Abstract
Ape chromosomes homologous to human chromosomes 14 and 15 were generated by a fission event of an ancestral submetacentric chromosome, where the two chromosomes were joined head-to-tail. The hominoid ancestral chromosome most closely resembles the macaque chromosome 7. In this work, we provide insights into the evolution of human chromosomes 14 and 15, performing a comparative study between macaque boundary region 14/15 and the orthologous human regions. We construct a 1.6-Mb contig of macaque BAC clones in the region orthologous to the ancestral hominoid fission site and use it to define the structural changes that occurred on human 14q pericentromeric and 15q subtelomeric regions. We characterize the novel euchromatin–heterochromatin transition region (∼20 Mb) acquired during the neocentromere establishment on chromosome 14, and find it was mainly derived through pericentromeric duplications from ancestral hominoid chromosomes homologous to human 2q14–qter and 10. Further, we show a relationship between evolutionary hotspots and low-copy repeat loci for chromosome 15, revealing a possible role of segmental duplications not only in mediating but also in “stitching” together rearrangement breakpoints.
Human chromosomes 14 and 15 were generated by fission of an ancestral chromosome composed of chromosome 14 attached, head-to-tail, to chromosome 15 around 25 million years ago (Ventura et al. 2003). The rearrangement occurred in the hominoid ancestor and involved a cluster of olfactory receptor (OR) genes (Rudd et al. 2009). At the point of fission, the 15q telomere and the neocentromere of chromosome 14 were seeded (Ventura et al. 2003). The 15q subtelomeric region experienced numerous structural rearrangements, including interstitial deletions and transfers of material to and from other subtelomeric regions (Rudd et al. 2009).
Low-copy repeats (LCR), also named segmental duplications (SDs) (Bailey et al. 2001; Cheung et al. 2001), mediate chromosomal rearrangements through non-allelic homologous recombination (NAHR) events (Hastings et al. 2009) and are enriched at pericentromeric and subterminal regions of the human genome (Bailey and Eichler 2006). LCR15 (low-copy repeats on human chromosome 15) loci have been widely studied (Pujana et al. 2001; Zody et al. 2006); those of primary focus are on 15q11–q13 because of their involvement in the Prader-Willi and Angelman syndromes (Pujana et al. 2002).
In this work, we provide insights into and highlight the role of LCR in the evolution of human chromosomes 14 and 15. We assembled a 1.6-Mb contig of macaque BAC clones encompassing the 14/15 boundary and tracked evolutionary changes initiated by the fission in the human lineage. We detected duplications from the newly seeded subtelomeric region on chromosome 15 and pericentromeric region on chromosome 14 to other existing subtelomeric and pericentromeric regions, respectively. Further, we traced the dynamics of the formation of the chromosome 14 neocentromere and demonstrated the recruitment of new pericentromeric sequences mainly from the pericentromeric regions of ancestral chromosomes orthologous to human 2q14–qter and 10. We found a correlation between clusters of LCR15 and evolutionary “hotspot” regions. (Note: For simplicity, we used the orthologous numbering of great ape and human chromosomes as proposed for great ape genome sequencing projects [Supplemental Fig. S1; McConkey 2004].)
Results
Evolutionary history of chromosomes 14 and 15
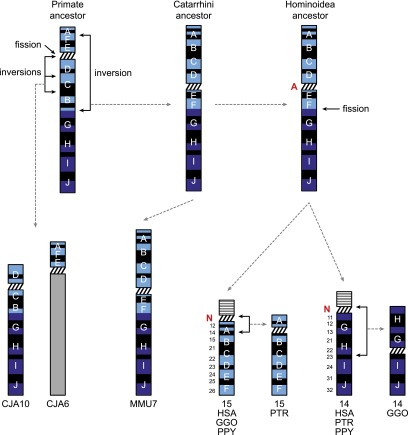
We reconstructed the evolutionary history of chromosomes 14 and 15 in the primate lineage from published data (Yunis et al. 1980; Yunis and Prakash 1982; Locke et al. 2003; Ventura et al. 2003; Zody et al. 2006; Stanyon et al. 2008; Locke et al. 2011; Ventura et al. 2011) and data from the marmoset genome sequencing project (Marmoset Genome Sequencing Consortium, unpubl.). The synteny of chromosomes 14/15 is ancestral to placental mammals (Wienberg and Stanyon 1995, 1997; Kent et al. 2002; Pontius et al. 2007; Wade et al. 2009; Meyer et al. 2013), as demonstrated by cat chromosome B3, horse chromosome 1, and rabbit chromosome 17 (UCSC Genome Browser alignment net, http://genome.ucsc.edu/). In the primate ancestor, chromosomes 14 and 15 were a submetacentric chromosome with a small p arm (Fig. 1). In the marmoset lineage, one fission and two inversions derived marmoset chromosomes 6p and 10, with 6p maintaining the ancestral association of A–F–E markers (Zody et al. 2006). A pericentric inversion in the Catarrhini ancestor generated a submetacentric chromosome with the same organization as the rhesus macaque chromosome 7 (MMU7). In the Hominoidea ancestor the submetacentric chromosome underwent a fission event and generated the acrocentric chromosomes 14 and 15. The ancestral centromere was inactivated and two neocentromeres emerged (Ventura et al. 2003). Finally, a pericentric inversion occurred in the gorilla and chimpanzee lineages, respectively on chromosomes 14 and 15 (Yunis et al. 1980; Yunis and Prakash 1982; Locke et al. 2003; Ventura et al. 2011).
Figure 1.
Evolutionary history of chromosomes 14 and 15. Ten segments (A–J) are used to track the chromosomal rearrangements and are defined as in Stanyon et al. (2008). The following abbreviations are used: A in red, ancestral centromere; N in red, evolutionary neocentromere; (HSA) Homo sapiens; (PTR) Pan troglodytes; (GGO) Gorilla gorilla; (PPY) Pongo pygmaeus; (MMU) Macaca mulatta; (CJA) Callithrix jacchus. In Figures 1, 2, and 5, chromosomes 14 and 15 are colored according to the color code of the UCSC Genome Browser (dark blue for chromosome 14 and light blue for chromosome 15). The NOR (nucleolus organizer region) bearing short arm is drawn with a horizontal black/white striped box.
