Abstract
Penicillium marneffei is an opportunistic human pathogen endemic to Southeast Asia. At 25° P. marneffei grows in a filamentous hyphal form and can undergo asexual development (conidiation) to produce spores (conidia), the infectious agent. At 37° P. marneffei grows in the pathogenic yeast cell form that replicates by fission. Switching between these growth forms, known as dimorphic switching, is dependent on temperature. To understand the process of dimorphic switching and the physiological capacity of the different cell types, two microarray-based profiling experiments covering approximately 42% of the genome were performed. The first experiment compared cells from the hyphal, yeast, and conidiation phases to identify “phase or cell-state–specific” gene expression. The second experiment examined gene expression during the dimorphic switch from one morphological state to another. The data identified a variety of differentially expressed genes that have been organized into metabolic clusters based on predicted function and expression patterns. In particular, C-14 sterol reductase–encoding gene ergM of the ergosterol biosynthesis pathway showed high-level expression throughout yeast morphogenesis compared to hyphal. Deletion of ergM resulted in severe growth defects with increased sensitivity to azole-type antifungal agents but not amphotericin B. The data defined gene classes based on spatio-temporal expression such as those expressed early in the dimorphic switch but not in the terminal cell types and those expressed late. Such classifications have been helpful in linking a given gene of interest to its expression pattern throughout the P. marneffei dimorphic life cycle and its likely role in pathogenicity.
Keywords: Penicillium marneffei, Talaromyces marneffei, microarray, hyphae, conidiation, yeast, dimorphic switch, cell wall
Human morbidity and mortality linked to fungal infections are increasing. This is a direct result of increasing numbers of immunocompromised patients associated with escalating HIV/AIDS incidence and increasing numbers of immunosuppressed patients because of medical intervention such as cancer treatment and organ transplantation (De Pauw 2001; Dixon et al. 1996; McNeil et al. 2001; Nielsen and Heitman 2007; Samonis and Bafaloukos 1992). Pathogenic fungi show a polyphyletic distribution and often have as their closest relatives nonpathogenic species, suggesting that the evolution of pathogenicity is likely to show species-specific characteristics (Bowman et al. 1992; Bowman et al. 1996; Nielsen and Heitman 2007). Despite this, a common theme in fungal pathogenesis is the ability to undergo developmental and morphological transitions during infection in response to the host environment. Dimorphism is one morphological alteration whereby certain fungi can undergo an environmentally induced morphological switch between a hyphal and a yeast growth form. In many dimorphic fungi, one of the strongest inducers is temperature. This switch allows for adaptation to different environmental conditions and has been correlated with the ability to infect, survive, and cause disease in a host (Gow et al. 2002; San-Blas et al. 2000). Blocking the ability of a fungus to undergo the switch has been shown to limit its virulence (Klein and Tebbets 2007; Lo et al. 1997). Examples of pathogenic dimorphic fungi include Blastomyces dermatitidis, Coccidioides immitis, Histoplasma capsulatum, Paracoccidioides brasiliensis, Sporothrix schenkii, and Penicillium marneffei. P. marneffei has recently been renamed Talaromyces marneffei as part of a kingdom-wide taxonomic reassessment (Samson et al. 2011).
P. marneffei is an opportunistic pathogen endemic to Southeast Asia and is strongly associated with AIDS (Cooper and Vanittanakom 2008). P. marneffei is the only known dimorphic species in the Eurotiales, which includes the large genera of Aspergillus and Penicillium (Andrianopoulos 2002). P. marneffei grows as multinucleate filamentous hyphae at 25° and can undergo asexual reproduction (conidiation) to produce the infectious asexual spores (conidia) (Vanittanakom et al. 2006). At 37° P. marneffei undergoes a morphological switch to grow as pathogenic uninucleate yeast cells that divide by fission. The dimorphic nature of P. marneffei provides an ideal opportunity to tease out key differences between nonpathogenic and pathogenic growth forms, all within the one organism (Andrianopoulos 2002). This process has been significantly aided with the recent completion of the P. marneffei genome sequence (Alex Andrianopoulos and William C. Nierman, unpublished data), highlighting gene groups common to dimorphic fungi as well as genes unique to P. marneffei.
In the past decade, DNA microarray analysis has proved to be a powerful and cost-efficient means of assaying differential gene expression on a large scale and has provided insight into the mechanisms that regulate processes such as growth, pathogenicity, and antifungal susceptibility (Black and Hare 2000; Garaizar et al. 2006; Kadosh and Johnson 2005; Saville et al. 2005). In fungi, microarray analysis has been successful in identifying cell-type–specific differential gene expression in dimorphic fungi such as H. capsulatum (Hwang et al. 2003) and Coccidioides posadasii (Johannesson et al. 2006). In the dimorphic pathogen P. brasiliensis, comparative microarray analysis of mycelial and yeast cells showed that genes upregulated in yeast cells encode proteins implicated in respiratory and metabolic processes as well as a range of transporters (Felipe et al. 2005). In contrast, genes involved in cell division and protein catabolism were notably associated with mycelial cells. Interspecies comparisons were also conducted to identify orthologous genes identified in P. brasiliensis from the genome sequence of H. capsulatum and C. posadasii in search of common dimorphic fungi-specific pathways. Common to the pathogenic forms of all three fungi was the expression of orthologous transporters, including calcium, copper, and drug resistance transporters (Monteiro et al. 2009).
To gain insight into the physiological capacity of the different growth forms in P. marneffei, phase or cell-type–specific gene expression patterns were examined using hyphal and yeast cells, as well as cells of the asexual development apparatus (conidiophore) that generate the infectious conidia. All other studies to date have only examined the hyphal and yeast vegetative growth forms in dimorphic fungi, including those in P. marneffei (Lin et al. 2012). Further, a second set of microarray experiments was also performed to examine gene expression during the early stages of the switch between the hyphal and yeast growth forms.
These data sets have identified several classes of genes that are likely to be involved in cell-type specification and maintenance, as well as those required for early and late events in cell-type specificity. The differentially expressed genes identified were organized into clusters based on expression patterns and predicted function. Detailed inspection of some of these genes has been helpful in linking temporal expression pattern to function throughout the dimorphic life cycle of P. marneffei and, in turn, in identifying the likelihood of a role in pathogenicity. For example, genes that were highly differentially expressed in yeast cells reflected the requirement for adaptation to a stressful environmental, with genes involved in iron acquisition and ergosterol biosynthesis upregulated throughout yeast cell morphogenesis and growth. Expression profiling of all the genes in the ergosterol biosynthetic pathway revealed a unique regulatory pattern for certain steps in the pathway that highlight differences between the cell types. Deletion of the highly differentially regulated ergM (encoding C-14 sterol reductase) resulted in severe growth defects, particularly at 37°, and an alteration in antifungal drug susceptibility. Genes upregulated late in yeast cell morphogenesis included those related to adhesion to host tissue, genes involved in the synthesis of core components of the fungal cell wall, and P. marneffei–specific genes of unknown function. One gene, ystA, was shown to encode a short 68-amino-acid protein that has rapidly diverged under selective pressure and to play a role in germination. A consistent difference between cell types in this study is the differential expression of genes involved in cell wall and membrane biogenesis, highlighting fundamental differences in these structures between the hyphal and yeast cell types.
Materials and Methods
Fungal strains and culture conditions
P. marneffei strains used in this study are described in Table 1 and were grown on Aspergillus-defined medium containing 1% glucose and 20 mM GABA (ANM) (Cove 1966). For microarray experiments, P. marneffei was grown at either 25° on ANM plates for asexual development (4 d) or shaken in liquid brain–heart infusion (BHI) (Oxoid) for hyphal growth at 25° (2 d) and for yeast growth at 37° (4 d of growth and then 10 ml was transferred to fresh medium for an additional 2 d). Auxotrophic supplements were added as required. For temperature switching microarray experiments, hyphal cells were transferred to 37° for 6 hr after 2 d of growth at 25°, or yeast cells were moved to 25° for 6 hr after 4 d of growth at 37°.
Table 1. P. marneffei strains.
| Name | Strain | Relevant Genotype | Origin |
|---|---|---|---|
| FRR2161 | Wild-type | ATCC 18224 or Borneman et al. 2000 | |
| G147 | SPM4 | niaD1 pyrG1 | Borneman et al. 2000 |
| G816 | ΔligD::pyrG− | ΔligD niaD1 pyrG1 | Bugeja et al. 2012 |
| G809 | ΔligD::pyrG+ | ΔligD::pyrG+ niaD1 pyrG1 | Bugeja et al. 2012 |
| G526 | ΔpkuA pyrG− | ΔpkuA areAΔDBD pyrG1 niaD1 | Bugeja et al. 2012 |
| G681 | ΔpkuA::pyrG+ | ΔpkuA::pyrG+ areAΔDBD niaD1 | Bugeja et al. 2012 |
| SBA001 | ΔystA pyrG− | ΔystA pyrG- areAΔDBD niaD1 | This study |
| SBA002 | ΔystA pyrG+ | ΔystA::pyrG+ areAΔDBD niaD1 | This study |
| SBA003 | ΔystA ystA+ | ΔystA::ystA+ pkuA::pyrG+ niaD1 | This study |
| SBA004 | ystAORF1-GFP | ΔystA pyrG+[pSB7326, barA+)] | This study |
| SBA005 | ystAORF2-GFP | ΔystA pyrG+[pSB7338, barA+] | This study |
| SBA006 | ΔystA, ystAM(ORF1)L | ΔystA pyrG+ [pSB7550, pyrG+] | This study |
| SBA007 | ΔystA, ystAM(ORF2)L | ΔystA pyrG+ [pSB7551, pyrG+] | This study |
| MPA001 | ΔergM pyrG+ | ΔergM::pyrG+ ΔligD niaD1 | This study |
| MPA002 | ΔergM ergM+ | ΔergM::ergM+ and ligD::pyrG+ niaD1 | This study |
Molecular techniques
For genomic DNA isolation, strains were grown in synthetic dextrose (SD) liquid medium supplemented with 10 mM (NH4)2SO4 for 4 d at 25° before mycelia was harvested by filtration through Miracloth, washed with 0.1 M MgSO4, dried, and frozen in liquid nitrogen. Genomic DNA was then isolated per standard protocols (Borneman et al. 2001). Genotypes were confirmed by Southern blot hybridization after transfer of DNA onto Amersham Hybond N+ membranes using 0.4 M NaOH according to manufacturer’s instructions. Radiolabeled probes (α-32P dATP) were added in hybridization solution (50% formamide, 4% SSPE, 1% SDS) at 37°, incubated overnight and washed with 2% SSC at 37° (low stringency) and 0.1% SSC at 65° (high stringency). Radiolabeled probes were made using the random hexamer primer method (Sambrook et al. 1989).
RNA for microarray analysis was prepared using the QIAGEN RNAeasy kit according to the manufacturer’s instructions, followed by a LiCl2 precipitation. RNA samples for northern blot analysis (10 µg) were separated on 1.2% agarose formaldehyde denaturing gels, transferred onto Amersham Hybond N+ membranes, and analyzed by northern blot hybridization (Sambrook et al. 1989).
