Abstract
Ubiquitination is a posttranslational modification that regulates protein degradation and signaling in eukaryotes. Although it is acknowledged that pathogens exploit ubiquitination to infect mammalian cells, it remains unknown how microbes interact with the ubiquitination machinery in medically relevant arthropods. Here, we show that the ubiquitination machinery is present in the tick Ixodes scapularis and demonstrate that the E3 ubiquitin ligase named x-linked inhibitor of apoptosis protein (XIAP) restricts bacterial colonization of this arthropod vector. We provide evidence that xiap silencing significantly increases tick colonization by the bacterium Anaplasma phagocytophilum, the causative agent of human granulocytic anaplasmosis. We also demonstrate that (i) XIAP polyubiquitination is dependent on the really interesting new gene (RING) catalytic domain, (ii) XIAP polyubiquitination occurs via lysine (K)-63 but not K-48 residues, and (iii) XIAP-dependent K-63 polyubiquitination requires zinc for catalysis. Taken together, our data define a role for ubiquitination during bacterial colonization of disease vectors.
Keywords: ticks, Rickettsia, Ehrlichia, insecta, ubiquitin
Ubiquitin is an evolutionarily conserved protein that carries 7 lysine (K) amino acids (K6, K11, K27, K29, K33, K48, and K63). Ubiquitin may form linkages with the K of a target protein or the K of another ubiquitin [1, 2]. This process is referred to as protein ubiquitination and involves a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3) [1, 2]. Ubiquitination has emerged as a key mechanism regulating pathogenesis and immunity in mammals [3]. Ubiquitination plays a central role in adaptive immunity, antigen presentation, and regulation of several immune signaling pathways. The nuclear factor (NF)-κB family of transcription factors, Toll-like (TLRs), Nod-like (NLRs) and RIG-like receptors (RLRs) have all been shown to be regulated by ubiquitination [3]. Hence, pathogens have evolved strategies to manipulate the ubiquitination machinery and colonize the mammalian host (reviewed in [3–5]). Bacterial factors target host proteins for degradation via the ubiquitin-proteasome system (reviewed in [3]). Salmonella enterica and Burkholderia pseudomallei interfere with antimicrobial pathways preventing or reversing ubiquitin modifications, whereas Shigella flexneri and Listeria monocytogenes prevent ubiquitination, thus, evading autophagy (reviewed in [3–5]).
Surprisingly, medically relevant arthropod vectors have not been studied in the context of ubiquitination, and ubiquitin dynamics during pathogen colonization has yet to be explored. This is a scientific constraint because understanding the ubiquitination machinery in disease vectors may pave the ground for the development of novel therapeutics that prevent or delay the onset of illnesses. Here, we begin to unravel how ubiquitination regulates microbial pathogenesis in ticks. By using the blacklegged tick and the rickettsial bacterium Anaplasma phagocytophilum, the agent of human granulocytic anaplasmosis [6], we first show that the ubiquitome is functional in Ixodes scapularis. Then, we characterize the tick E3 ubiquitin ligase named x-linked inhibitor of apoptosis protein (XIAP) via biochemical and molecular assays. We demonstrate that XIAP polyubiquitination is dependent on the really interesting new gene (RING) domain, requires zinc for catalysis, and occurs via lysine (K)-63 but not K-48 residues. Finally, we use RNA interference (RNAi) to show that xiap silencing significantly increases colonization of I. scapularis ticks by A. phagocytophilum. Altogether, our findings shed some light onto microbial colonization of ticks and may serve as a prelude for discussions in terms of ubiquitin-pathogen interactions in disease vectors.
MATERIALS AND METHODS
Ethics Statement
Experiments were approved by the Institutional Animal Care and Use Committee (IACUC number A-20110030BE). C57BL/6 mice (6–10 weeks) were purchased from Jackson Laboratories. I. scapularis nymphs were obtained from Oklahoma State University and reared at 23°C with 85% relative humidity and 14 hour light/10 hour dark cycle. Experimentation with A. phagocytophilum (HZ strain) was approved by the Biological Use Authorization Committee (BUA number 20120020). A. phagocytophilum was grown in HL-60 cells, as described elsewhere [7].
I. scapularis siRNA Microinjection
The siRNA synthesis, details about I. scapularis ISE6 cells, and viability assays are available in supplemental materials and methods. In total, 10–15 I. scapularis nymphs were held with forceps and microinjected (Nanoject II, Drummond Scientific, Broomall, PA) in the abdomen at 45 degrees and a 46 ηL/second injection rate, with 9.2 ηL containing 1 × 1013 molecules/µL of xiap or scrambled siRNAs. I. scapularis were left to rest for 30 minutes to 2 hours and allowed to feed for 72 hours on A. phagocytophilum-infected C57BL/6 mice. Nymphs were then dissected under the microscope and salivary glands or midguts were processed either individually or in pools of 2 for analysis.
