Abstract
Most speciation events probably occur gradually, without complete and immediate reproductive isolation, but the full extent of gene flow between diverging species has rarely been characterized on a genome-wide scale. Documenting the extent and timing of admixture between diverging species can clarify the role of geographic isolation in speciation. Here we use new methodology to quantify admixture at different stages of divergence in Heliconius butterflies, based on whole-genome sequences of 31 individuals. Comparisons between sympatric and allopatric populations of H. melpomene, H. cydno, and H. timareta revealed a genome-wide trend of increased shared variation in sympatry, indicative of pervasive interspecific gene flow. Up to 40% of 100-kb genomic windows clustered by geography rather than by species, demonstrating that a very substantial fraction of the genome has been shared between sympatric species. Analyses of genetic variation shared over different time intervals suggested that admixture between these species has continued since early in speciation. Alleles shared between species during recent time intervals displayed higher levels of linkage disequilibrium than those shared over longer time intervals, suggesting that this admixture took place at multiple points during divergence and is probably ongoing. The signal of admixture was significantly reduced around loci controlling divergent wing patterns, as well as throughout the Z chromosome, consistent with strong selection for Müllerian mimicry and with known Z-linked hybrid incompatibility. Overall these results show that species divergence can occur in the face of persistent and genome-wide admixture over long periods of time.
Ongoing hybridization between closely related species appears to be common in nature (Mallet 2005; Rieseberg 2009) and theoretical work has demonstrated a diversity of scenarios whereby species can emerge without complete geographical isolation (Kirkpatrick and Ravigné 2002; Gavrilets 2004; van Doorn et al. 2009). Despite widespread interest in these scenarios, there remains little consensus among speciation biologists regarding the extent to which ongoing gene flow actually plays a role during speciation. This is partly because it is challenging to reconstruct ancestral ranges and therefore almost impossible to know for sure the extent of historical contact between species. Fortunately, genomic approaches are now beginning to allow us to address these long-standing questions from a different angle, by documenting the extent of admixture between species on a genome-wide scale (Kulathinal et al. 2009; Ellegren et al. 2012; Garrigan et al. 2012; The Heliconius Genome Consortium 2012; Nosil et al. 2012). Speciation genomics therefore offers an opportunity to address long-standing questions regarding the extent to which divergence and speciation occurs in the face of ongoing gene flow.
One prediction of models of speciation with gene flow is that the level of divergence should be heterogeneous across the genome. Some loci are likely to be shared between incipient species, while selection maintains divergence at others (Turner et al. 2005; Nosil et al. 2009). Recently, considerable progress has been made in documenting patterns of genomic divergence between incipient species (Hohenlohe et al. 2010; Lawniczak et al. 2010; Michel et al. 2010; Ellegren et al. 2012; Nosil et al. 2012; Gagnaire et al. 2013). Genome-wide studies of threespine sticklebacks (Hohenlohe et al. 2010, 2012) and Ficedula flycatchers (Ellegren et al. 2012) revealed patterns of divergence consistent with a model of “islands” of divergence amidst a sea of gene flow. In contrast, analyses of Anopheles gambiae subspecies (Lawniczak et al. 2010) and Rhagoletis host races (Michel et al. 2010) reported widespread divergence throughout the genome. One problem is that patterns of divergence are typically noisy, reflecting the complex interactions of selection, drift, migration, recombination, mutation, and ancestral polymorphism, all of which can lead to heterogeneity in divergence (Noor and Bennett 2009; Michel et al. 2010; Nadeau et al. 2012). A key challenge is therefore to distinguish the signal of gene flow from background noise.
Analyses of genomic divergence therefore need to be complemented with more sensitive tests for gene flow between populations (Kulathinal et al. 2009; Ellegren et al. 2012; Garrigan et al. 2012; The Heliconius Genome Consortium 2012; Nosil et al. 2012). A widely used approach is to fit coalescent models (Pinho and Hey 2010), but this can be computationally prohibitive for genomic data sets and requires strong assumptions about population parameters. A simpler method is to compare the extent of shared variation between sympatric and allopatric populations (Grant et al. 2005). Recent gene flow should result in reduced differentiation and an excess of shared variation between sympatric populations compared with allopatric populations. This logic has been applied on a genomic scale to test for gene flow in Drosophila (Kulathinal et al. 2009) and hominids (Green et al. 2010). However, this approach does not account for the age of shared variation, such that recent admixture may be confounded with ancestral geographic structure (Green et al. 2010; Durand et al. 2011; Eriksson and Manica 2012). It is therefore best used in combination with other methods that can distinguish recent gene flow from ancient shared variation.
In this paper, we focus on the closely related neotropical butterfly species Heliconius melpomene, Heliconius cydno, and Heliconius timareta (Fig. 1). These species are distasteful to predators and often involved in Müllerian mimicry with other species. All three comprise multiple distinct wing pattern races that have been considered as an early stage in speciation (Jiggins 2008). Indeed there is strong evidence that selection for Müllerian mimicry can lead to wing pattern divergence and assortative mating without the need for geographic separation (Chamberlain et al. 2009). Heliconius cydno and H. timareta together form a clade that is sister to H. melpomene, estimated about two million years divergent (Bull et al. 2006; Salazar et al. 2008). Heliconius melpomene and H. cydno have distinct wing patterns and other ecological differences, and display strong assortative mating (Merrill et al. 2011a). Hybrids occur at low frequency (<1/1000) (Mallet et al. 2007), and are female-sterile (Naisbit et al. 2002), as well as being preferentially attacked by predators due to their non-mimetic wing patterns (Merrill et al. 2012). Unlike H. cydno, several H. timareta races have H. melpomene-like patterns (Giraldo et al. 2008; Mérot et al. 2013) and similarly show differences in host plant use and mating preferences (Giraldo et al. 2008). Recent genomic studies have begun to dissect the genetic variation underlying color pattern diversity in this genus (The Heliconius Genome Consortium 2012; Nadeau et al. 2012; Supple et al. 2013). One important insight is that the shared color patterns between H. melpomene and H. timareta appear to have resulted from introgression (The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012). There is also evidence for exchange of other loci between H. melpomene and the H. cydno/timareta clade (Bull et al. 2006; Kronforst et al. 2006; The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012; Nadeau et al. 2013). RAD-tag analyses of Peruvian races of H. melpomene and H. timareta suggest that at least ∼2%–5% of the genome is admixed (The Heliconius Genome Consortium 2012). The recent completion of the H. melpomene genome now allows investigation of genome-wide patterns of divergence and gene flow within and between these species.
Figure 1.
Populations sampled and their phylogenetic relationships. The entire distribution of H. melpomene is shown in gray. The entire distribution of the H. cydno/timareta clade is shown with dots (Rosser et al. 2012). Colors depict distributions of races used in this study, with dots indicating the sampling locations, and correspond to the colored dots on the tree. The tree is a compressed version of the whole-genome ML tree (Supplemental Fig. S1). The three general sampling locations, Panama, Peru, and French Guiana, are indicated. The scale bar refers to the number of substitutions per site.
Here we take advantage of the geographic distribution of H. melpomene, with some populations many thousands of kilometers from the current range of the H. cydno/timareta clade (Fig. 1), and carry out a much more powerful genome-wide test for gene flow than was possible with the sequenced fragments hitherto studied. We analyzed 31 resequenced individuals (30 of which were newly sequenced in this study) from replicate sympatric species pairs of the two clades in Peru, where they are convergent in wing pattern, and Panama, where they are divergent. We also sampled an allopatric H. melpomene population from French Guiana (Fig. 1). Four species of the silvaniform clade of Heliconius were included as outgroups. Our new methods allowed us to investigate the extent and time course of genomic admixture, both before speciation and during different time periods after speciation.