Comparative analysis of the human and macaque genomic regions orthologous to the hominoid 14/15 fission site
MMU7 has a chromosomal organization resembling the ancestral hominoid 14/15 association and shows perfect marker order conservation with respect to the two human-derivative chromosomes (Ventura et al. 2003). To gain insights into the genomic organization of the hominoid fission region, we constructed a 1.6-Mb contig of 61 macaque BAC clones on MMU7 spanning the 14/15 boundary orthologous to human 14q11 and 15q26.3 (Fig. 2; Supplemental Table S1). We screened the macaque genomic library, CHORI-250, using STS derived from the human 15q subterminal (Hsa15a–d) and 14q pericentromeric (Hsa14a–c) regions (hg18/build36) together with macaque-specific STS derived from CH250 BAC end sequences (BES). The corresponding region was single copy in macaque, except for a 134-kb segment duplicated in tandem. We FISH- (fluorescence in situ hybridization) mapped all clones on both macaque and human chromosome metaphases and grouped them accordingly by FISH pattern (F1–F3 clones map to human 15q26.3; G1–G4 clones map to human 14q11); most of the clones were single copy in macaque and duplicated intra- and/or interchromosomally in human (Supplemental Note). We generated 91 BES and mapped them on the macaque (rheMac2) and human (hg18/build36) reference genome assemblies. We found consistent placements on rheMac2, except for those partly covering assembly gaps. In the human reference assembly, several BACs were one-end anchored and some end sequences showed multiple matches due to the presence of SDs in the human genome (Supplemental Table S1).
Figure 2.
Contig of CH250 BAC clones on MMU7. The schematic shows features of the macaque genomic region spanned by the contig (chr7:81552000–83132000, rheMac2). In particular, gap and STS locations, BAC clones, FISH mapping data, chained alignments with hg18 reference, WSSD (whole-genome shotgun sequence detection) depth-of-coverage data (the green color corresponds to single copy; the red color to duplication; Marques-Bonet et al. 2009), macaque RefSeq genes, and human RefSeq genes in the orthologous human regions are shown. STS names are color-coded: STS used in radioactive hybridization (orange), STS used in PCR amplification (blue), and STS used in both assays (green). FISH results on MMU7 and human chromosomes 14/15 are shown for representative BAC clones marked by names and arrowheads in red. Seven different FISH patterns are identified: BAC clones of Groups F1–F3 map to the homologous region on chromosome 15 (light blue); BAC clones of Groups G2–G4 map to the homologous region on chromosome 14 (dark blue); and gray-colored BAC clones (Group G1) showed absent or weak signal on human chromosome 14 and correspond to the fission breakpoint. An asterisk in front of the BAC name indicates BAC clones belonging to the second copy of the macaque duplication. A yellow arrowhead marks BES not anchored to the human reference hg18. Additional FISH signals on other human chromosomes due to the presence of interchromosomal SDs (further indicated by the chained alignments) are shown for the BAC clones of Groups F3, G2, and G3. An empty black box marks the human GOLGA2–ITSN2 core duplicon. Both end segments of the contig (Groups F1 and G4) are in single copy and direct orientation in macaque and human.
Extreme groups (F1 and G4) are made up of clones spanning single copy and directly oriented segments in human and macaque. For the homologous region on chromosome 15, we identified an LCR15 copy in macaque and human genomes (Group F2) and human subterminal duplications of the last segment preserving the same centromere–telomere orientation except for 1p (Group F3) (Supplemental Note). We detected clones (Groups F3 and G1) covering the evolutionary fission breakpoint as revealed by the BAC end mapping (one-end anchored on human chromosomes 14 or 15) and absence of signal on human metaphases for three clones. For the homologous region on chromosome 14, we detected three human interchromosomal pericentromeric duplications (15q11, 18p11, and 21q11; Groups G2 and G3) dated to have duplicated prior to human–orangutan divergence (Supplemental Note). Furthermore, we identified a hominoid-specific inversion and macaque tandem duplication with 99.85% identity (Fig. 2) terminally flanked by human and macaque single copy as well as human/macaque direct-oriented regions. This suggests that the hominoid inversion and 14–15 duplication, as well as the macaque tandem duplication, end at the same position. Gene content analysis showed the presence of OR genes restricted to the duplicated regions in human and macaque (Fig. 2; Supplemental Table S1; Supplemental Note).
Relationships between the LCR15 and chromosome 15 evolutionary hotspots
Since we found an LCR15 copy in proximity to the fission point, we investigated the relationships between the LCR15 (ITSN2/DNM1–GOLGA2 duplications) (Supplemental Note) and chromosome 15 evolutionary breakpoints in the primate lineage using molecular cytogenetic and phylogenetic analyses. We performed three-color FISH experiments on human, macaque, and marmoset chromosome metaphases of 10 RP11 BAC triplets designed at 15q11.2–q14 and 15q24.2–q25.2 in addition to a CH250 BAC triplet for LCR15 at 15q26.3 (Fig. 3). We identified six LCR15 loci corresponding to evolutionary hotspots (named AB, D1, D2, DE1, DE2, and F) and the LCR15 cluster at the novel pericentromeric region (15q11.2, locus A). (Note: the loci nomenclature links up the evolutionary history markers in Fig. 1.) Loci AB and F are orthologous to the evolutionary Catarrhini inversion breakpoints and show orthologous LCR15 duplications in human and macaque, but not in marmoset, whereas locus DE1 is orthologous to the marmoset fission breakpoint and has orthologous LCR15 copies in all three species (Figs. 1, 3). BAC clones at 15q24–q25 (loci D1, D2, DE1, and DE2) mapped to the pericentromeric region of both the macaque and marmoset chromosomes, orthologous to the ancestral hominoid pericentromeric region. Noteworthy, BAC clones spanning LCR15 at loci D1 and D2 exclusively showed a tandem duplicated signal in all three species, comprising the marmoset 10p pericentromeric region. Alternatively, three-color FISH experiments using BACs for the duplication clusters at the remaining loci did not show any break of synteny or pericentromeric localization in the three species analyzed (data not shown).