XIAP Cloning and Expression
Details about XIAP bioinformatics, an expanded version of the cloning and expression procedures, are available in supplemental materials and methods. We used the TOPO cloning strategy to clone I. scapularis XIAP and XIAP-ΔRING. OligoPerfect (Invitrogen, Grand Island, NY) was used to design primers (Table S1). Amplicons were ligated into pCR 2.1-TOPO and Escherichia coli TOP10 strain (Invitrogen, Grand Island, NY) was transformed. EcoRI and NotI restriction sites were added to I. scapularis XIAP and ΔRING-XIAP amplicons for subcloning. Amplicons were digested with EcoRI and NotI high-fidelity restriction enzymes (New England BioLabs, Ipswich, MA) and ligated into the digested EcoRI and NotI PGEX-6P-2 plasmid (GE Healthcare, Pittsburg, PA). XIAP and ΔRING-XIAP PGEX-6P-2 constructs were then used to transform the E. coli BL21 Gold (DE3) strain (Agilent, Santa Clara, CA). Expression was induced with isopropylthio-β-galactoside (IPTG; 0.1 mM) at 18°C for 20 hours. Purification and solubilization were performed, as described elsewhere [8]. Briefly, E. coli BL21 Gold (DE3) strain induced with IPTG was pelleted by centrifugation (3220 × g, 15 minutes, 4°C) and washed with sodium chloride and Tris-ethylenediaminetetraacetic acid (EDTA) (STE) buffer (150 mM NaCl, 10 mM Tris, pH 8.0, 1 mM EDTA). Pellets were resuspended in 1 mg/mL lysozyme in STE buffer and incubated for 1 hour at 4°C with rotation. Dithiothreitol (DTT) was added to a final concentration of 5 mM. Bacteria were lysed by the addition of 1.5% N-laurylsarcosine (sarkosyl: final concentration 1.5%). After sonication on ice, lysates were obtained by centrifugation (3220 × g, 4°C, 20 minutes). Supernatant was taken and Triton X-100 added to a concentration of 2%. Glutathione-S-transferase (GST) beads (BD Biosciences, Franklin Lakes, NJ) were added and incubated on a rotator at 4°C overnight. Beads were washed with cleavage buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM, EDTA, 1 mM DTT), and the GST fusion XIAP was cleaved with PreScission protease (GE Healthcare, Pittsburgh, PA) for 4 hours at 4°C.
Ubiquitination Assays
Ubiquitination assays were performed by combining 3 µg of I. scapularis GST-XIAP, XIAP, or XIAP-ΔRING with 0.3 µg E1, 0.1 µg of E2 enzymes, 5 µg of ubiquitin, 1.5 µL of 10× energy regeneration solution (ERS) and 2.5 µL of polyubiquitination buffer (50 mM Tris-HCl, pH 7.4, 1 mM DTT, 200 µM ZnCl2; Boston Biochem, Cambridge, MA). E2 enzymes were selected because they are commercially available (Boston Biochem) and commonly used. Reactions were carried out for 2 hours at 35°C. Samples were heated at 95°C in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) 4× sample buffer (Bio-Rad Hercules, CA) before loading onto 10% SDS-PAGE. Proteins were blotted onto 0.2 µm nitrocellulose membrane (Bio-Rad, Hercules, CA). Immunoblots were probed with primary antibodies at 4°C overnight (1:2500 Ubpan dilution, 1:1000 UbK48 dilution, 1:250 UbK63 dilution; Millipore, Billerica, MA). Antibody specificity was assessed by preincubating the antibodies with 4 µg of either TetraK48 or TetraK63-linked ubiquitin for 1 hour. Custom-made I. scapularis XIAP antibodies were obtained (Thermo Scientific, Lafayette, CO). XIAP (1:500 dilution) and GST (1:500 dilution; Calbiochem, Millipore, Billerica, MA) antibodies were used for autoubiquitination studies. Secondary antibodies were used at a 1:8000 dilution. Blot was covered with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Lafayette, CO). Immunoblots were stripped using Multi-Western Stripping Buffer (Bioland Scientific, Paramount, CA). For Zn chelation, 2.5 µg of XIAP were incubated with 2 mM tetrakis-(2-pyridylmethyl) ethylenediamine (TPEN) (Sigma-Aldrich, St. Louis, MO) or 0.5% ethanol (mock) overnight at 4°C. Samples were then treated with indicated amounts of ZnCl2 for 45 minutes at room temperature. For alkylation experiments, 3 µg of XIAP were treated with indicated concentrations of N-ethylmaleimide (NEM; Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline (PBS) for 30 minutes at room temperature. Details related to the confocal microscopy are available in supplemental materials and methods.
Statistical Analysis
Data were expressed as means ± standard errors of the mean (SEM). We used D'Agostino-Pearson omnibus test, unpaired Student t test and 1-way analysis of variance (ANOVA), followed by Bonferroni post hoc multiple-comparison tests. Analyses were performed using GraphPad Prism 5.04. P≤ .05 was considered statistically significant.
RESULTS
The I. scapularis Ubiquitome
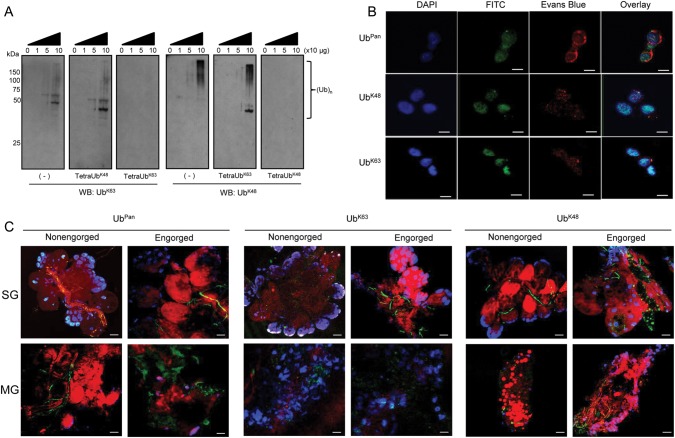
K48 (UbK48) and K63 (UbK63)-ubiquitination are the most widely studied ubiquitin chains [9]. Thus, we used antibodies specific for these linkages to determine whether UbK48- and/or UbK63-polyubiquitination are present in I. scapularis. We established that both UbK48 and UbK63 linkages are present in protein lysates of the ISE6 cell line (Figure 1A, left panels). To confirm specificity, we performed antibody-competition assays with linkage-specific tetraubiquitins. TetraUbK63 and tetraUbK48 are the minimum recognition units by the polyubiquitin antibodies [9]. Coincubation of UbK63- or UbK48-specific antibodies with their respective tetraubiquitin units abolished recognition of polyubiquitination (Figure 1A, right panels). Conversely, tetraUbK48 coincubated with the UbK63-specific antibody or tetraUbK63 coincubated with the UbK48-specific antibody did not affect polyubiquitination recognition (Figure 1A, center panels).
