Abstract
Background. Staphylococcus aureus produces numerous virulence factors but little is known about their in vivo regulation during an infection.
Methods. The production of capsule and α-toxin, and the expression of their respective genes, cap5 and hla, were analyzed by comparing CYL11481 (derivative of Newman) and its isogenic regulatory mutants in vitro. The temporal expression of cap5 and hla and the regulatory genes in vivo was carried out using a rat infective endocarditis model.
Results. In vitro analyses showed that capsule was positively regulated by MgrA, Agr, Sae, ArlR, and ClpC, and negatively by CodY and SbcDC. The α-toxin was positively regulated by MgrA, Agr, Sae, ArlR, and SbcDC but negatively by ClpC and CodY. In vivo analyses showed that cap5 expression correlated best with mgrA expression, whereas hla expression correlated best with sae expression. Mutation in mgrA drastically reduced cap5 expression in vivo.
Conclusions. Our results suggest that, in vitro, Agr is the most important regulator for capsule and α-toxin production, as well as for cap5 transcription, but SaeR is the most critical for hla transcription. However, in vivo, MgrA is the major transcriptional regulator of capsule, but not α-toxin, whereas saeR expression correlates best with hla expression.
Keywords: Staphylococcus aureus, virulence, infective endocarditis model, mgrA, capsule, α-toxin
Staphylococcus aureus causes many diseases, ranging from superficial skin infections to severe and life-threatening infections such as endocarditis, septic arthritis, and sepsis [1]. Endovascular infections caused by S. aureus are often associated with implanted medical devices, such as prosthetic heart valves and vascular catheters [2]. The pathogenicity of S. aureus is a very complex process involving coordinated expression of numerous secreted enzymes, toxins, antiphagocytic capsular polysaccharides (CPs) and adhesins [1]. These virulence factors are coordinately regulated by a network of regulatory genes, which include 2-component systems (TCSs) and various transcriptional regulators [3, 4]. The accessory gene regulator (agr) and staphylococcal accessory regulator (sarA) represent the most well-characterized global regulatory elements in S. aureus, which coordinate synthesis of multiple virulence factors [3, 5]. MgrA is a multiple gene regulator belonging to the SarA family, which has been shown to directly regulate several virulence genes by binding to their respective promoter regions [6–8]. Several virulence factors have been shown to be upregulated by MgrA, such as capsular polysaccharides, sortase A, serine proteases, leukotoxins, and α-toxin. In contrast, several cell wall–associated or surface-associated proteins are downregulated by MgrA [9, 10]. MgrA has also been shown to regulate genes involved in bacterial metabolism, antibiotic resistance, and autolysis [7–9]. Regulation by MgrA is affected by the redox and phosphorylation state of the regulator [11, 12]. MgrA is critical for S. aureus pathogenesis, as shown in a murine abscess model and a murine sepsis arthritis model [11, 13].
Capsule is one of several virulence factors that contribute to S. aureus pathogenicity, which has been shown to attenuate virulence in experimental infective endocarditis (IE) [14, 15]. This cell surface molecule is produced by most S. aureus strains, with type 5 (CP5) or type 8 (CP8) being the dominant serotypes. The cap5 and cap8 operons involved in the biosynthesis of CP5 and CP8, respectively, are allelic, and each contains 16 genes in which 4 serotype-specific genes are flanked by common genes [16]. The nearly identical promoter regions of the cap5 and cap8 operons indicate that their mechanisms of regulation are similar. Staphylococcal capsule has been shown to be affected by various environmental conditions such as carbon dioxide, iron limitation, pH, oxygen tension, and in vivo environmental cues [14, 15]. We have previously identified several genes that regulate capsule expression in vitro. Among these, Agr, MgrA, and ArlR are activators, whereas ClpC, Sae, SbcDC, and CodY are repressors of capsule production [6, 9, 17–22]. ArlR is the response regulator of the ArlRS TCS that activates the expression of capsule primarily through an MgrA-dependent pathway in vitro in strain Newman [16]. SbcDC has been shown to be involved in DNA repair in Escherichia coli [23]; however, in S. aureus, SbcDC represses capsule by negatively regulating the arl locus [19]. CodY, found in Bacillus subtilis and many low G + C gram-positive bacteria, is a pleiotropic regulator sensing nutrient limitation [24] that represses capsule in S. aureus [18, 20]. Recently, we reported that ClpC divergently regulated capsule via sae-dependent repression or via codY-dependent activation [21].
Staphylococcus aureus α-toxin, encoded by the hla gene, is a pore-forming extracellular protein that targets a variety of host cell types, and is tightly regulated by multiple genes in S. aureus [25]. The regulation of hla has been extensively studied in vitro. These studies suggest that agr exerts a direct positive impact on hla expression, whereas SarA positively affects hla expression indirectly by both agr-dependent and agr-independent pathways [26–28]. The TCS encoded by saeS and saeR positively regulates the expression of hla at the transcriptional level [29, 30]. MgrA has been shown to activate hla by direct promoter binding or through activation of agr [10].
