G., Caldwell Z., Yu X., Wood B., Self S., Kalams S., Stamatatos L. (2009) Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gray E. S., Madiga M. C., Hermanus T., Moore P. L., Wibmer C. K., Tumba N. L., Werner L., Mlisana K., Sibeko S., Williamson C., Abdool Karim S. S., Morris L. (2011) The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85, 4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mikell I., Sather D. N., Kalams S. A., Altfeld M., Alter G., Stamatatos L. (2011) Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7, e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwong P. D., Mascola J. R. (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klein F., Diskin R., Scheid J. F., Gaebler C., Mouquet H., Georgiev I. S., Pancera M., Zhou T., Incesu R. B., Fu B. Z., Gnanapragasam P. N., Oliveira T. Y., Seaman M. S., Kwong P. D., Bjorkman P. J., Nussenzweig M. C. (2013) Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao H. X., Lynch R., Zhou T., Gao F., Alam S. M., Boyd S. D., Fire A. Z., Roskin K. M., Schramm C. A., Zhang Z., Zhu J., Shapiro L., NISC Comparative Sequencing Program, Mullikin J. C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S. M., Montefiori D. C., Parks R., Lloyd K. E., Scearce R. M., Soderberg K. A., Cohen M., Kamanga G., Louder M. K., Tran L. M., Chen Y., Cai F., Chen S., Moquin S., Du X., Joyce M. G., Srivatsan S., Zhang B., Zheng A., Shaw G. M., Hahn B. H., Kepler T. B., Korber B. T., Kwong P. D., Mascola J. R., Haynes B. F. (2013) Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parren P. W., Burton D. R., Sattentau Q. J. (1997) HIV-1 antibody–debris or virion? Nat. Med. 3, 366–367 [DOI] [PubMed] [Google Scholar]
- 12. Ofek G., Guenaga F. J., Schief W. R., Skinner J., Baker D., Wyatt R., Kwong P. D. (2010) Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. U.S.A. 107, 17880–17887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Azoitei M. L., Correia B. E., Ban Y. E., Carrico C., Kalyuzhniy O., Chen L., Schroeter A., Huang P. S., McLellan J. S., Kwong P. D., Baker D., Strong R. K., Schief W. R. (2011) Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334, 373–376 [DOI] [PubMed] [Google Scholar]
- 14. Stanfield R. L., Julien J. P., Pejchal R., Gach J. S., Zwick M. B., Wilson I. A. (2011) Structure-based design of a protein immunogen that displays an HIV-1 gp41 neutralizing epitope. J. Mol. Biol. 414, 460–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McLellan J. S., Correia B. E., Chen M., Yang Y., Graham B. S., Schief W. R., Kwong P. D. (2011) Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. Mol. Biol. 409, 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Batista F. D., Neuberger M. S. (1998) Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8, 751–759 [DOI] [PubMed] [Google Scholar]
- 17. Berzofsky J. A. (1985) Intrinsic and extrinsic factors in protein antigenic structure. Science 229, 932–940 [DOI] [PubMed] [Google Scholar]
- 18. Shih T. A., Meffre E., Roederer M., Nussenzweig M. C. (2002) Role of BCR affinity in T cell dependent antibody responses in vivo. Nat. Immunol. 3, 570–575 [DOI] [PubMed] [Google Scholar]
- 19. Yang G., Holl T. M., Liu Y., Li Y., Lu X., Nicely N. I., Kepler T. B., Alam S. M., Liao H. X., Cain D. W., Spicer L., VandeBerg J. L., Haynes B. F., Kelsoe G. (2013) Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J. Exp. Med. 210, 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiesa M. D., Martensen P. M., Simmons C., Porakishvili N., Justesen J., Dougan G., Roitt I. M., Delves P. J., Lund T. (2001) Refocusing of B-cell responses following a single amino acid substitution in an antigen. Immunology 103, 172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zlatkovic J., Stiasny K., Heinz F. X. (2011) Immunodominance and functional activities of antibody responses to inactivated West Nile virus and recombinant subunit vaccines in mice. J. Virol. 85, 1994–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geysen H. M., Tainer J. A., Rodda S. J., Mason T. J., Alexander H., Getzoff E. D., Lerner R. A. (1987) Chemistry of antibody binding to a protein. Science 235, 1184–1190 [DOI] [PubMed] [Google Scholar]
- 23. Rubinstein N. D., Mayrose I., Halperin D., Yekutieli D., Gershoni J. M., Pupko T. (2008) Computational characterization of B-cell epitopes. Mol. Immunol. 45, 3477–3489 [DOI] [PubMed] [Google Scholar]
- 24. Salzwedel K., West J. T., Hunter E. (1999) A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73, 2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cardoso R. M., Zwick M. B., Stanfield R. L., Kunert R., Binley J. M., Katinger H., Burton D. R., Wilson I. A. (2005) Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 [DOI] [PubMed] [Google Scholar]
- 26. Huang J., Ofek G., Laub L., Louder M. K., Doria-Rose N. A., Longo N. S., Imamichi H., Bailer R. T., Chakrabarti B., Sharma S. K., Alam S. M., Wang T., Yang Y., Zhang B., Migueles S. A., Wyatt R., Haynes B. F., Kwong P. D., Mascola J. R., Connors M. (2012) Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muster T., Guinea R., Trkola A., Purtscher M., Klima A., Steindl F., Palese P., Katinger H. (1994) Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68, 4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson J. D., Brunel F. M., Jensen R., Crooks E. T., Cardoso R. M., Wang M., Hessell A., Wilson I. A., Binley J. M., Dawson P. E., Burton D. R., Zwick M. B. (2007) An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81, 4033–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun Z. Y., Oh K. J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J. H., Wagner G., Reinherz E. L. (2008) HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63 [DOI] [PubMed] [Google Scholar]