Figure 1.
Polyubiquitination in Ixodes scapularis. A, Protein lysates were obtained from I. scapularis ISE6 cells and aliquots (10–100 µg) resolved in 10% SDS-PAGE. Dashes represent immunoblots treated with UbK63 and UbK48 antibodies. Antibody specificity was assessed by preincubating the antibodies with either TetraUbK48 or TetraUbK63 for 1 h prior to immunoblotting. B, ISE6 cells, (C) tick salivary glands (SG), and midguts (MG) were fixed with paraformaldehyde and stained with DAPI (blue), Evans blue (red), and FITC (green) UbPan, UbK63, or UbK48 antibodies. The scale represents 10 µm in (B) and 20 µm in (C). Original magnification at 63× (B) and 20× (C). These experiments were repeated twice. Abbreviations: DAPI, 4′, 6-diamidino-2-phenylindole; FITC, fluoresceine-isothiocyanate; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Confocal microscopy with an antibody that recognizes a wide range of ubiquitin chains, here described as a pan ubiquitin (UbPan) antibody, showed wide polyubiquitination distribution across ISE6 cells (Figure 1B, upper panel). However, foci of UbK48 polyubiquitination in ISE6 cells revealed a pattern within the nuclear and perinuclear cellular region (Figure 1B, middle panel). Seemingly denser, UbK63 polyubiquitination foci patterns were observed in ISE6 cells (Figure 1B, lower panel). Because ticks experience a dramatic change in physiology during blood feeding [10], we addressed polyubiquitination in vivo. We focused our studies on salivary glands and midguts of nonengorged and engorged ticks because these organs are targeted by pathogens during a blood meal [10, 11]. It was difficult to estimate the extent to which differences observed were due to feeding or tissue reorganization because engorgement affected UbPan, UbK63 and UbK48 polyubiquitination dynamics in tick midguts and salivary glands (Figure 1C). As in ISE6 cells, UbK63 polyubiquitination was present in the nuclei of non-engorged tick salivary glands (Figure 1C, SG nonengorged, middle panel). Conversely, we did not detect UbK48 polyubiquitination in the nuclei of nonengorged tick salivary glands (Figure 1C, SG nonengorged, right panel). A more widespread distribution of UbK48 polyubiquitination was seen after tick engorgement in both salivary glands and midguts (Figure 1C, engorged, right panels). Less UbK63 ubiquitination was observed in blood-fed midguts when compared to UbK48 and UbPan, but it is unclear if this effect is due to engorgement or blood derived from mice. Nevertheless, UbK63 can still be seen in the nuclear area of the midgut cells (Figure 1C). Overall, our data support a functional ubiquitome in I. scapularis ticks.
I. scapularis XIAP Is an E3 Ubiquitin Ligase
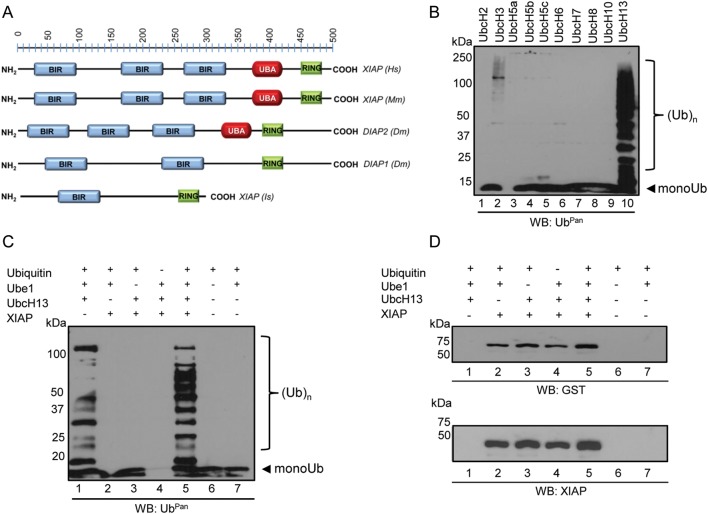
The human XIAP is an important E3 ubiquitin ligase involved in neutrophil infection by the tick-borne rickettsial agent A. phagocytophilum [12]. The tick XIAP sequence suggested similarities with mammalian XIAPs and related proteins in Drosophila (Figure 2A). However, I. scapularis XIAP is substantially shorter when compared to the mammalian and Drosophila proteins and does not carry 2 baculoviral IAP repeat (BIR) or the ubiquitin-associated (UBA) domains [13]. To address the role of I. scapularis XIAP in the context of ubiquitination, we expressed and purified this protein and performed assays with commercially available ubiquitin, E1 (Ube1) and E2 (UbcH) enzymes. We used recombinant I. scapularis XIAP derived from Escherichia coli because this system has been used for polyubiquitination assays [14–16]. We detected low levels of polyubiquitination when XIAP was incubated with the E2 enzyme UbcH3 (Figure 2B, lane 2) and high levels of polyubiquitination when XIAP was combined with UbcH13 (Figure 2B, lane 10). Addition of ubiquitin in the absence of E1 (Ube1), E2 (UbcH13), and XIAP yielded only monoubiquitination (Figure 2C, lane 6). As previously observed, UbcH13 alone is capable of producing polyubiquitin due to autoubiquitination (Figure 2C, lane 1) [17]. However, addition of I. scapularis XIAP revealed increased quantity and diversity of polyubiquitin in the 50–100 KDa range, as judged by Ubpan immunoblots (Figure 2C, lane 5).