Virulence gene regulation in S. aureus has mainly been dissected only under in vitro conditions, and little is known regarding gene regulation under in vivo conditions. An in vivo study on hla gene regulation in endovascular infection by Xiong et al [31] suggests that hla expression is more dependent on Sae than on Agr and/or SarA. The results of a transcriptional analysis using a soft-tissue guinea pig infection model by Goerke et al [27] also supports the notion that Sae is the key regulator of hla expression in strain Newman and RN6390. Using a rabbit IE model, van Wamel et al [32] showed that Agr, but not SarA, significantly affected capsule gene transcription in vivo. However, these in vivo studies focused on the effect of 1 or 2 regulatory genes at 1 fixed time-point during infection. To better understand virulence gene regulation, we investigated the effect of a large cadre of regulators known to affect capsule in vitro on cap5 gene expression in a rat IE model at 3 distinct time-points during disease development. We found that, despite its strong effect in vitro, Agr was not a key regulator of cap5 gene expression in vivo; instead, MgrA was pivotal for cap gene transcription in vivo. We also analyzed hla gene expression in our study.
MATERIALS AND METHODS
Bacterial Strain and Plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. The S. aureus strain CYL11481, in which the proline at the 18th codon of saeS in strain Newman—that is, Newman saeS(P18L)—has been replaced with leucine, was used as the parental strain (21). Transduction between S. aureus strains was carried out by phage 52A or 80α. The isogenic mutants of CYL11481 were constructed by phage transduction from their respective parents (Table 1). All the mutants were verified by polymerase chain reaction (PCR) amplification, and complemented strains were verified by plasmid digestion.
Table 1.
Strains and Plasmids Used in the Study
| Strain or Plasmid | Description | Reference |
|---|---|---|
| Staphylococcus aureus | ||
| Newman | Wild-type CP5 strain | T. Foster |
| CYL11481 | Newman saeS(P18L) | [21] |
| CYL12659 | CYL11481 ΔmgrA | This study |
| CYL12676 | CYL11481 ΔmgrA (pCL52.2) | This study |
| CYL12678 | CYL11481 ΔmgrA (pTL2989) | This study |
| CYL12661 | CYL11481 Δagr | This study |
| CYL12752 | CYL11481 ΔarlR | This study |
| CYL12723 | CYL11481 ΔcodY | This study |
| CYL11489 | CYL11481 ΔclpC | This study |
| CYL12750 | CYL11481 ΔsbcDC | This study |
| CYL12367 | Newman ΔsaePQRS | [22] |
| Plasmids | ||
| pCL 52.2 | Cloning vector | [6] |
| pTL2989 | pCL 52.2 with mgrA | [6] |
Rat IE Model
All experiments involving animals followed protocols that have been reviewed and approved by the institutional animal care and use committee at the University of Arkansas for Medical Sciences. Rats were purchased from Harlan Inc, and were housed in Institutional Animal Care and Use Committee–accredited animal care facilities at the University of Arkansas for Medical Sciences. The surgical procedures were carried out under full anesthesia, as well as local delivery of lidocaine. Rats were euthanized by exposure to 100% carbon dioxide in a closed chamber.
A well-characterized experimental rat IE model was used in the present study [33]. Sprague-Dawley female rats (220–240 g each) were anesthetized with an isoflurane-oxygen gas mixture (1:1 ratio) during the surgery. In brief, an indwelling polyethylene catheter was positioned in the left ventricle of each animal via the retrograde transcarotid artery approach, with the tip passing across the aortic valve, to induce sterile vegetations. At 48 hours after catheterization, animals were infected iv through the tail vein with an inoculum of 1 × 104 colony-forming units (CFU)/rat of strain CYL11481 or its isogenic mgrA mutant (this inoculum was chosen after extensive pilot studies identified an appropriate ID90). Animals were sacrificed at days 1, 2, and 3 after infection. Vegetations were collected immediately in sterile tubes and frozen in dry ice until RNA isolation (see below). Only samples from rats with proper catheter placement and macroscopic vegetations were used for the study. Groups of 3 rats per time-point were used for the study, and each experiment was repeated in duplicate or triplicate.