Cloning and plasmid construction
The propagation of plasmid DNA was performed by transformation of Escherichia coli TOP10 cells (Life Technologies). Cells were grown overnight at 37° on Luria-Bertani broth plates supplemented with 50 μg/ml ampicillin. The ystA coding sequence with 5′ and 3′ flanking regions was amplified by PCR (primers AA50 and AA51). The ΔystA construct, in which the coding region was replaced with the pyrG selectable marker gene, was generated by fusion PCR (primers AA52, AA53, AA62, AA63, AA64, AA65; pSB7593). The ergM coding sequence with 5′ and 3′ flanking regions was amplified by PCR (primers KK1 and KK2). The PCR product was then ligated into the plasmid pALX223 at the EcoRV restriction site, resulting in plasmid pMP7459. The ΔergM construct was generated by inverse PCR using primers KK25 and KK26 on plasmid pMP7459. This PCR removed the coding region of the gene. The pyrG selectable marker was ligated into the PCR products to generate the deletion constructs (pMP7507). Linearized deletion constructs of ystA and ergM were transformed into the P. marneffei ΔpkuA pyrG- and ΔligD pyrG strains, respectively, as previously described (Borneman et al. 2001) and selected on uracil free medium.
The ystA locus was PCR-amplified from FRR2161 genomic DNA and used to construct two plasmids. A 2.6-kb PCR product incorporating the predicted ORF1 (primers GG73 and EE71) and a 2.8-kb PCR product incorporating ORF2 (primers GG73 and GG72) were separately ligated into pGEM-T Easy. After sequencing, the inserts were excised by a ClaI/HindIII digest and ligated into pALX190 to generate in-frame fusions with GFP. These fused products were then ligated into pMT1612 RV SK+, containing the selectable marker barA. These ystA-GFP fusion constructs, ystAORF1-GFP (pSB7326) and ystAORF2-GFP (pSB7338), were targeted to the 5′ upstream region of ΔystA pyrG+, with the presence of barA allowing for candidate transformant screening on glufosinate-supplemented medium. Constructs of ystA with a mutated ATG in either ORF1 [ystAM(ORF1)L] or ORF2 [ystAM(ORF2)L] were generated by inverse PCR (ORF1 invPCR primers GG67 and GG68; ORF2 invPCR primers GG69 and GG70) on ystA::pyrG targeting vector. Circular mutant constructs were transformed into P. marneffei and selected on uracil free medium.
Library construction
Two independent genomic DNA libraries were constructed to ensure maximal representation of the P. marneffei genome. Fungal genomic DNA was partially digested with either Sau3AI or TaqI. DNA fragments were separated on a 1.5% agarose gel and those ranging from 400 to 600 bp were purified. The size-selected genomic DNA was ligated into pALX223 cut with either BamHI (for the Sau3AI library) or ClaI (for the TaqI library) and chemically competent E. coli cells transformed with these libraries. Transformants were selected on ampicillin/X-gal/IPTG plates and screened for white colonies.
Microarray construction
Independent colonies from both libraries were selected for the construction of the microarrays at the Australian Genome Research Facility (http://www.agrf.org.au/). Colonies (5376) were picked using the BioRobotics BioPick machine and individually placed into 384-well plates containing Luria-Bertani broth. Colonies were then grown overnight and DNA was prepared from a 2-μl aliquot of each culture and used to PCR-amplify the gDNA inserts with M13 forward and reverse primers. The resulting PCR products were purified using Millipore Multiscreen filter plates via a manifold vacuum. The quality and quantity of the purified amplified product were ascertained through agarose gel electrophoresis. Amplified DNA inserts were adjusted to 250 ng/μl in 50% DMSO buffer for printing. The amplified DNA inserts were arrayed in duplicate onto Corning GAPS II aminosilane glass slides using the ESI Virtek Arraying Robot. DNA was spotted using Point Technology split pins. Slides were baked and UV-irradiated to crosslink the DNA.
cDNA synthesis, labeling, and hybridization
First-strand cDNA synthesis was achieved using the SuperScript Indirect cDNA Labeling System (Invitrogen) according to the manufacturer’s instructions. Total RNA (50 µg) was used in each labeling reaction. Cy3 and Cy5 dyes (Amersham) were coupled to the amino-allyl groups in 0.1 M sodium bicarbonate pH 9.0 buffer for 1 hr in the dark. Unincorporated dyes were removed using QIAquick PCR Purification Kit (Qiagen). Samples were heated at 100° for 2 min in hybridization buffer (25% formamide, 2.5× SSC, 0.01% SDS, 0.32 mg/ml tRNA, and 0.8 mg/ml herring sperm DNA). Samples labeled with Cy3 and Cy5 were then competitively hybridized on microarray slides prehybridized with prehybridization buffer (25% formamide, 5× SSC, 0.1% SDS, and 10 mg/ml BSA). Hybridizations were performed at 42° for 16–20 hr. Slides were washed four times in wash buffer containing decreasing concentrations of SSC and SDS. Dye switch was performed for every pairwise comparison.
Microarray data analysis
Microarray slides were scanned on either a GenePix 4000B or a 4100A scanner (Axon Instruments), and the resulting images were analyzed with GenePix Pro 4 or 6.1 and Acuity 4.0 to obtain foreground and background intensity estimates for each spot. Statistical analysis was performed using the limma software package (Smyth 2005a). Intensities were background-corrected by the “normexp” method with offset equal to 16 to stabilize the log-ratios of low-intensity probes (Ritchie et al. 2007) and then normalized using a global loess curve for each array (Smyth and Speed 2003). The first microarray experiment with eight two-color arrays provided four replicates of conidiating samples and six replicates of hyphal and yeast cells. The second microarray experiment provided four replicates of each temperature switch. Comparisons between the three cell states or responses to temperature switching were extracted from the competitive hybridizations by linear modeling, as previously described (Smyth 2005a). An intercept term was included in the probe-wise linear models to account for probe-specific dye effects (Holloway et al. 2006). The duplicate spots for each probe, printed on the top and bottom halves of each slide, were combined using the common correlation method and using an estimated duplicate correlation of 0.50 for the first experiment and 0.72 for the second (Smyth et al. 2005b). Differential expression between the three states was assessed using an empirical Bayes moderated analysis of variance (Smyth 2004). The false discovery rate (FDR) was controlled using the Benjamini and Hochberg method. The ternary diagram was produced using the compositions software package (http://www.r-project.org) after first converting the log-fold changes between the states into compositional values representing relative expression. Microarray data are available from the NCBI Gene Expression Omnibus database under accession numbers GSE51109 and GSE51110.
DNA sequencing and analysis
Sequences of the probes representing genes with differential expression were obtained by sequencing the corresponding plasmid using the M13 forward or reverse primer. A pipeline was created for automated vector sequence trimming and database searches (blastn and blastx) for the sequences of the top 500 differentially expressed clones in the phase-specific microarray and those uniquely differentially expressed in the switching microarray experiment. Blast outputs were then evaluated for hits with an E-value more than 0.001 and manually curated. Clones with ambiguous gene assignment or high E-values were examined further by obtaining the sequence of both flanking DNA regions from the P. marneffei genome. Clones showing low-quality sequence or more than one ORF within the insert region were discarded from further clustering analysis (Supporting Information, Table S2 and Table S3).
Microarray data clustering and gene ontology analysis
Clustering analysis was performed with Multiexperiment Viewer (MeV) 3.1 (Saeed et al. 2006) using k-means clustering and Pearson correlation (Table S4 and Table S5). Gene ontology (GO) terms were allocated to each gene represented by the sequenced microarray probes using Blast2GO (http://www.blast2go.com/). These GO assignments were then used to examine significant gene relationships within each cluster using GoTermFinder (http://go.princeton.edu/cgi-bin/GOTermFinder) and statistical significance determined by calculating p-values (p-value < 0.05) using the hypergeometric distribution described by Boyle et al. (2004) that takes into account bias in the sample population. The FDR was also calculated (Table S6 and Table S7).
Gene expression analysis
RNA was extracted using TRIzol Reagent (Invitrogen) and the MP FastPrep-24 bead beater according to the manufacturer’s instructions. RNA was DNase-treated (Promega) before RT-PCR and cDNA synthesis using the DyNaMo cDNA synthesis kit. An increasing number of cycles was used to ensure the amplification was in the exponential phase, and H3 (histone H3) or benA (β-tubulin) were used as a loading control. Microarray analysis results were validated by performing RT-PCR on a subset of differentially expressed genes from independently isolated RNA produced under the same conditions as those for the microarray experiments (PMAA_076130, PMAA_071450, PMAA_065680, PMAA_053710, PMAA_082030, PMAA_082040, PMAA_010220, PMAA_097290, PMAA_0572450, PMAA_040300, PMAA_018640, and PMAA_091310) (Figure S1) using primers listed in Table S1.
Western blot analysis
Strains were grown at 37° in liquid BHI medium for 6 d. Total protein extraction was performed as described previously (Todd et al. 2005) and concentrations were determined using a protein assay reagent (Bio-Rad). Proteins were fractionated using a SDS-PAGE gel and transferred to Hybond ECL. Blots were probed with polyclonal rabbit anti-GFP (1/5000) followed by anti-rabbit IgG HRP (1/4000) (Promega).
Microscopy
For yeast cells, P. marneffei strains were grown on a thin layer of solid heart infusion medium (HI) covering a glass slide resting at an angle in a sterile Petri dish, with one end in liquid HI, and incubated at 37° for 5 d (Borneman et al. 2000). Cells were fixed in 4% paraformaldehyde in PME [50 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 1 mM MgSO4, 20 mM EGTA pH 6.7] for 30 min at room temperature followed by two washes with PME. Cells were then stained with either fluorescent brightener 28 (calcofluor; final concentration 0.014 mg/ml) or Hoescht 33258 nuclear stain (final concentration 0.14 μg/ml) in Tween 80. The same procedure was followed for hyphal cells, except ANM medium was used and growth occurred for 3 d at 25°. Slides were examined using differential interference contrast and fluorescent optics on a Reichart Jung Polyvar II microscope. Images were captured using a SPOT CCD camera (Diagnostic Instruments) and processed with Adobe Photoshop.
Germination assay
Conidia (1×106) were added to 500 μl liquid SD [with 10 mM (NH4)2SO4] and allowed to germinate at 25° or 37° for 0, 12, 16, or 24 hr. One hundred cells were counted from each sample and the cell type was recorded as either ungerminated or germinated. Three biological repeats were performed for quantification and error bars represent the SEMs. Data were analyzed using Prism 4.0 (GraphPad Software) and statistical analysis was performed using the R statistics package (http://www.r-project.org/).
Antifungal drug assay
Minimum inhibitory concentration was determined for the antifungal drugs voriconazole, fluconazole, and amphotericin B for the ΔergM strain and controls using e-test strips (bioMérieux). Agarose medium (4 ml of 0.7%) containing 105 conidia was poured onto a Petri dish containing a 2% agarose solution of the same medium. The e-test strips were then placed on top of the medium and the plates were incubated for 5 d (for control strains) or 10 d (for ΔergM strain) at 25° and 37°. At 25° ANM medium was supplemented with 10 mM (NH4)2SO4, whereas at 37° HI medium was used. Minimum inhibitory concentration was determined from the zone of inhibition according to the manufacturer’s instructions.