Figure 2.
Ixodes scapularis XIAP is an E3 ubiquitin ligase. A, I. scapularis (Is) XIAP domains [(BIR: 61–135 aa., blue); (RING: 255–289 aa., green)] compared to related proteins in humans (Hs), mice (Mm), and Drosophila (Dm). UBA, ubiquitin associated domain (red). B–C, Ubiquitination assays followed by Western blot. Polyubiquitination assays were performed in the presence of ubiquitin, an E1 ubiquitin-activating enzyme (Ube1), E2 ubiquitin-conjugating enzymes (UbcH), and the tick recombinant E3 ubiquitin ligase XIAP expressed in Escherichia coli. Ten different E2 enzymes were used in (B), and UbcH13 was used as the E2 in (C and D). Aliquots were resolved in 12% SDS-PAGE and probed with an UbPan ubiquitin. Experiments were repeated at least twice. D, Polyubiquitination assays were performed in the presence of XIAP expressed in E. coli tagged with GST. Aliquots were resolved in 12% SDS-PAGE and probed for GST (upper panel) and XIAP (lower panel). Abbreviations: GST, glutathione-S-transferase; RING, really interesting new gene; XIAP, x-linked inhibitor of apoptosis protein.
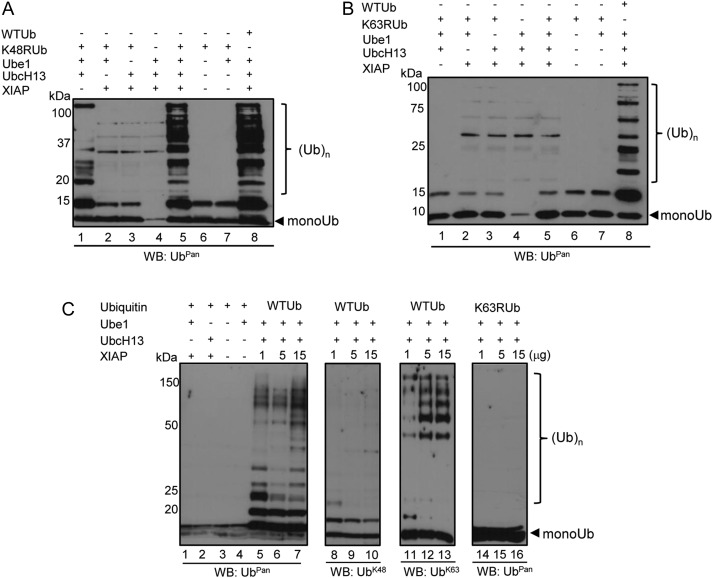
To demonstrate that the results obtained were not an UbcH13 artifact, we performed more stringent experiments. First, we used Ubpred [18] to predict XIAP autoubiquitination sites. Although Ubpred predicted that XIAP may be autoubiquitinated (Supplementary Figure 1A), we did not observe any autoubiquitination activity (Figure 2D). Immunoblots using 2 independent antibodies (GST tag or XIAP) showed that XIAP did not autoubiquitinate under our experimental conditions. Next, we used ubiquitins with lysine 63 (UbK63R) or lysine 48 mutated to arginine (UbK48R) to determine the type of linkages the I. scapularis XIAP is involved. Incubating XIAP with UbK48R did not show any alteration in activity (Figure 3A). On the other hand, polyubiquitination was not observed when XIAP was incubated with UbK63R ubiquitin (Figure 3B). As expected, incubation of XIAP and the wild-type ubiquitin (UbWT) showed polyubiquitination (Figure 3). Third, a dose-dependent polyubiquitination assay indicated that increased levels of I. scapularis XIAP enhanced polyubiquitination (Figure 3C, lanes 5–7). These results were confirmed with subsequent immunoblotting with UbK63 and UbK48 antibodies. UbK48 polyubiquitination was not observed when UbK48 immunoblots were performed (Figure 3C, lanes 8–10), whereas UbK63 polyubiquitination increased with higher amounts of XIAP (Figure 3C, lanes 11–13). Importantly, ubiquitin chains were not observed when the UbK63R mutant was used, despite increased levels of XIAP (Figure 3C, lanes 14–16). Altogether, our findings provide strong evidence that I. scapularis XIAP carries out UbK63-linked polyubiquitination.
Figure 3.
Ixodes scapularis XIAP promotes K63-linkage polyubiquitin chains. Ubiquitination assays were performed and followed by Western blot. A, K48RUb and (B) K63RUb were included in polyubiquitination assays. Wild-type (WT) ubiquitin was used as a positive control. C, WT and K63R ubiquitins were used. Aliquots were resolved in 12% SDS-PAGE and then probed with UbPan ubiquitin (lanes 1–7, 14–16), UbK48 (lanes 8–10), and UbK63-specific antibodies (lanes 11–13). These experiments were repeated at least twice. Abbreviation: XIAP, x-linked inhibitor of apoptosis protein.