Results
Phylogenomic analysis
Five populations of H. melpomene, one population of H. cydno, one population of H. timareta, and four outgroup species were sequenced (Fig. 1; Supplemental Table S1). Populations were represented by four wild-caught individuals (eight haploid genomes) except H. m. melpomene from Panama, for which three individuals were sampled (Supplemental Table S1). All individuals were wild-caught except for H. m. melpomene specimen no. 1, which was from the inbred reference genome strain. Whole-genome shotgun sequencing using the Illumina GA IIx and HiSeq 2000 technology gave an average coverage per individual of 15–62× (Supplemental Table S1). Sequences were aligned to the H. melpomene reference genome (The Heliconius Genome Consortium 2012) (version 1.1), including the complete mitochondrial scaffold. Genotyping and quality filtering (see Methods for details) produced an average of 190 million high-quality genotype calls per individual (69% of the genome). Proportions of variant sites were similar across all wild-caught individuals, and the ratio of transitions to transversions with respect to the reference was similar across all taxa, indicating that there was no systematic bias in the distribution of sequencing errors among taxa.
Maximum likelihood (ML) phylogenetic reconstruction using all sites with high-quality genotype calls for all 31 individuals (60 Mb of sequence, ∼25% of the genome) confirmed that the H. melpomene and the H. cydno/timareta clades are reciprocally monophyletic (Fig. 1; Supplemental Fig. S1; The Heliconius Genome Consortium 2012; Nadeau et al. 2013). We here term this topology “the species tree.” A tree generated from complete mitochondrial sequences produced a similar topology (Supplemental Fig. S1).
Phylogenetic discordance across the genome
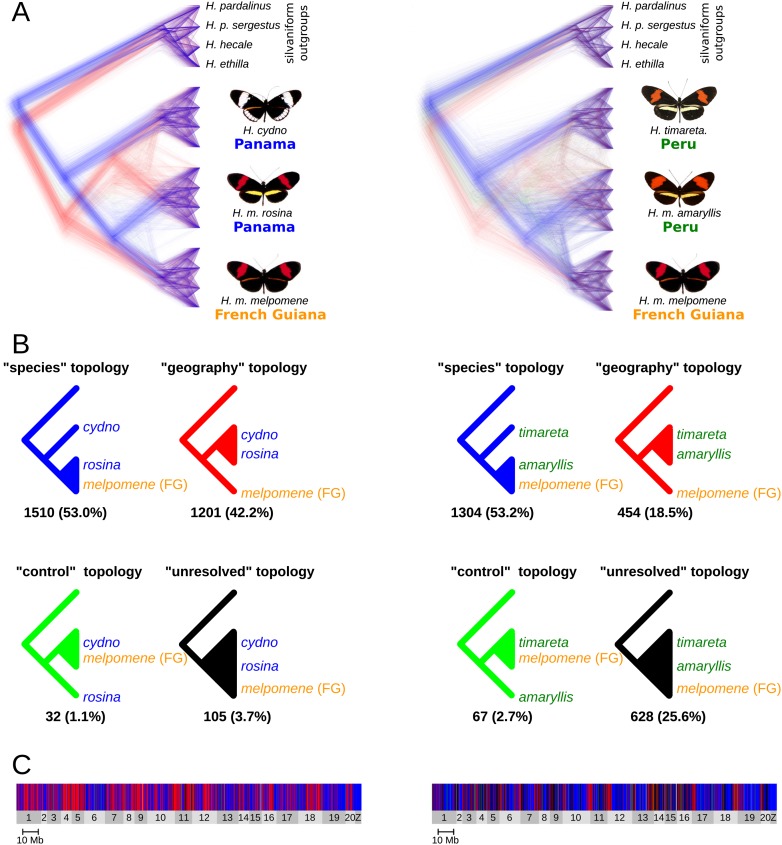
Although the genome-wide ML tree revealed strong support for the expected “species tree,” most speciation scenarios predict discordant coalescent histories among genomic regions (Garrigan et al. 2012). To investigate this, we generated maximum-likelihood trees for non-overlapping 100-kb windows throughout the genome. To simplify the hypotheses being tested, we analyzed two sets of four taxa separately, each representing a sympatric species pair and an allopatric “control” population. The cydno/melpomene set consisted of H. cydno and H. m. rosina (both from Panama), H. m. melpomene from French Guiana and outgroups, while the timareta/melpomene set consisted of H. timareta and H. m. amaryllis (both from Peru), with H. m. melpomene from French Guiana and outgroups. We summed the frequency of four possible topologies: species, geography, control, and unresolved (Fig. 2B). Three of these we considered “resolved,” meaning that two of the ingroup populations (eight individuals) formed a monophyletic clade, while the four individuals comprising the third ingroup population formed a distinct monophyletic sister clade (see Supplemental Fig. S3 for examples). For both data sets, the majority of genomic windows (53% and 53.2%, respectively) supported a resolved “species tree” topology in which the H. melpomene populations are monophyletic (Fig. 2). Under a bifurcating topology, incomplete lineage sorting should result in similar numbers of two alternative resolved topologies, which we term the “geography tree” (sympatric populations of different species cluster together) and the “control tree” (allopatric populations of different species cluster together). The final possibility is an “unresolved tree,” in which the three ingroup populations are not neatly partitioned into two monophyletic clusters (Fig. 2; see Supplemental Fig. S3 for examples).
Figure 2.
Four-taxon ML trees for 100-kb windows. (A) Trees were superimposed using DensiTree (Bouckaert 2010). There were 2848 trees for the H. cydno–H. melpomene data set (left) and 2453 for the H. timareta–H. melpomene data set (right). Tree lengths were equalized so that all trees could be superimposed, and then a random jitter was added to all branch lengths to show density. Trees supporting each of the four possible topologies are colored accordingly: blue for the species tree, red for the geography tree, green for the control tree, and black for unresolved trees. (B) The four topologies scored, along with the number and percentage of trees supporting each. See Supplemental Figure S3 for examples of trees assigned to each topology. (C) The distribution of the four topologies across the genome. Chromosomes are shaded light and dark gray. See Supplemental Figure S4 for an enlarged version.
As expected under admixture, we found that the geography tree was far more prevalent than the control tree in both data sets: 42.2% versus 1.1% for the cydno/melpomene set and 18.5% versus 2.7% for the timareta/melpomene set; and widely distributed across the genome (Fig. 2C; Supplemental Fig. S4). While only 3.7% of trees were unresolved in the cydno/melpomene set, 25.6% were unresolved in the timareta/melpomene set. The greater fraction of unresolved trees in the second case is expected given greater shared ancestral polymorphism due to the more recent divergence between H. m. amaryllis and H. m. melpomene from French Guiana (Fig. 1; Supplemental Fig. S1). These findings show that there is not only a large amount of phylogenetic discordance across the genome, but that it is strongly structured by geography, consistent with gene flow between these clades where their ranges overlap. Here we report results for 100-kb windows because linkage disequilibrium (LD) tends to break down completely within 100 kb in Heliconius genomes (Supplemental Fig. S2), making each 100-kb block effectively independent from its neighbors. However, we also repeated the tests at various window sizes between 10 and 200 kb (Supplemental Table S2). Although the number of unresolved trees increases at smaller window sizes, the relative ratios of resolved trees are robust to window size variation (Supplemental Table S2).