Results and Discussion
Identification of trends in cell-type specific differential gene expression
At 25° P. marneffei grows as multinucleate filamentous hyphal cells. It undergoes asexual reproduction (conidiation) in response to specific environmental cues via a series of cellular differentiation steps culminating in the production of asexual conidia (Boyce and Andrianopoulos 2013). On exposure to 37°, P. marneffei cells undergo a morphological switch to uninucleate yeast cells. The hyphal, yeast, and conidial cells are distinct and stable morphological cellular states that represent saprophytic, pathogenic, and infectious cells, respectively, but they also have the capacity to interconvert in response to environmental stimuli. To examine the transcriptional profile of each of these cell states, RNA from three biological replicates was isolated and used in pairwise combinations to probe a custom-generated microarray. This microarray was produced by spotting inserts from two size-selected, random fragment genomic libraries containing short (400-600 bp) inserts from the type strain of P. marneffei (FRR2161) (see Materials and Methods section). The rationale for this approach was that given there was no P. marneffei genome sequence available at the time, the genome size is small (estimated to be approximately 26–30 Mb) (Yuen et al. 2003) and gene-rich [fungal genomes from related species showed that intergenic regions are relatively short, genes do not have many introns, and those introns that exist are generally short (50–200 bp)] (Galagan et al. 2005; Kupfer et al. 1997), then most short random fragments would cover transcribed regions and rarely overlap two independent transcriptional units. This was preferred over a cDNA-based approach to probe design because it was subject to less bias. Such an approach has been used previously for other microorganisms (DeRisi et al. 1997; Hwang et al. 2003; Monteiro et al. 2009).
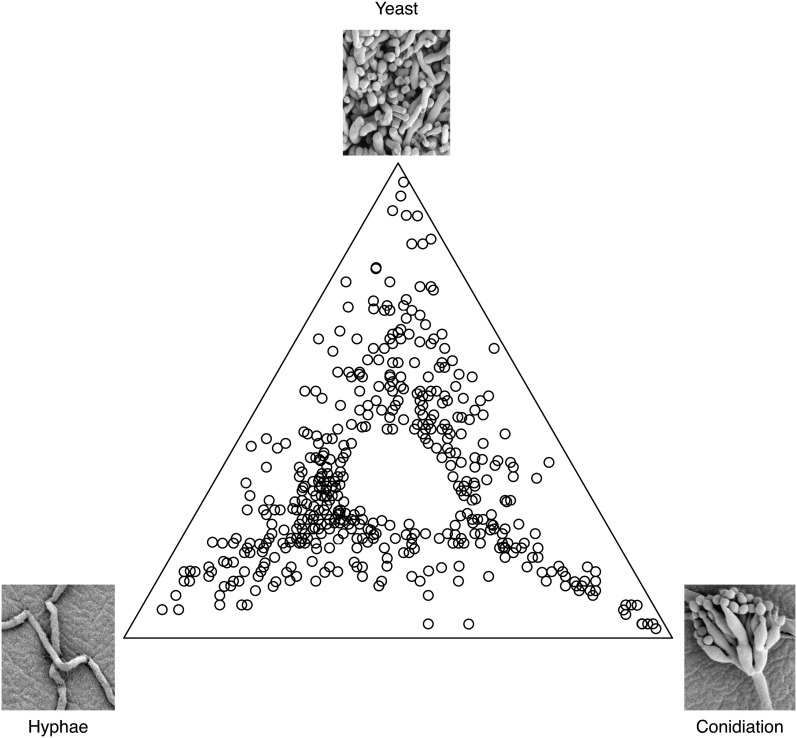
One of the goals of this study was to identify a set of genes that represent cell-type–specific markers to be used for genetic screens and studies of cell function. A second goal was to identify potentially interesting genes for further characterization. Random probes from the libraries (5376) along with 38 probes for 34 previously characterized genes, spotted in duplicate, were arrayed and used in these experiments. More than 38% of the microarray probes were found to show statistically significant differences in expression levels between the three cell states (FDR < 0.05). The 500 most significant clones were selected for further study. Of the top 500 probes, 31.2% were found to be hyphal-specific, 18.2% were conidiation-specific, and 20.2% were yeast-specific (Figure 1). Here, we define hyphal-specific probes to be those with at least 50% higher expression in hyphal cells compared to either of the other two states (similarly for conidiation-specific and yeast-specific probes). The data successfully show dynamic changes in gene expression profiles among the three states and clearly identify probes expressed predominantly in one cell state. Such probes correspond to genes with possible roles in cell-type maintenance, whether they are morphological or physiological.
Figure 1.
Cell-state–specific differential gene expression in P. marneffei. Microarray analysis was conducted with wild-type P. marneffei under three growth conditions, 25° on ANM plates for asexual development (4 d), agitated in liquid BHI for hyphal growth at 25° (2 d), and yeast growth at 37° (4 d of growth followed by 10 ml transferred to fresh medium for an additional 2 d). The data set represents the top 500 differentially expressed genes. Points closest to the top corner represent genes with maximum log expression in the yeast phase. Points in the bottom right corner are genes most highly expressed during conidiation, and points at the bottom left corner of the triangle represent genes differentially expressed highest in the hyphal phase. Points between two corners, along the sides of the triangle, depict clones upregulated in two conditions with respect to the one in the opposite corner. Probes expressed evenly among the three states were omitted from the data set used to generate this representation, accounting for the absence of spots in the center of the triangle.
Clones representing the top 500 differential expressed probes were sequenced and used in BLAST searches (blastn and tblastn) against the available fungal sequence databases in Genbank (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Subsequently, blastn searches against the P. marneffei FRR2161 genome sequence were performed when it became available. Microarray clones were omitted from the data set if BLAST searches resulted in no significant P. marneffei hits using an E-value cut-off <1×10−6. These clones represent either missing sequences from the P. marneffei genome project or contaminating clones in the library. In cases in which multiple clones identified the same gene, duplicates were removed. This resulted in a set of 400 unique differentially expressed P. marneffei genes under the three tested conditions. Based on this frequency of “false” or duplicate clones, the entire microarray is predicted to represent 4301 unique P. marneffei genes, which is 42% of the predicted genes in the genome. Microarray analysis results were validated by RT-PCR on a subset of differentially expressed genes (Figure S1). The trends in expression between the microarray data and the RT-PCR were highly correlative for the genes identified as yeast-enriched and conidiation-enriched, but less so for the genes identified as hyphal-enriched (Figure S1).
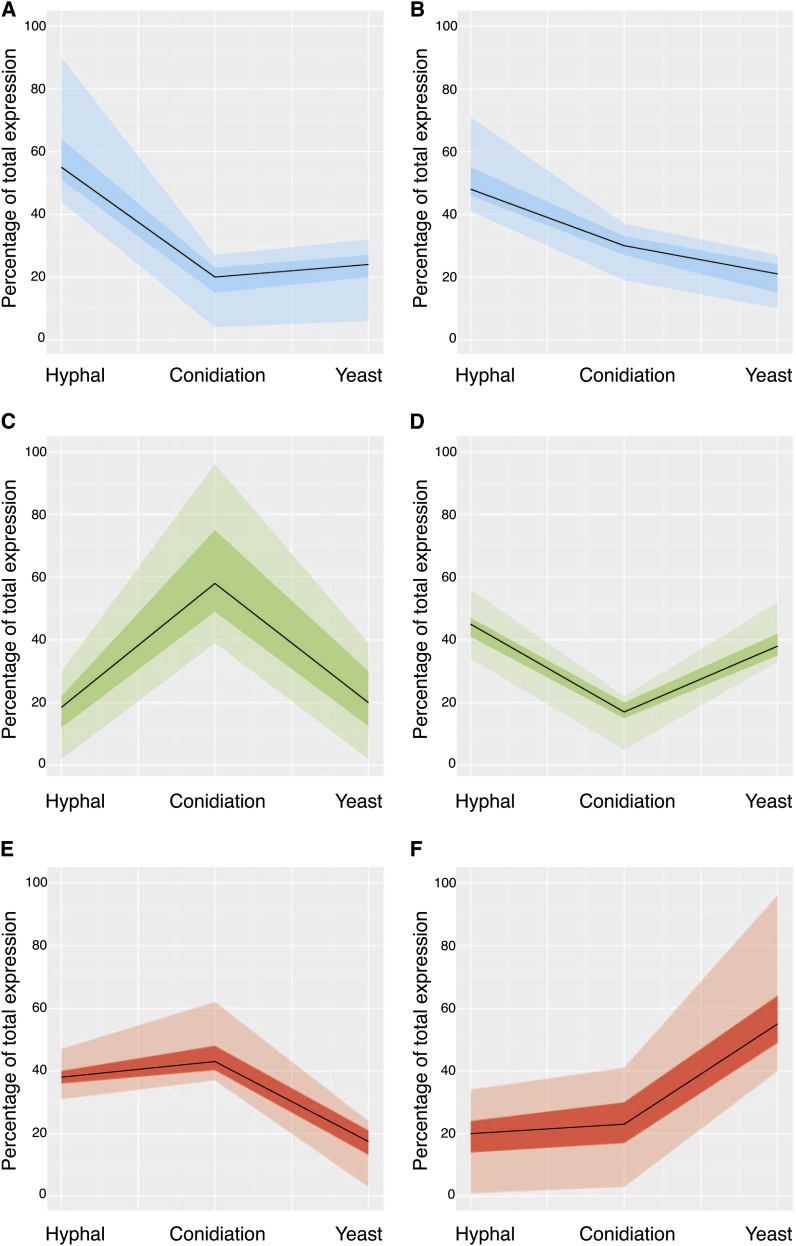
Trends in gene expression were examined by k-means clustering using MeV (Saeed et al. 2006). The data were clustered into six general expression profiles using Pearson correlation (Figure 2). Gene clusters reflect genes that are exclusively expressed or upregulated in one state, as well as those coregulated in two states (represented as percentages of total expression). Because both hyphal growth and conidiation occur at 25°, and because conidiation begins only after a period of hyphal growth, the presence of genes commonly upregulated in both states is expected. Genes that showed increased expression in hyphal and yeast cells relative to conidiation are likely to represent general cellular processes required for vegetative growth as opposed to those specific for the differentiation processes involved in conidiation. Interestingly, there is an underrepresentation of genes downregulated in hyphal cells and upregulated during both conidiation and in yeast cells. This may be indicative of the difference in specificity involved in reproduction by fission in yeast cells and differentiation during conidiation. Binary fission involves the segregation of replicated genetic material to the poles of a cell that then splits into two identical cells. However, asexual reproduction during conidiation is performed in a budding fashion with the specialized differentiation of uninucleate cells (Borneman et al. 2000; Clutterbuck 1969). Genes that have opposing expression in these cell states may provide information regarding what gene groups govern these differences in cell division. As a way of addressing what categories of genes are within each cluster and whether any of these are enriched for a particular state, GO terms were assigned to each gene represented by the microarray probes, and these were curated further by a qualitative literature-based approach. These GO assignments were then used to examine significant gene relationships within each cluster (p-value < 0.05) (http://go.princeton.edu/) (Figure S2), and they highlighted, in particular, the enrichment of gene groups during conidiation (Table 2).
Figure 2.