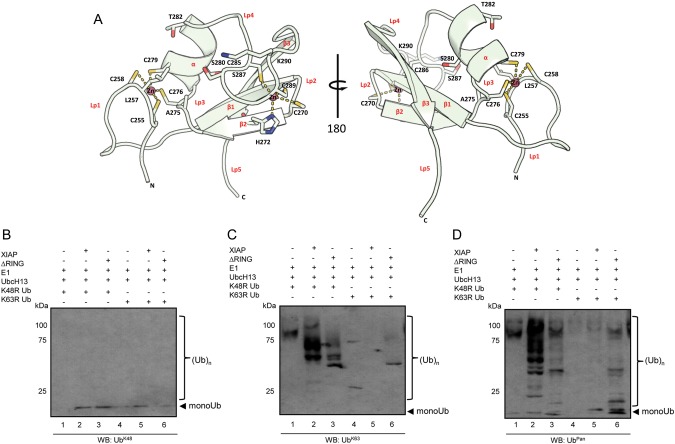
I. scapularis XIAP Requires the RING Domain for Polyubiquitination
To gain additional insight into I. scapularis XIAP function, we modeled its catalytic RING domain based on the E3 ubiquitin ligase MDMX, a negative regulator of the tumor suppressor protein p53 [19]. The I. scapularis XIAP RING domain consisted of 1 α-helix, 3 β-sheets, and 5 loops that accommodated 2 structural zinc ions folding in a “cross-brace” fashion (Figure 4A). Of the residues in the RING domain, cysteine and histidine amino acids were the most evolutionarily conserved (Supplementary Figure 1B). From the consensus amino acids [20], leucine, alanine, threonine, lysine, and isoleucine were retained, but one hydrophobic amino acid was replaced by a serine (Supplementary Figure 1C, in purple). Using antibodies specific for UbK48- and UbK63, we showed that XIAP polyubiquitination occurs via UbK63 but not UbK48 residues (Figure 4B and 4C). To assess the role of the RING domain in polyubiquitination, we expressed I. scapularis XIAP without this domain (XIAP-ΔRING) and performed polyubiquitination assays. When the RING domain was deleted from I. scapularis XIAP, polyubiquitination activity was greatly diminished when compared to the wild-type XIAP (Figure 4C and 4D, lane 3). Importantly, we noticed the residual autoubiquitination activity of UbcH13 (Figure 4C and 4D, lane 1), and the UbK48R mutant did not influence polyubiquitination by the wild-type XIAP (Figure 4C and 4D, lane 2). Conversely, wild-type I. scapularis XIAP catalysis was greatly influenced by the UbK63R mutant. I. scapularis XIAP catalysis did not occur when the UbK63R mutant was used (Figure 4C, lane 5).
Figure 4.
XIAP requires RING domain for polyubiquitination. A, Two views (180° rotated angle) of a ribbon diagram for the Ixodes scapularis XIAP RING domain based on protein threading with the published MdmX (2vjf:D) structure. This model shows the characteristic cysteine-histidine “cross-brace” conserved motif where cysteines and histidines provide zinc (Zn) coordination sites for maintenance of the protein structure. Side chains of some amino acid residues are shown in stick format with oxygen labeled orange, nitrogen labeled blue, and sulfur labeled yellow; α -helix, β-sheets, and secondary loop structures are labeled in red, whereas conserved and consensus amino acids are written in black. Hydrogen bonds are shown as dashed yellow lines. B–D, Polyubiquitination assays were performed using 3 µg XIAP and XIAP-ΔRING as ubiquitin ligases. Ubiquitin was replaced by UbK48R (lanes 1–3) or UbK63R (lanes 4–6) mutants. Reactions were immunoblotted (WB) with (B) UbK48, (C) UbK63, and (D) Ubpan antibodies. These experiments were repeated at least twice. Abbreviations: RING, really interesting new gene; XIAP, x-linked inhibitor of apoptosis protein.
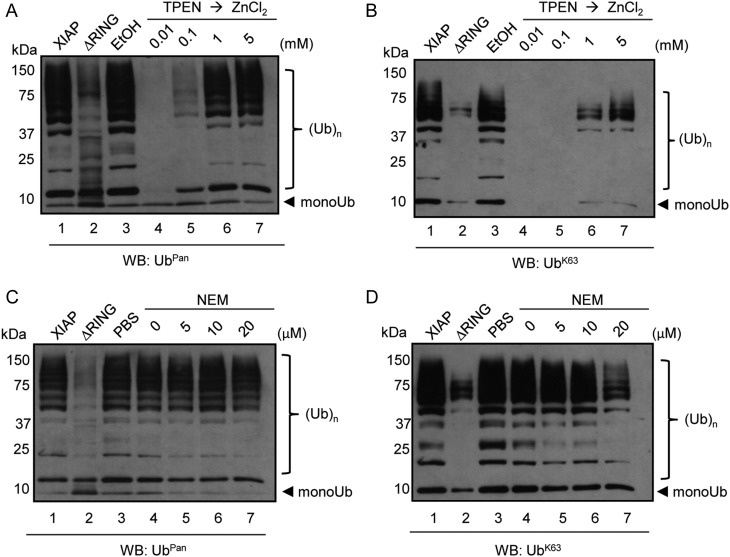
RING domains have previously been shown to require 2 zinc cations to provide a stable structure for E3 ligases [14]. To determine whether I. scapularis XIAP was sensitive to zinc depletion, XIAP was incubated with the zinc chelator TPEN [14] and subsequently rescued by the addition of ZnCl2. When I. scapularis XIAP was subsequently probed with an antibody that recognizes UbPan or UbK63, polyubiquitination activity was abrogated (Figure 5A and 5B, lane 4). Interestingly, XIAP-dependent polyubiquitination was readily restored in a ZnCl2 concentration-dependent manner (Figure 5A and 5B, lanes 5–7). We then tested the sensitivity of I. scapularis XIAP to the alkylating agent NEM. NEM interacts with the sulfhydryl group of cysteine residues in certain E3 ligases. However, RING-type E3 ligases are relatively insensitive to NEM activity [14, 21, 22]. We only observed an effect of NEM on XIAP at very high concentrations (Figure 5D, lane 7), perhaps, due to the alkylation of cysteine residues within the RING finger [21]. Overall, these results suggest that I. scapularis XIAP requires zinc cations for polyubiquitination activity, and the RING domain is essential for UbK63-type polyubiquitination.