Evidence of recent gene flow
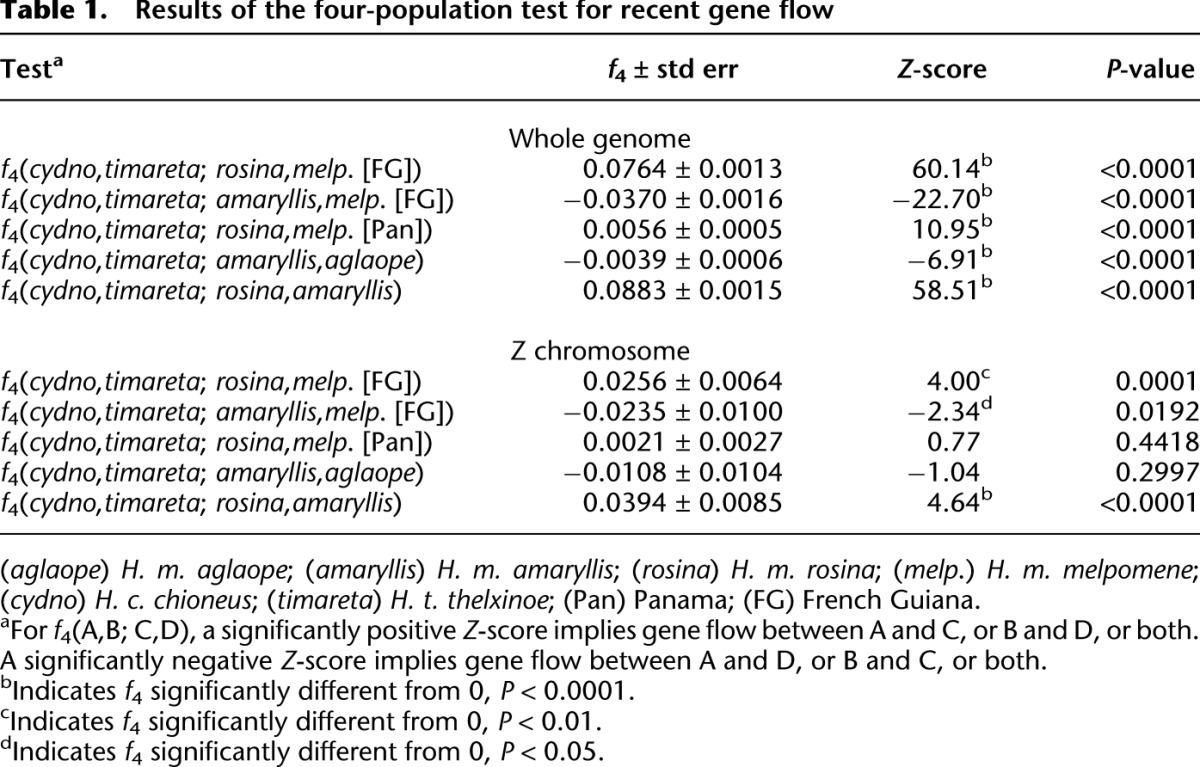
Allele frequency correlations provide further evidence for recent interspecific gene flow. Our geographically structured sampling design allowed us to distinguish between ancient and recent admixture using a sensitive “four-population” test (Reich et al. 2009, 2012) for geographical correlations in allele frequencies. In the absence of admixture, allele frequency changes due to drift in disparate populations should not be correlated. Across all tests, there was a highly significant allele frequency correlation between H. m. rosina and H. cydno from Panama, and between H. m. amaryllis and H. timareta from Peru (Table 1). These correlations indicate recent gene flow between these species where they occur in sympatry.
Table 1.
Results of the four-population test for recent gene flow
Gene flow has occurred at multiple points since early in speciation
Evidence for recent gene flow does not necessarily imply that gene flow has persisted throughout speciation. Secondary contact after allopatric speciation might be characterized by a burst of recent gene flow, while sympatric speciation should leave a signature of continuous gene flow during speciation. We estimated admixture along different branches of the phylogeny using a method devised by Green et al. (2010), which compares two classes of shared derived alleles, termed ABBAs and BABAs. For three populations and an outgroup, with the relationship {[(P1,P2),P3],O}, we can test for differential admixture between P3 and either of P1 or P2 by examining the numbers of shared derived alleles between P3 and P2 (ABBAs) and between P3 and P1 (BABAs). We calculated two statistics: “D” used to test for a significant imbalance of ABBAs and BABAs, indicative of admixture; and “f,” the estimated fraction of the genome that has been shared between populations (Green et al. 2010; Durand et al. 2011). These measures are robust to variation in effective population size (Durand et al. 2011).
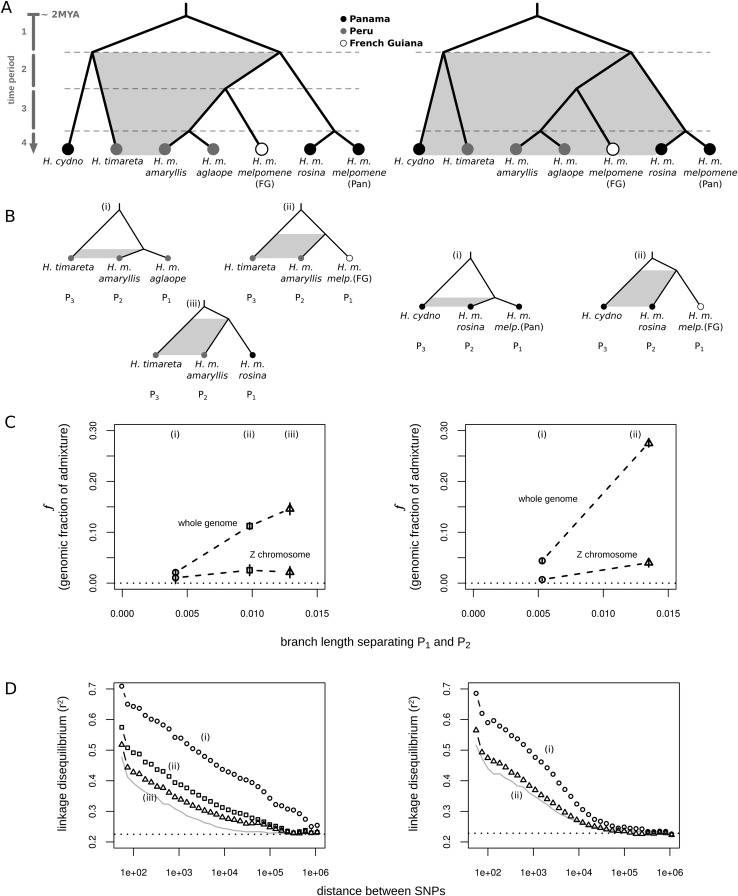
We examined rates of gene flow between H. timareta and H. m. amaryllis across three time periods (Fig. 3): a short, recent period, subsequent to the divergence between H. m. amaryllis and H. m. aglaope (period 4 of Fig. 3A); an intermediate period, subsequent to the divergence of the Peruvian populations from French Guianan H. m. melpomene (periods 3, 4 of Fig. 3A); and a long period, subsequent to the divergence between Peruvian and Panamanian populations (periods 2, 3, 4 of Fig. 3A). Across these comparisons there was a strong trend of increasing f with time (Fig. 3C; Table 2). Similarly, for H. m. rosina and H. cydno, over two time periods, f was again much greater for the longer period (Fig. 3C; Table 2). Because this method assumes unidirectional gene flow from P3, and complete isolation between P3 and P1, the actual fraction of the genome that has been shared may be greater than estimated here. What is important is that the relative values of f increase with the length of the period examined, which is consistent with gene flow having occurred during time periods 2, 3, and 4. One potential caveat is that the extent of isolation between P3 and P1 could differ between these tests, accounting for some of the variation in f. We therefore investigated linkage disequilibrium among these shared derived sites as an additional signal to differentiate between recent and long-term gene flow.
