Abstract
Genome sequencing has provided us with gene lists but cannot tell us where and how their encoded products work together to support life. Complex organisms rely on differential expression of subsets of genes/proteins in organs and tissues, and, in concert, evolved to their present state as they function together to improve an organism's overall reproductive fitness. Proteomics studies of individual organs help us understand their basic functions, but this reductionist approach misses the larger context of the whole organism. This problem could be circumvented if all the organs in an organism were comprehensively studied by the same methodology and analyzed together. Using honey bees (Apis mellifera L.) as a model system, we report here an initial whole proteome of a complex organism, measuring 29 different organ/tissue types among the three honey bee castes: queen, drone, and worker. The data reveal that, e.g., workers have a heightened capacity to deal with environmental toxins and queens have a far more robust pheromone detection system than their nestmates. The data also suggest that workers altruistically sacrifice not only their own reproductive capacity but also their immune potential in favor of their queen. Finally, organ-level resolution of protein expression offers a systematic insight into how organs may have developed.
Honey bees epitomize hard work and evoke thoughts of honey, stings, and their telltale black and gold coloration. Apis mellifera L. (the western or European honey bee) is one of the few insects that humans have domesticated, and now they contribute tens of billions of dollars per year to the value of agricultural crops. Bees have been dying at unprecedented rates across much of the world over the past six years, leading to an increased public awareness of their importance. They are one of the few eusocial animals known; they are organized into a three-caste system where reproduction is delegated specifically to the single queen and hundreds of drones (males), while caring for young, foraging, and defense fall to the female workers. Pheromones, especially those emitted by the queen, control the physiology and behavior of hive members. Despite all hive members having the same genome, workers live only for several weeks in the summer, while the queen can live for years. While we partially understand how bees' social behaviors manifest (Robinson et al. 2008), we have little mechanistic understanding of what allows the castes to carry out their individual duties.
Completion of the honey bee genome sequence in 2006 constituted a quantum leap forward (The Honey Bee Genome Sequencing Consortium 2006) in our molecular understanding of this insect. Comparative genomics revealed that many gene families had expanded (e.g., of genes coding for brood food and odorant reception) or contracted (e.g., of immunity and detoxification genes) relative to those of Drosophila melanogaster. These are merely a few examples of the insight that a sequenced genome can provide, in addition to providing a genetic “parts list” that can be used by the research community: “with the genome at hand, prospects are bright for elucidating the molecular and genetic bases of many complex traits…” (The Honey Bee Genome Sequencing Consortium 2006). While an organism's chromosomes can now be sequenced from telomere to telomere, defining the complete transcriptome or proteome of any life-form remains beyond our capabilities, especially for multicellular organisms where transcriptomes and proteomes will be organ if not cell specific. Transcript atlases based on microarrays exist for some species (Edwards et al. 2010; Huttlin et al. 2010) and RNA-seq data for specific tissues are becoming available (Krupp et al. 2012). For multicellular organisms, however, our view of the proteome is largely confined to one or a few organs, with just two exceptions: The antibody-based Protein Atlas is examining several different human proteins (Uhlen et al. 2010), while a very recent report has cataloged tissue expression across 28 mouse tissues (Geiger et al. 2013). Here we present the protein expression profiles across all three castes and across essentially all organs in adult bees. These data reveal the remarkable specialization required of the proteome to generate distinct organs: major caste-related trends observed for enzymes responsible for digestion, immunity, and detoxification, to name a few. This represents one of the few whole-body, cross-gender, and cross-caste proteome maps for any multicellular organism, and is a comprehensive resource not only for bee researchers but also in comparative functional genomics.
Results and Discussion
Honey bee proteins
We dissected 29 organs from adult worker, drone, and queen honey bees, generating biological triplicates of all and pooling specimens where necessary to collect enough material. Bees were taken from three different colonies, all of which are first- or second-generation descendants from a single mother queen. Samples were analyzed by liquid chromatography mass spectrometry (LC-MS/MS), and all raw spectral data are available in the honey bee PeptideAtlas (Chan et al. 2011b) along with all the bee-specific peptide spectra our group has published previously, which to date represent the largest repository of honey bee fragment mass spectra. This freely accessible resource allows the growing community of bee mass spectrometrists to plan targeted proteomic studies for their protein of interest, a major time-saving improvement from the trial-and-error work that is usually associated with target selection. Furthermore, Gene Ontology annotation by Blast2GO has already been performed for each protein hit, and other helpful parameters such as peptide hydrophobicity and frequency of observation.
In total, 2288 honey bee proteins were identified with at least two peptides with a protein false discovery rate of 1.2%. The number of identifications initially surprised us, but bees have a considerably smaller genome than mammals; even so, we typically identify relatively (to number of annotated genes) fewer proteins in a bee sample compared with an equivalent amount of human protein. This may suggest that there are still many unannotated or incorrectly annotated genes in bee, that the distribution of protein abundances in bee are more skewed toward lower copy numbers, or that there are a large number of polymorphisms that are not considered in database searching. Thus, the profiles for each organ here are most certainly incomplete, but organ-specific patterns can already be distinguished despite the number of proteins identified, similar to how organ-specific patterns can also be observed with the 100 most abundant proteins in mouse organs (Geiger et al. 2013). Likewise, while we used every organ we could to maximize protein identifications, there were still several identified structures that could not be obtained in sufficient quantities to allow effective analysis. Another factor that limited the depth of coverage of each organ was the difficulties in raising (particularly queens) and dissecting enough bees to obtain sufficient amounts of protein for analysis (e.g., thoracic glands and heart). Here we are able to identify about 2000 proteins, which is about one-fifth of the predicted ∼10,000 genes in the bee genome (Elsik et al. 2007), but even still there is more that could be learned with more exhaustive sampling and improved mass spectrometry technology.