Figure 5.
XIAP is resistant to NEM but sensitive to TPEN. A–B, 2.5 µg of full length XIAP or XIAP-ΔRING were incubated with 2 mM TPEN at 4°C overnight. 0.5% ethanol was used as a mock control. Samples were then incubated with increasing amounts of ZnCl2 (0.01 mM, 0.1 mM, 1 mM, and 5 mM) for 45 minutes at room temperature. The total mixtures were used in polyubiquitination assays, resolved in 10% SDS-PAGE and immunoblotted with (A) Ubpan and (B) UbK63 antibodies. C–D, 3 µg of XIAP or XIAP-ΔRING was incubated with increasing amounts of NEM (5 µM, 10 µM, and 20 µM) for 30 min at room temperature. Samples were then used in polyubiquitination assays. Reactions were immunoblotted with (C) Ubpan and (D) UbK63 antibodies. These experiments were repeated at least twice. Abbreviations: NEM, N-ethylmaleimide; RING, really interesting new gene; XIAP, x-linked inhibitor of apoptosis protein.
XIAP Restricts A. phagocytophilum Colonization of I. scapularis
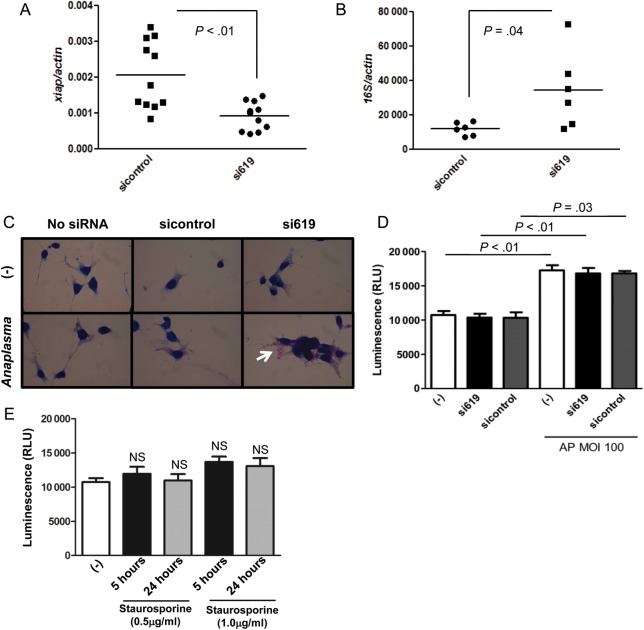
Because human XIAP was previously associated with A. phagocytophilum infection [12], we designed siRNA to determine whether the tick XIAP had any role in microbial pathogenesis. From 2 siRNAs designed, siRNA targeting the nucleotide positions 619–639 (si619) was proven to be the most successful (Supplementary Figure 2). We then compared A. phagocytophilum load in xiap silenced (si619) vs nonsilenced ISE6 cells (sicontrol; Figure 6A). A. phagocytophilum infection increased upon xiap silencing in ISE6 cells, as indicated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Romanowsky staining (Figure 6B and 6C). XIAP in mammals has been associated with apoptosis [12, 23]. Thus, we silenced xiap in ISE6 tick cells and verified cell death. We used adenosine triphosphate (ATP) quantification as a read-out for metabolically active cells because measuring cell death with standard mammalian lactate dehydrogenase (LDH) assays was hampered by high background levels—most likely due to the complexity of the ISE6 cell culture media. Importantly and similar to mammalian cells [6], A. phagocytophilum inhibited cell death in tick cells at multiplicity of infection (MOI) 100 (Figure 6D). We also observed that xiap silencing did not affect cell death in I. scapularis ISE6 cells (Figure 6D), suggesting that this gene may not have an apoptotic role in I. scapularis. Alternatively, the redundancy of the I. scapularis genome [24] could have “masked” the phenotype. This is reasonable because stimulation of I. scapularis ISE6 cells with different concentrations of staurosporine, a common trigger for mammalian cell apoptosis [25], did not induce cell death at 5 and 24 hours poststimulation (Figure 6E).
Figure 6.
Xiap silencing facilitates Anaplasma phagocytophilum colonization of ISE6 cells and does not affect cell death. A, ISE6 cells (1 × 105) were transfected with xiap (si619) or scrambled siRNA (sicontrol) and xiap expression analyzed by qRT-PCR to confirm silencing. B, Xiap (si619) or scrambled siRNA (sicontrol) transfected ISE6 cells (n = 6) were infected with A. phagocytophilum (MOI 100) for 24 h and A. phagocytophilum load was measured by qRT-PCR using the ΔΔCt method. C, ISE6 cells (2 × 104) were stained by using kwik-diff, a commercial Romanowsky variant stain. A. phagocytophilum morulae are shown in purple (white arrow), whereas I. scapularis cells are shown in dark blue. D, ISE6 cells (2 × 104) were transfected with siRNA 619 (600 ng) or siRNA control (600 ng) and infected with A. phagocytophilum (MOI 100) for 24 h posttransfection. ATP presence signals cell viability and was measured as relative luminescence units (RLU). These experiments were repeated twice. E, ISE6 cells (2 × 104) were treated with staurosporine at indicated concentrations and cell viability (as judged by ATP presence) was measured at described time points. Error bars (D and E) represent standard error of the mean. Statistical analysis (P < .05) was performed using Student t test (A, B, D) and ANOVA (Bonferroni) (E). Abbreviations: ANOVA, analysis of variance; ATP, adenosine triphosphate; MOI, multiplicity of infection; qRT-PCR, quantitave reverse transcription polymerase chain reaction.