Figure 3.
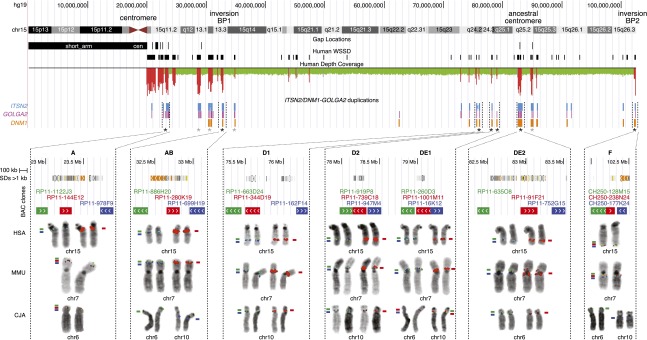
Relationships between the ITSN2/DNM1–GOLGA2 duplications and chromosome 15 evolutionary hotspots. Gap locations, WSSD-positive regions, depth-of-coverage data, and locations (BLAT search; Kent 2002) of the ITSN2, DMN1, and GOLGA2 duplications are shown for the sequence of chromosome 15 (hg19/build37). (Note, only a few LCR15 copies lack one of the three components.) Asterisks indicate the 11 loci analyzed through FISH. The BAC triplets were chosen as follows: one BAC (in red) spanning LCR15 and the other two at the single-copy regions immediately upstream (in green) and downstream (in blue) of the duplication cluster. SDs (hg19) and FISH mapping data on human, macaque, and marmoset chromosomes are shown for seven loci (named A, AB, D1, D2, DE1, DE2, and F, and indicated by a black asterisk). For BAC clones spanning LCR15 sequences (red-colored) only signals co-mapping with the single-copy clones (green and blue colored) are shown. BAC clones designed in single-copy regions of loci AB and F co-hybridized in human and macaque, whereas in marmoset they hybridized on chromosomes 6 and 10, respectively, for green- and blue-colored clones; no signal is on marmoset chromosomes for the red-colored BAC. Locus DE1 single-copy BAC clones mapped to marmoset chromosomes 10 and 6, respectively; the red-colored BAC clone showed signal on both chromosomes. Locus DE2 single-copy BAC clones hybridized to the macaque 7p and 7q pericentromeric regions, respectively.
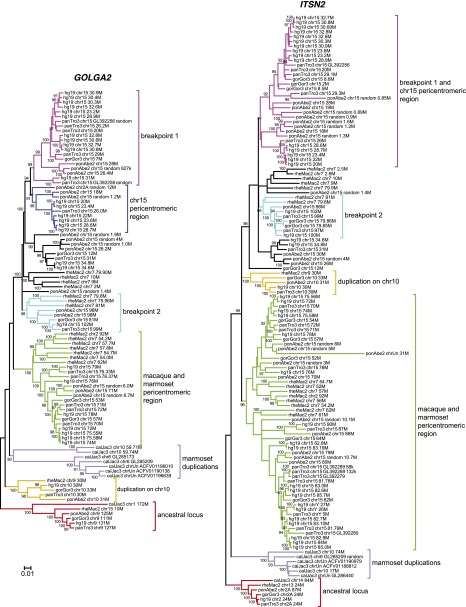
GOLGA2 and ITSN2 neighbor-joining trees reported monophyletic clades of duplications mapping to orthologous locations of the ape pericentromeric region (human 15q11.1–q13.1), the macaque and marmoset pericentromeric region (human 15q24–q25), and the Catarrhini inversion breakpoints (human 15q11.2–q13.3 and 15q26.3) (Fig. 4). Marmoset LCR15 copies map to chromosomes 6 and 10, as well as to unassembled scaffolds, and are monophyletic (bootstrap values equal to 80 and 100), likely because they homogenized. The ortholog to the human chromosome 10 copy is annotated in ape and macaque genomes; however, no copy is annotated in the marmoset genome though the assembly of the region appears complete (Supplemental Fig. S2).
Figure 4.
Primate phylogeny of GOLGA2 and ITSN2 duplicons. Neighbor-joining phylogenetic trees were constructed using a 2.5-kb region of the GOLGA2 module and a 3.3-kb region of the ITSN2 module. Sequences were retrieved from human (hg19), chimpanzee (panTro3), gorilla (gorGor3), orangutan (ponAbe2), macaque (rheMac2), and marmoset (calJac3) reference genomes. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site and bootstrap values (1000 replicates) shown. Monophyletic clades of duplicons mapping to the ancestral loci (9q34.11 and 2p23.3 for GOLGA2 and ITSN2, respectively), chromosome 10, marmoset duplications, macaque and marmoset pericentromeric region, inversion breakpoints, and the pericentromeric region of ape chromosome 15 are highlighted in color code. The copies at the proximal inversion breakpoint and ape pericentromeric region homogenized, especially in the ITSN2 phylogenetic tree, do not group into two different branches.
Study of human chromosome 14 newly minted pericentromeric region
To analyze human 14q pericentromeric sequence acquired after the fission event, we followed two different approaches: (1) screening of macaque high-density filters using three STS at chr14:19 Mb (hg18) (Supplemental Table S3); and (2) FISH mapping comparison in human, orangutan, and macaque of an RP11 BAC clone tiling path spanning the entire assembled sequence. In the first approach, we mapped macaque positive BAC clones on both human and macaque by FISH, BES alignment on reference genome assemblies (rheMac2 and hg18), and sequencing of representative clones using Illumina technology (Supplemental Table S2). We arranged positive clones into six groups (I–VI) according to the main FISH signals on macaque chromosomes and FISH-mapped one clone per group on chimpanzee, gorilla, and orangutan chromosomes (Supplemental Fig. S7; Supplemental Table S3). The majority of macaque clones (Groups I–IV) mapped in all primate species to human 2q21 orthologous location, an ancestral primate pericentromeric region (Yunis and Prakash 1982; Wienberg et al. 1994; Roberto et al. 2008), and showed interchromosomal pericentromeric duplication in apes. Of note, Group IV reported a tandem duplicated signal at the macaque 9q pericentromeric region (orthologous to human chromosome 10), but orthologous FISH localization was identified only in orangutan chromosomes. The end sequences of this group had inconsistent or no homologous placement on both the macaque and human genome references, suggesting that the macaque 9q pericentromeric sequence is incomplete in the macaque reference and differs significantly from the human 10q pericentromeric sequence. This finding was confirmed by the Illumina sequencing of two clones (CH250-48F1 and CH250-34C6). On the other hand, Group V and VI clones mapped to a nonpericentromeric region (human 10q22.3 orthologous location) and to a different pericentromeric region (human 12p11 orthologous location), respectively. At human 14q11, clones showed signal (Groups II, III, and IV), faint signal (Groups I and V), or no signal (Group VI).