Gene expression clustering pattern in P. marneffei cell states. Clustering analysis was performed using the top 400 unique genes differentially expressed in hyphal, yeast, or conidiation conditions with MeV Six clusters were created using k-means clustering with Pearson correlation. The median of gene expression under each condition is represented as a continuous black line. The dark shaded region in each graph represents the differential expression values that lie within the first and third quartiles of the cluster. The lighter shading extends the spread of data from zero to the fourth quartile. (A) Genes upregulated in hyphal cells. (B) Genes upregulated in hyphal cells, followed by conidiation, and genes downregulated in yeast cells. (C) Genes upregulated during conidiation. (D) Genes upregulated in both hyphal and yeast cells with respect to conidiation. (E) Genes upregulated in hyphae and during conidiation, with respect to yeast cells. (F) Genes upregulated in yeast cells.
Table 2. Gene groups upregulated during conidiation.
| GO ID (cluster uncorrected p-value) and description | Gene ID | Percentage of Total Expression |
BLASTX Description and Role | Reference | ||
|---|---|---|---|---|---|---|
| Hyphal | Conidiation | Yeast | ||||
| GO:0070317 (0.003): negative regulation of G0-to-G1 transition | PMAA_096670 (smD3 ortholog) | 16 | 56 | 29 | Small nuclear ribonucleoprotein: part of the heteroheptameric complex that is essential for spliceosomal U1, U2, U4, and U5 snRNPs. Involved in nuclear mRNA splicing | Roy et al. (1995); Bordonne (2000) |
| PMAA_042130 (nimE) | 28 | 52 | 20 | G2/M-specific cyclin: regulates activity of cyclin-dependent protein kinase processes | O’Connell et al. (1992) | |
| PMAA_017210 (erg13 ortholog) | 20 | 43 | 37 | Hydroxymethylglutaryl-CoA synthase: catalyzes the second step of mevalonate biosynthesis | Parks and Casey (1995) | |
| GO:0008643 (0.008): carbohydrate transport | PMAA_049040 | 17 | 66 | 17 | MFS monosaccharide transporter | |
| PMAA_047000 | 12 | 78 | 10 | Sugar transporter | ||
| PMAA_049550 | 21 | 60 | 19 | Sugar transporter | ||
| PMAA_059270 | 29 | 50 | 22 | MFS alpha-glucoside transporter | ||
| GO:0019954 (0.035): asexual reproduction | PMAA_081680 (wetA) | 24 | 54 | 22 | Developmental regulatory protein: activates conidiation-specific gene expression | Boylan et al. (1987); Marshall and Timberlake (1991); |
| PMAA_082040 (arpA) | 3 | 94 | 3 | Scytalone dehydratase: involved in conidial pigment biosynthesis | Tsai et al. (1999) | |
| PMAA_075300 (brlA) | 3 | 91 | 7 | C2H2 type conidiation transcription factor: regulation of conidiophore development | Adams et al. (1988) | |
| PMAA_006690 (dfgE) | 21 | 56 | 23 | Cell wall glycosyl hydrolase: required for cell wall synthesis in bud formation and filamentous growth | Kitagaki et al. (2002) | |
Transcription profiles of nonpathogenic hyphal and pathogenic yeast cells during early and late cell morphogenesis
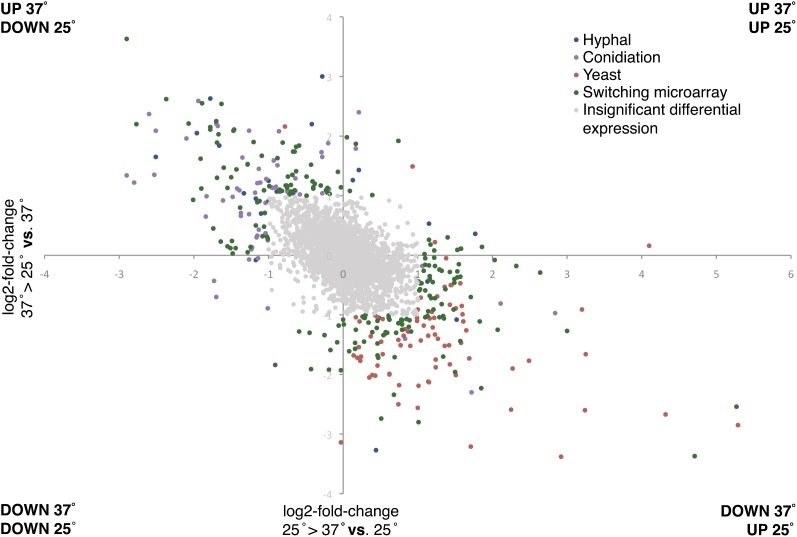
Microarray analyses of hyphal, yeast, and conidiation cell states reflect gene expression differences late in morphogenesis and development and are not necessarily representative of the transcriptional events during the switch from one temperature to another and the initial events of cell-type establishment. To examine differential gene expression early in the transition between hyphal and yeast cells, microarray analysis was performed using RNA from cells grown at 25° or 37° and then either maintained at this temperature or switched to the alternate temperature for an additional 6 hr (see Materials and Methods section). The resulting data were converted to log2 ratios of expression after temperature switching vs. maintenance at a constant temperature (Figure 3). The initial data set showed substantial changes in gene expression across many probes (2.4% upregulated and 2.1% downregulated by two-fold or more during 25° to 37° switch, 2.5% upregulated and 2.7% downregulated by two-fold or more during 37° to 25° switch). From this larger data set, clones that showed less than a two-fold increase or decrease in expression were omitted from further analysis, as were known duplicates, and the remaining values were then compared to expression later during that state. After which, clones representing differentially expressed probes unique to the switching microarray (73 genes) were sequenced and identified in the same manner described in the three-state microarray. From this new subset of 473 genes, 31.6% of the probes were differentially regulated in both the three-state microarray and the switching microarray experiments. Results indicate that 43% of the genes are upregulated and 57% are downregulated by two-fold or more during the 25° to 37° switch, and 42% are upregulated and 58% are downregulated by two-fold or more during the 37° to 25° switch.
Figure 3.
Differential gene expressions during a temperature-induced morphological switch. Temperature switching microarray experiments involved the transfer of hyphal cells to 37° for 6 hr after 2 d of growth at 25° and the transfer of yeast cells to 25° for 6 hr after 4 d of growth at 37°. Log2 fold change during 25° to 37° vs. 25° represents the expression of genes at 25° compared to those transferred to 37° for 6 hr and Log2 fold change during 37° to 25° vs. 37° represents the expression of genes at 37° compared to those transferred to 25° for 6 hr. Color-coding is based on comparison to three-phase–specific microarray (Figure 1). Blue probes identified the highest expression during late hyphal cell growth compared to other cell states and red probes identified the highest expression during late yeast cell growth. Green clones represent probes that showed significantly different expression between hyphal and yeast cells during the first 6 hr and not during later development. Probes shaded gray were omitted from subsequent analysis and showed no significant difference between the cell states in expression during the first 6 hr of growth.
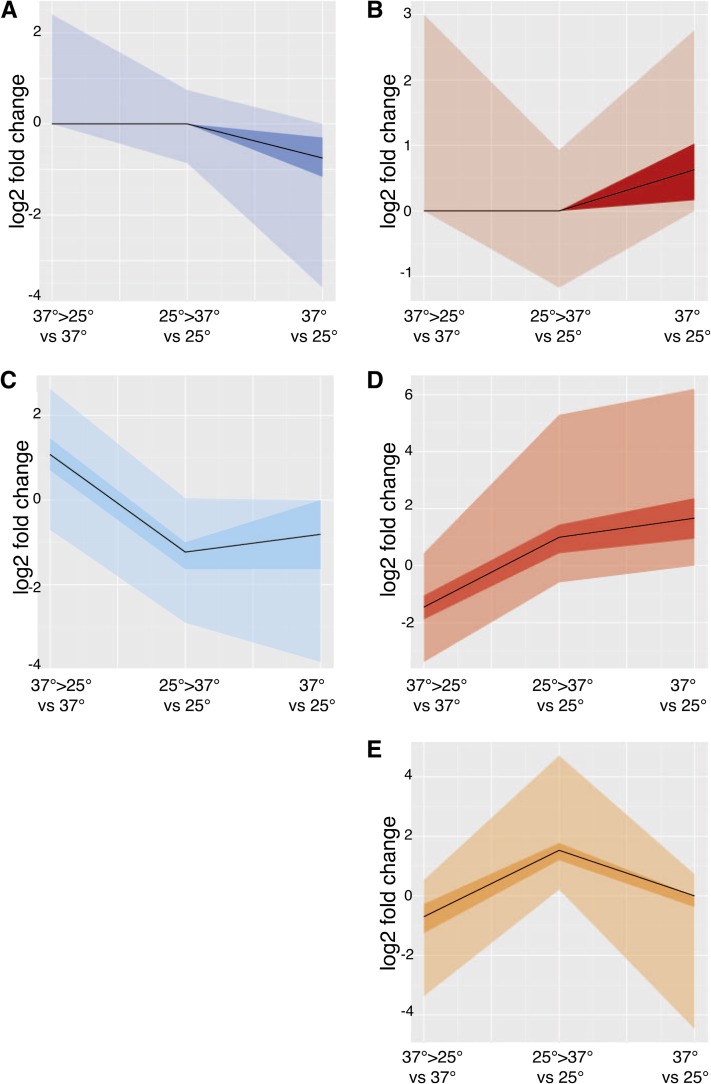
To examine the overlap between the expression patterns for the cell-state–specific genes and those identified during the switch from 25° to 37° and vice versa, k-means Pearson correlation testing was performed in MeV using log2-fold change values for all data sets. This analysis allowed for the identification of early and late expressed genes in hyphal and yeast cells. The data were clustered into six expression profiles, with two similar clusters showing genes with increased expression in early and late hyphal cells being manually combined (Figure 4), and GO terms were allocated in a manner previously described (see Materials and Methods section). Genes upregulated solely during the switch from 37° to 25° clustered with those that were differentially upregulated late in hyphal growth compared to late yeast cell expression, together accounting for 20% of all the genes (Figure 4C). The clustering showed that 36% of the genes were highly expressed only late in hyphal growth (Figure 4A) and 4% were upregulated only during the switch from 25° to 37° (Figure 4E). An additional cluster of 16% of the genes was upregulated during the switch to 37° and remained upregulated late in yeast cell growth (Figure 4D). Finally, the 24% of genes upregulated only late in yeast cell growth grouped together (Figure 4B). A consequence of clustering the data set into six expression groups, as opposed to binning probes in all possible patterns of gene expression, is that some genes designated as outliers, in the zero to fourth quartiles, may in fact represent genes with underrepresented rare patterns of gene expression (Figure 4).
Figure 4.