Figure 3.
Measuring admixture at different phylogenetic scales. (A) We can distinguish between admixture in different time periods as follows. If gene flow was ancient only (i.e., period 1), then H. m. amaryllis and H. m. rosina should both be equally admixed with H. timareta and H. cydno. However, if gene flow is more recent (i.e., period 2, 3, or 4), then H. m. amaryllis should be more admixed with Peruvian H. timareta, and H. m. rosina should be more admixed with Panamanian H. cydno. The same logic applies when quantifying admixture for a specific branch: If H. timareta shares more derived alleles with H. m. amaryllis than with H. m. aglaope, this skew must reflect gene flow between H. timareta and H. m. amaryllis that is more recent than the coalescence between H. m. amaryllis and H. m. aglaope (i.e., during period 4). (B) Our sampling allowed us to quantify admixture at three time scales between H. timareta and H. m. amaryllis, and two time scales between H. cydno and H. m. rosina. (C) The estimated fraction of admixture (f), plotted for the whole genome and the Z chromosome specifically against the estimated length of the time period being analyzed, calculated as the average branch length separating populations P1 and P2 in the genomic ML phylogeny (Supplemental Fig. S1). Vertical lines depict standard errors. (D) LD (r2) between shared-derived alleles in the P2 population (left, H. m. amaryllis; right, H. m. rosina), plotted as a function of distance on a logarithmic scale. The SNPs used to estimate LD were those carrying a shared derived allele in P2 and P3, while P1 was fixed for the ancestral state (i.e., an ABBA pattern, where the B alleles are not necessarily fixed). The gray line represents the average genomic LD level, and the dashed line shows the average LD among unlinked sites.
Table 2.
Results of ABBA BABA tests to quantify gene flow over specific time periods

Linkage disequilibrium between shared derived alleles
The extent of LD between introgressed alleles carries information about the age of admixture. Recently introgressed haplotypes have had insufficient time to be broken down by recombination, and therefore closely linked introgressed alleles should occur in LD with one another (Machado et al. 2002; Sankararaman et al. 2012). In contrast, anciently introgressed alleles should display levels of LD similar to the average genomic level. We tested for this signal by examining LD at sites carrying shared derived variants (i.e., ABBA SNPs) in H. m. amaryllis and H. m. rosina. For H. m. amaryllis, three sets of SNPs carrying shared variants could be examined, corresponding to the three time periods described above (Fig. 3B). Likewise, in H. m. rosina, two sets of SNPs could be examined.
We found that the extent of LD differed dramatically between the time periods (Fig. 3D). Variants shared between H. timareta and H. m. amaryllis but absent from H. m. aglaope displayed the strongest LD, extending up to a megabase. This is consistent with the existence of large introgressed haplotypes that have yet to be fully broken down. Variants shared between H. timareta and H. m. amaryllis but absent from French Guianan H. m. melpomene displayed weaker LD, while those shared between H. timareta and H. m. amaryllis but absent from H. m. rosina displayed the weakest LD, declining with distance at a similar rate to the genomic average (Fig. 3D). Thus, these two latter comparisons include variation that appears to have been shared more anciently, giving sufficient time for introgressed haplotypes to be broken down. Similar differences were observed in the extent of LD among variants shared between H. cydno and H. m. rosina at the two time intervals examined. By exploiting a different aspect of the data, these results provide an independent line of evidence that both recent and ancient admixture has occurred between these species pairs.
Patterns of genomic divergence along the speciation continuum
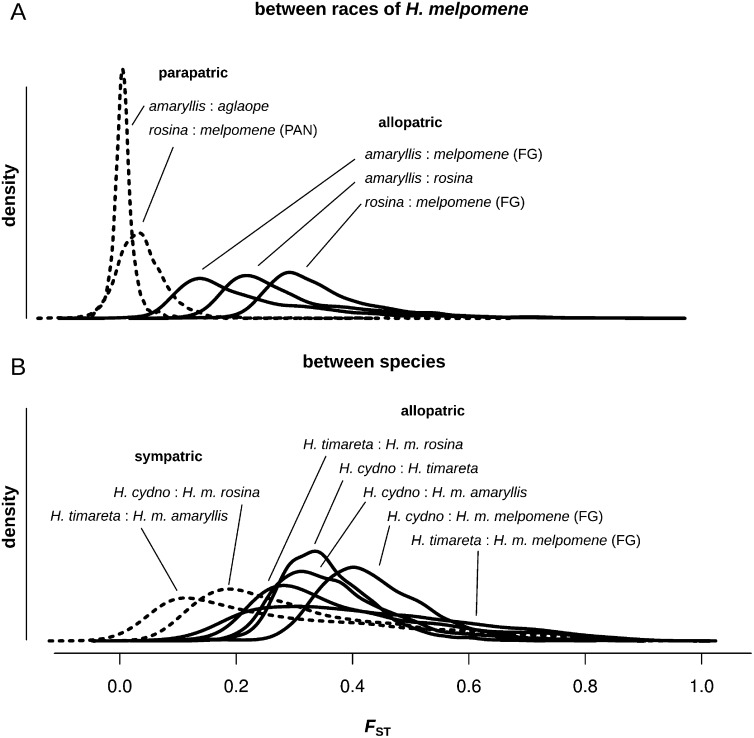
We characterized patterns of divergence across the genome between populations at various levels of divergence and geographic separation using the fixation index, FST. At the earliest stage of divergence, between parapatric races, FST was low throughout the genome with just a few narrow peaks (Fig. 4), which are partly explained by known wing pattern divergence. Between H. m. aglaope and H. m. amaryllis from Peru, only two pronounced divergence peaks were present, corresponding to the known pattern loci HmB (red elements) and HmYb (yellow elements) (Baxter et al. 2010; Nadeau et al. 2012). For the Panamanian races, the level of FST was noisier but there was a small FST peak at the HmYb locus (Fig. 4). There was no peak at the HmB locus, consistent with the fact that the Panamanian races share the same red mimetic patterns. Between allopatric races, background FST was significantly higher and more heterogeneous, and color pattern loci no longer appeared as clear outliers. Patterns of FST between species were broadly similar in mean and variance to those between allopatric races of H. melpomene (Figs. 4, 5).
Figure 4.
Genomic divergence along the speciation continuum. FST values were calculated for 100-kb windows sliding in increments of 20 kb. Chromosomes are shown with alternating light and dark shading. Point colors reflect the absolute level of FST to allow for comparison between plots. The locations of the wing pattern loci HmYb and HmB are indicated by arrows. (amaryllis) H. m. amaryllis; (rosina) H. m. rosina; (melpomene) H. m. melpomene; (cydno) H. c. chioneus; (timareta) H. t. thelxinoe; (Pan) Panama; (Per) Peru; (FG) French Guiana.