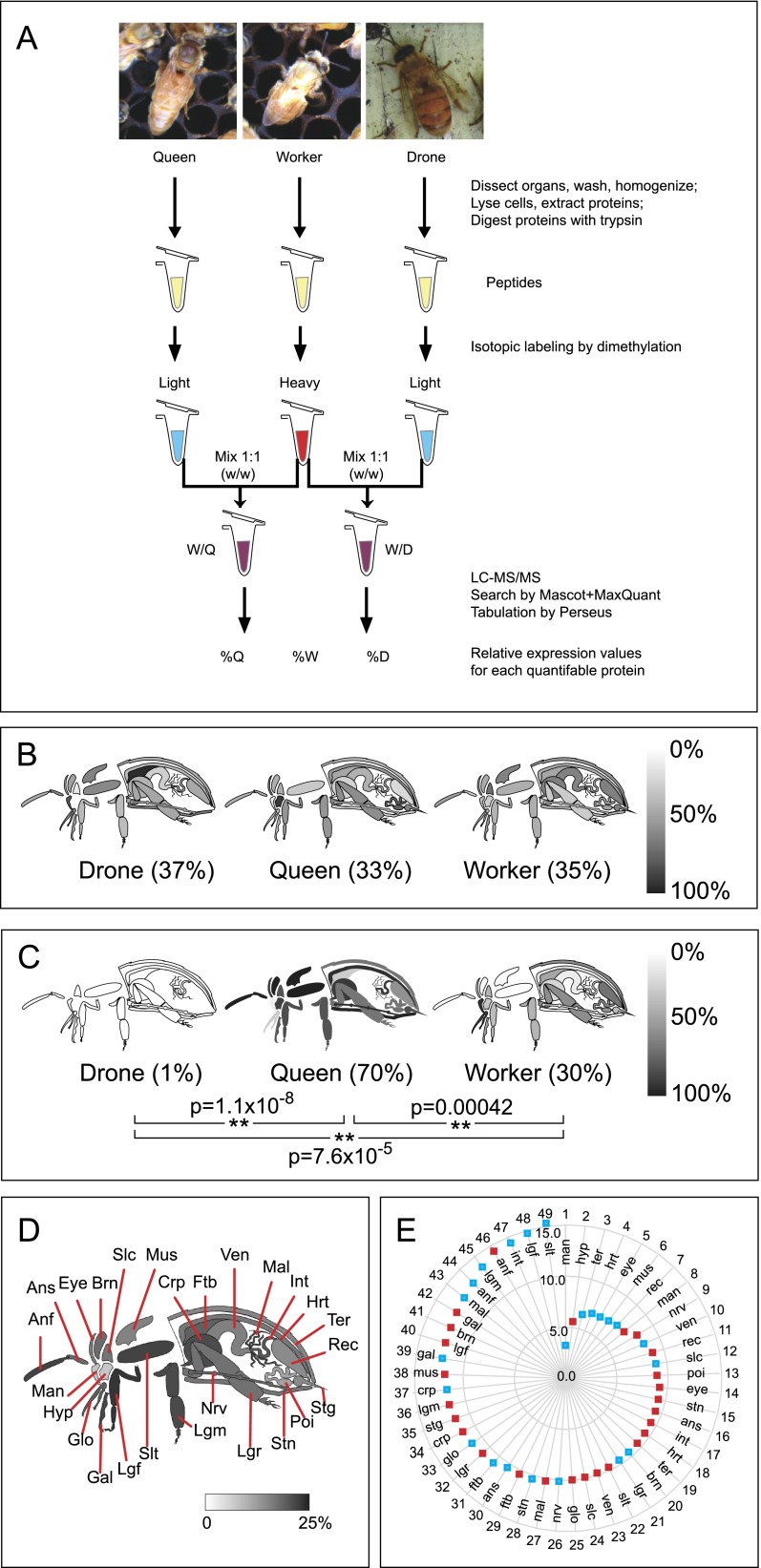
With the exception of three caste-specific organs (spermatheca, testes, and mucus gland), peptides derived from all organs and all castes were isotopically labeled for relative quantitation by LC-MS/MS (summarized in Fig. 1A). This approach has been widely used by us (Chan et al. 2009) and others (Kovanich et al. 2012) and provides more accurate data than label-free approaches. This analysis resulted in 31,669 individual measurements of relative protein levels, with drone or queen levels being measured relative to the worker level in all cases. In order to express values in such a way as to allow comparisons across castes without having to always think of the worker level as equal to 1.0, the ratios were converted into percent values, where the expression levels for a given protein in drone, queen, and worker sum to 100%. Classic “housekeeping” proteins such as actin (Fig. 1B) showed nearly uniform expression across all organs and castes. In contrast, the largely female-specific protein vitellogenin (gi:58585104) (Fig. 1C), particularly important for reproductive females (i.e., queens), is clearly most abundant throughout the queen's body, with lower levels detected in workers. The data for all quantifiable proteins can be browsed and searched in an interactive graphical format (http://www.chibi.ubc.ca/faculty/leonard-foster/foster-lab/data/ under Bee Protein Atlas).
Figure 1.
An overview of the Honey Bee Protein Atlas. (A) Sample handling flowchart. Worker proteins from each organ served as a reference against which queen and drone organs were compared. The relative abundance of actin (B) and vitellogenin (C) in the organs of each caste are shown on a grayscale. The absence of an organ indicates that either the organ does not exist (e.g., stinger in drones) or the protein was not detected there. Whole-body averages are shown in parentheses; (**) P < 0.01. (D) A diagrammatic representation of each tissue, and the percentage of quantified proteins in queens and/or drones whose expression level is significantly different from the worker at P < 0.05. Individual percentage differences in queen–worker (red squares) and drone–worker (blue squares) comparisons for the tissues are shown as a radar graph (0% at the origin up to 15%) in E, arranged from the least different (wheel position 1) to most different (wheel position 49) tissue. (anf) flagellum (antenna); (ans) scape (antenna); (brn) brain; (crp) crop (forgut); (ftb) fat body; (gal) galea (mouthpart); (glo) glossa (mouthpart); (hrt) heart; (hyp) hypopharngeal gland (queen and worker only); (int) intestine; (lgf) front leg; (lgm) middle leg; (lgr) rear leg; (mal) Malpighian tubules; (man) mandibular gland; (mus) thorax muscle; (nrv) nerve; (poi) poison sac (queen and worker only); (rec) rectum; (slc) post-cerebral salivary gland; (slt) thoracic salivary gland; (stg) sting (queen and worker only); (stn) sternite; (ter) tergite; (ven) ventriculus (midgut).
Caste-specific differences in honey bee organs
The quantitative aspects of the data set presented here cover two dimensions of bee biology: (1) across dissectible bee organs and (2) across castes. Anywhere from 3% to 16% of the quantified proteins in each organ were significantly different among castes at the P < 0.05 level (Fig. 1D), implying that some organs have more caste-specific functions, or specialization, than others. By ranking each worker–drone and worker–queen organ according to the percentage of proteins that were significantly different (Fig. 1E) it is possible to visualize how similar the drone or queen organ is to the equivalent baseline worker organ. Near the center of the wheel (wheel position 1, 3–7) are a number of drone organs responsible for what would be expected to be caste-independent, basic life functions (e.g., heart, eyes). Such similarity was not the case for the same comparisons between the worker and queen. For example, the drone's mandibular glands (wheel position 1) are much more similar to worker than are the queen's (wheel position 8). The secretions from the queen's mandibular glands have an enormous impact on colony behavior and physiology, and the dissimilarity to worker glands may reflect this critical specialization.
Prior to starting this analysis, we had assumed that tissues such as the legs would show minimal caste differences. Worker hind legs are the only ones to carry pollen sacs, but the basic function of all legs in all castes is the same: locomotion. Intriguingly, however, the profiles of drone or queen to worker legs (wheel positions 21, 32, 36, 40, 45, 48) were actually quite different. Other organs such as the galea (wheel positions 39, 42) and flagella (distal portion of the antenna, wheel positions 44, 46) were also among the most caste diverse. The most peculiar is the drone thoracic salivary gland, the most divergent organ from the worker above all others. Very little is known of its function other than to moisten food before it enters the alimentary canal. These observations hint at more substantial variations in the function and/or efficiency of organs among the castes than was previous appreciated. In humans, there are measurable differences between males and females with regard to lipid metabolism (Sugiyama and Agellon 2012), the handling of oxidative stress (Vina et al. 2011), and the immune system (Oertelt-Prigione 2012), to name a few. Hence, it is reasonable that honey bees should have diverse caste differences in their organs, and we explore this phenomena in more detail below.