Early and late hyphal and yeast gene expression clustering. Clustering analysis was performed on genes expressed during early and/or late cell-state specific growth with MeV using k-means clustering. Six clusters were created using Pearson correlation, two of which were manually merged because they displayed similar trends. The median of gene expression under each condition is represented as a continuous black line. The dark shaded region in each graph represents the differential expression values that lie within the first and third quartiles of the cluster. The lighter shading extends the spread of data from zero to fourth quartile. X-axis condition 37° to 25° vs. 37° shows cells that were transferred to 25° from 37° for 6 hr and compared to 37°. Condition 25° to 37° vs. 37° shows cells that were transferred to 37° for 6 hr and compared to 25° gene expression. Finally, condition 37° vs. 25° shows cells grown at 37° for 4 d (yeast cells) compared to cells grown at 25° for 2 d (hyphal cells). (A) Genes upregulated in late hyphal cells. (B) Genes upregulated in late yeast cells. (C) Genes upregulated in the switch to 25° and that stayed upregulated in late hyphal cells or remained evenly expressed in late hyphal and yeast cells. (D) Genes upregulated during the switch to 37° and that remained upregulated in late yeast cells. (E) Genes that are upregulated during the switch to 37° and upregulated in late hyphal cells.
Genes upregulated during early and late hyphal cell growth are involved in apical growth and cell wall integrity
Despite being clustered with genes upregulated primarily in established hyphal cells, the homolog of the fungal-specific Ras small monomeric GTPase, rasB, is upregulated during the switch to hyphal cells and throughout hyphal development. Ras GTPases have characterized roles in cell polarization and morphology in numerous pathogenic and nonpathogenic fungi as well as other eukaryotes (Boyce et al. 2005; Lengeler et al. 2000). Deletion of the rasB ortholog in Aspergillus fumigatus, a prominent monomorphic opportunistic fungus that grows filamentously, results in decreased germination and growth rates, increased hyphal branching, and a loss of virulence (Fortwendel et al. 2005). In Neurospora crassa, the rasB homolog, NC-ras2, regulates cell wall synthesis, apical growth of hyphal cells, and conidial formation (Kana-uchi et al. 1997).
Another gene highly expressed during early and late hyphal cell growth encodes an ortholog of the GPI-anchored protein Ecm33, which is involved in apical growth and cell wall integrity in A. fumigatus and S. cerevisiae, among others (Latgé 2007; Pardo et al. 2004). S. cerevisiae Ecm33 is important for cell wall organization and deletion results in a weakened cell wall and associated glycosylation defects. Ecm33 mutants have defects in the construction of the cell wall mannoprotein outer layer and secrete increased levels of 1,6-β-glucan linked proteins into the growth medium (Pardo et al. 2004). Another gene in this group is an ortholog of the S. cerevisiae BUD4 gene. The GTP binding protein Bud4 is important for apical bud growth and is essential for axial budding pattern and septin organization. A Bud4 ortholog also has morphogenetic roles in Aspergillus nidulans, where it is involved in septum formation in hyphae and conidiophores (Si et al. 2012).
Genes upregulated during conidiation play integral roles in asexual development
Conidiation in many filamentous fungi begins with the establishment of a specialized foot cell from which a differentiated aerial stalk extends. Conidia are then produced either directly or after the differentiation of one or more layers of sterigmata (metula and phialide) cells (Cole 1986). In P. marneffei, a metula buds off the aerial stalk, phialides bud from the metula, and then a chain of uninucleate conidia sequentially bud off each phialide (Borneman et al. 2000; Boyce and Andrianopoulos 2013). Genes identified as upregulated in the conidiation samples are consistent with what is known about the molecular control of conidiation in P. marneffei and reinforce the similarities with monomorphic filamentous fungi, such as A. nidulans. GO term clustering of these genes highlighted gene groups previously characterized with roles in asexual reproduction (Table 2). For example, orthologuous genes (smdC, nimE, and erg13) involved in the negative regulation of the G0-to-G1 transition, which coincides with conidial dormancy and cell-cycle arrest in the G1 phase (Roy et al. 1995; Bordonne 2000; O’Connell et al. 1992; Parks and Casey 1995), were upregulated. Important to the long-term survival and dormancy of asexual propagules in filamentous fungi is their pigmentation via dihydroxynaphthalene (DHN) melanin biosynthesis. The conidial pigment biosynthesis gene arpA, encoding scytalone dehydratase, showed the second highest conidiation-specific gene expression relative to hyphal and yeast cells and clustered with other “asexual reproduction” genes in the GO term clustering (p-value = 0.035) (Table 2). In A. fumigatus, arp1 is a member of a developmentally regulated pigment biosynthesis gene cluster and shows sequence similarity to scytalone dehydratase of the DHN-melanin cluster identified in other fungi (Tsai et al. 1998), and arp1 mutants produce conidia with a pigmentation that is red-pink rather than blue-green (Tsai et al. 1997). Similar to A. fumigatus, the P. marneffei ortholog of arpA is located in a putative DHN-melanin pigment biosynthesis cluster. The biosynthesis of melanin has been associated with increased conidial survival and plays a role in pathogenicity, aiding the fungus in survival and in escaping host defenses (Kwon-Chung et al. 1982; Wang et al. 1995; Wheeler and Bell 1988).
In A. nidulans, A. fumigatus, and Aspergillus oryzae, the wetA and brlA genes are required for conidiation (Mah and Yu 2006; Ogawa et al. 2010; Yu 2010). Together with abaA, deletion of these genes blocks conidiation at different stages of development but leaves hyphal growth unaffected. Induction of brlA alone is sufficient for rudimentary conidiation to occur and brlA mutants are able to form stalks, but these fail to undergo any further differentiation (Adams et al. 1988). The abaA mutants produce aberrant phialides that fail to produce conidia, whereas wetA mutant strains produce conidia, but these fail to mature and eventually autolyze (Adams et al. 1988; Clutterbuck 1969; Ogawa et al. 2010; Timberlake et al. 1987). Similarly, the P. marneffei brlA, abaA, and wetA orthologs control various stages of differentiation during asexual development, supporting the conserved roles of these key central regulatory pathway genes (Borneman et al. 2000; Anthony R. Borneman and Alex Andrianopoulos, unpublished data).
Genes upregulated during late stages of hyphal growth are involved in reproduction, hyphal morphogenesis, and cell polarity
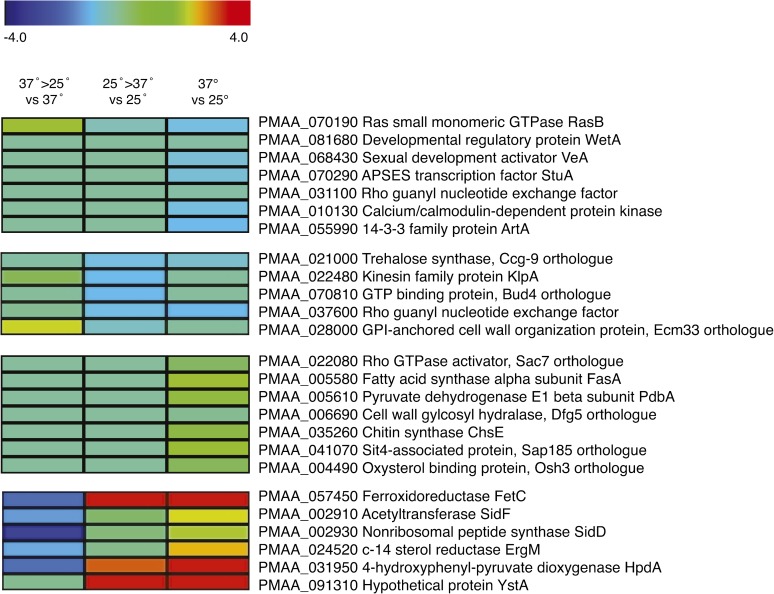
At 25°, hyphal growth of P. marneffei begins with the germination of dormant conidia. During germination isotropic growth is activated, followed by the establishment of a polarized axis and germ tube extension. Apical growth of the germ tube culminates in the formation of radiating septate hyphae that branch subapically (Andrianopoulos 2002). Comparing expression profiles of genes differentially expressed in the phase-specific microarray and the switching microarray revealed a cluster of genes that were upregulated in late hyphal growth relative to all other growth conditions (Figure 4). GO analysis grouped a subset of these genes as being involved in developmental processes of reproduction and the generation of precursor metabolites and energy (Figure S2). Further analysis highlighted additional genes fundamental to hyphal morphogenesis and cell polarity (Figure 5).
Figure 5.
Cell-state–specific expressed genes of interest. Differential expression heat maps of representative genes selected from each cluster, most of which have characterized roles in other fungi. Expression of genes from P. marneffei grown at 37° then transferred to 25° for 6 hr compared to 37° (37°>25° vs 37°) or grown at 25° then transferred to 37° for 6 hr compared to 25° (25°>37° vs 25°). In addition, gene expression from cells grown at 37° for 6 d (yeast cells) compared to cells grown at 25° for 2 d (hyphal cells) is shown.
As alluded to earlier, it is not unexpected that genes upregulated in hyphal cells relative to yeast cells have roles in asexual (conidiation) and sexual reproduction. The gene encoding the APSES transcriptional regulator of mating and asexual reproduction StuA was upregulated late during hyphal cell growth. In A. nidulans, stuA acts as a modifier of the central regulatory pathway affecting conidiophore patterning and differentiation and is required for sexual reproduction (Clutterbuck 1969). In P. marneffei, stuA has been shown to be required for the correct spatial organization of conidiophores, with deletion of stuA resulting in spore production despite a lack of distinct metula and phialide cell differentiation (Borneman et al. 2002).
Appropriate polarity establishment is fundamental to normal cell differentiation and, in turn, hyphal morphology. Present in this cluster is the 14-3-3 family protein encoding gene artA (Figure 5). In A. nidulans, ArtA plays a role in hyphal morphogenesis, with overexpression causing delays in conidial germination and germ tube establishment (Kraus et al. 2002). Also present in this cluster are genes encoding a kinesin family protein and dynactin. Polarized growth requires the delivery of secretory vesicles via microtubules to the hyphal apex and back to the cell center, and vesicle delivery acts as a rate-limiting factor for hyphal tip extension. The microtubule motors kinesin and dynein cooperatively deliver secretory vesicles via microtubules and are likely to be important for maintaining continuous growth (Steinberg 2011). Additionally, the hyphae-specific upregulation of a calcium/calmodulin-dependent protein kinase–encoding gene is consistent with the requirements of cell polarity and apical growth. Large quantities of calcium are essential at the apex of growing hyphal cells, and the calmodulin-calcineurin-Crz1 signaling pathway has been shown to upregulate an array of target genes as a response to increased cytosolic calcium levels (Yoshimoto et al. 2002). In A. fumigatus, the calcium/calmodulin-dependent kinases and calcineurin catalytic subunit cnaA mutant showed dramatic hyphal defects, including decreased apical extension and polarized growth (Steinbach et al. 2006).
Genes upregulated early during yeast cell morphogenesis and throughout yeast growth include those involved in nutrient assimilation and membrane fluidity
The cluster of greatest interest with respect to pathogenicity defines genes that, compared to other cell stages, are highly expressed during the switch to 37° and persist into the yeast cell stage. This cluster, including a series of iron acquisition and assimilation genes, is expressed in the initiation and establishment of yeast cells and, in turn, is likely to have roles in the pathogenic potential of P. marneffei (Figure 5).