Figure 5.
Density plots of pairwise FST values for non-overlapping 100-kb windows. All pairwise comparisons, corresponding to the plots in Figure 4, between races of H. melpomene (A), and between species (B).
Reduced interspecific divergence in sympatry
Gene flow between sympatric populations should lead to reduced FST as compared with that between allopatric populations. Consistent with phylogenetic evidence for gene flow in sympatry, FST between sympatric species pairs in both Panama and Peru was significantly lower than that between either H. timareta or H. cydno and the allopatric H. m. melpomene from French Guiana (Table 3). Each of the 21 chromosomes independently showed the same trend of significantly lower FST in sympatry than in allopatry (Supplemental Fig. S5B). This trend is also robust to the use of different allopatric populations. Peruvian H. m. amaryllis can be considered as allopatric to Panamanian H. cydno (separated by the Andes). Likewise, H. m. rosina can be considered allopatric to H. timareta. These allopatric comparisons both displayed significantly higher average FST than the sympatric comparisons, although not quite as high as when the French Guianan population was used (Supplemental Table S3). This variation may be partly due to differences in the extent of isolation, but probably also reflect differences in effective population size. Nevertheless no inter-species allopatric comparison showed anything close to the reduced FST observed between the species in sympatry (Fig. 5).
Table 3.
FST between sympatric and allopatric populations

Plotted across individual chromosomes, the pattern of FST was highly heterogeneous in both sympatry and allopatry (Supplemental Figs. S5C, S6, S7). As admixture between species is expected to be non-uniformly distributed across the genome, we predicted that there would be greater heterogeneity in FST in sympatry. Indeed the coefficient of variation was significantly greater for FST between sympatric pairs than allopatric pairs (Table 3).
Comparison of FST in sympatry relative to that in allopatry may be useful in identifying regions subject to divergent selection and hence reduced gene flow. When plotted across individual chromosomes, the trend of lower FST in sympatry was widespread but punctuated by narrow regions at which FST between the sympatric populations approached and occasionally exceeded that between allopatric populations (Supplemental Figs. S5C, S6, S7). Assuming that the allopatric population pair provides a reference for expected FST in the absence of gene flow, these regions indicate putative loci at which selection has acted to eliminate introgressed alleles. This hypothesis is supported by the wing pattern loci. H. cydno and H. m. rosina have divergent color patterns, and FST at both the HmB and HmYb patterning loci was similar in sympatry and allopatry (Supplemental Fig. S5C). In contrast, between H. timareta and H. m. amaryllis, which have convergent wing patterns due to recent introgression (The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012), there were narrow regions of reduced FST between the sympatric pair at both patterning loci.
Enhanced reproductive isolation of the Z chromosome
Multiple lines of evidence suggest that gene flow has been reduced throughout the Z chromosome compared with the rest of the genome. Estimates of the fraction of introgression for the Z chromosome were strikingly reduced compared with genome-wide estimates (Fig. 3C; Table 2). In fact, there is no evidence for significant recent Z chromosomal admixture between H. timareta and H. m. amaryllis (Tables 1, 2).
Patterns of genomic differentiation were also consistent with reduced gene flow across the Z chromosome. For most population pairs, the Z chromosome had a significantly elevated level of FST compared with autosomes (Supplemental Table S3; Supplemental Fig. S5B,C). Higher FST on this chromosome compared with autosomes is expected given its lower effective population size. However, the ratio of sympatric/allopatric FST was closer to one on the Z chromosome (∼0.9) than on autosomes (∼0.65) (Table 3). This further supports the hypothesis that admixture on the Z is significantly reduced compared with that on autosomes.
Discussion
The dominant paradigm among evolutionary biologists has recently shifted from widespread belief in the virtually universal importance of allopatric speciation toward increasing acceptance that speciation may occur in the presence of some gene flow. However, despite plenty of evidence for hybridization and gene flow between good species, we remain largely ignorant of the extent to which speciation involves ongoing gene flow, both across the genome and through time. If gene flow is indeed common and persistent, then theoretical models of sympatric speciation might be very widely applicable, justifying the recent shift in emphasis (Wu 2001; Pinho and Hey 2010; Smadja and Butlin 2011; Feder et al. 2012). Using whole-genome resequencing combined with structured geographic sampling we now have much greater power to answer these questions. Our data indicate strong signals of admixture between species across a surprisingly large fraction of the genome. This has occurred either continuously or during multiple periods since their initial divergence. Taken together, our results indicate that species divergence can occur in the face of persistent and genome-wide admixture over long periods of time.
Quantifying gene flow through time
It has long been recognized that both incomplete lineage sorting and hybridization can lead to discordant genealogical histories across the genome. By using an allopatric population of Heliconius melpomene from French Guiana for comparison, we provide evidence that 20%–40% of the genome in H. melpomene shows admixture with H. cydno or H. timareta in sympatry. The window-based phylogenetic approach, using 100-kb regions, averages over large numbers of sites but ensures that each 100-kb tree is statistically well-supported. Furthermore, this result was highly robust to variation in window size.
We extended the site-based ABBA/BABA method to quantify gene flow through time. A previous analysis of H. melpomene and H. timareta indicated that ∼2%–5% of the genome was influenced by gene flow (The Heliconius Genome Consortium 2012), but this comparison could detect admixture that occurred only over a short, recent time period. Our sampling design here allowed us to vary the choice of ingroup populations and examine gene flow over different time scales. Estimates of admixture increased with increasing length of the time period examined, implying continued gene flow during speciation as opposed to a recent burst. Furthermore, LD between derived alleles that were shared during the recent time period was strongest, indicating the existence of introgressed haplotype blocks that are yet to be broken down fully by recombination. This signal was most extensive for alleles shared between H. timareta and H. m. amaryllis but absent from H. m. aglaope, with LD extending up to 1 Mb. This is consistent with extremely recent gene flow, as H. m. amaryllis and H. m. aglaope coalesce very recently. In contrast, LD between variants shared over longer time periods was weaker and declined with physical distance at a rate similar to the genome-wide average, implying that most of these admixed variants were shared very long ago. Thus two independent lines of evidence suggest that gene flow extends from early in speciation to the present. While we cannot rule out periods of allopatry during this time, particularly very early during the species divergence, our results imply that admixture has been a major influence on the genome throughout most of the speciation process.
Genomic divergence through time and space
There has been mixed support for the verbal model of islands of divergence amidst a sea of gene flow (Noor and Bennett 2009; Nosil et al. 2009; Feder et al. 2012). Here we examined this model by comparing patterns of genomic divergence at different stages of speciation and different levels of geographical separation. Parapatric races that are known to hybridize in nature, and in particular H. m. amaryllis and H. m. aglaope from Peru, displayed patterns of differentiation strongly congruent with this islands of divergence model, with strong differentiation at known wing patterning loci. Nonetheless, patterns of divergence are likely to be heterogeneous regardless of gene flow (Noor and Bennett 2009; Michel et al. 2010). For example, between allopatric populations of H. melpomene, subject to isolation by distance and biogeographic barriers such as the Andes, there is a higher average FST but also considerable heterogeneity across the genome, including divergence peaks at the color pattern loci. This probably reflects the fact that strong selection, and various other demographic factors, can cause localized reductions in effective population size, such that certain regions appear as outliers for population differentiation, even in the absence of homogenizing gene flow at other loci (Charlesworth 1998; Turner and Hahn 2010). The presence of FST outliers alone does not provide sufficient evidence that divergence occurred with ongoing gene flow.