Divergent metabolic capacity of castes
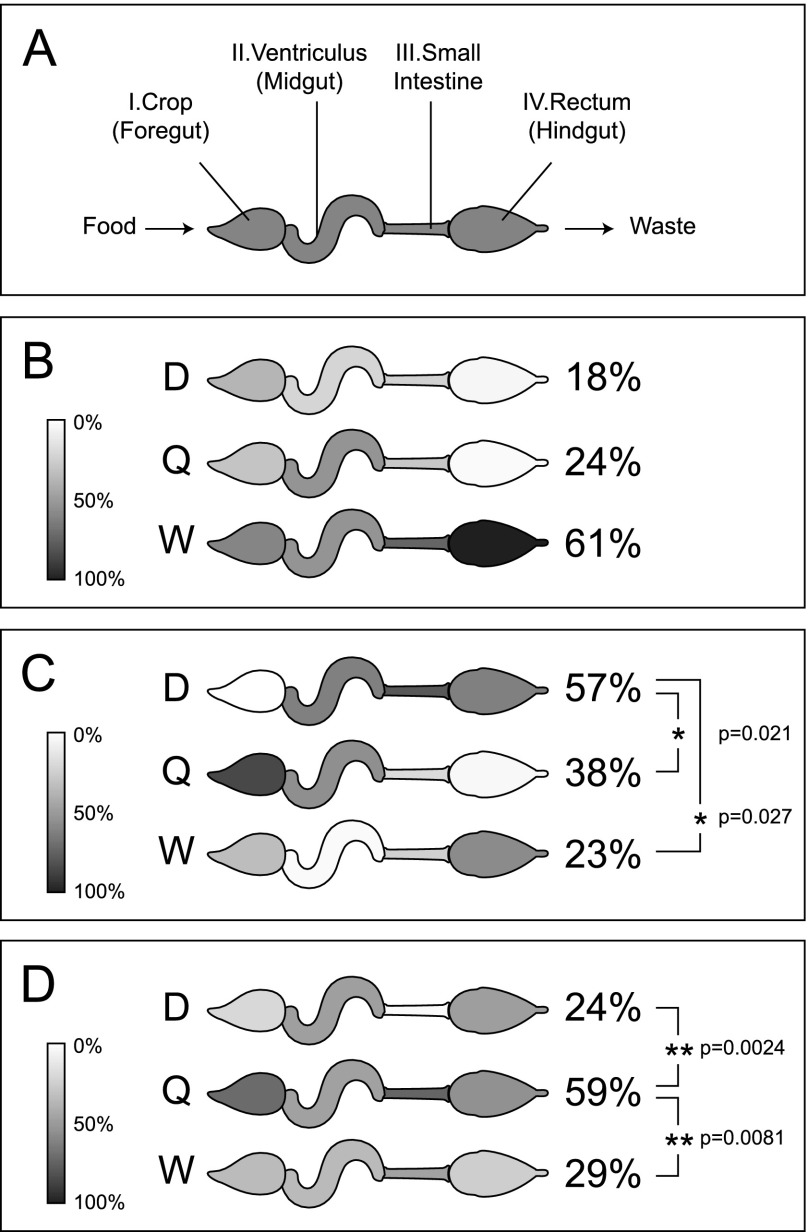
Males and females of most animal species perform gender-specific roles and display gross anatomical differences, but the mechanisms underlying these differences are ill defined. Queens, drones, and workers have distinct dietary needs, which should be reflected in their metabolic capacity, but this notion has never been explored at the molecular level. The exact composition of their food is unknown, but generally workers are fueled by carbohydrates from honey, queens rely on lipids and proteins in the form of royal jelly fed to them by workers, and drones fall somewhere in between (Winston 1987a). After the food travels through the esophagus, it proceeds through four major organs in the abdomen before waste is eliminated (Fig. 2A): (1) the crop, or foregut, an expandable sac that holds nectar collected from flowers; (2) the ventriculus, or midgut, where digestion is mainly thought to occur; (3) the small intestine, where nutrients are absorbed; and (4) the rectum, or hindgut, which stores waste until it can be expelled outside of the hive.
Figure 2.
Caste-dependent differences in major digestive system enzymes. (A) The abdominal sections of the digestive tract. Relative expression values of major enzymes for digestion of mono- and polysaccharides (B), proteins (C), and lipid transport (D) are shown on a grayscale. Averages among the four organs are shown in parentheses; (*) P < 0.05, (**) P < 0.01.
Differences in the castes' diets are reflected in gut enzymes. Workers express threefold more alpha-glucosidases (gi:94158854, gi:94400901, gi:58585164) and alpha-amylase (gi:58585144) across the entire digestive tract (but does not meet significance at the P = 0.05 level) (Fig. 2B) than queens or drones. A worker's diet is rich in carbohydrates, given their need for easily accessible energy during frequent movement and flight activity. On the other hand, drones clearly are better equipped to use protein as an energy source, since they expressed higher average levels (P < 0.05) of two trypsins (gi:328783370, gi:110762227), two carboxypeptidases (gi:328782267, gi:328782015), one chymotrypsin (gi:110762229), and aminopeptidase (gi:66512450). Notably, the worker midgut appeared comparatively devoid of these enzymes (Fig. 2C). To round out the three primary energy sources, four lipid binding and transport proteins (gi:328780884, gi:328785302, gi:328787757, gi:328790232) were more highly expressed in the digestive tract of queens (Fig. 2D), in keeping with their royal jelly diet, which is relatively high in lipids (Howe et al. 1985).
Robust detoxification machinery in workers
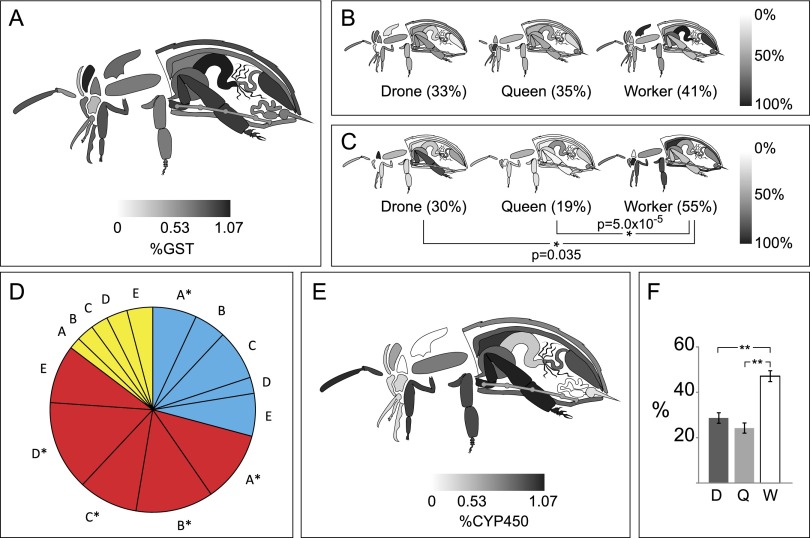
Despite identical genomes, bees of each caste have very different life spans, ranging from a few weeks for an adult worker to a few years for a queen. These differences are thought to be at least partially attributable to their varied ability to cope with oxidative stress (Finkel and Holbrook 2000). However, our data indicate that bees express several glutathione-S-transferases (Fig. 3A), glutathione peroxidases, peroxiredoxins, superoxide dismutases, and thioredoxin proteins ubiquitously with little or no organ or caste bias (Fig. 3B). This is not surprising, given that endogenous metabolic processes generate free radicals that can damage proteins, lipids, and DNA, and all three castes could expect roughly equivalent exposure to these. Catalase, which reduces hydrogen peroxide to water and oxygen, is heavily expressed in workers, followed by drones then queens (Fig. 3C). This enzyme is concentrated in the worker fat body, consistent with the liver-like functions of this organ. Malpighian tubules are another important site for detoxification because they selectively remove unneeded metabolites from the hemolymph and surrounding organs (Dow 2009). Multidrug resistant proteins (MDRPs) are rarely found in most organs, but at least five distinct isoforms are expressed in Malpighian tubules. More surprising, however, is the queen's uniformly high expression of all five of them compared with the other castes (Fig. 3D).