Iron is essential for the survival of all eukaryotes and most prokaryotes. On microbial infection, the host withholds free iron through sequestration, reducing available free iron within the blood and extracellular fluids of the host (Philpott 2006; Weinberg 2009). Thus, the challenges for invading pathogens become finding ways to obtain and assimilate iron from its host (Dancis et al. 1990). In iron-limiting conditions, such as within macrophages, fungi use high-affinity uptake systems, including reductive iron assimilation and siderophore-mediated iron uptake system (Johnson 2008; Kosman 2003). Multicopper ferrioxidase, FetC acting in conjunction with permease FtrA in the reductive iron assimilation pathway to take-up ferrous iron, shows higher gene expression throughout yeast cell initiation and growth compared to hyphal cells (Figure 5). This was also the case with siderophore biosynthetic genes sidF and sidD. In conditions of low iron, many fungi secrete siderophores, low-molecular-mass ferric iron–specific chelators, to mobilize and transport iron (Schrettl et al. 2004; Timmerman and Woods 1999), resulting in siderophore function being considered an important virulence factor in other pathogenic fungi (Haas 2003) and a potential iron-specific target for antifungal drugs (Miethke and Marahiel 2007; Roosenberg et al. 2000).
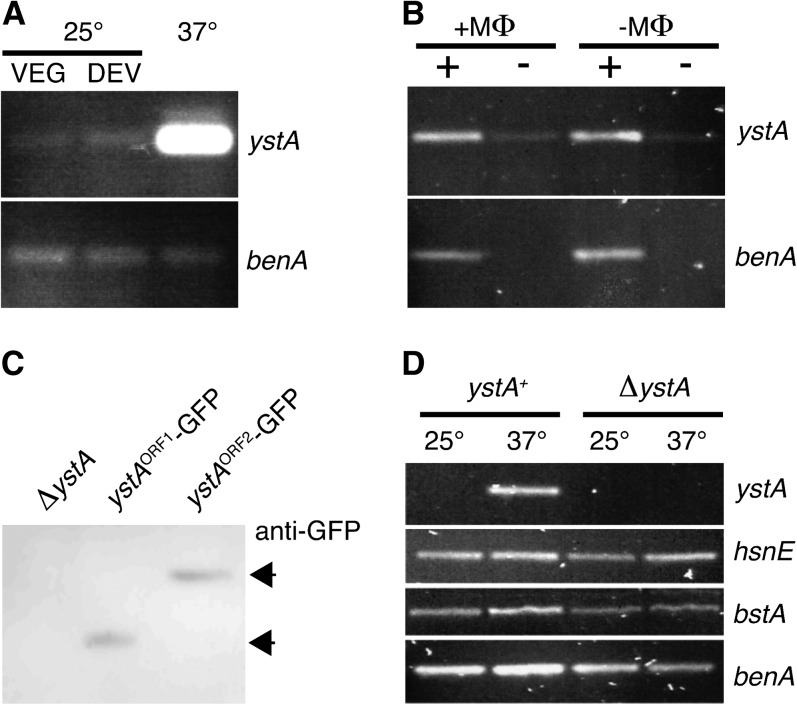
Although most of the probes on the microarray identified genomic sequences with readily identifiable gene annotations, several were either intergenic or had low coding potential, making gene annotation difficult. Such probes might represent genes that encode novel short proteins or long noncoding RNAs. One of the most differentially expressed probes was 1E11, which showed very high levels of expression throughout yeast cell growth but no expression during the hyphal phase (Figure 6A). The 1E11 probe is between a gene encoding a second histone H4 (hsnE) and a gene encoding a GPI maturation protein (bstA) (Figure S2). Northern blot analysis using RNA isolated from hyphal cells grown at 25° and yeast cells at 37° identified ∼850 nt RNA that is transcribed from the opposite strand to hsnE (Figure S3). A combination of 3′ RACE and RT-PCR identified the boundaries of this transcript, which contained three short ORFs (encoding 68, 66, and 69 aa) and potentially a long 3′ UTR if any of these ORFs encode a protein (Figure S3). RT-PCR using RNA derived from P. marneffei isolated from macrophages showed that this transcript was as abundant under these conditions as in yeast grown in vitro. As such, the gene was named ystA (yeast-specific transcript A) (Figure 6B).
Figure 6.
Expression from the 1E11 region in P. marneffei. (A) RT-PCR using P. marneffei RNA extracted from vegetative hyphal cells growing at 25° for 2 d (VEG), cells undergoing asexual development at 25° after 7 d (DEV), and yeast cells growing at 37° for 2 d using ystA-specific primers. Primers for the β-tubulin–encoding benA gene were used as a control. (B) RT-PCR using P. marneffei RNA extracted from yeast cells growing in J774 mouse macrophages (+MΦ) or in macrophage cell culture medium (−MΦ) for 24 hr using ystA-specific primers. To assess the amount of contaminating genomic DNA in the samples, both reverse-transcriptase (+) and no reverse-transcriptase (−) reactions were performed; benA was used as a control. (C) Western blot analysis using total cell extracts from P. marneffei yeast cells of the ystA deletion strain (ΔystA) and strains containing either the ystA ORF1 or the ORF2 GFP fusions. Cells were grown at 37° for 2 d and then extracts were fractionated on an SDS-PAGE gel, blotted, and Western blotted with a polyclonal anti-GFP primary antibody and an anti-rabbit HRP secondary antibody. Polyclonal anti-β-tubulin was used as a control. (D) RT-PCR expression analysis of ystA and the flanking hsnE and bstA genes in the wild-type and ystA deletion strain using RNA extracted from vegetative hyphal cells growing at 25° for 2 d or yeast cells growing at 37° for 2 d.
Sequence comparisons identified ystA homologous sequences in the closely related monomorphic fungus T. stipitatus, but not in other Eurotiales fungi such as Penicillium chrysogenum or A. fumigatus. Using the flanking genes, this region was cloned from another closely related fungus, Penicillium funiculosum, and sequencing showed that ystA was also conserved. Consistent with this, genes across this region are syntenic in the closely related species and disrupted in P. chrysogenum and A. fumigatus (Figure S2). Alignment of the ystA region showed that only ORF2 was conserved. To assess if any of these ORFs are translated, gene fusions with the GFP-encoding gene were generated and integrated into the P. marneffei genome either at the ystA locus or at the pyrG ectopic locus (Materials and Methods section). Western blot analysis using a monoclonal anti-GFP antibody identified a single fusion protein of the correct estimated size for each ORF1 and ORF2 in protein extracts from P. marneffei grown at 37°, suggesting that both the trans-species–conserved ORF2 and ORF1 encode proteins (Figure 6C).
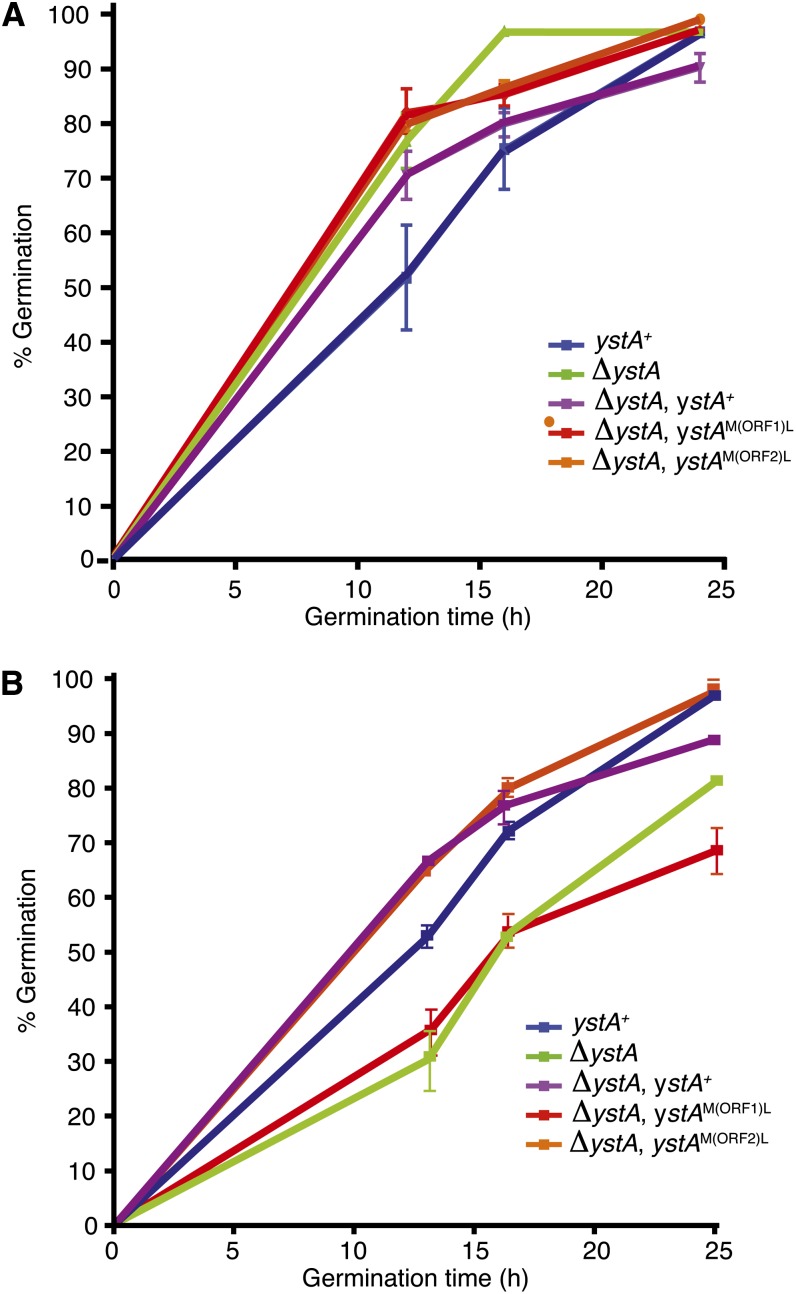
To understand the function of ystA, a deletion construct was generated that removes the entire transcribed region and transformation of the G526 strain yielded a number of transformants in which the locus had been deleted (as verified by Southern blot hybridization; data not shown). Phenotypic analysis showed that ΔystA strains were not substantially different from the ystA+ controls for germination at 25° (Figure 7A) or for hyphal growth and asexual development (data not shown). In contrast, ΔystA strains showed a delay in germination at 37° in vitro (Figure 7B). The ystA locus was deleted in a second strain of P. marneffei (G147) and the delayed germination phenotype was consistent with the previous transformants (data not shown). Despite this delay, normal yeast cell morphogenesis was evident at 37°. In addition, ΔystA germination and growth in macrophages displayed no clear difference compared to the wild-type (data not shown), suggesting that host-derived signals can override the observed delay. Because ystA has one short translated ORF (ORF1) that is not conserved in other species and one ORF (ORF2) that is conserved in closely related species, alleles were generated that mutated the initiator ATG codon of either ORF1 [ystAM(ORF1)L] or ORF2 [ystAM(ORF2)L], and these were used to transform a ΔystA strain. Phenotypic examination of these strains showed that the allele containing the ATG mutation in ORF1 failed to complement the germination delay of the ΔystA strain, whereas the allele with the ATG mutation in ORF2 fully complemented this phenotype (Figure 7B).
Figure 7.