FST between sympatric species was highly heterogeneous, and was not congruent with an idealized scenario of islands of divergence against an otherwise homogenized genome. Nevertheless, interspecific FST between sympatric species was generally lower, and more variable (Table 3) than between the corresponding allopatric populations, as expected under a model of admixture with variable selection against introgressing alleles. The trend of lower FST in sympatry was widespread across all chromosomes, consistent with pervasive admixture across the whole genome. Despite this widespread signal, the rate of effective gene flow between H. melpomene and the H. cydno/timareta clade is apparently insufficient to completely abolish differentiation across most of the genome (Nosil et al. 2009; Feder et al. 2012).
Comparisons of sympatric and allopatric populations also permit detection of outlier loci using the joint distribution of FST in sympatry and allopatry. Loci at which interspecific FST is similar in sympatry and allopatry could indicate putative targets of divergent selection where the effective rate of gene flow is reduced. In effect, the allopatric population provides a reference for the expected divergence value in the absence of gene flow, controlling for the inherent heterogeneity in rates of divergence across the genome. This is conceptually similar to an approach applied in hybrid zones, where allopatric populations are used as a control to detect introgressed loci (Gompert and Buerkle 2010). Loci known to be under selection offer a test of this logic. H. cydno and H. melpomene from Panama have distinct wing patterns and both the HmB and HmYb pattern loci fall under peaks at which FST is similar in sympatry and allopatry. The Peruvian pair has convergent wing patterns, and narrow tracts of the genome have here introgressed at both color pattern loci (The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012). Indeed, at both loci, there is a narrow trough of low FST between these populations. The relatively high levels of FST surrounding these troughs may be remnants of hitchhiking following initial divergence in wing pattern. Although we are here mostly interested in the genome-wide patterns of divergence and admixture, we believe that in the future such joint distributions of FST are likely to provide a powerful method for detection of genomic regions subject to selection.
The Z chromosome is at a more advanced stage of speciation
There is both theoretical and empirical evidence for a disproportionate role of the sex chromosomes in speciation (Qvarnström and Bailey 2009). Sex-linked genes are expected to diverge more rapidly (Coyne and Orr 2004), and in the Lepidoptera species differences have been shown to map disproportionately to the Z chromosome (Prowell 1998). In our data there was a significantly reduced signal of admixture on the Z chromosome compared with autosomes. The discrepancy between the Z and autosomal FST was also considerably greater in sympatry than in allopatry (Table 3). Thus the difference cannot be explained solely by reduced effective population size of sex chromosomes. Numbers of shared derived alleles suggest that ancient gene flow did occur on the Z, but that the contemporary migration rate for this chromosome is very low. This can be explained, in part, by Z-autosome incompatibilities known to cause female hybrid sterility (Jiggins et al. 2001; Naisbit et al. 2002). These sex chromosome versus autosome patterns are similar to those seen in the genomes of the Drosophila simulans group and in Ficedula flycatchers (Ellegren et al. 2012; Garrigan et al. 2012), providing general support for the hypothesis that sex chromosomes play a major role in speciation.
Conclusions
Genomic methods offer the opportunity to address the ongoing debate between recent proponents of sympatric speciation and the classical wisdom of ubiquitous allopatric speciation. It is unlikely that genomic data from extant species will ever rule out brief periods of allopatry during speciation. Nonetheless, it is clear from our results that admixture between H. melpomene and the H. cydno/timareta lineage has taken place on a large scale throughout much of their divergence history. To some extent, our findings fit with verbal ideas of speciation with gene flow (Wu 2001; Feder et al. 2012), in which a progression from narrow islands leads to more genomically widespread divergence. Indeed, despite increasing genome-wide divergence later on, the effects of gene flow remain pervasive throughout the genome. Up to 40% of the genome shows a discordant phylogenetic pattern consistent with admixture in sympatry. Our results imply that the recent focus on mechanisms that permit speciation-with-gene-flow in the literature is not misguided (Servedio et al. 2011; Smadja and Butlin 2011). In the case of H. melpomene and H. cydno, wing patterns have a relatively simple genetic basis (Naisbit et al. 2007), and the loci that affect male mate preference and hybrid sterility are associated with color pattern loci (Merrill et al. 2011b), both of which should make speciation easier. Genomics therefore has provided empirical data that help answer thorny questions about the relative importance of allopatric isolation in speciation, which have hitherto proved to be among the most intractable debates in evolutionary biology.
Methods
Whole-genome resequencing and genotype calling
Samples were preserved in NaCl-saturated DMSO solution at −20°C and DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). Illumina paired-end libraries were generated according to the manufacturer's protocol (Illumina Inc.). These were shotgun sequenced on either Illumina's Genome Analyzer IIx system or Illumina's HiSeq 2000 system, according to the manufacturer's protocol (Illumina Inc.).
Quality-filtered, paired-end sequence reads were mapped to the H. melpomene genome scaffolds (version 1.1) (The Heliconius Genome Consortium 2012) using Stampy v1.0.13 (Lunter and Goodson 2011). Defaults were used for all parameters with the exception of the expected substitution rate, which was set to 0.03 for H. melpomene samples (0.001 for the individual from the reference genome strain), 0.04 for H. cydno/timareta samples, and 0.05 for outgroup silvaniform samples to allow mapping of reads from divergent species. To minimize false SNPs due to inconsistent mapping around indels, base alignment quality (BAQ) was considered during mapping, and then local realignment around indels was performed using the Genome Analysis Tool Kit (GATK) v1.6 (DePristo et al. 2011). SAM/BAM file conversion, analysis, and filtering were performed using SAMtools (Li et al. 2009) and Picard (http://picard.sourceforge.net). PCR-duplicate reads were removed using Picard.
Genotypes were called using the GATK v1.6 UnifiedGenotyper (DePristo et al. 2011). Individuals from the same population were genotyped simultaneously. Default parameters were used, except expected heterozygosity was set to 0.01, and BAQ calculation was performed where necessary to optimize calls around indels. For a genotype call to be considered high quality, it had to meet the following criteria: Quality (QUAL) ≥ 30, 10 ≤ depth ≤ 200 (the upper bound was imposed to avoid false SNPs due to mis-mapping in repetitive regions), and for variant (non-reference) calls, genotype quality (GQ) ≥ 30. Only these “high-quality” genotype calls were used in downstream analyses. Genotyping summary statistics for each sample are provided in Supplemental Table S1.
Assigning scaffolds to chromosomes
Several analyses involved comparisons among chromosomes. Scaffolds were assigned to chromosomes based on the Heliconius melpomene linkage map (The Heliconius Genome Consortium 2012), version 1.1, which has ∼80% of the genome assigned to chromosomes. An important focus of this study was the comparison between autosomal and Z-linked regions. We therefore performed extra tests to confirm Z-linkage of mapped scaffolds and identify additional Z-linked scaffolds among those previously unmapped (see Supplemental Appendix A for details). This procedure also identified several mis-assembled scaffolds that were Z/autosome chimeras. Using the most likely breakpoints identified, we removed Z-linked regions from autosomes and also removed autosomal regions from the Z-linked scaffolds.