RNA Isolation and Quantitative Reverse Transcription PCR
For in vivo transcriptional analysis, vegetations in each tube were suspended in 1 mL of sterile phosphate-buffered saline (pH7.4) and homogenized with a mortar and pestle, and serial dilutions of 100 µL were plated on tryptic soy agar plates and incubated at 37°C for 24 hours for quantitative cultures (expressed as mean log10 CFU/g tissue ± SD). The remaining sample was processed for RNA isolation as described previously [34] using RNAzol-RT (Molecular Research Center, Inc) with some modifications. After homogenization, tissues were pelleted at 500g for 5 minutes, and supernatants containing bacterial cells were mixed with an equal volume of an ice-cold 1:1 mixture of ethanol-acetone and kept at −20°C for 2 hours or overnight. Cells were pelleted, washed 2 times with TNE buffer (50 mM Tris [pH 7.6], 150 mM NaCl, and 5 mM EDTA) and suspended in 50 µL of TNE buffer with 2.5 M NaCl. Cells were digested with 0.8 µg/µL of lysostaphin at 37°C for 5 minutes and immediately lysed with 1 mL RNAzol-RT, and RNA was isolated according to the manufacturer's instructions. Total RNA was further purified from the host tissue RNA using the MICROBEnrich Kit (Ambion AM1901). Purified bacterial RNA was checked for DNA contamination by running a no–reverse transcription (RT) PCR using hu gene primers for 25 cycles in a thermocycler. RNA was stored in −80°C until used. For day 1 sample, vegetations from 2 to 3 animals were pooled to obtain adequate quantity of RNA for analysis. For in vitro expression analysis of the cap5 and hla genes, RNA was isolated as described previously [34] using RNAzol-RT from 1.5 mL overnight cultures adjusted to an OD660 of 2.0. The quantitative RT-PCR (qRT-PCR) was performed as described previously [9] using the primers listed in Supplementary Table 1.
In Vitro Capsule and α-Toxin Production Assays
Capsule quantification was done as described previously [6]. The semiquantitative assay of α-toxin was performed by spotting the filtered (0.22 µm) supernatants of overnight cultures onto 5% sheep blood agar plates (Remel, Lenexa), with the zones of clearance measured with calipers.
Statistical Analysis
Relative expression and bacterial burden data were analyzed by the GraphPad Prism version 4.00 using a Student t test (P < .05 considered as significant). Correlation coefficient (r2) among the genes was analyzed by linear regression with best fit model in GraphPad Prism software.
RESULTS
Regulation of Capsule and α-Toxin In Vitro
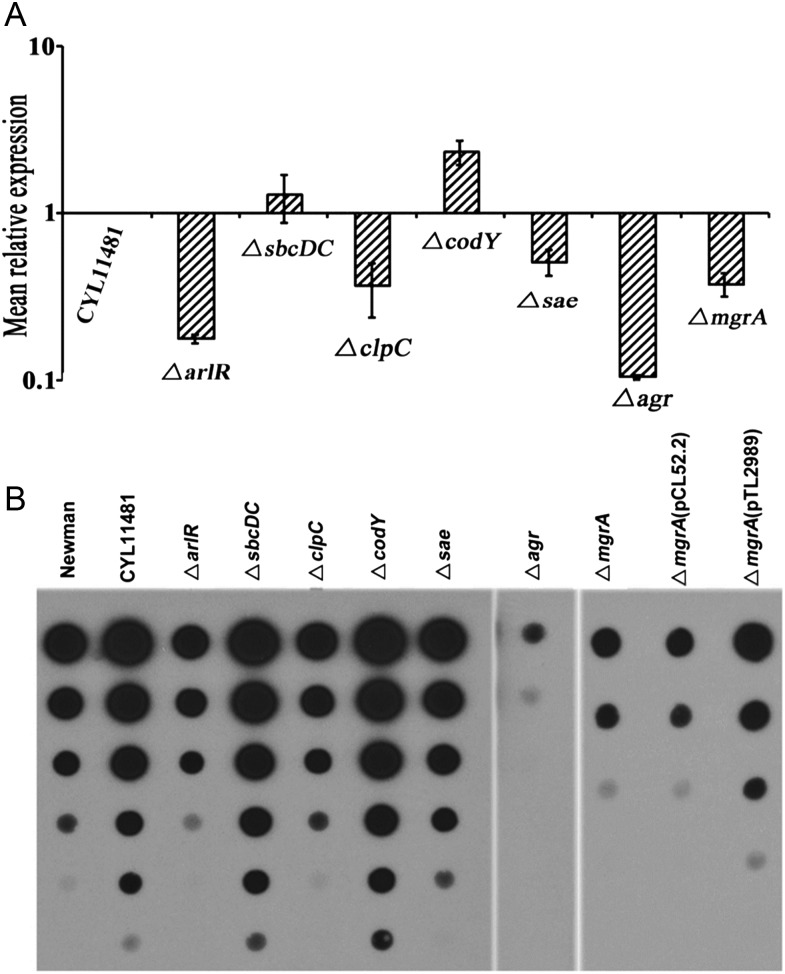
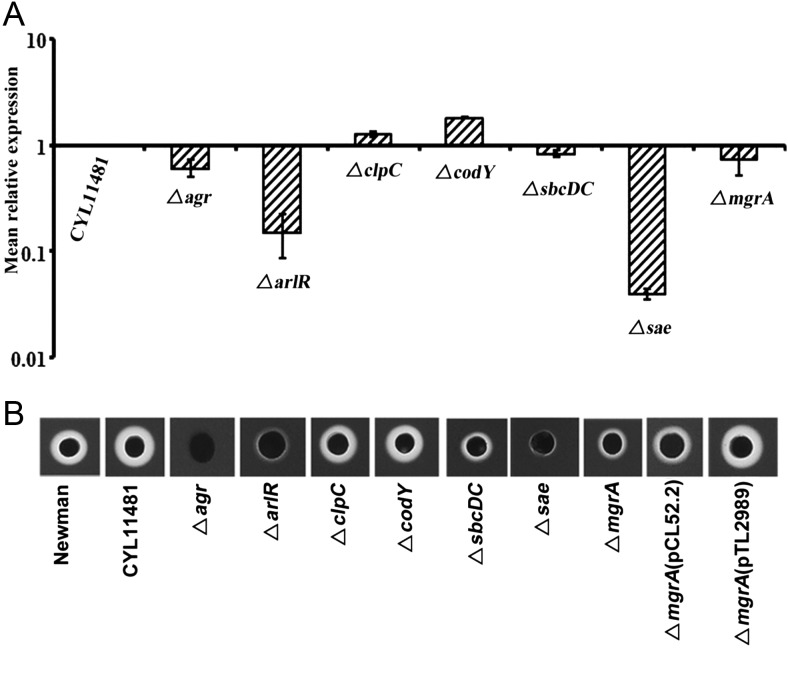
Strain Newman has often been used in virulence regulation studies. However, genome sequencing revealed a single nucleotide variation in the saeS gene that resulted in unusually high expression of sae. Restoration of this point mutation resulted in strain CYL11481, ie, Newman saeS(P18L) [21]. Because Sae has been shown to affect many virulence regulators, we first assessed the effect of regulatory gene mutations in the saeS-corrected strain CYL11481 background on capsule and α-toxin production at the transcriptional as well as posttranscriptional levels. Capsule expression in CYL11481 was positively regulated (in decreasing order of relative impact) by Agr, ArlR, MgrA, ClpC, Sae, and negatively regulated by CodY and SbcDC (Figure 1A and 1B). For α-toxin, we found that (in decreasing order of effect) Sae, ArlR, Agr, MgrA, and SbcDC were positive regulators, and CodY and ClpC were repressors at the transcriptional level. Interestingly, the agr mutation has a strongest effect on α-toxin activity, suggesting that Agr is a key regulator of α-toxin at both the transcriptional and the posttranscriptional levels (Figure 2A and 2B).