Figure 3.
Caste and organ distribution of detoxification enzymes. (A) GST proteins as a fraction of all quantified proteins across castes. (B) Peroxiredoxin (gi:328787790) and (C) catalase expression levels. Whole-body averages are shown in parentheses; (*) P < 0.05, (**) P < 0.01. (D) In the Malpighian tubules, MDRPs were collectively highly expressed in queens (red) compared with drones (blue) and workers (yellow). The size of each slice is determined by the relative proportion of a given protein in each caste; (*) P < 0.05 compared with the worker. A (gi:328789595), B (gi:328787148), C (gi:328784175), D (gi:328791429), E (gi:328777607). (E) P450 proteins as a fraction of all proteins across castes, and (F) among the P450s, a caste breakdown of their average P450 expression (±SEM, n = 85).
The enzyme family responsible for detoxification and xenobiotic breakdown is the cytochrome P450 monooxygenases (P450). These enzymes are concentrated in organs one might expect to be exposed to toxins directly, in the outer extremities, including the antennae, legs, and exoskeleton, as well as along parts of the digestive tract (Fig. 3E). Of the three castes, workers spend by far the most time outside the hive so would be most likely to encounter insecticides, defensive phytochemicals, and pollutants; indeed, they express significantly higher levels of P450s compared with queens and drones (Fig. 3F).
Our data do not support a link between detoxification enzymes and life span. A queen's longevity cannot be solely attributed to her heightened expression of MDRPs since workers are enzymatically better equipped to minimize the effect of xenobiotics but their life span remains relatively short. In keeping with the principles of natural selection, bees have evolved such that each caste can neutralize the toxins their bodies typically generate or encounter.
Queens may have the best sense of smell
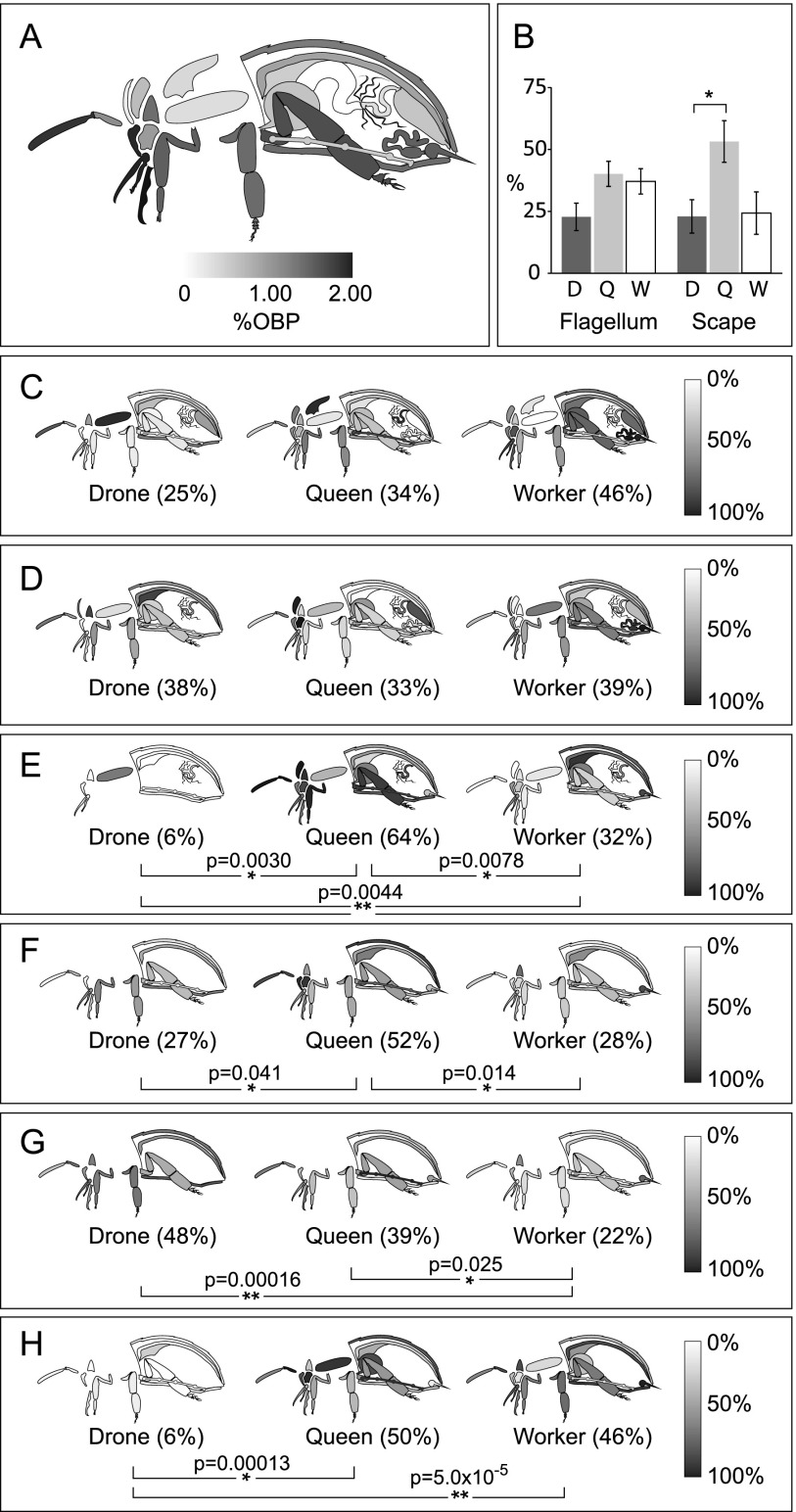
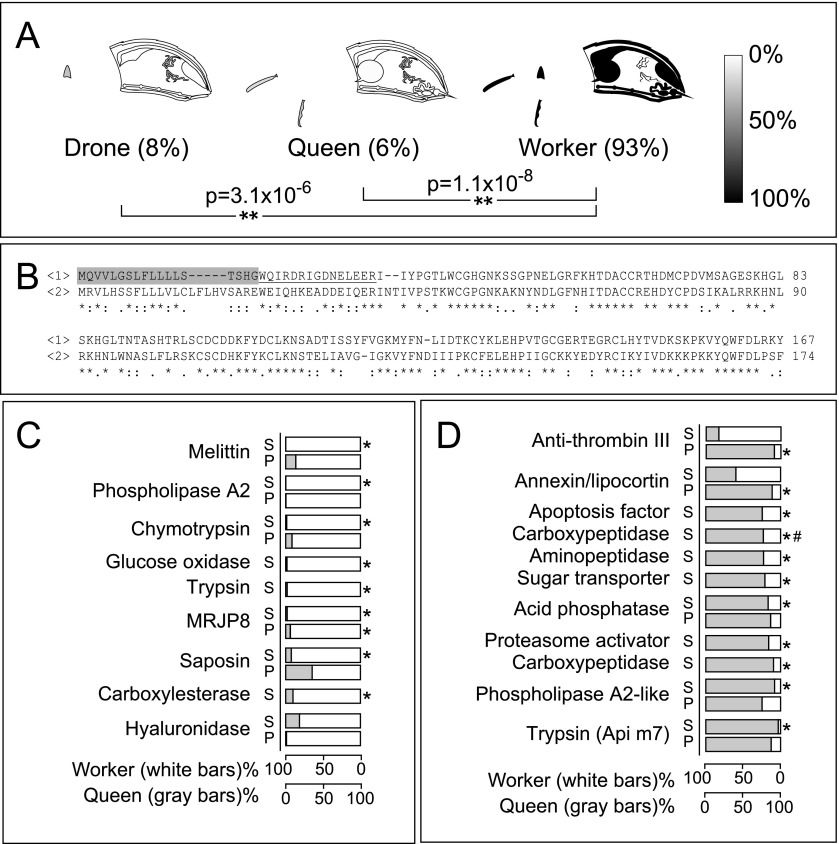
Scent is a vital means of communication among bees, with pheromones affecting the behavior and physiology of every member in the hive. The largely insoluble odorant molecules used by bees are carried through the aqueous lymph by soluble odorant binding proteins (OBPs). Despite the importance of OBPs, their specific substrates are unknown, although several expression studies exist for some organs and developmental stages (Foret and Maleszka 2006; Dani et al. 2010). OBPs are expressed in nearly all regions of the body but are most concentrated in tissues exposed to the external environment (Fig. 4A). Antennae have the richest variety of OBPs, expressing at least 18, plus two chemosensory proteins (CSPs), while queens had more than other castes, shown most clearly in the short proximal segment of the antenna called the scape (P = 0.038 and P = 0.093 compared with the drone and worker, respectively) (Fig. 4B).