We found a similar pattern by a reciprocal approach using human BAC clones (Supplemental Fig. S8). In human, 14q pericentromeric sequence (tandem duplicated signal at this locus) showed interchromosomal pericentromeric duplication mainly with chromosome 22. Of note, all clones hybridized to human 2q21 and the orangutan orthologous locus along with the macaque 9q pericentromeric region. In orangutan they showed interchromosomal pericentromeric duplicated signals (in two cases at 10q); in addition, four clones showed a tandem duplicated signal on macaque chromosome 10.
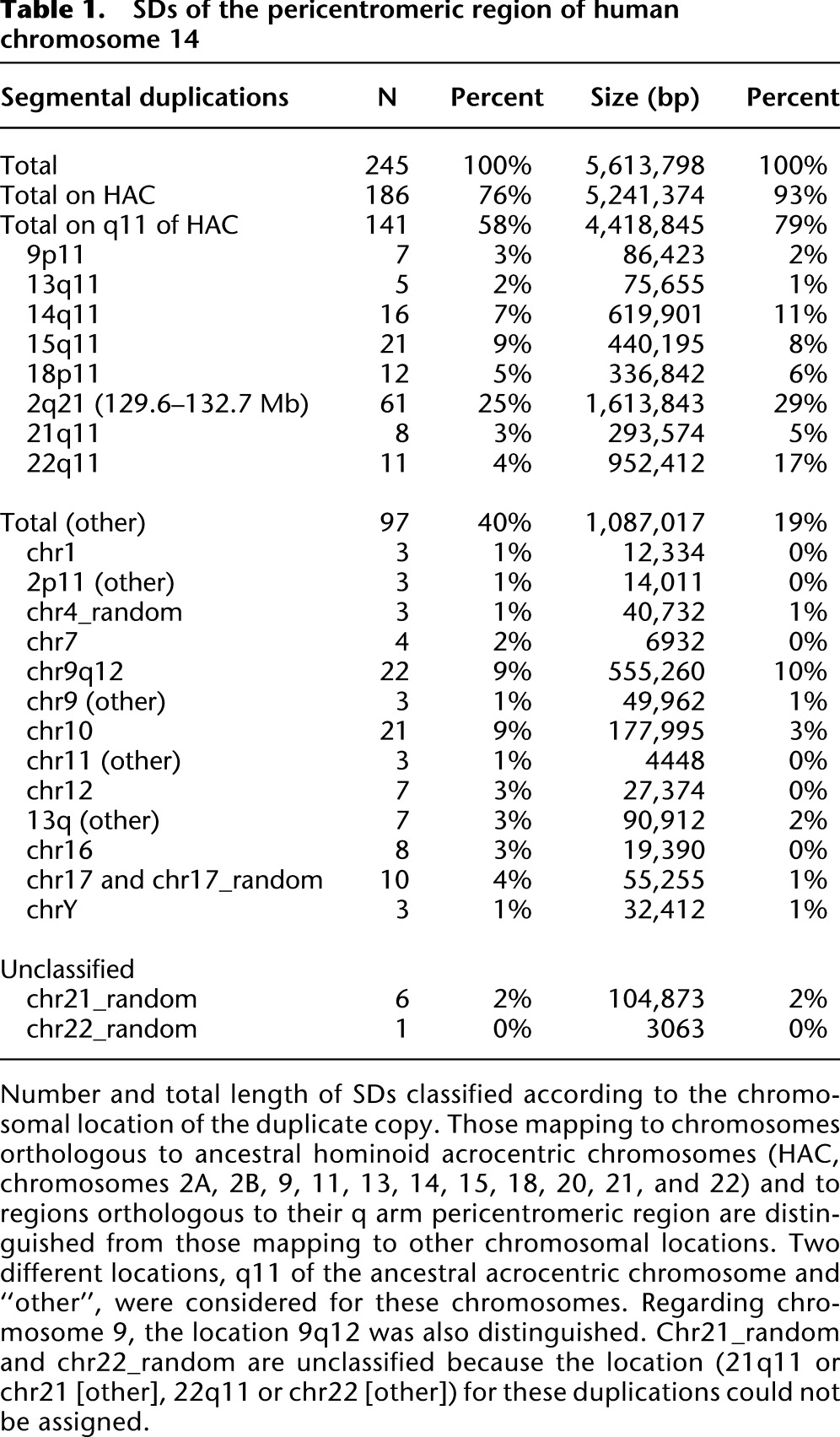
SD data (Bailey et al. 2001, 2002a) for this region (Table 1) reported that (1) 76% of the duplications (93% of the total length) are shared with ancestral hominoid acrocentric chromosomes (13, 14, 15, 21, and 22 together with 2A, 2B, 9, 11, and 18, which are no longer acrocentric in human; Supplemental Note); and (2) 58% of sites (79% of the total length) map to the q arm pericentromeric region of these chromosomes (13q11, 14q11, 15q11, 21q11, and 22q11 together with 2q21, 9p11, and 18p11, which are orthologous to ancestral q arm pericentromeric regions). The 2q21 locus shows the greatest homology in terms of both the number and the sequence length of shared duplications. Among the other chromosomal positions, the highest values of SD are with 9q12 (chromosome 9 heterochromatin) and chromosome 10.