Figure 1.
In vitro analysis of cap5 gene expression and capsule production. A, The expression of cap5D gene was analyzed by quantitative reverse transcription polymerase chain reaction for 18-hour cultures of CYL11481 (parent strain) and its isogenic mutants of arlR (CYL12752), sbcDC (CYL12750), clpC (CYL11489), codY (CYL12723), sae (CYL12367), agr (CYL12661), and mgrA (CYL12659). Expression levels are expressed relative to that of CYL11481, which was arbitrarily set at 1. Data represent the mean relative expression of duplicate experiments with standard deviation. B, Capsule production was assayed for 18-hour cultures of the strains listed above; strain Newman, and the complemented mgrA mutant. The crude capsule preparations were serially diluted 3-fold and assayed by immunoblotting with anti–type 5 capsule antibody.
Figure 2.
In vitro analysis of hla expression and α-toxin activity. A, The expression of the hla gene was analyzed by quantitative reverse transcription polymerase chain reaction as described in the legend of Figure 1A with the same set of strains. Data represent the mean relative expression of duplicate experiments with standard deviation. B, α-Toxin activity was evaluated by spotting 15 µL of filtered culture supernatants of the strains listed in Figure 1B on 5% sheep blood agar plates followed by incubation for 24 hours at 37°C. Zones of clearance on the blood agar plate indicate Hla activity.
Regulation of Capsule and α-Toxin In Vivo
To study gene regulation in vivo, we chose to focus on capsule because regulation of capsule has been studied extensively in vitro. A total of 7 regulatory genes, as well as the cap5 and hla genes, were analyzed for their expression longitudinally during the temporal progression of experimental IE. The inclusion of the hla gene served as an appropriate comparison, as regulation of α-toxin has also been well studied in vitro, and regulation of hla in vivo has already been reported [27, 31]. As shown in Figure 3, we found in the parental strain that most of the genes of interest were strongly induced in vivo. The exceptions were the clpC and codY genes, which were both reduced, especially the clpC gene. Interestingly, RNAIII, the effector of agr, was reduced at days 1 and 3 but was highly increased at day 2. The expression of cap5 and mgrA were at a relatively constant level over time, whereas hla and saeR were more variable, peaking at day 2. Overall, the expression of cap5 correlated best with that of mgrA (r2 = 0.99) and poorly with other regulators (r2 = 0.007–0.82). The expression of hla matched best with that of saeR (r2 = 0.88). These results suggest that MgrA is the major cap5 regulator in vivo in this model, and that Sae is a key regulator of hla under these in vivo conditions.