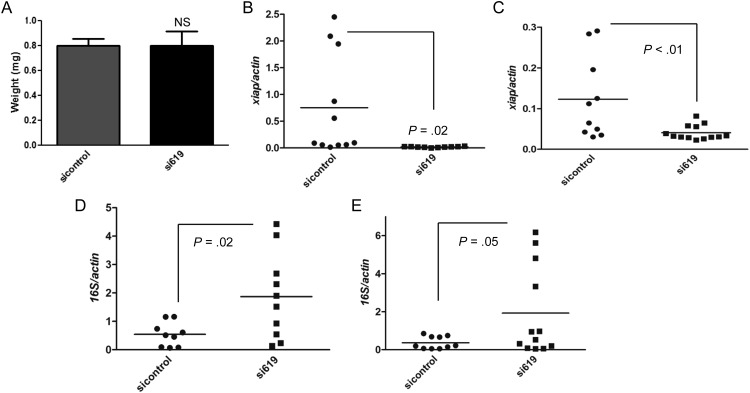
I. scapularis nymphs were also microinjected with siRNAs. No difference in feeding (as judged by tick engorgement) was observed between ticks injected with xiap (si619) and control siRNAs (Figure 7A). This is important because it suggested that both groups of ticks were feeding similarly and xiap silencing did not influence engorgement. Silencing was obtained in tick salivary glands (Figure 7B) and midguts (Figure 7C). A. phagocytophilum load was also found to be higher in I. scapularis upon xiap silencing (Figure 7D and 7E). These findings suggest that xiap restricts A. phagocytophilum colonization of I. scapularis ticks.
Figure 7.
XIAP restricts Anaplasma phagocytophilum colonization of Ixodes scapularis. A–E, I. scapularis nymphs were body injected with 9.2 ηL containing 1 × 1013 molecules/µL of xiap siRNA (si619) or scrambled siRNA (sicontrol) and allowed to feed for 72 h on A. phagocytophilum infected C57BL/6 mice. A, Average weight of ticks (n = 30) treated with sicontrol and si619 is shown. Xiap is silenced in (B) salivary glands (SG) and (C) midguts (MG). D and E, A. phagocytophilum load was measured in the (D) SG and (E) MG by qRT-PCR using the ΔΔCt method for the 16S gene relative to tick β-actin expression. Each dot indicates individual or pools of 2 ticks (n = 15 per group). Experiments were repeated twice. Error bars in (A) represent standard error of the mean. Statistical analysis was performed using the Student t test (P≤ .05). Abbreviations: qRT-PCR, quantitave reverse transcription polymerase chain reaction; XIAP, x-linked inhibitor of apoptosis protein.
DISCUSSION
How polyubiquitination regulates pathogen colonization of medically relevant arthropods has not yet been determined. Here, we describe a tick E3 ubiquitin ligase, named XIAP, restricting bacterial colonization of an arthropod vector. Polyubiquitination has been widely demonstrated to regulate microbial pathogenesis and immunity [26, 27]. For example, NF-κB activation is controlled by polyubiquitination in MyD88-dependent pathways following exposure to pathogens [27]. The E3 ubiquitin ligase TRAF6 is also recruited when TLRs are activated, leading to UbK63 polyubiquitination of kinases [26]. This mechanism appears evolutionarily conserved because UbK63 polyubiquitination of the DREDD caspase and the immunodeficiency (IMD) molecule requires DIAP2 during infection of Drosophila [28]. I. scapularis XIAP and DIAP2 share similarities, and our results indicate that the tick XIAP catalyzes UbK63 polyubiquitination via the RING domain, despite the absence of an UBA domain. This corroborates with findings that illustrated that the mammalian XIAP does not require the UBA domain for E3 ligase activity [29].
We posit that XIAP-mediated UbK63 polyubiquitination may regulate immune signaling during A. phagocytophilum colonization of ticks. This hypothesis is supported by increased A. phagocytophilum acquisition of I. scapularis after xiap silencing. Interaction between XIAP and UbcH13, a protein that shares strong similarities with bendless in I. scapularis (E value <2e-96) and a modulator of the IMD pathway in arthropods [30], reiterates our reasoning. It is unclear how RING domains of E3 ubiquitin ligases transfer ubiquitin to substrate proteins. It is suggested that a dimeric XIAP RING domain is necessary for polyubiquitination activity [31]. Though not yet proved, we have some evidence of XIAP dimerization during A. phagocytophilum infection of ISE6 cells. XIAP dimerization appears very strong because our attempts to rupture this dimer under different conditions were unsuccessful.
I. scapularis XIAP was not found to be autoubiquitinated. This is contrary to reports observed for XIAP homologues in mammals, where autoubiquitination and proteasomal degradation seems to be a requirement for apoptosis [13]. Although we did not observe any effect of I. scapularis XIAP on cell death, we do not exclude the possibility that I. scapularis XIAP may perform autocatalytic functions under physiological conditions, as many E3 ligases require accessory proteins for their activity [5, 13, 32, 33]. As previously shown, I. scapularis XIAP share similarity with MdmX, and this protein interacts with another E3 ubiquitin ligase named Mdm2 through its RING domain [19]. Undoubtedly, clarifying the physiological role of XIAP during pathogen infection of ticks will be important. However, this endeavor is not currently possible because the technology to insert or delete genes in ticks is not available.
