J. Clin. Invest. 106:1039–1052 (2000)
During the final stages of producing this manuscript for publication, errors were introduced in Figures 2 and 6. The correct versions, accompanied by the legends, appear below
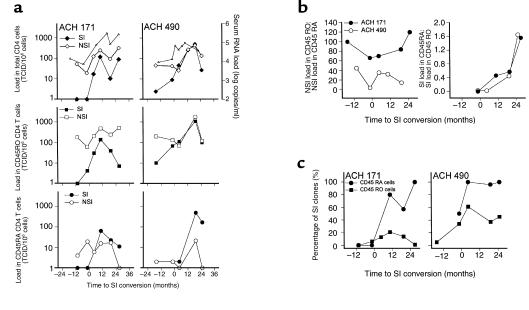
Figure 2.
Tropism for CD45RA+ and CD45RO+ CD4+ T cells during NSI to SI HIV-1 conversion. Patient PBMCs were FACS sorted in CD45RA+ CD45RO– CD4+ and CD45RA– CD45RO+ CD4+ T cells, and clonal virus isolation was performed on these T-cell subsets. (a) The frequency of infected cells with either NSI or SI HIV-1 variants (NSI or SI load) in total (top), CD45RO+ (middle), and CD45RA+ (bottom) CD4+ T cells relative to SI conversion in patient ACH171 (left panels) and 490 (right panels). Filled symbols indicate SI clones, and open symbols indicate NSI clones. In the top panel, serum RNA load is indicated by triangles. (b) Ratio of NSI load in CD45RO+ to NSI load in CD45RA+ CD4+ T cells (left) and ratio of SI load in CD45RA+ to SI load in CD45RO+ CD4+ T cells (right) of patient ACH171 (filled circles) and ACH490 (open circles). (c) Prevalence of SI phenotype among HIV-1 clones isolated from CD45RA+ (circles) or CD45RO+ (squares) CD4+ T-cell subset in ACH171 and ACH490.
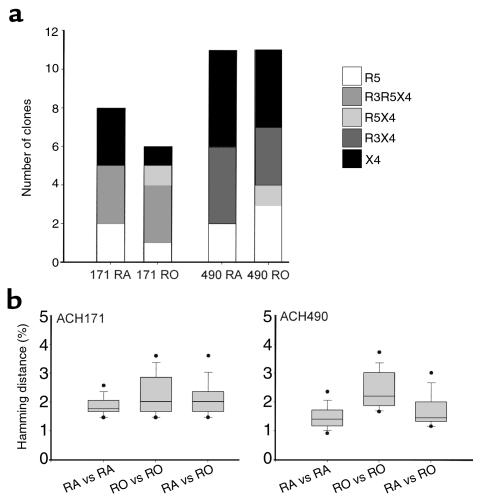
Figure 6.
SI HIV-1 clones isolated from CD45RA+ and CD45RO+ CD4+ T cells are phenotypically and evolutionarily indistinguishable. (a) Coreceptor usage of SI clones isolated from CD45RA+ and CD45RO+ CD4+ T cells from ACH171 and ACH490. Virus clones were pooled from the time points in which SI clones were obtained from both subsets (10.5, 19.5, and 27 months for ACH171 and 4.4, 19.2, and 25.1 months for ACH490). Bar descriptions are as in Figure 3. (b) Pairwise hamming distances were calculated between clones obtained from CD45RA+ CD4+ T cells (RA vs. RA), between clones obtained from CD45RO+ CD4+ T cells (RO vs. RO), and between clones isolated from CD45RA+ to clones from CD45RO+ CD4+ T cells (RA vs. RO). Uncorrected hamming distances between each pair of sequences were calculated and are depicted in a box plot as described in Figure 5.