Figure 3.
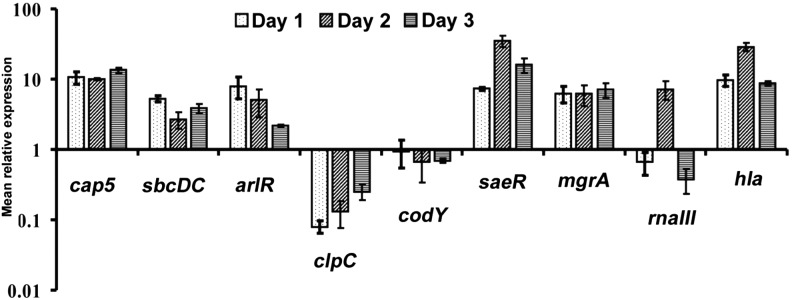
Gene expression in CYL11481 during infective endocarditis infection. RNA was isolated from groups of 3 rats infected with CYL11481 at days 1, 2, and 3 after infection, and each experiment was repeated 2–3 times. The expression of the genes indicated in the figure was assayed by quantitative reverse transcription polymerase chain reaction as described in the legend of Figure 1A.
Role of MgrA in cap5 and hla Expression In Vivo
To further study the role of MgrA in the regulation of capsule and α-toxin in vivo, we constructed an mgrA mutant of strain CYL11481 and performed qRT-PCR to measure the expression profile of the cap5 and hla genes during the progression of disease in the rat IE model. The mutant was verified by complementation to ensure that no second site mutation occurred during strain construction (Figure 1B and 2B). We also included RNAIII expression in the experiments, as MgrA has been shown to affect agr in vitro [10]. Our results showed that cap5 gene expression in the mgrA mutant was highly reduced during the 3 days of infection suggesting that MgrA is a major regulator of capsule in vivo. On the other hand, the expression of hla and RNAIII was reduced at day 2 but slightly increased in day 3. Comparing these results to those of the wild type shown in Figure 3, we concluded that the mgrA mutation had a negative effect on both agr and hla at day 2, but its effect at day 3 was unclear. Based on the expression profiles, we suggest that MgrA may have only a marginal and possibly an indirect effect on the expression of agr and hla in the rat IE model (Figure 4).
Figure 4.
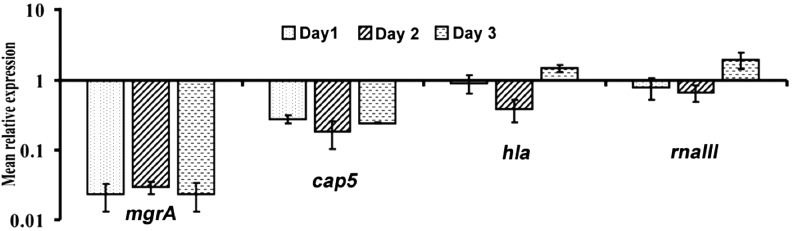
Gene expression in mgrA mutant during infective endocarditis infection. RNA was isolated from groups of 3 rats infected with CYL12659 at days 1, 2, and 3 after infection, and each experiment was repeated 2 times. The expression of the genes indicated in the figure was assayed by quantitative reverse transcription polymerase chain reaction as described in the legend of Figure 1A.