Phylogenomic analysis
A whole-genome ML tree was generated using only sites in the genome with high-quality genotype calls for all 31 individuals, resulting in an alignment of 60 Mb. RAxML (Stamatakis 2006; Ott et al. 2007; Stamatakis et al. 2008) was used with the GTRGAMMA model, and 100 bootstrap replicates were performed. A separate tree was constructed for the mitochondrial genome (alignment of 9.5 kb), using the same procedure, but with 1000 bootstraps. To investigate phylogenetic discordance across the genome, independent ML trees were generated for non-overlapping 100-kb windows. To minimize artifacts of data quality, only sites with a high-quality genotype call for all 31 genomes were used, and windows that contained <10000 sites were rejected.
Four-population tests for admixture
To test for admixture between pairs of heterospecific populations, we used the four-population test (Reich et al. 2009, 2012). This test is based on the fact that genetic drift should be uncorrelated in unadmixed populations. Given the populations A, B, C, and D, with the unrooted relationship [(A,B),(C,D)], the f4 statistic, f4 (A,B;C,D), allows a test for whether allele frequency differences between A and B are correlated with differences between C and D, thus indicative of admixture (either between A and C, or between B and D, or both). We calculated the f4 statistic (Equation S6.1 of Reich et al. 2012) using all informative sites, i.e., biallelic sites at which both pairs of populations differ in allele frequency. The mean and variance in f4 were then estimated using a block jack-knifing approach (Reich et al. 2009), which controls for LD among sites. We used a block size of 1 Mb, far greater than the extent of LD in the Heliconius genomes studied here (Supplemental Fig. S2; The Heliconius Genome Consortium 2012). This allowed us to test whether f4 deviated significantly from zero. Such deviations would indicate that the allele frequency differences between the two population pairs are significantly correlated, indicating gene flow.
Quantifying gene flow over specific time periods
To quantify gene flow along a specific branch of the phylogeny, we used a method based on the relative abundance of two classes of polymorphic sites called “ABBAs” and “BABAs” (Green et al. 2010; Durand et al. 2011). Given four populations, P1, P2, P3, and an outgroup O, with the relationship {[(P1,P2),P3],O}, ABBAs are SNPs at which P2 and P3 share a derived allele “B,” while P1 retains the ancestral allele “A,” as inferred from the outgroup (i.e., P2 = P3 ≠ P1 = O). Similarly, BABAs are SNPs at which P1 and P3 share a derived allele “B,” while P2 retains the ancestral allele “A” (i.e., P1 = P3 ≠ P2 = O). Under the null hypothesis of no gene flow, ABBA and BABA patterns can only arise via incomplete lineage sorting, and should be equally infrequent (assuming no recurrent mutation and random mating in the ancestral population). However, if there has been gene flow between P3 and P2 since the split between P1 and P2, there should be an overrepresentation of ABBA patterns. The relative abundance of ABBA and BABA patterns throughout the genome was compared using the D statistic (Equation 2 of Durand et al. 2011), based on allele frequencies at each SNP. Only sites at which the four outgroup genomes were homozygous for the same allele were considered to ensure confident assignment of the ancestral and derived states. We used a 1-Mb block jack-knifing approach to calculate the mean and variance of D, allowing a test for whether D differed significantly from zero.
We then estimated f, the fraction of the genome that is admixed. In the example described above, the fraction of the genome that is admixed between P3 and P2 subsequent to the split between P1 and P2 can be estimated by comparing the observed difference in abundance of ABBA and BABA patterns with that which would be expected under a scenario of 100% admixture between P3 and P2 (Equation 8 of Durand et al. 2011). As above, we used a 1-Mb block jack-knife approach to calculate the mean and variance of the f value.
Estimating the extent of linkage disequilibrium (LD)
Linkage disequilibrium (LD) was estimated using all pairs of biallelic sites with high-quality genotype calls in all 31 genomes and a minor allele count of at least five. We estimated r2 within H. melpomene populations using the ML estimator (Clayton and Leung 2007), implemented in the R package “snpstats,” which does not require phased haplotypes. To investigate how LD breaks down with distance, r2 values were binned according to distance in logarithmically increasing bin sizes, to account for small numbers of SNP pairs at large distances. Only SNP pairs on the same scaffold were considered. To obtain an estimate of background LD between unlinked sites, subsets of 500 SNPs were randomly selected and r2 was estimated for all pairs for which the two SNPs were on separate chromosomes. This procedure was repeated 100 times and a 95% confidence interval was calculated.
We investigated the rate of decline in LD between shared derived alleles in H. m. amaryllis and H. m. rosina. Following the definition of an ABBA site above, all sites at which P1 was fixed for the ancestral state while P2 and P3 carried a derived allele were considered. r2 values were binned according to distance as described above.
Patterns of genetic differentiation between populations
We estimated levels of genetic differentiation between populations by calculating FST for 100-kb genomic windows. Nadeau et al. (2012) showed that averaging over large numbers of sites in this way provides highly repeatable FST estimates from small samples. FST was calculated using the EggLib Python module (De Mita and Siol 2012). To minimize variation due to stochasticity and genotyping errors, windows were rejected if they contained <2500 variant sites genotyped with high quality for all individuals from the two populations being analyzed. Windows were restricted to single scaffolds (i.e., they did not cross scaffold boundaries). To plot FST across chromosomes, scaffolds were arranged according to the H. melpomene linkage map (The Heliconius Genome Consortium 2012), version 1.1, having corrected for the several Z/autosome chimeric scaffolds identified as described in Supplemental Appendix A.
Data access
Whole-genome shotgun sequencing paired-end FASTQ files have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena/) under accession number ERP002440. The following files have been deposited in the Data Dryad repository (http://datadryad.org/resource/doi:10.5061/dryad.dk712): all processed VCF files, site-based allele frequency data used for the four-population tests and ABBA BABA analyses, all pairwise FST values for 100-kb windows, maximum-likelihood trees for each 100-kb window for the two four-taxon data sets analyzed (Newick format), data files providing the topology supported by each window, a list of scaffolds and scaffold regions designated as Z-linked, and custom Python and R scripts used for data analyses.
Acknowledgments
We thank Richard Merrill and Simon Baxter for field work. Judith Mank and Jamie Walters kindly contributed funds from their Fell Fund grant. We also thank Anders Eriksson for his input in discussions of this study. Lastly, we are most grateful to the three anonymous referees, whose thoughtful and in-depth reviews greatly improved the standard of this work. This study was funded by the BBSRC (G006903/1, G008841/1, and H01439X/1), the Leverhulme Trust (F/09364/E), and The Fell Fund.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.159426.113.