Table 1.
SDs of the pericentromeric region of human chromosome 14
Discussion
Reshuffling of the hominoid OR gene repertoire after the 14/15 fission
An ancestral hominoid fission event gave rise to human chromosomes 14 and 15 and converted a single-copy euchromatic region from an interstitial to a terminal location. Since the genomic organization of the macaque region orthologous to the fission site closely resembles the ancestral hominoid one, we constructed a 1.6-Mb macaque BAC clone contig and compared the content and organization of human and macaque. Sequences from the novel 15q subtelomeric and 14q pericentromeric regions were extensively and specifically duplicated to other subtelomeric and pericentromeric regions, respectively, probably as a direct consequence of the evolutionary change in chromosomal location and consistent with the SD enrichments observed for these regions in the human genome (Bailey et al. 2001; Linardopoulou et al. 2005; Bailey and Eichler 2006). Notably, we found that 14q pericentromeric sequences were duplicated to the q arm pericentromeric region of other acrocentric (15 and 21) or ancestrally acrocentric (18) (Dennehey et al. 2004) chromosomes and subsequently experienced duplication exchanges mainly with acrocentric or ancestrally acrocentric chromosomes (Table 1). These observations suggest that, first, the pericentromeric region of acrocentric chromosomes is rather preferentially duplicated with other acrocentric chromosomes and, second, these exchanges occur primarily between the q arms possibly because the presence of the rDNA and other repetitive sequences prevent the “invasion” of the p arm, as previously reported in chimpanzee and gorilla telomeric ends (Ventura et al. 2012).
Clones positive to the STS 265F14t7 showed weak FISH signals and single-end sequence placement to human 15q26.3 or 14q11 (Fig. 2; Supplemental Table S1), consistent with the occurrence of the fission event and sequence loss at this site, as previously shown for clone CH250–246C20 (Rudd et al. 2009). The fission and consequent chromosomal rearrangements shuffled the OR gene repertoire of hominoids. In particular, the fission caused the loss of OR gene copies at the breakpoint (Rudd et al. 2009) while the consequent duplication events created additional copies in other subterminal and pericentromeric regions. We observed a correlation between the presence of OR genes and the involvement in chromosomal rearrangements (deletion, duplication, and inversion)—all regions that underwent rearrangement in human or in both human and macaque genomes embedded OR genes (besides the ITSN2/DNM1–GOLGA2 duplicon) whereas the flanking single-copy regions contained few OR genes and genes other than OR. OR genes form the largest mammalian gene family and have been subjected to extensive pseudogenization particularly in the primate lineage. Not surprisingly, they have been continuously reshuffled during mammalian evolution by frequent duplication and deletion events (Niimura and Nei 2007). In this regard, their presence at the novel subtelomeric and pericentromeric regions and subsequent duplications unlikely affected the species' survival, and the fission event and its consequences were likely neutral.
ITSN2/DNM1–GOLGA2 duplication clusters map to evolutionary hotspots
Multiple studies demonstrated the association of SDs and evolutionary rearrangements in primates (Armengol et al. 2003; Locke et al. 2003; Bailey et al. 2004; Murphy et al. 2005). In this study, we further prove this association, specifically between LCR15 and regions involved in evolutionary rearrangements. We show that LCR15 sequences cluster around (1) the evolutionary new centromere (15q11.2, ∼3 Mb away from the centromere); (2) the breakpoints of the pericentric inversion that occurred in the Catarrhini ancestor (15q13 and 15q26.3); and (3) the ancestral centromere (15q24–q25, <1 Mb from the ancestral centromere locus). Interestingly, golgin repeats were also found at the breakpoints of pathogenic chromosome 15 deletions (F Antonacci, MY Dennis, J Huddleston, PH Sudmant, K Meltz Steinberg, TA Graves, L Vives, M Malig, CT Amemiya, A Stuart, et al., unpubl.), emphasizing and confirming the dual effect of SD genomic instability in both evolution and disease.
LCR15 copies are present on the macaque and marmoset orthologous regions of 15q11.2 and, therefore, were likely present in the Haplorhini ancestor, but expanded in human, possibly as a result of the neocentromere emergence. LCR15 sequences are present on human and macaque yet absent from marmoset regions orthologous to the inversion breakpoints that occurred in the Catarrhini ancestral submetacentric chromosome (loci AB and F), revealing these copies may have emerged as a result of the inversion or may have driven the event.
There is additional evidence of LCR15 evolutionary instability. The same site at 15q11 was shown to be involved in the pericentric inversion of chimpanzee 15p and in the insertion of a novel SD within the human lineage (Locke et al. 2003). Here, we found the occurrence of two independent chromosomal rearrangements in proximity to one site (human 15q26.3) after its acquisition of an LCR15 copy—the chromosomal fission in the hominoid lineage and the duplicative insertion from chromosome 13 in the macaque lineage. This suggests that LCR15 may not only serve as preferential sites of chromosomal breakage and rearrangement, like the chimpanzee inversion, but they may be subject to further expansion and duplication associated with recurrent events, such as the duplicate copies mapping to the inversion breakpoints in the Catarrhini ancestor.
In FISH experiments, LCR15 signals observed on marmoset metaphases mapped mainly to the pericentromeric regions of chromosomes 6p and 10p—particularly 10p—orthologous to human 15q24–q25 (loci D1, D2, DE1, and DE2), suggesting that the ancestral pericentromeric region was probably the seed of LCR15 and the first chimeric ITSN2/DNM1–GOLGA2 unit likely formed at this locus in the Haplorhini ancestor. Moreover, since no copy is present in the marmoset region orthologous to human chromosome 10, this copy emerged in the Catarrhini ancestor; therefore, it was unlikely the site where the first combined unit formed as previously suggested (Zody et al. 2006).