Mutation in mgrA Attenuated S. aureus Virulence in Experimental IE
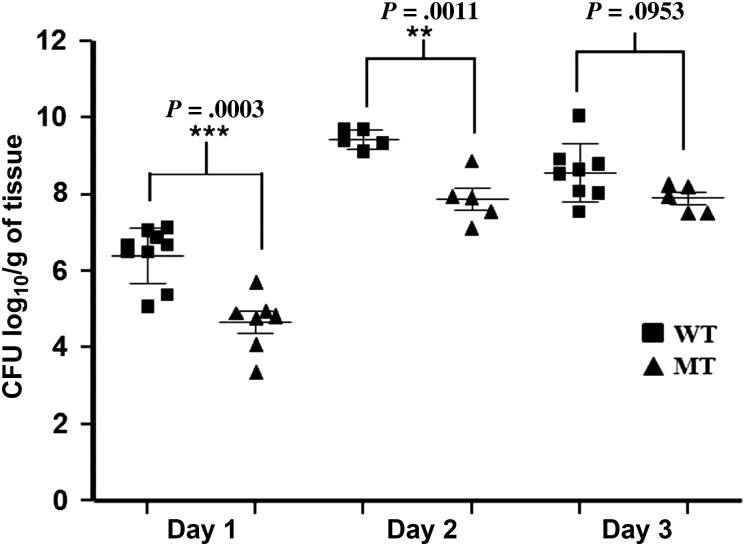
The role of MgrA in virulence in endocarditis has not been studied. Here, we quantified the temporal bacterial burden within cardiac vegetations in animals with IE infected with either the parent strain or its isogenic mgrA mutant strain. As shown in Figure 5, animals infected with the wild-type S. aureus strain, CYL11481, had significantly higher vegetation bacterial counts than in animals infected with the mgrA mutant strain at both days 1 and 2 of infection. At day 3, the mgrA mutant strain had slightly lower bacterial counts than the parent strain but the difference was not statistically significant. These results suggest that MgrA plays an important role in virulence in the rat IE model.
Figure 5.
Bacterial burden in wild-type CYL11481 (WT) and its isogenic mgrA mutant (MT) in rat cardiac vegetations. Homogenized heart vegetation (100 µL) from each time point of the WT and mgrA mutant strains were quantitatively cultured onto tryptic soy agar plate and incubated at 37°C for 24 hours. Data represent colony-forming units (CFU) log10/g of tissue from 5 to 9 rats per time-point with P value. Statistical significance was analyzed by Student t test. ***Highly significant; **Significant.
DISCUSSION
The ability of S. aureus to cause a multitude of life-threatening human diseases suggests that the pathogenesis of S. aureus infections is highly complex. The expression of virulence genes is expected to be influenced by various host signals and thus likely to be different between in vivo and in vitro conditions. However, little is known about virulence gene regulation in vivo. In this study, we chose capsule as the model virulence factor in an in vivo virulence gene regulation study using a well-characterized rat IE model, because (1) regulation of capsule has been studied extensively in vitro; and (2) capsule production appears to be a key virulence factor [14, 15]. Our strategy was to correlate the temporal expression of this target virulence gene with that of its known regulators, during the infection course. We reasoned that the expression of the primary regulatory gene should correlate well with that of its target gene over time. To our knowledge, this is the first study of such temporal expression of a virulence gene, as well as its regulatory modulators in S. aureus in vivo. Our results suggest that, among all regulators tested, MgrA is the key regulator affecting capsule regulation in the rat IE model. This conclusion was further supported by the result that mgrA deletion abolished cap5 gene expression during the course of infection. This is in contrast to the in vitro results wherein Agr had the strongest effect on capsule expression. We have also previously shown that Agr strongly affected capsule over time in different growth phases in vitro [35]. Our results therefore suggest that capsule regulation is affected by host factors and regulated differently in vitro and in vivo; this supports the notion that S. aureus encounters different environmental signals in vivo than in vitro, and modulates its expression of virulence genes accordingly. The sharp increase in RNAIII expression in vivo at day 2 is noteworthy. The increase is consistent with a report that agr promoter activity in vegetations increased at 48 hours of infection in a rabbit IE model [36]. Interestingly, our results showed a drastic decrease of RNAIII at day 3. This spike in expression may indicate that Agr could still have an effect on capsule at that time point, which may explain the earlier study by van Wamel et al, who showed that the cap5 promoter in Newman was strongly affected by Agr in a rabbit IE model [32].
It is interesting that clpC expression in vivo was greatly reduced, especially in day 1 of infection. ClpC is an ATP-dependent chaperone involved in protein quality control by refolding or degrading misfolded proteins [37]. The reduction may reflect a drastic change in physiology involving protein synthesis of the bacteria at an early stage of infection. In parental strain CYL11481, we found that ClpC promoted capsule by repressing codY expression in vitro. These results are opposite to the results in vivo where we found increased expression of cap5, but decreased expression of clpC, whereas codY remained largely unchanged. These results provide further evidence that regulation of gene expression is different between in vitro and in vivo conditions.
Detection of capsule in vivo depends on the specific animal models under study, and particularly the principal site of infection [14, 15, 32]. Production of CP8 has been shown to be increased in a rabbit IE model after 4 days of infection [38], consistent with our present study. However, paradoxically, capsule has been shown to attenuate S. aureus virulence possibly by masking adhesins required for colonization [39, 40]. How can capsule be induced in vivo, and yet attenuate virulence? One possible explanation is that S. aureus cells covered with capsule are less able to colonize the damaged heart tissue; however, after establishing colonization at such sites, induced capsule could then facilitate evasion of localized phagocytosis from host cells (eg, neutrophils [41–44]).