Understanding the polyubiquitination machinery may allow for the development of innovative strategies to treat vector-borne illnesses. It would be fascinating to apply chemical screening assays with the intent of modulating the arthropod ubiquitome. This approach would be a first step toward the development of structure-based molecules that target vector-pathogen interactions. This is not unreasonable as pharmacological inhibitors named second mitochondria-derived activator of caspase (SMAC) mimetics have been successfully used in Drosophila [34]. Hence, SMAC mimetics may provide novel therapeutic opportunities for the treatment of vector-borne diseases. In summary, the results presented here promote a significant advancement in ubiquitin biology in the context of pathogen colonization of medically relevant arthropod vectors.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Ulrike G. Munderloh, Jason Stajich, Sukanya Narasimhan, and Kathleen DePonte for excellent technical assistance; invaluable colleagues for intellectual input and manuscript comments; and the Institute for Integrative Genome Biology Core Facilities at the University of California-Riverside.
Financial support. This work was supported by the Centers for Disease Control and Prevention (K01 CK000101 to J. H. F. P.); the National Institutes of Health (R01 AI093653 to J. H. F. P.); the Initial Complement provided by the University of California to J. H. F. P.; and by an International Fellowship from the American Association of University Women to M. S. S.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 2.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annual Review of Biochemistry. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steele-Mortimer O. Exploitation of the ubiquitin system by invading bacteria. Traffic. 2011;12:162–9. doi: 10.1111/j.1600-0854.2010.01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins CA, Brown EJ. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends in Cell Biology. 2010;20:205–13. doi: 10.1016/j.tcb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Severo MS, Stephens KD, Kotsyfakis M, Pedra JH. Anaplasma phagocytophilum: deceptively simple or simply deceptive? Future Microbiology. 2012;7:719–31. doi: 10.2217/fmb.12.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Severo MS, Sakhon OS, et al. Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infection and Immunity. 2012;80:3194–3205. doi: 10.1128/IAI.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Analytical Biochemistry. 1993;210:179–87. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 9.Newton K, Matsumoto ML, Wertz IE, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–78. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 10.Piesman J, Eisen L. Prevention of tick-borne diseases. Annual Review of Entomology. 2008;53:323–43. doi: 10.1146/annurev.ento.53.103106.093429. [DOI] [PubMed] [Google Scholar]
- 11.Bowman AS, Sauer JR. Tick salivary glands: function, physiology and future. Parasitology. 2004;129(Suppl):S67–81. doi: 10.1017/s0031182004006468. [DOI] [PubMed] [Google Scholar]
- 12.Ge Y, Rikihisa Y. Anaplasma phagocytophilum delays spontaneous human neutrophil apoptosis by modulation of multiple apoptotic pathways. Cellular Microbiology. 2006;8:1406–16. doi: 10.1111/j.1462-5822.2006.00720.x. [DOI] [PubMed] [Google Scholar]
- 13.Beug ST, Cheung HH, Lacasse EC, Korneluk RG. Modulation of immune signalling by inhibitors of apoptosis. Trends Immunol. 2012;33:535–45. doi: 10.1016/j.it.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 15.Mace PD, Linke K, Feltham R, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–40. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 16.Grant K, Grant L, Tong L, Boutell C. Depletion of intracellular zinc inhibits the ubiquitin ligase activity of viral regulatory protein ICP0 and restricts herpes simplex virus 1 replication in cell culture. J Virol. 2012;86:4029–33. doi: 10.1128/JVI.06962-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doss-Pepe EW, Chen L, Madura K. α-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J Biol Chem. 2005;280:16619–24. doi: 10.1074/jbc.M413591200. [DOI] [PubMed] [Google Scholar]
- 18.Radivojac P, Vacic V, Haynes C, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Jiang X. Mdm2 and MdmX partner to regulate p53. FEBS Letters. 2012;586:1390–6. doi: 10.1016/j.febslet.2012.02.049. [DOI] [PubMed] [Google Scholar]
- 20.Ying M, Huang X, Zhao H, et al. Comprehensively surveying structure and function of RING domains from Drosophila melanogaster. PLoS One. 2011;6:e23863. doi: 10.1371/journal.pone.0023863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seol JH, Feldman RM, Zachariae W, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Development. 1999;13:1614–26. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–80. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagel Van Zee J, Geraci NS, Guerrero FD, et al. Tick genomics: the Ixodes genome project and beyond. Int J Parasitol. 2007;37:1297–305. doi: 10.1016/j.ijpara.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Luhrmann A, Roy CR. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun. 2007;75:5282–9. doi: 10.1128/IAI.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandenabeele P, Bertrand MJ. The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling. Nat Rev Immunol. 2012;12:833–844. doi: 10.1038/nri3325. [DOI] [PubMed] [Google Scholar]
- 28.Silverman N, Paquette N, Aggarwal K. Specificity and signaling in the Drosophila immune response. Invertebrate Surviv J. 2009;6:163–174. [PMC free article] [PubMed] [Google Scholar]
- 29.Gyrd-Hansen M, Darding M, Miasari M, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-κB as well as cell survival and oncogenesis. Nat Cell Biol. 2008;10:1309–17. doi: 10.1038/ncb1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meinander A, Runchel C, Tenev T, et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–83. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feltham R, Khan N, Silke J. IAPS and ubiquitylation. IUBMB Life. 2012;64:411–8. doi: 10.1002/iub.565. [DOI] [PubMed] [Google Scholar]
- 32.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–44. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 34.Chew SK, Chen P, Link N, Galindo hour KA, Pogue K, Abrams JM. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature. 2009;460:123–7. doi: 10.1038/nature08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.