Figure 4.
Caste and organ distribution of OBPs. (A) OBPs as a fraction of all quantified proteins across castes. (B) The percent expression of all quantified OBPs in each caste, averaged across the flagellum (the long distal portion of the antenna, n = 16) and the scape (the short proximal portion, n = 11) (±SEM). The relative abundance of OBP14 (C), CSP3 (D), OBP3 (E), OBP17 (F), OBP18 (G), and OBP21 (H) in the organs of each caste is shown on a grayscale. Whole-body averages are shown in parentheses; (*) P < 0.05, (**) P < 0.01.
Expression levels of OBPs were roughly in-line with a semi-quantitative analysis of transcript abundance (Foret and Maleszka 2006), especially with regard to the widespread presence of OBP14 (Fig. 4C). This protein and CSP3 (Fig. 4D) were the most ubiquitous and most evenly distributed among all the castes, suggesting that they may share a ligand and/or that their ligands may have very broad functions, such as maintaining hive unity. All other OBPs demonstrated a strong organ bias and typically were highest in queens (Fig. 4E,F,H).
The overall observation that queens express the most OBPs seems to correlate inversely with the number of olfactory organs, the sensilla placodea, on the antenna surface (queens have ∼1600, workers ∼3000, and drones ∼30,000) (Winston 1987b). One hypothesis to explain the inverse correlation between OBP variety and sensilla placodea is that drones may have the most acute sense of smell, but only to very few pheromones. Drones do not have many functions, but they do have to be able to track down a virgin queen in flight to mate with her. In contrast, the queen is surrounded at all times by tens of thousands of workers and larvae, each emitting pheromones, so she may have evolved to have fewer antennal pores to avoid being overwhelmed. Her heightened expression of many different OBPs may indicate that she is capable of sensing numerous types of odorant signals. It implies that she is also capable of responding to cues from workers and other sources, in addition to being the chief pheromone producer in the colony.
Queen immune system linked to longevity
The recent unsustainable losses of managed bee colonies to infectious diseases (Evans and Schwarz 2011) underlines our poor understanding of innate immunity in bees. Initial indications from the genome sequence suggested that bees have far fewer immunity genes than other insects (The Honey Bee Genome Sequencing Consortium 2006), and our data suggest that even the few genes that the species does have are not protecting all three castes equally. The major melanization pathway enzyme, prophenoloxidase (PPO, gi:58585196), was nearly ubiquitous across organs but was highly biased toward queens, followed by workers and then drones (Fig. 5A). Interestingly, the only known enzyme that can activate PPO (gi:328783409) was expressed solely in the heart of queens and workers (81%–19%, P < 0.05).
Figure 5.
Caste and organ distribution of immunity proteins. (A) PPO, (B) PGRP-S3, and (C) GNBP 1-2 expression is significantly higher in queens compared with the other castes. Overall expression levels of the antimicrobial peptides hymenoptaecin (D) and defensin (E) are not different (P > 0.1), except that drones express far less defensin than queens or workers. Relative levels across each organ shown on a grayscale. Whole-body averages are shown in parentheses; (*) P < 0.05, (**) P < 0.01.
This queen-biased expression also holds true for two pathogen-binding proteins that were quantified: Peptidoglycan recognition protein S3 (PGRP-S3, gi:254910928) (Fig. 5B) and Gram-negative bacteria-binding protein 1-2 (GNBP 1-2, gi:254911140) (Fig. 5C). Trends for antimicrobial peptides were not as clear-cut; e.g., hymenoptaecin (gi: 58585174) (Fig. 5D), trended toward higher expression in queens but was not statistically significant. On the other hand, workers express much more defensin than other castes (Fig. 5E), although this is almost exclusively in the head and thoracic glands, as well as the hypopharyngeal glands which synthesize royal jelly for larvae and queens to consume. As such, the defensin workers do produce is probably not used for their own protection but, in true altruistic fashion, goes toward protecting their queen. Overall, queens appear to have the most robust immune system, even to the point of the other castes being completely expendable.
Queen versus worker venom
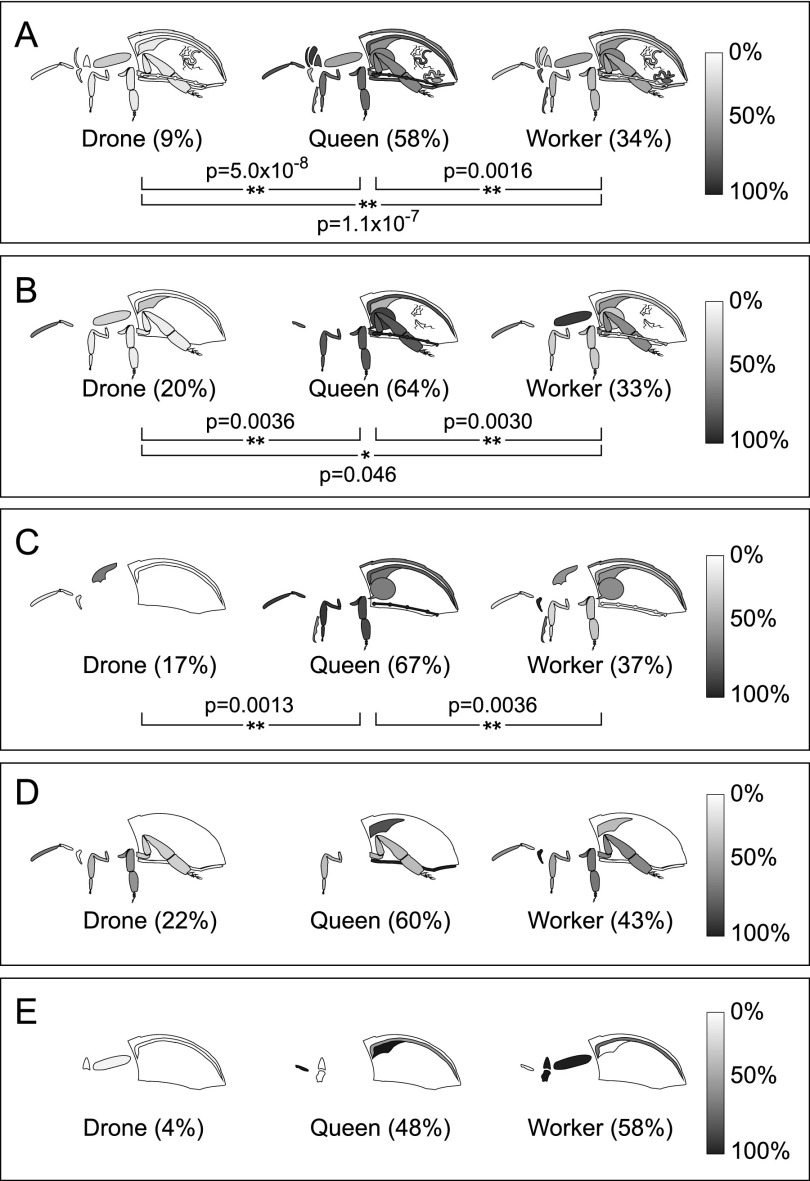
The stinger apparatus (i.e., the poison sac plus the stinger) is a bee's primary weapon and is exclusive to the female castes. Workers use it to defend the colony, using a ratchet mechanism in the barbed stingers to help it initially lodge in and then work its way further into thick tissues, even as it means death for the worker. Queens use their barbless stingers exclusively to kill rival queens. Previous analyses of venom composition have identified certain proteins specific to worker or queen venom, as might be expected given the contrasting end goals of their stinging behavior. The differences between queen and worker venom are even more dramatic than previously thought, however. Phospholipase A2 (gi:58585172) is a well-known venom protein, and it is heavily expressed in the worker throughout the body (Fig. 6A) and particularly in the stinger and poison sac (Fig. 6C; and as shown before, Marz et al. 1981). However, a homolog (gi:110758297, 49% identical) is more abundant in the queen stinger (Fig. 6D). Differences in their signal peptide and propeptide regions (Fig. 6B) suggest they are sorted and regulated differently. Queens also have more lipocortin (gi:328790767, also called annexin) (Fig. 6D), a known suppressor of phospholipase A2 activity. On the other hand, workers express a stinger-specific trypsin (gi: 328778042) (Fig. 6C), which may cleave the phospholipase A2 propeptide with arginine at the C terminus.