Our findings here that MgrA was the major activator of capsule in vivo suggests that MgrA may contribute to S. aureus resistance to phagocytosis and intracellular killing, in part, through its effect on capsule. Indeed, it was suggested that MgrA endows the bacteria with the ability to evade the host defense based on the finding that an mgrA mutation attenuated virulence in septic arthritis animal models [13]. Our results here also showed that mgrA mutation resulted in significant reduced bacterial burden by 1.5 to 2 log CFU/g vegetation at days 1 and 2 after infection. However, whether capsule is the key virulence factor through which MgrA affects virulence requires further studies.
Several regulators included in this study (Agr, Sae, MgrA, Arl, and CodY) have been shown to affect α-toxin expression in S. aureus [6, 18, 20, 16–30, 45, 46]. Agr activates hla through its effector RNAIII. The 5′ end of RNAIII directly interacts with the hla mRNA and unblocks the ribosome binding site of the hla gene [47], whereas the 3′ half of RNAIII inhibits rot translation by an antisense mechanism to relieve the repression of hla transcription by Rot [48, 49]. SaeR activates the transcription of hla by direct promoter binding [50], whereas MgrA activates hla transcription by binding to the promoter and by modulating Agr through SarS but not Sae [9]. The upstream region of the hla gene contains a CodY box suggesting that repression of hla transcription is by direct CodY binding. CodY can also repress hla via Agr [18, 20]. Although the results from these reports have been derived from different strains, they are largely consistent with our in vitro studies presented here using CYL11481. However, we found Sae had the strongest effect at the transcriptional level, although Agr had the most effect on hemolytic activity, implying potential posttranscriptional regulation. The expression of hla in vivo has been studied in 2 reports. A study by Xiong et al [31] investigated the role of agr, sarA, and sae in controlling hla expression in S. aureus RN6390 and SH1000 backgrounds in a rabbit IE model; they found that hla expression in cardiac vegetations was moderately reduced in the agr and sarA single mutants, but was markedly reduced in the sae mutant [31]. In another study using a guinea pig device–related infection model, Goerke et al [27] demonstrated that Sae, but not Agr or SarA, was the key regulator of hla in vivo. These results are consistent with our finding that the sae expression correlated best with that of the hla in the rat IE model. Interestingly, we also found that the mgrA mutation in our study resulted in noticeable change in the hla and rnaIII expression patterns in the rat IE model, suggesting that MgrA has an effect, perhaps indirect, on both hla and agr expression in vivo.
In summary, the present investigation established the disparity of in vitro and in vivo regulation of capsule, and strongly underscored that MgrA is a major positive regulator of capsule gene expression in vivo in experimental S. aureus IE. The expression of the hla gene correlated best with the saeR gene expression in vivo, although MgrA may also have an effect on the regulation of hla gene expression.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank David Cue and Mei Lei for critical review of the manuscript and helpful suggestions.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grants AI037027 to C. Y. L., AI-39108 to A. S. B., and AI097657 to Y. Q. X.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lowy FD. How Staphylococcus aureus adapts to its host. N Engl J Med. 2011;364:1987–90. doi: 10.1056/NEJMp1100251. [DOI] [PubMed] [Google Scholar]
- 2.El-Ahdab F, Benjamin DK, Wang A, et al. Risk of endocarditis among patients with prosthetic valves and Staphylococcus aureus bacteremia. Am J Med. 2005;118:225–9. doi: 10.1016/j.amjmed.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Junecko J, Zielinska AK, Mrak LN, et al. Transcribing virulence in Staphylococcus aureus. World J Clin Inf Disease. 2012;2:63–76. [Google Scholar]
- 4.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 5.Cheung AL, Zhang G. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front Biosci. 2002;7:d1825–42. doi: 10.2741/A882. [DOI] [PubMed] [Google Scholar]
- 6.Luong TT, Newell SW, Lee CY. Mgr, a novel global regulator in Staphylococcus aureus. J Bacteriol. 2003;185:3703–10. doi: 10.1128/JB.185.13.3703-3710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingavale SS, Van Wamel W, Cheung AL. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol. 2003;48:1451–66. doi: 10.1046/j.1365-2958.2003.03503.x. [DOI] [PubMed] [Google Scholar]
- 8.Truong-Bolduc QC, Zhang X, Hooper DC. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J Bacteriol. 2003;185:3127–38. doi: 10.1128/JB.185.10.3127-3138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol. 2006;188:1899–910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun. 2005;73:1423–31. doi: 10.1128/IAI.73.3.1423-1431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen PR, Bae T, Williams WA, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–5. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 12.Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J Bacteriol. 2010;192:2525–34. doi: 10.1128/JB.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonsson IM, Lindholm C, Luong TT, Lee CY, Tarkowski A. mgrA regulates staphylococcal virulence important for induction and progression of septic arthritis and sepsis. Microbes Infect. 2008;10:1229–35. doi: 10.1016/j.micinf.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–34. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CY, Lee JC. Staphylococcal capsules. In: Fischetti V, Novick RP, Ferretti J, Portnoy D, Rood J,, editors. Gram-positive pathogens. 2. Washington, DC: American Society for Microbiology; 2006. pp. 456–63. [Google Scholar]