- 30. Haran G., Cohen R., Bar L. K., Barenholz Y. (1993) Transmembrane ammonium sulfate gradients in liposomes produce efficient and stable entrapment of amphipathic weak bases. Biochim. Biophys. Acta 1151, 201–215 [DOI] [PubMed] [Google Scholar]
- 31. Song L., Sun Z. Y., Coleman K. E., Zwick M. B., Gach J. S., Wang J. H., Reinherz E. L., Wagner G., Kim M. (2009) Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U.S.A. 106, 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 33. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu X., McGraw P. A., House F. S., Crowe J. E., Jr. (2008) An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J. Immunol. Methods 336, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim M., Sun Z. Y., Rand K. D., Shi X., Song L., Cheng Y., Fahmy A. F., Majumdar S., Ofek G., Yang Y., Kwong P. D., Wang J. H., Engen J. R., Wagner G., Reinherz E. L. (2011) Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat. Struct. Mol. Biol. 18, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bershteyn A., Chaparro J., Yau R., Kim M., Reinherz E., Ferreira-Moita L., Irvine D. J. (2008) Polymer-supported lipid shells, onions, and flowers. Soft Matter 4, 1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lazarski C. A., Chaves F. A., Jenks S. A., Wu S., Richards K. A., Weaver J. M., Sant A. J. (2005) The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity 23, 29–40 [DOI] [PubMed] [Google Scholar]
- 38. Torchilin V. P., Omelyanenko V. G., Papisov M. I., Bogdanov A. A., Jr., Trubetskoy V. S., Herron J. N., Gentry C. A. (1994) Poly(ethylene glycol) on the liposome surface: on the mechanism of polymer-coated liposome longevity. Biochim. Biophys. Acta 1195, 11–20 [DOI] [PubMed] [Google Scholar]
- 39. Haynes B. F., Fleming J., St Clair E. W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R. M., Plonk K., Staats H. F., Ortel T. L., Liao H. X., Alam S. M. (2005) Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906–1908 [DOI] [PubMed] [Google Scholar]
- 40. Dennison S. M., Sutherland L. L., Jaeger F. H., Anasti K. M., Parks R., Stewart S., Bowman C., Xia S. M., Zhang R., Shen X., Scearce R. M., Ofek G., Yang Y., Kwong P. D., Santra S., Liao H. X., Tomaras G., Letvin N. L., Chen B., Alam S. M., Haynes B. F. (2011) Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One 6, e27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watson D. S., Szoka F. C., Jr. (2009) Role of lipid structure in the humoral immune response in mice to covalent lipid-peptides from the membrane proximal region of HIV-1 gp41. Vaccine 27, 4672–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., Hendrickson W. A. (1984) The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature 312, 127–134 [DOI] [PubMed] [Google Scholar]
- 43. Westhof E., Altschuh D., Moras D., Bloomer A. C., Mondragon A., Klug A., Van Regenmortel M. H. (1984) Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature 311, 123–126 [DOI] [PubMed] [Google Scholar]
- 44. Clackson T., Wells J. A. (1995) A hot spot of binding energy in a hormone-receptor interface. Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 45. Jackson R. M. (1999) Comparison of protein-protein interactions in serine protease-inhibitor and antibody-antigen complexes: implications for the protein docking problem. Protein Sci. 8, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gras-Masse H. S., Jolivet M. E., Audibert F. M., Beachey E. H., Chedid L. A., Tartar A. L. (1986) Influence of CONH2 or COOH as C terminus groups on the antigenic characters of immunogenic peptides. Mol. Immunol. 23, 1391–1395 [DOI] [PubMed] [Google Scholar]
- 47. Chua M. M., Goodgal S. H., Karush F. (1987) Germ-line affinity and germ-line variable-region genes in the B cell response. J. Immunol. 138, 1281–1288 [PubMed] [Google Scholar]
- 48. Clarke S. H., Staudt L. M., Kavaler J., Schwartz D., Gerhard W. U., Weigert M. G. (1990) V region gene usage and somatic mutation in the primary and secondary responses to influenza virus hemagglutinin. J. Immunol. 144, 2795–2801 [PubMed] [Google Scholar]
- 49. Dal Porto J. M., Haberman A. M., Shlomchik M. J., Kelsoe G. (1998) Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J. Immunol. 161, 5373–5381 [PubMed] [Google Scholar]
- 50. Kocks C., Rajewsky K. (1988) Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 85, 8206–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takahashi Y., Dutta P. R., Cerasoli D. M., Kelsoe G. (1998) In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 187, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foote J., Milstein C. (1991) Kinetic maturation of an immune response. Nature 352, 530–532 [DOI] [PubMed] [Google Scholar]
- 53. Nakra P., Manivel V., Vishwakarma R. A., Rao K. V. (2000) B cell responses to a peptide epitope. X. Epitope selection in a primary response is thermodynamically regulated. J. Immunol. 164, 5615–5625 [DOI] [PubMed] [Google Scholar]
- 54. Vijayakrishnan L., Sarkar S., Roy R. P., Rao K. V. (1997) B cell responses to a peptide epitope: IV. Subtle sequence changes in flanking residues modulate immunogenicity. J. Immunol. 159, 1809–1819 [PubMed] [Google Scholar]
- 55. Guenaga J., Dosenovic P., Ofek G., Baker D., Schief W. R., Kwong P. D., Karlsson Hedestam G. B., Wyatt R. T. (2011) Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One 6, e16074. [DOI] [PMC free article] [PubMed] [Google Scholar]