Deletion of ystA delays germination at 37°. Time course measuring conidial germination of the wild-type (ystA+), ystA deletion (ΔystA), and complemented ystA deletion (ΔystA, ystA+) strains, as well as two strains carrying point mutations of the predicted initiator ATG of either ORF1 [ΔystA, ystAM(ORF1)L] or ORF2 [ΔystA, ystAM(ORF1)L] at 25° (A) and 37° (B). Conidia (1×106) from each strain were inoculated into SD medium at 25° or BHI at 37° in a 24-well microtiter plate and incubated at the appropriate temperature for 0, 12, 16, or 24 hr. One hundred cells at each time point were microscopically examined to assess their germination status. The experiment was performed in triplicate and the error bars represent SEM.
The high conservation of ORF2, but not of the functional ORF1, among these closely related species was unexpected. To examine this further, the ystA region was PCR-amplified from three clinical isolates of P. marneffei (FRR3840, 3841, and 4059), cloned, and sequenced. DNA sequence alignment identified a single nonsynonymous base substitution difference between FRR2161/FRR3841 and FRR3840/FRR4059 over the 206 bp of ORF1. Alignment of a 206-bp region of the adjacent bstA gene identified a single nonsynonymous nucleotide difference between FRR2161/FRR3840/FRR4059 and FRR3841. Thus, ystA produces ∼850 base transcript with a long 3′ UTR and encodes a short 68 aa protein unique to P. marneffei that is involved in germination at 37°. The biochemical activity of YstA is currently obscure.
Genes predominantly expressed in yeast cells include those involved in interactions with the host cell and fundamental to infection
The cell wall of fungi plays a vital role in the survival and growth of the cell. Its composition and structure have been shown to affect adhesion and virulence in a number of pathogens (Latgé 2007; Lesage and Bussey 2006; Sosinska et al. 2009). The cell wall typically comprises cross-linked glycans, chitins, and cell wall proteins. Chitin is a homopolymer of 1,4-N-acetylglucosamine and is found in all fungi. In C. albicans, four chitin synthases are responsible for the production of chitin, which has a vital role in cell shape and viability and is responsible for creating layers in the inner walls of all cell types (Munro and Gow 2001; Munro et al. 2003). Chitin is used to mask C. albicans from the host pathogen recognition receptor Dectin-1 (Mora-Montes et al. 2011). The chitin synthase encoded by chsE shows upregulated expression in P. marneffei yeast cells compared to hyphae. The ortholog in P. brasiliensis also has higher expression in yeast cells compared to hyphae, and this correlates with an increase in chitin production (Barreto et al. 2009).
Additionally involved in the cell wall glucan/chitin matrix is the ortholog of the glycosyl hydrolase Dfg5, which in N. crassa contributes to the covalent cross-linking of glycoproteins into the cell wall (Latgé 2007). Cell wall glycoproteins are structural components vital for cell wall biogenesis and the glycoprotein content of cell walls varies between particular cell types, allowing specialized adaptation to specific environmental stresses (Latgé 2007; Sosinska et al. 2009). In N. crassa, dfg5 mutant cells have irregular hyphal diameters, exhibit dichotomous branching, secrete higher levels of cell wall proteins, and are sensitive to various stress reagents (Maddi et al. 2012). Finally, the ability of fungal cells to adhere to various host tissues is fundamental to successful infection. Another gene upregulated in established yeast cells encodes E1 beta subunit pyruvate dehydrogenase. Apart from its major metabolic roles in converting pyruvate into acetyl-CoA, pyruvate dehydrogenase and its complex have been associated with adhesion to host tissue, particularly to fibronectin in Mycoplasma mycoides and Mycoplasma pneumonia (Dallo et al. 2002; Jores et al. 2009).
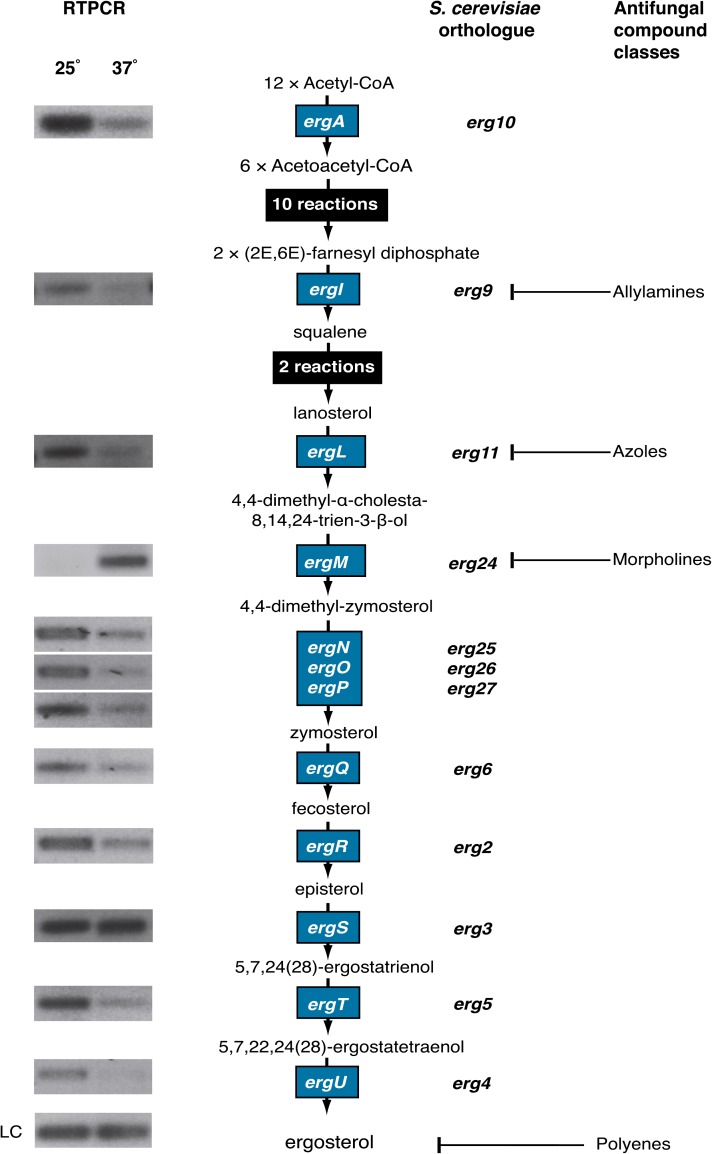
Ergosterol is a key component of the fungal cell membrane, shown to control fluidity and formation of lipid rafts (Brown and London 2000; Lees et al. 1980; Sharma 2006). Lipid rafts allow the stable association of membrane-bound proteins to the cell surface, thus ergosterol production has been linked to many important cellular functions from nutrient uptake and drug efflux to polarity determination (Alvarez et al. 2007; Iwaki et al. 2008; Pasrija et al. 2008). The biochemistry and genetics involved in ergosterol biosynthesis and sterol biosynthesis more broadly is well-characterized in several species and has become the target of several antifungal compound classes (Nes 2011). The C-14 sterol reductase encoding gene ergM, active in the ergosterol biosynthetic pathway, showed differentially upregulated expression throughout yeast cell growth in P. marneffei (Figure 5 and Figure 8).
Figure 8.
Ergosterol pathway expression analysis. The ergosterol biosynthesis pathway from acetyl-CoA to ergosterol including the chemical intermediates and genes encoding the enzymes for each step. Expression of P. marneffei genes indicated by blue boxes were examined by RT-PCR using RNA from vegetative hyphal cells grown at 25° for 2 d, yeast cells grown at 37° for 2 d, and primers specific for each gene. S. cerevisiae orthologs are shown adjacent to the P. marneffei genes. The proteins targeted by various classes of antifungal compound are also indicated. LC, loading control (benA).
Ergosterol biosynthesis is dependent on oxygen and iron. Regulation of the ergosterol biosynthetic pathway in the variable environment of the host, where both of these may be limiting, is therefore vital (Jones 1986; Weinberg 1986). RT-PCR validation of ergM expression confirmed that it was upregulated at 37°. Although it was not surprising that ergosterol biosynthesis may differ between hyphal cells growing at 25° and yeast cells at 37°, to compensate for membrane fluidity differences at the two temperatures, ergM was the only upregulated gene in the ergosterol biosynthesis pathway (Figure 8). This suggests that the reduction of C-14 on the sterol backbone was more important for sterol function at 37° than at 25°. Alternatively, the biochemical activity of ErgM, C-14 reduction, may be the limiting step in ergosterol biosynthesis at 37° and ergM is upregulated to increase flux through the pathway.
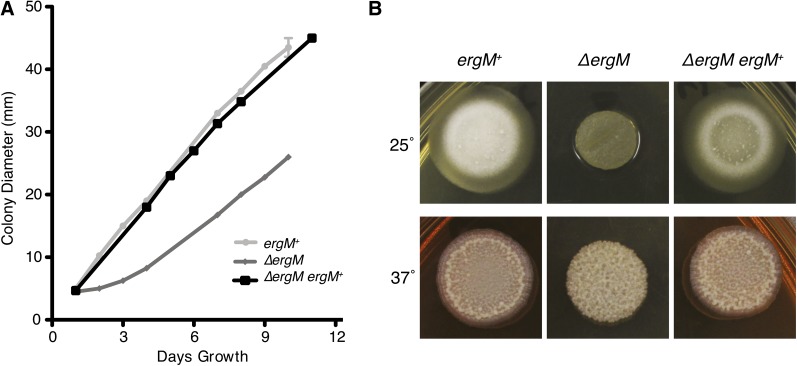
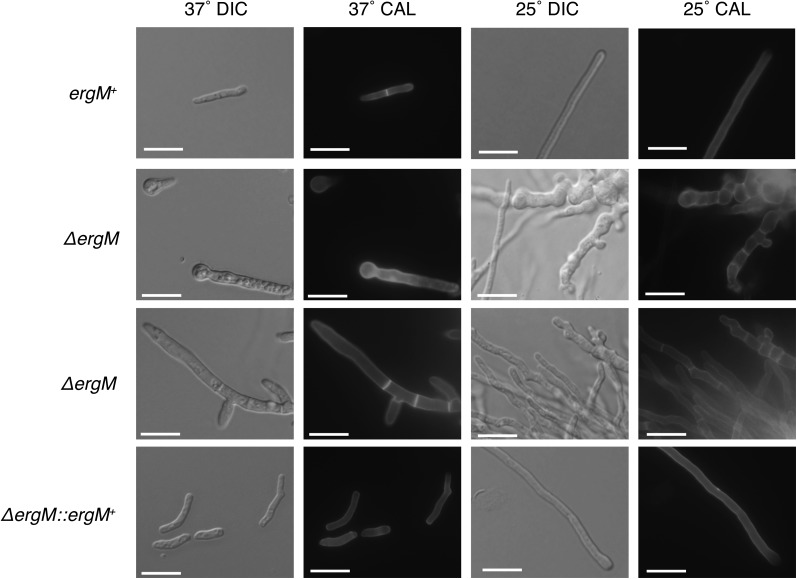
To assess the role of ergM during growth and morphogenesis in P. marneffei, the ergM coding sequence was deleted by homologous gene replacement (see Materials and Methods section). Deletion of ergM led to significant and pleiotropic effects, including swollen and highly branched cells, slow growth, delayed conidiation, and altered antifungal sensitivity. The most obvious of these was a severe reduction in growth rate and altered colony morphology at both 25° and 37° (Figure 9). Microscopic analysis showed normal levels of progression past the germling stage but severe polarity defects in a proportion of cells at 25° (Figure 10), whereas at 37° a large proportion of the conidia failed to produce more than a small germ tube. The remaining cells grew at a reduced rate with very few yeast cells produced (Figure 10). The more severe phenotype at 37° confirmed the importance of ergM at this temperature. The growth reduction of the ergM deletion strain at both temperatures was initially severe; however, there was a marked recovery in growth over time (Figure 10). This recovery was not attributable to the accumulation of suppressor mutations because conidia from the deletion strain exhibited the severe growth phenotype of the original transformant at both temperatures (data not shown). The phenotypic change was therefore a biochemical or regulatory response to the altered sterol production. This phenomenon might be akin to a response observed in S. cerevisiae, in which other membrane components such as phospholipids were altered in ergosterol biosynthesis mutants, allowing growth (Sharma 2006).