Figure 6.
Proteome of the poison and sting apparatus: select proteins. (A) Relative abundance of phospholipase A2 (gi:58585172) in the organs of each caste is shown on a grayscale, and the whole-body average is shown in parentheses. Worker expression is significantly different from the others; (**) P < 0.01). (B) Multiple sequence alignment of <1> phospholipase A2 and <2> phospholipase A2-like (gi:110758297). The signal peptide (gray) and propeptide (underlined) are as given in Kuchler et al. (1989). Select proteins that tend to be more highly expressed in (C) workers (white bars) and (D) queens (gray bars) in the poison sac (P) and stinger (S) are shown; (*) P < 0.05, (#) the protease also found in the digestive tract.
Melittin (gi:58585154), a pore-forming peptide that causes cell leakage (Raghuraman and Chattopadhyay 2007), is already known to make up 50% of the worker venom by weight (Habermann 1972). In our data, workers have >1000-fold more melittin than queens in the stinger (Fig. 6C). Given that it can also synergistically activate phospholipase A2, leading to release of pro-inflammatory arachidonic acid in the victim, it appears that worker venom is designed to cause quick and localized damage. Furthermore, the extracellular matrix–degrading enzyme hyaluronidase (gi:58585182), the hydrogen peroxide–producing glucose oxidase (gi:58585090), and the lysosomal lipase–activating enzyme saposin (gi:328782499) are all much higher in the worker poison sac and stinger (Fig. 6C). Given that workers need to defend the colony against predators that are often hundreds of thousands of times their own size (e.g., bears, humans), killing the antagonist would be a tall order, but inflicting enough pain to drive it away achieves the same goal.
On the other hand, queen venom is used exclusively to kill rival queens. Queen venom contains higher levels of an apoptosis-inducing factor (gi:328783113), a proteasome activator (gi:328788437), and several peptidases (Fig. 6C), including a trypsin-like enzyme with unknown substrates (gi:58585116) (Georgieva et al. 2010), better known as the venom allergen Api m7. Thus, queen venom appears to be optimally suited to damaging cells and tissues. Even the anti-coagulant anti-thrombin III (gi:328782084) (Fig. 6C) helps to ensure that this toxic concoction circulates thoroughly in the victim queen's body.
Insights into organ differentiation
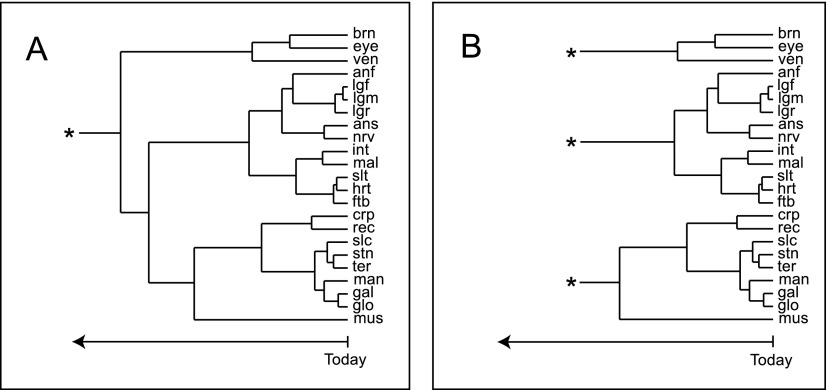
The body of data generated here can be used to explore how the proteomes among the castes differ, but just as interesting is its capacity to help us understand how each organ is similar or divergent from one another. To this end, we identified the proteins for each organ that were in approximately equal levels among all three castes (minimum 20% in each, thereby excluding heavily caste-specific proteins), averaged the number of peptides we were able to find for each protein between the worker-drone and worker-queen samples, and divided this value by the total number of peptides found in a given organ; this effectively corrected for the incongruent numbers of peptides (and proteins) found in each organ, and with these values we performed complete linkage clustering for the top 500 most abundant proteins (Fig. 7A).
Figure 7.
Evidenced-based insight into organ differentiation and specialization within castes. Dendrograms generated from the top 500 most common proteins among the tested tissues propose two models of organogenesis: (A) a common, single ancestral organ, or (B) multiple organogenesis events. The time scale and its linearity are uncertain but likely go back to the appearance of early bilaterans.
Numerous insights emerged from this analysis, but first we observed that some patterns are consistent with existing knowledge. For example, the front, middle, and rear legs have effectively identical functions (i.e., locomotion) and formed the tightest cluster. Other organs that have similar functions also have similar expression patterns, such as the major exoskeletal plates of the abdomen (ternite, sternite) and the mouthparts (galea, glossae). Not everything is so obvious, however. The nerve chord and the short antennal base (scape) are quite similar, perhaps related to their roles in transducing nerve signals. The small intestine, which is poorly understood in the bee, may have similar functions to the Malpighian tubules, which play a major role in osmoregulation and ion transport. Due to the physical proximity of these two organs, cross-contamination during dissection cannot be ruled out, but is unlikely to have had a major contribution since the two organs are morphologically quite different and easy to distinguish under a dissecting microscope. The heart and thoracic salivary gland are physically a good distance apart so cross-contamination cannot be a factor, yet the two form a tight cluster. This observation cannot be reasonably explained at this point—both are thought to be highly dissimilar (anatomically and physiologically) but are also not well studied.