The chromosome 14 pericentromeric region consists of SDs from the pericentromeric region of ancestral hominoid chromosomes 2B and 10
After the fission event, human chromosome 14 acquired ∼19 Mb of sequence over the last 25 million years. It is largely composed of three blocks: (1) a 1.3-Mb transition region between the q arm euchromatin and the centromere; (2) a 3-Mb centromere composed of alphoid DNA; and (3) a 15-Mb heterochromatic short arm containing arrays of ribosomal RNA genes, satellite sequences, and other repetitive elements (Kehrer-Sawatzki et al. 1998). We analyzed the first of these three blocks and showed that it was mainly formed through pericentromeric duplicative transpositions from ancestral hominoid chromosomes 2B and 10 (homologous to human 2q14–qter and 10, respectively). We note that the centromere of chromosome 2B inactivated in both the macaque (Ventura et al. 2007) and human lineages—the latter after a fusion event occurred 3–5 million years ago and mapping to 2q21 (IJdo et al. 1991). The pericentromeric sequences on chromosome 10 were lost in human and African ape, and evidence of their ancestral location on this chromosome still exists in orangutan and macaque. Notably, chromosomes 2B and 10 have been partners in another process during great ape evolution—the formation of subterminal heterochromatin in the chimpanzee and gorilla lineages (Ventura et al. 2012)—but the genomic regions involved in the process were different.
STS derived from the new human chromosome 14 pericentromeric sequence and probed against the macaque genomic library retrieved clones mapping to either pericentromeric or euchromatic regions. Among the latter, the euchromatic SD-rich region at human 10q22.3 is associated with chromosomal rearrangement and human disease (Melberg et al. 1999; Quintero-Del-Rio et al. 2002; Fallin et al. 2003; Faraone et al. 2006; Hofmann et al. 2008; Kuhl et al. 2008; Venken et al. 2008; O'Meara et al. 2012); in the gorilla genome, one breakpoint of the inversion of chromosome 10q is 1 Mb proximal to this locus (Carbone et al. 2002; Ventura et al. 2011).
In summary, we propose a model that describes the evolution of ape chromosomes 14 and 15 (Fig. 5). After the fission, the resulting two new chromosomal ends achieved different functional roles with respect to the chromosome. The chromosome 14 portion acquired the centromeric functionality necessary for daughter cell segregation, resulting in a telocentric chromosome, while the chromosome 15 end became a telomere participating in interchromosomal DNA exchanges with other subtelomeric regions. Both novel chromosome ends earned telomeric repetitive sequences (TTAGGG) to be protected from degradation and to prevent chromosomal end-to-end fusions. The original centromere, located on chromosome 15, became inactive and a neocentromere was seeded at the chromosome tip not derived by the fission, also making chromosome 15 a telocentric chromosome. The neocentromeres rapidly acquired the complex organization characteristic of primate centromeres—a large core of alphoid sequences surrounded by SDs (Eichler et al. 1999). SDs mainly from the long arm pericentromeric region of the acrocentric chromosome 2B and from the pericentromeric region of chromosome 10 contributed to the formation of the 14q pericentromeric region. SDs involving this genomic region continue to accumulate in the human lineage, mainly within the long arm pericentromeric region of chromosome 22 (Bailey et al. 2002b). It is known that the juxtaposition of constitutive heterochromatin and transcriptionally active genes induces position effects, effectively silencing transcription (Dillon and Festenstein 2002). The SDs served the potentially important role of buffering chromosome 14 euchromatin and the novel centromeric heterochromatin, possibly protecting and maintaining the transcription regulatory landscape of the 14q11 genes suddenly found in a pericentromeric region. Chromosome 15 experienced further LCR15 duplications that generated new copies, particularly within the novel pericentromeric region. In the human karyotype, the neocentromere of chromosome 14 acquired alphoid DNA of the suprachromosomal families 2, 4, and 5; the one of chromosome 15 acquired alphoid DNA of the suprachromosomal families 2 and 4 (Alexandrov et al. 2001). Finally, duplications from the p arm of acrocentric chromosomes created the heterochromatic p arm of ribosomal RNA genes, turning chromosomes 14 and 15 from telocentric to acrocentric chromosomes. We note that in human, chimpanzee, and orangutan, only acrocentric chromosomes bear the rDNA on the p arm, implying that the acquisition of the rDNA short arm was an obligated fate for chromosomes 14 and 15. Yet, in the gorilla genome only two acrocentric chromosomes, 21 and 22, carry the rDNA while the short arms of the remaining acrocentric chromosomes are composed mainly of satellite III sequences (Ventura et al. 2012).
Figure 5.
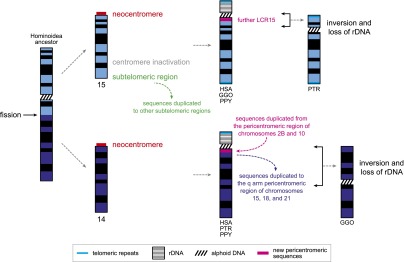
Model of the evolution of ape chromosomes 14 and 15. The picture highlights the centromere repositioning on chromosome 15, the neocentromere formation on chromosome 14, the sequence duplications from and to the novel pericentromeric and subtelomeric regions, and the acquisition of telomeric repeats, rDNA, and alphoid DNA that occurred in the last 25 million years of human evolution. The pericentric inversions in the chimpanzee and gorilla lineages caused the loss of the rDNA from the inverted chromosomes.
This work highlights the dynamic and intimate relationship between duplication and rearrangement in the evolution of human chromosomes 14 and 15. It raises the possibility that not only duplication triggers chromosomal rearrangement through NAHR but also that chromosomal breakage gives rise to duplication events (Kehrer-Sawatzki and Cooper 2008; Girirajan et al. 2009). We propose a possible additional role of SDs in genome evolution where the occurrence of duplication events in response to chromosomal breakage might represent a possible “feedback mechanism” that produces additional DNA material to recover the damage caused by the breakage.
Methods
Hybridization of high-density filters
Radioactive genomic hybridization of the sixfold coverage CHORI-250 (rhesus macaque) segment 1 BAC library was carried out according to the protocol available at CHORI BACPAC resources (http://bacpac.chori.org/highdensity.htm). Probes were obtained by means of PCR amplification of macaque genomic DNA (Supplemental Table S4). The number of positive clones provided estimation of the copy number in the macaque genome. BES were repeatmasked (Smit and Hubley 2010) and mapped on macaque (rheMac2) and human (hg18) reference genomes using the BLAT tool at the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat).