References
- Baxter SW, Nadeau NJ, Maroja LS, Wilkinson P, Counterman BA, Dawson A, Beltran M, Perez-Espona S, Chamberlain N, Ferguson L, et al. 2010. Genomic hotspots for adaptation: The population genetics of Müllerian mimicry in the Heliconius melpomene clade. PLoS Genet 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR 2010. DensiTree: Making sense of sets of phylogenetic trees. Bioinformatics 26: 1372–1373 [DOI] [PubMed] [Google Scholar]
- Bull V, Beltrán M, Jiggins CD, McMillan WO, Bermingham E, Mallet J 2006. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biol 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain NL, Hill RI, Kapan DD, Gilbert LE, Kronforst MR 2009. Polymorphic butterfly reveals the missing link in ecological speciation. Science 326: 847–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol 15: 538–543 [DOI] [PubMed] [Google Scholar]
- Clayton D, Leung H-T 2007. An R package for analysis of whole-genome association studies. Hum Hered 64: 45–51 [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA 2004. Speciation. Sinauer, Sunderland, MA [Google Scholar]
- De Mita S, Siol M 2012. EggLib: Processing, analysis and simulation tools for population genetics and genomics. BMC Genet 13: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol 28: 2239–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756–760 [DOI] [PubMed] [Google Scholar]
- Eriksson A, Manica A 2012. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci 109: 13956–13960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J, Egan S, Nosil P 2012. The genomics of speciation-with-gene-flow. Trends Genet 28: 342–350 [DOI] [PubMed] [Google Scholar]
- Gagnaire P-A, Pavey SA, Normandeau E, Bernatchez L 2013. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by rad-sequencing. Evolution 67: 2483–2497 [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton K, Presgraves DC 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 22: 1499–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. 2004. Fitness landscapes and the origin of species. Princeton University Press, Princeton, NJ. [Google Scholar]
- Giraldo N, Salazar C, Jiggins CD, Bermingham E, Linares M 2008. Two sisters in the same dress: Heliconius cryptic species. BMC Evol Biol 8: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Buerkle AC 2010. Introgress: A software package for mapping components of isolation in hybrids. Mol Ecol Resour 10: 378–384 [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Petren K 2005. Hybridization in the recent past. Am Nat 166: 56–67 [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Currey M, Cresko WA 2012. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos T Roy Soc B 367: 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C 2008. Ecological speciation in mimetic butterflies. Bioscience 58: 541–548 [Google Scholar]
- Jiggins CD, Linares M, Naisbit RE, Salazar C, Yang ZH, Mallet J 2001. Sex-linked hybrid sterility in a butterfly. Evolution 55: 1631–1638 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Ravigné V 2002. Speciation by natural and sexual selection: Models and experiments. Am Nat (Suppl) 159: S22–S35 [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Young LG, Blume LM, Gilbert LE 2006. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution 60: 1254–1268 [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF 2009. The genomics of speciation in Drosophila: Diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet 5: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, Redmond S, Fulton L, Appelbaum E, Godfrey J, et al. 2010. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330: 512–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M 2011. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J 2002. Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Mol Biol Evol 19: 472–488 [DOI] [PubMed] [Google Scholar]
- Mallet J 2005. Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237 [DOI] [PubMed] [Google Scholar]
- Mallet J, Beltrán M, Neukirchen W, Linares M 2007. Natural hybridization in heliconiine butterflies: The species boundary as a continuum. BMC Evol Biol 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot C, Mavárez J, Evin A, Dasmahapatra KK, Mallet J, Lamas G, Joron M 2013. Genetic differentiation without mimicry shift in a pair of hybridizing Heliconius species (Lepidoptera: Nymphalidae). Biol J Linn Soc Lond 109: 830–847 [Google Scholar]
- Merrill RM, Gompert Z, Dembeck LM, Kronforst MR, McMillan WO, Jiggins CD 2011a. Mate preference across the speciation continuum in a clade of mimetic butterflies. Evolution 65: 1489–1500 [DOI] [PubMed] [Google Scholar]
- Merrill RM, Van Schooten B, Scott JA, Jiggins CD 2011b. Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc Roy Sci B 278: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill RM, Wallbank RWR, Bull V, Salazar PC, Mallet J, Stevens M, Jiggins CD 2012. Disruptive ecological selection on a mating cue. Proc Roy Sci B 279: 4907–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel APA, Sim S, Powell THQ, Taylor MS, Nosil P, Feder JL 2010. Widespread genomic divergence during sympatric speciation. Proc Natl Acad Sci 107: 9724–9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW, Quail MA, Joron M, Ffrench-Constant RH, Blaxter ML, et al. 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci 367: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW, Baxter SW, Blaxter ML, Mallet J, Jiggins CD 2013. Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol Ecol 22: 814–826 [DOI] [PubMed] [Google Scholar]
- Naisbit RE, Jiggins CD, Linares M, Salazar C, Mallet J 2002. Hybrid sterility, Haldane's rule and speciation in Heliconius cydno and H. melpomene. Race 1526: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit RE, Jiggins CD, Mallet J 2007. Mimicry: Developmental genes that contribute to speciation. Evol Dev 5: 269–280 [DOI] [PubMed] [Google Scholar]
- Noor MAF, Bennett SM 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol 18: 375–402 [DOI] [PubMed] [Google Scholar]
- Nosil P, Gompert Z, Farkas TE, Comeault AA, Feder JL, Buerkle CA, Parchman TL 2012. Genomic consequences of multiple speciation processes in a stick insect. Proc Roy Sci B 279: 5058–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Zola J, Stamatakis A, Aluru S 2007. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. In Proceedings of the 2007 ACM/IEEE conference on supercomputing, SC '07. Reno, Nevada [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Mérot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho C, Hey J 2010. Divergence with gene flow: Models and data. Annu Rev Ecol Evol Syst 41: 215–230 [Google Scholar]
- Prowell DP. 1998. Sex linkage and speciation in Lepidoptera. In Endless forms: Species and speciation (ed. Howard DJ, Berlocher SH), pp. 309–319. Oxford University Press, New York. [Google Scholar]
- Qvarnström A, Bailey RI 2009. Speciation through evolution of sex-linked genes. Heredity 102: 4–15 [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L 2009. Reconstructing Indian population history. Nature 461: 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. 2012. Reconstructing Native American population history. Nature 488: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH 2009. Evolution: Replacing genes and traits through hybridization. Curr Biol 19: R119–R122 [DOI] [PubMed] [Google Scholar]
- Rosser N, Phillimore AB, Huertas B, Willmott KR, Mallet J 2012. Testing historical explanations for gradients in species richness in heliconiine butterflies of tropical America. Biol J Linn Soc Lond 105: 479–497 [Google Scholar]
- Salazar C, Jiggins CD, Taylor JE, Kronforst MR, Linares M 2008. Gene flow and the genealogical history of Heliconius heurippa. BMC Evol Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Patterson N, Li H, Pääbo S, Reich D 2012. The date of interbreeding between Neandertals and modern humans. PLoS Genet 8: e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servedio MR, Van Doorn GS, Kopp M, Frame AM, Nosil P 2011. Magic traits in speciation: “Magic” but not rare? Trends Ecol Evol 26: 389–397 [DOI] [PubMed] [Google Scholar]
- Smadja CM, Butlin RK 2011. A framework for comparing processes of speciation in the presence of gene flow. Mol Ecol 20: 5123–5140 [DOI] [PubMed] [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Supple MA, Hines HM, Dasmahapatra KK, Lewis JJ, Nielsen DM, Lavoie C, Ray DA, Salazar C, McMillan WO, Counterman BA 2013. Genomic architecture of adaptive color pattern divergence and convergence in Heliconius butterflies. Genome Res 23: 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW 2010. Genomic islands of speciation or genomic islands and speciation? Mol Ecol 19: 848–850 [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol 3: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Edelaar P, Weissing FJ 2009. On the origin of species by natural and sexual selection. Science 326: 1704–1707 [DOI] [PubMed] [Google Scholar]
- Wu C 2001. The genic view of the process of speciation. Science 14: 851–865 [Google Scholar]