As far as we can ascertain, this clustering approach has never been attempted before for a large number of organs for a whole organism, likely because no comprehensive assessment of gene expression across organs has been attempted. The relationships among organs shown in Figure 7A is obviously correlated to their individual functions. Visually, it echoes the dendrograms shown for the evolution of gene families, and for that reason, it may be possible to use these diagrams to draw parallels to organ evolution. When bilaterians first appeared in evolution, they would have had only one or two rudimentary organs, such as the precursor to the vertebrate heart and the dorsal vessel of arthropods (Xavier-Neto et al. 2007), so the organ systems of complex animals must have been the result of several divergent steps in evolution, much the same way as one gene might duplicate and then each copy evolves independently. Such a process is suggested in the organization of various animals that we see today, but there is little evidence of this in the fossil record since soft tissues are very poorly preserved. However, given that sequence homology between genes implies a common evolutionary origin (Koonin 2005), expression similarity at the protein level of different organs should reflect their ancestry. However, the dendrogram in Figure 7A suggests an evolutionary path each organ may have followed from an undefined common ancestral organ to get to its modern state. While all the organs may have evolved from a single ancestor, it seems more likely that independent organogenesis events probably occurred throughout history. If so, then depicting all the organs as belonging to a single supercluster would be artificial; the real situation may have been more like that presented in Figure 7B. Clearly this is an entirely speculative interpretation of the data, and to the best of our knowledge, such a molecular approach to define organ evolution is completely novel, even among the vast microarray literature. The lack of an appropriate fossil record of arthropod organs, or for any other phyla, makes verification of the relationships implied in Figure 7, A and B, impossible. Only as more pan-organism organ-level protein expression profiles become available will its validity be established, but at least the data set presented here allows us to systematically address how organs may have evolved, at least in insects.
Conclusion
Of the three levels of information in the Central Dogma, proteins are most closely linked to phenotype. Mass spectrometry–based proteomics thus enables a functional genomics view of gene expression in a tissue and how that may be linked to the function(s) of that tissue. Despite having nearly complete genomes for many multicellular organisms, no comprehensive, organ/tissue-level proteomic description of a multicellular organism has been attempted previously, although the 28 mouse tissues reported by Geiger et al. (2013) comes closest. The 29 organs that we could obtain in sufficient quantities to analyze have enabled us to construct a detailed protein expression atlas for the adult stage of all three castes. The data are generally consistent with known bee biology but have revealed several novel insights into specific metabolic and sensory capabilities of individual organs and of specific castes. In this article we have merely selected a few highlights to discuss, but the vast wealth of data pertaining to other aspects of bee proteins function are available online in a searchable format that includes illustrated caste and organ comparisons for all quantifiable proteins (http://www.chibi.ubc.ca/faculty/leonard-foster/foster-lab/data/ under Bee Protein Atlas). This resource should serve as a catalyst for studying the finer details of biological regulation, such as post-translational modifications and protein–protein interactions.
Perhaps the most profound and novel finding here is the first hint of how animal organs may have evolved and, indeed, even the realization that questions along this line can be answered. Comparative genomics helps us to understand the evolutionary relationship between organisms. The fossil record is also useful in this regard and can explain how certain structures like limbs evolved, but neither fossils nor genomics are useful for softer structures. By looking at protein expression profiles across many organs, however, a picture of which individual organs may share common ancestral structures begins to emerge. As with fossils and genomes, however, data from a single organism are not sufficient to build an accurate picture; our understanding of how organs may have evolved will become clearer when data from multiple species become available.
Methods
Reagents
All salts and chemicals were of analytical grade or better and were obtained from Sigma-Aldrich unless otherwise indicated. All solvents were of HPLC grade and were obtained from ThermoFisher Scientific. The following materials were obtained as indicated: endopeptidase Lys-C, Wako Chemicals; porcine-modified trypsin, Promega; loose ReproSil-Pur 120 C18-AQ 3 μm, Dr. Maisch (Ammerbuch-Entringen, Germany); 96-well full skirt PCR plates, Axygen; fused silica capillary tubing, Polymicro; soft forceps for holding bees, BioQuip; protease inhibitor mixture, Roche Applied Science.
Apiculture and bee dissection
A randomly selected, naturally mated queen was initially chosen as the initial source of all bee organ. This queen's colony and all her progeny colonies were maintained at an apiary in Langley, British Columbia from 2007 to 2010, during which time adult bees were collected as needed. Dissections were all performed as described (Dade 1994), and all organs were washed with phosphate-buffered saline (PBS) three times and frozen until proteins could be extracted. Three biological replicates of each organ were dissected, and material from multiple animals (up to 30 in some cases) was pooled to obtain enough protein material for each replicate.
Protein extraction and digestion
Exoskeleton-containing hard tissues were homogenized in a bead mill using a single tungsten bead in each 2-mL self-locking tube (Eppendorf). Tissues were pulsed at 30 H for 5 min in 50 μL of PBS containing a protease inhibitor cocktail (Roche) at 8× the suggested concentration. Soft tissues were placed on a glass plate and cut into smaller pieces before collecting them in a microfuge tube containing 50 μL of PBS. All samples were centrifuged for 10 min at 16,100 relative centrifugal force (RCF) at 4°C to pellet cells and debris, and the supernatant was removed. Lysis buffer (100 μL of 1% NP-40, 150 mM NaCl, 20 mM Tris at pH 7) was added to the pellet, and the contents were passed through a 25 G needle 10 times. The sample was clarified for 10 min at 16,100 RCF at 4°C, and the pelleted debris was discarded. Proteins were precipitated for 3 h through the addition of 1 mL of ethanol (100%) with 20 μL of 3 M sodium acetate (pH 5) and 20 μg of glycogen. Protein pellets were collected by centrifugation for 10 min at 16,100 RCF, briefly dried for 5 min in a vacuum centrifuge, and resolubilized in as little solubilization buffer (6 M urea, 2 M thiourea in 10 mM HEPES at pH 8) as was required to fully resolubilize the pellet (as low as 2 μL). The Coomassie Plus Protein Assay (Pierce) was used to determine protein concentrations of the tissue lysates according to the manufacturer's instructions before they were stored at −20°C until downstream analysis. Proteins were then digested as described (Chan et al. 2011a), desalted, isotopically labeled by dimethylation, and desalted (Chan and Foster 2008). Labeled proteins were mixed prior to the second desalting, with the worker sample acting as the reference in mixtures of worker–queen, worker–drone, and worker–queen–drone. Caste-specific organs (i.e., reproductive organs) were analyzed without isotopic labeling. Samples that contained a total of 7 μg of protein or more were analyzed in the mass spectrometer in duplicate, or step-gradient-fractionated by strong cation exchange STAGE tips (Ishihama et al. 2006). Liquid chromatography–tandem mass spectrometry (LC-MS/MS) was conducted as described (Chan et al. 2011a) using a 1100 Series nanoflow high-performance liquid chromatography system (Agilent) with a C18 column, directly coupled through a nanoelectrospray ion source (Proxeon) to a linear trapping quadrupole Orbitrap XL (LTQ Orbitrap XL, Thermo Scientific).