Fluorescence in situ hybridization (FISH)
Metaphase spreads and interphase nuclei were prepared from lymphoblastoid or fibroblast cell lines of Homo sapiens (HSA), Pan troglodytes (PTR), Gorilla gorilla (GGO), Pongo pygmaeus (PPY), Macaca mulatta (MMU), Cercopithecus aethiops (CAE), Trachypithecus cristatus (TCR), and Callithrix jacchus (CJA). FISH experiments were performed using BAC clones (300 ng) directly labeled by nick-translation with Cy3-dUTP, Cy5-dUTP, and fluorescein-dUTP as previously described (Lichter et al. 1990) with minor modifications. Hybridization was performed at 37°C in 2× SSC, 50% (v/v) formamide, 10% (w/v) dextran sulfate, 3 μg C0t-1 DNA, and 3 μg sonicated salmon sperm DNA, in a volume of 10 μL. Post-hybridization washing was at high stringency (60°C in 0.1× SSC, three times) or at low stringency for cross-species hybridization (37°C in 2× SSC, 50% formamide, three times, and 42°C in 2× SSC, three times). Nuclei and chromosome metaphases were DAPI-stained. Digital images were obtained using a Leica epifluorescence microscope equipped with a cooled CCD camera. Fluorescence signals detected with Cy3, Fluorescein, and Cy5 filters and chromosomes and nuclei images detected with DAPI filter were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Phylogenetic analysis
Duplicate copies of a 3-kb region from ITSN2 and a 2.5-kb region from GOLGA2 were retrieved from the hg19, panTro3, gorGor3, ponAbe2, rheMac2, and calJac3 reference genomes through BLAT search (http://genome.ucsc.edu/cgi-bin/hgBlat) (Kent 2002). The hg19 reference was used instead of hg18 because in the most recent release of the human genome (hg19), all LCR15 duplicons are placed on the chromosome 15 sequence, and there are no chr15_random locations. Multiple sequence alignments were performed using Clustal W (Thompson et al. 1994) (match 5, mismatch −2, match with N 0, gap opening 25, gap extension 0.05 for the ITSN2 region; match 4, mismatch −2, match with N 0, gap opening 15, gap extension 0.05 for the GOLGA2 region). Neighbor-joining phylogenetic trees based on the Kimura 2-parameter model (Kimura 1980) (pairwise deletion option) were constructed in MEGA5 (Tamura et al. 2011). A bootstrap test with 1000 replicates was conducted to evaluate the statistical significance of each node (Felsenstein 1985).
Sanger sequencing
Ten STS regions were PCR-amplified from positive BAC clones and PCR products were sequenced using PCR primers (Supplemental Table S4). DNA from BAC clones were extracted using the BAC Prep Protocol (Schein et al. 2004), and BAC ends were sequenced (if not available in the NCBI Trace Archive) using SP6 and T7 primers. Sequencing was performed on an ABI PRISM 3100 Genetic Analyzer using the BigDye Terminator v3.1 Chemistry (Applied Biosystems) according to manufacturer instructions.
Illumina sequencing and analysis of BAC clones
BAC clone DNA from the CH250 macaque library was isolated by a modified alkaline lysis miniprep procedure and paired-end sequenced on an Illumina HiSeq 2000 (Supplemental Note). Short Illumina reads (50 bp) were mapped to the hard-masked references hg18 and rheMac2 with mrsFAST (Hach et al. 2010) using an edit distance of three and single-end mapping mode. We determined the best location for each clone using the coverage of reads across each genome. For each clone we calculated the mean and standard deviation of its coverage and selected regions for which the coverage was greater than the mean plus two standard deviations and the total size of the region was >1 kbp. We manually investigated each of the resulting regions in the UCSC Genome Browser to determine the best location and whether any regions should be merged. We determined the repeat content of short reads by aligning reads to all known RepeatMasker and Tandem Repeat Finder (TRF) sequences in hg18 and rheMac2. Repetitive coordinates were obtained from UCSC Genome Browser tracks (rmsk.txt.gz for RepeatMasker and chromTrf.tar.gz for TRF) and corresponding sequences were extracted from each reference to create a “repeat” reference. We mapped short reads to these repeat references using BWA (Burrows-Wheeler Alignment) in single-end mapping mode and counted the total number of reads that mapped to each reference with at least a quality of 1. To determine the amount of vector sequence or bacteria contamination present in the BAC short reads, we aligned these reads to a reference consisting of the vector pTARBAC2.1 and the Escherichia coli strain DH10B using mrsFAST with an edit distance of three and single-end mapping mode. We counted the number of reads that mapped to the vector and the E. coli genome with a quality of at least 1 to determine the proportion of vector or E. coli reads in the complete set of reads.
Data access
BAC end sequences are available in the Genome Survey Sequence division of GenBank (http://www.ncbi.nlm.nih.gov/nucgss) under accession numbers KG088791–KG088955. CHORI-250 BAC clone sequence data are available in the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP028268.
Acknowledgments
We acknowledge “Futuro in Ricerca 2010” for funding. We thank Tonia Brown for manuscript editing.
Author contributions: G.G. and M.V. designed the study. G.G. and M.P. performed BAC library hybridizations and FISH experiments. G.G. performed PCR product sequencing and phylogenetic analysis. F.A. constructed shotgun sequencing libraries and J.H. analyzed sequencing reads. M.M. and L.V. performed BAC end sequencing. G.G. and M.V. analyzed the data. G.G., E.E.E., and M.V. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.156240.113.