- 16.Sau S, Bhasin N, Wann ER, Lee JC, Foster TJ, Lee CY. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143(Pt 7):2395–405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 17.Luong TT, Lee CY. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology. 2006;152:3123–31. doi: 10.1099/mic.0.29177-0. [DOI] [PubMed] [Google Scholar]
- 18.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–65. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Luong TT, Lee CY. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J Bacteriol. 2007;189:7343–50. doi: 10.1128/JB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majerczyk CD, Dunman PM, Luong TT, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–77. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luong TT, Sau K, Roux C, Sau S, Dunman PM, Lee CY. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain Newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol. 2011;193:686–94. doi: 10.1128/JB.00987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhuber A, Goerke C, Bayer MG, Döring G, Wolz C. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol. 2003;185:6278–86. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–46. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–7. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Moreillon P, Entenza JM, Francioli P, et al. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–43. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arvidson S, Janzon L, Lofdahl S. The role of the d-lysin gene (hld) in the agr-dependent regulation of exoprotein synthesis in Staphylococcus aureus. In: Novick RP, editor. Molecular biology of the staphylococci. New York: VCH; 1990. pp. 419–31. [Google Scholar]
- 27.Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol. 2001;40:1439–47. doi: 10.1046/j.1365-2958.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 28.Vandenesch F, Kornblum J, Novick RP. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991;173:6313–20. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraudo AT, Cheung AL, Nagel R. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol. 1997;168:53–8. doi: 10.1007/s002030050469. [DOI] [PubMed] [Google Scholar]
- 30.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–9. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis. 2006;194:1267–75. doi: 10.1086/508210. [DOI] [PubMed] [Google Scholar]
- 32.van Wamel W, Xiong YQ, Bayer AS, Yeaman MR, Nast CC, Cheung AL. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb Pathog. 2002;33:73–9. doi: 10.1006/mpat.2002.0513. [DOI] [PubMed] [Google Scholar]
- 33.Xiong YQ, Willard J, Kadurugamuwa JL, Yu J, Francis KP, Bayer AS. Real-time in vivo bioluminescent imaging for evaluating the efficacy of antibiotics in a rat Staphylococcus aureus endocarditis model. Antimicrob Agents Chemother. 2005;49:380–7. doi: 10.1128/AAC.49.1.380-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol. 2011;193:5231–41. doi: 10.1128/JB.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luong T, Sau S, Gomez M, Lee JC, Lee CY. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect Immun. 2002;70:444–50. doi: 10.1128/IAI.70.2.444-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong YQ, Van Wamel W, Nast CC, Yeaman MR, Cheung AL, Bayer AS. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J Infect Dis. 2002;186:668–77. doi: 10.1086/342046. [DOI] [PubMed] [Google Scholar]
- 37.Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–99. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 38.Lee JC, Takeda S, Livolsi PJ, Paoletti LC. Effects of in vitro and in vivo growth conditions on expression of type 8 capsular polysaccharide by Staphylococcus aureus. Infect Immun. 1993;61:1853–8. doi: 10.1128/iai.61.5.1853-1858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baddour LM, Lowrance C, Albus A, Lowrance JH, Anderson SK, Lee JC. Staphylococcus aureus microcapsule expression attenuates bacterial virulence in a rat model of experimental endocarditis. J Infect Dis. 1992;165:749–53. doi: 10.1093/infdis/165.4.749. [DOI] [PubMed] [Google Scholar]
- 40.Nemeth J, Lee JC. Antibodies to capsular polysaccharides are not protective against experimental Staphylococcus aureus endocarditis. Infect Immun. 1995;63:375–80. doi: 10.1128/iai.63.2.375-380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson PK, Wilkinson BJ, Kim Y, Schmeling D, Quie PG. Influence of encapsulation on staphylococcal opsonization and phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1978;19:943–9. doi: 10.1128/iai.19.3.943-949.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voyich JM, Braughton KR, Sturdevant DE, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–19. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 43.Luong TT, Lee CY. Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect Immun. 2002;70:3389–95. doi: 10.1128/IAI.70.7.3389-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson IM, Lee JC, Bremell T, Rydén C, Tarkowski A. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect Immun. 1997;65:4216–21. doi: 10.1128/iai.65.10.4216-4221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang X, Yu C, Sun J, et al. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun. 2006;74:4655–65. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955–64. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–77. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–48. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 49.Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111–27. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.