Data processing
Fragment spectra were searched against the Apis Official Gene Set (v2, 22,037 sequences) using MaxQuant (v1.2.0.13) (Cox and Mann 2008) with default parameters. Lys and N-terminal dimethylation with the three isotopologs of dimethyl groups were considered, but otherwise default parameters were used for both identification and quantitation. For a protein to be considered identified, two or more unique peptides were required. Quantitative data extracted by MaxQuant were further processed by Perseus, converting peak volume data for all isotope labels into natural logarithm-transformed ratios of Worker/Drone (W/D) and Worker/Queen (W/Q). Statistical significance for these relative quantitation values were obtained by calculating Significance B in the Perseus module of MaxQuant (Cox and Mann 2008) at P < 0.05, after correction for multiple hypothesis testing. The W/D and W/Q ratios, where available for each protein, are converted into percent values, with the total expression of W + D + Q assigned as 100%. These percent expression values are represented on an inverted grayscale (i.e., 100% expression is converted to 0 or black, and 0% expression is converted to 100 or white) for each organ of each caste (e.g., Fig. 1B). Missing organs in the diagram indicate that the organ does not exist or that the protein did not meet the limit of quantitation there. Whole-body expression values for each protein are calculated by averaging the percent expression values across each organs for each caste (each organ treated as one n), and two-tailed t-tests were used to evaluate whether a caste is significantly different from the other two. For the digestive system, similar calculations were conducted to generate the diagram and expression values, but only limited to carbohydrate and peptide degradation and lipid transport proteins (see the body of the text for their identities) and only to the crop, ventriculus, small intestine, and rectum.
For particular protein families of interest, the numbers of isoforms expressed in each organ that fall within the limit of quantitation were added (regardless of caste) and divided by the total number of quantified proteins in that organ (as seen in Figs. 4A,E, 5A). For Figure 4E, each measurement of each P450 subtype level across each organ where detectable across all three castes is considered one n, for a total of n = 85. For Figure 5A, all detectable OBPs or CSPs were considered in the average values as one n, equating to n = 16 for the flagellum and n = 11 for the scape.
Data access
The mass spectrometry proteomics raw data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaino et al. 2010) with the data set identifier PXD000084. The spectra are publicly available at PeptideAtlas (http://www.peptideatlas.org/builds/honeybee/). Interpreted data for all quantifiable proteins can be browsed and searched in an interactive graphical format (http://www.chibi.ubc.ca/faculty/leonard-foster/foster-lab/data/ under Bee Protein Atlas).
Acknowledgments
This work was supported by an NSERC Discovery Grant to L.J.F. Q.W.T.C. was supported by an NSERC post-graduate scholarship. Y.F. was supported by a Genome Sciences and Technologies studentship. L.J.F. is the Canada Research Chair in Quantitative Proteomics. We extend our gratitude to the staff at PRIDE for helping to make these data publicly available.
Author contributions: H.H. raised and managed the bee colonies and performed the majority of the organ dissections. M.Y.C., M.L., and Y.F. dissected bees, processed samples for MS analysis, and analyzed data. Q.W.T.C. dissected bees, processed samples for MS analysis, analyzed data, and wrote the paper. L.J.F. designed the study and wrote the paper.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.155994.113.
References
- Chan QW, Foster LJ 2008. Changes in protein expression during honey bee larval development. Genome Biol 9: R156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Melathopoulos AP, Pernal SF, Foster LJ 2009. The innate immune and systemic response in honey bees to a bacterial pathogen, Paenibacillus larvae. BMC Genomics 10: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Mutti NS, Foster LJ, Kocher SD, Amdam GV, Wolschin F 2011a. The worker honeybee fat body proteome is extensively remodeled preceding a major life-history transition. PLoS ONE 6: e24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Parker R, Sun Z, Deutsch EW, Foster LJ 2011b. A honey bee (Apis mellifera L.) PeptideAtlas crossing castes and tissues. BMC Genomics 12: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dade H. 1994. Anatomy and dissection of the honeybee. Alden Press, Oxford, UK. [Google Scholar]
- Dani FR, Iovinella I, Felicioli A, Niccolini A, Calvello MA, Carucci MG, Qiao H, Pieraccini G, Turillazzi S, Moneti G, et al. 2010. Mapping the expression of soluble olfactory proteins in the honeybee. J Proteome Res 9: 1822–1833 [DOI] [PubMed] [Google Scholar]
- Dow JA 2009. Insights into the Malpighian tubule from functional genomics. J Exp Biol 212: 435–445 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Bombarely A, Story GW, Allen F, Mueller LA, Coates SA, Jones L 2010. TobEA: An atlas of tobacco gene expression from seed to senescence. BMC Genomics 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM 2007. Creating a honey bee consensus gene set. Genome Biol 8: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Schwarz RS 2011. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol 19: 614–620 [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Foret S, Maleszka R 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res 16: 1404–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Velic A, Macek B, Lundberg E, Kampf C, Nagaraj N, Uhlen M, Cox J, Mann M 2013. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol Cell Proteomics 12: 1709–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva D, Greunke K, Betzel C 2010. Three-dimensional model of the honeybee venom allergen Api m 7: Structural and functional insights. Mol Biosyst 6: 1056–1060 [DOI] [PubMed] [Google Scholar]
- Habermann E 1972. Bee and wasp venoms. Science 177: 314–322 [DOI] [PubMed] [Google Scholar]
- The Honey Bee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe S, Dimick P, Benton A 1985. Composition of freshly harvested and commercial royal jelly. J Apic Res 24: 52–61 [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M 2006. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res 5: 988–994 [DOI] [PubMed] [Google Scholar]
- Koonin EV 2005. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39: 309–338 [DOI] [PubMed] [Google Scholar]
- Kovanich D, Cappadona S, Raijmakers R, Mohammed S, Scholten A, Heck AJ 2012. Applications of stable isotope dimethyl labeling in quantitative proteomics. Anal Bioanal Chem 404: 991–1009 [DOI] [PubMed] [Google Scholar]
- Krupp M, Marquardt JU, Sahin U, Galle PR, Castle J, Teufel A 2012. RNA-Seq Atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28: 1184–1185 [DOI] [PubMed] [Google Scholar]
- Kuchler K, Gmachl M, Sippl MJ, Kreil G 1989. Analysis of the cDNA for phospholipase A2 from honeybee venom glands. The deduced amino acid sequence reveals homology to the corresponding vertebrate enzymes. Eur J Biochem 184: 249–254 [DOI] [PubMed] [Google Scholar]
- Marz R, Mollay C, Kreil G, Zelger J 1981. Queen bee venom contains much less phospholipase than worker bee venom. Insect Biochem 11: 685–690 [Google Scholar]
- Oertelt-Prigione S 2012. The influence of sex and gender on the immune response. Autoimmun Rev 11: A479–A485 [DOI] [PubMed] [Google Scholar]
- Raghuraman H, Chattopadhyay A 2007. Melittin: A membrane-active peptide with diverse functions. Biosci Rep 27: 189–223 [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF 2008. Genes and social behavior. Science 322: 896–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama MG, Agellon LB 2012. Sex differences in lipid metabolism and metabolic disease risk. Biochem Cell Biol 90: 124–141 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. 2010. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250 [DOI] [PubMed] [Google Scholar]
- Vina J, Gambini J, Lopez-Grueso R, Abdelaziz KM, Jove M, Borras C 2011. Females live longer than males: Role of oxidative stress. Curr Pharm Des 17: 3959–3965 [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L 2010. The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston ML. 1987a. Biology of the honey bee, pp. 60–62. Harvard University Press, Cambridge, MA. [Google Scholar]
- Winston ML. 1987b. Table 3.2. Some structural differences between worker, queen, and drone castes in honey bees. In Biology of the honey bee, p. 40. Harvard University Press, Cambridge, MA. [Google Scholar]
- Xavier-Neto J, Castro RA, Sampaio AC, Azambuja AP, Castillo HA, Cravo RM, Simoes-Costa MS 2007. Parallel avenues in the evolution of hearts and pumping organs. Cell Mol Life Sci 64: 719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]