References
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y 2001. α-satellite DNA of primates: Old and new families. Chromosoma 110: 253–266 [DOI] [PubMed] [Google Scholar]
- Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X 2003. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet 12: 2201–2208 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE 2006. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res 11: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE 2002a. Recent segmental duplications in the human genome. Science 297: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE 2002b. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am J Hum Genet 70: 83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE 2004. Hotspots of mammalian chromosomal evolution. Genome Biol 5: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Ventura M, Tempesta S, Rocchi M, Archidiacono N 2002. Evolutionary history of chromosome 10 in primates. Chromosoma 111: 267–272 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, Kim UJ, Kuo WL, Olivier M, et al. 2001. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409: 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehey BK, Gutches DG, McConkey EH, Krauter KS 2004. Inversion, duplication, and changes in gene context are associated with human chromosome 18 evolution. Genomics 83: 493–501 [DOI] [PubMed] [Google Scholar]
- Dillon N, Festenstein R 2002. Unravelling heterochromatin: Competition between positive and negative factors regulates accessibility. Trends Genet 18: 252–258 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M 1999. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res 9: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, Liang KY, Pulver AE 2003. Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet 73: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Hwu HG, Liu CM, Chen WJ, Tsuang MM, Liu SK, Shieh MH, Hwang TJ, Ou-Yang WC, Chen CY, et al. 2006. Genome scan of Han Chinese schizophrenia families from Taiwan: Confirmation of linkage to 10q22.3. Am J Psychiatry 163: 1760–1766 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Girirajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK, et al. 2009. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res 19: 178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F, Hormozdiari F, Alkan C, Birol I, Eichler EE, Sahinalp SC 2010. mrsFAST: A cache-oblivious algorithm for short-read mapping. Nat Methods 7: 576–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, Schurmann M, Muller-Quernheim J, Krawczak M, Rosenstiel P, et al. 2008. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet 40: 1103–1106 [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA 1991. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci 88: 9051–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN 2008. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res 16: 41–56 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Wohr G, Schempp W, Eisenbarth I, Barbi G, Assum G 1998. Mapping of members of the low-copy-number repetitive DNA sequence family chAB4 within the p arms of human acrocentric chromosomes: Characterization of Robertsonian translocations. Chromosome Res 6: 429–435 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Kuhl A, Melberg A, Meinl E, Nurnberg G, Nurnberg P, Kehrer-Sawatzki H, Jenne DE 2008. Myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy 7: Corroboration and narrowing of the critical region on 10q22.3. Eur J Hum Genet 16: 367–373 [DOI] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC 1990. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247: 64–69 [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Archidiacono N, Misceo D, Cardone MF, Deschamps S, Roe B, Rocchi M, Eichler EE 2003. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol 4: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey EH 2004. Orthologous numbering of great ape and human chromosomes is essential for comparative genomics. Cytogenet Genome Res 105: 157–158 [DOI] [PubMed] [Google Scholar]
- Melberg A, Oldfors A, Blomstrom-Lundqvist C, Stalberg E, Carlsson B, Larrson E, Lidell C, Eeg-Olofsson KE, Wikstrom G, Henriksson G, et al. 1999. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy linked to chromosome 10q. Ann Neurol 46: 684–692 [DOI] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2013. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617 [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2: e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara E, Stack D, Lee CH, Garvin AJ, Morris T, Argani P, Han JS, Karlsson J, Gisselson D, Leuschner I, et al. 2012. Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J Pathol 227: 72–80 [DOI] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, et al. 2007. Initial sequence and comparative analysis of the cat genome. Genome Res 17: 1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Gratacos M, Peral B, Csiszar K, Gonzalez-Sarmiento R, Sumoy L, Estivill X 2001. Additional complexity on human chromosome 15q: Identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res 11: 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X 2002. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet 10: 26–35 [DOI] [PubMed] [Google Scholar]
- Quintero-Del-Rio AI, Kelly JA, Kilpatrick J, James JA, Harley JB 2002. The genetics of systemic lupus erythematosus stratified by renal disease: Linkage at 10q22.3 (SLEN1), 2q34-35 (SLEN2), and 11p15.6 (SLEN3). Genes Immun (Suppl 1) 3: S57–S62 [DOI] [PubMed] [Google Scholar]
- Roberto R, Misceo D, D'Addabbo P, Archidiacono N, Rocchi M 2008. Refinement of macaque synteny arrangement with respect to the official rheMac2 macaque sequence assembly. Chromosome Res 16: 977–985 [DOI] [PubMed] [Google Scholar]
- Rudd MK, Endicott RM, Friedman C, Walker M, Young JM, Osoegawa K, de Jong PJ, Green ED, Trask BJ 2009. Comparative sequence analysis of primate subtelomeres originating from a chromosome fission event. Genome Res 19: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein J, Kucaba T, Sekhon M, Smailus D, Waterston R, Marra M 2004. High-throughput BAC fingerprinting. Methods Mol Biol 255: 143–156 [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, 2010. RepeatMasker Open-3.2.9. http://www.repeatmasker.org/
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N 2008. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosome Res 16: 17–39 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken T, Alaerts M, Souery D, Goossens D, Sluijs S, Navon R, Van Broeckhoven C, Mendlewicz J, Del-Favero J, Claes S 2008. Chromosome 10q harbors a susceptibility locus for bipolar disorder in Ashkenazi Jewish families. Mol Psychiatry 13: 442–450 [DOI] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res 13: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246 [DOI] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. 2011. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res 21: 1640–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Sajjadian S, Vives L, Sudmant PH, Marques-Bonet T, Graves TA, Wilson RK, Eichler EE 2012. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res 22: 1036–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R 1995. Chromosome painting in mammals as an approach to comparative genomics. Curr Opin Genet Dev 5: 792–797 [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R 1997. Comparative painting of mammalian chromosomes. Curr Opin Genet Dev 7: 784–791 [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Ludecke HJ, Senger G, Horsthemke B, Claussen U, Cremer T, Arnold N, Lengauer C 1994. The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosome Res 2: 405–410 [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525–1530 [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Sawyer JR, Dunham K 1980. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science 208: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Zody MC, Garber M, Sharpe T, Young SK, Rowen L, O'Neill K, Whittaker CA, Kamal M, Chang JL, Cuomo CA, et al. 2006. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature 440: 671–675 [DOI] [PubMed] [Google Scholar]