Figure 9.
Growth rate defect of the ΔergM strain. (A) Radial growth rate (colony diameter) was measured every 24 hr for the control (ergM+), deletion (ΔergM), and complemented (ΔergM ergM+) strains at 25° on ANM medium. (B) Colony morphology after 4 d of growth on BHI medium at 25° and 37°.
Figure 10.
Microscopic examination of ΔergM strain. Control (ergM+), deletion (ΔergM), and complemented (ΔergM ergM+) strains were grown on 2% BHI for 4 d at the indicated temperature and examined at 100× magnification. DIC, differential interference contrast microscopy; calcofluor (CAL), 0.1 mg/ml calcofluor staining under UV filter; scale bars, 20 µm.
Ergosterol biosynthesis is the target of several classes of antifungal compounds, including azoles (ergL) and morpholines (ergM) (Figure 8). Amphotericin B (an allylamine) is commonly used to treat P. marneffei infections and also binds ergosterol itself to effect cell death. Therefore, it might be expected that deletion of an enzyme in the ergosterol biosynthesis pathway would result in resistance to these antifungals. Although resistance to amphotericin B was increased in the ergM deletion strain, this strain showed increased sensitivity to the azoles fluconazole and voriconazole (Table 3). This implies that either ergM or ergL was required for the production of the functional sterol because the loss of ErgL and ErgM activity had a combinatorial rather than epistatic effect on phenotype. In other species, the deletion of some ergosterol biosynthesis genes results in a modified sterol profile (Alcazar-Fuoli et al. 2008; Geber et al. 1995; Ruan et al. 2002). As such, deletion of biosynthetic genes does not eliminate sterol production but does modify the type of sterol produced. This modification may lead to reduced binding of amphotericin B to the sterol and could explain the resistance of the ergM deletion to this compound. These experiments suggest that combinatorial treatment of P. marneffei infections with recently developed compounds targeting ergM (Hata et al. 2010; Liang et al. 2009) and azoles would be effective, with a loss of ErgL and ErgM activity having a combinatorial effect on growth. Conversely, combination of compounds targeting ErgM with allylamines may not be effective because a loss of ergM leads to resistance to the allylamine amphotericin B.
Table 3. ΔergM sensitivity to antifungal drugs measured as minimum inhibitory concentrations using e-test strips.
| 25° |
37° |
|||||
|---|---|---|---|---|---|---|
| ergM+ | ΔergM | ΔergM ergM+ | ergM+ | ΔergM | ΔergM ergM+ | |
| Voriconazole | 0.032 | 0.001 | 0.032 | 0.006 | <0.001 | 0.006 |
| Fluconazole | 24 | 0.5 | 24 | 3 | 0.125 | 3 |
| Amphotericin B | >32 | >32 | >32 | 1.5 | 4 | 1.5 |
Minimum inhibitory concentration values are presented as µg/ml concentrations.
In a recent microarray study, Lin et al. (2012) compared the hyphal-specific and yeast-specific gene expression profiles of P. marneffei clinical isolate B-6323, rather than the type strain used here. Although the microarray design encompassed most annotated gene models, the study was restricted to 1884 P. marneffei genes orthologous to S. cerevisiae genes with identified GO terms. Despite the different strains and substantially different growth conditions that were used, 76% of the top 400 differentially expressed genes with respect to hyphal cells and yeast cells identified in the present study were also identified by Lin et al. (2012). Of these genes, 73% were consistent with respect to which cell type displayed higher gene expression. Numerous cell-type–specific gene expression differences identified in the present study are also evident in similar studies of other dimorphic fungi such as P. brasiliensis and H. capsulatum. For example, when comparing the differentially expressed genes identified in H. capsulatum by Hwang et al. (2003) and those identified in P. marneffei (Figure 5) (37° vs. 25°), 55% show similar patterns of expression between hyphal cells and yeast cells. Interestingly, of the 27% of genes that were inconsistent between this study and that of Lin et al. (2012) in terms of the cell type with the greater expression, 83% showed consistent cell-type expression between this study and the study by Hwang et al. (2003) using H. capsulatum. Overall, the gene expression profiles described in these studies highlight key similarities and also differences in the metabolic and reproductive modes of yeast and hyphal cells.
Conclusions
Cell-state–specific gene expression is one of the most informative strategies for understanding the physiological properties of different types of cells in multicellular organisms. This study effectively identified genes differentially expressed in hyphal, yeast, and conidiating cultures, and those expressed early and late in hyphal and yeast cell growth of the dimorphic opportunistic pathogen P. marneffei. P. marneffei hyphal cells behave like those of many other filamentous fungi at 25°, focusing on maintaining growth through hyphal tip extensions and tightly regulated asexual reproduction. Compared to hyphal cells, the selective pressures imposed on yeast cells are very different. Survival of yeast cells in the host is dependent on their ability to survive within the host macrophage, avoiding destruction, while scavenging limited nutrients, such as iron. Genes important for high-affinity iron assimilation were upregulated throughout yeast cell growth in P. marneffei. A cell wall specialized for host environments is also necessary for P. marneffei yeast cells with possibly different chitin and cell wall protein requirements than those of hyphal cells. Similarly, the ability to adhere to the host cell is considered a pathogenicity factor in many fungi, making the increased expression of these genes in yeast cells an area of great interest. Identification and characterization of ergM showed that this is a gene fundamental to cell wall maintenance, nutrient uptake, and polarity determination, and loss of function results in severe growth defects in P. marneffei and important differences in susceptibility to antifungal agents. Other genes such as ystA are specific to the yeast cell state and are highly restricted taxonomically, highlighting the differences between the dimorphic fungi and possibly their evolved mechanisms for growth in a host. There is much to be discovered about the specific roles of genes identified in this study; further functional analyses are necessary. What can be elucidated is that temporal and cell-specific transcriptional differences identified in this study reflect different modes of reproduction, requirements throughout development, and changing environmental pressures throughout the life cycle of P. marneffei.
Supplementary Material
Acknowledgments
We thank Luke Noble, Alisha McLauchlan, and Harshini Weerasinghe for assistance with some of the aspects of the experimentation and analysis, and the Australian Genomic Research Facility for assistance with development of the custom microarrays. This work was supported by grants from the National Health and Medical Research Council and the Howard Hughes Medical Institute (to A.A.), a European Union Marie-Curie fellowship (to D.C.), and the Thailand Research Fund (TRF) Royal Golden Jubilee Scholarship (to N.N.).
Footnotes
Communicating editor: J. C. Dunlap
Literature Cited
- Adams T., Boylan M., Timberlake W., 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54: 353–362 [DOI] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Lopez J. F., Grimalt J. O., et al. , 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73: 339–347 [DOI] [PubMed] [Google Scholar]
- Alvarez F. J., Douglas L. M., Konopka J. B., 2007. Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell 6: 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianopoulos A., 2002. Control of morphogenesis in the human fungal pathogen Penicillium marneffei. Int. J. Med. Microbiol. 292: 331–347 [DOI] [PubMed] [Google Scholar]
- Barreto L., Sorais F., Salazar V., San-Blas G., Nino-Vega G. A., 2009. Expression of Paracoccidioides brasiliensis CHS3 in a Saccharomyces cerevisiae chs3 null mutant demonstrates its functionality as a chitin synthase gene. Yeast 27: 293–300 [DOI] [PubMed] [Google Scholar]
- Black T., Hare R., 2000. Will genomics revolutionize antimicrobial drug discovery? Curr. Opin. Microbiol. 3: 522–527 [DOI] [PubMed] [Google Scholar]
- Bordonné R., 2000. Functional characterization of nuclear localization signals in yeast Sm proteins. Mol. Cell. Biol. 20: 7943–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Hynes M. J., Andrianopoulos A., 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol. Microbiol. 38: 1034–1047 [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Hynes M. J., Andrianopouls A., 2001. An STE12 homolog from the asexual dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 157: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Hynes M. J., Andrianopoulos A., 2002. A basic helix–loop–helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol. Microbiol. 44: 621–631 [DOI] [PubMed] [Google Scholar]
- Bowman B. H., Taylor J. W., White T. J., 1992. Molecular evolution of the fungi: human pathogens. Mol. Biol. Evol. 9: 893–904 [DOI] [PubMed] [Google Scholar]
- Bowman B. H., White T. J., Taylor J. W., 1996. Human pathogeneic fungi and their close nonpathogenic relatives. Mol. Phylogenet. Evol. 6: 89–96 [DOI] [PubMed] [Google Scholar]
- Boyce K. J., Andrianopoulos A., 2013. Morphogenetic circuitry regulating growth and development in the dimorphic pathogen Penicillium marneffei. Eukaryot. Cell 12: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce K. J., Hynes M. J., Andrianopoulos A., 2005. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 55: 1487–1501 [DOI] [PubMed] [Google Scholar]
- Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., et al. , 2004. GO:TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinform 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., London E., 2000. Structure and function of sphingolipid-and cholesterol-rich membrane rafts. J. Biol. Chem. 275: 17221–17224 [DOI] [PubMed] [Google Scholar]
- Bugeja H. E., Boyce K. J., Weerasinghe H., Beard S., Jeziorowski A., et al. , 2012. Tools for high efficiency genetic manipulation of the human pathogen Penicillium marneffei. Fungal Genet. Biol. 49: 772–778 [DOI] [PubMed] [Google Scholar]
- Clutterbuck A., 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. T., 1986. Models of cell differentiation in conidial fungi. Microbiol. Rev. 50: 95–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C. R., Vanittanakom N., 2008. Insights into the pathogenicity of Penicillium marneffei. Future Microbiol. 3: 43–55 [DOI] [PubMed] [Google Scholar]
- Cove D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56 [DOI] [PubMed] [Google Scholar]
- Dallo S. F., Kannan T. R., Blaylock M. W., Baseman J. B., 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G., 1990. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pauw B. E., 2001. Treatment of documented and suspected neutropenia-associated invasive fungal infections. J. Chemother. 13: 181–192 [DOI] [PubMed] [Google Scholar]
- DeRisi J., Iyer V., Brown P., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Dixon D. M., McNeil M. M., Cohen M. L., Gellin B. G., La Montagne J. R., 1996. Fungal infections: a growing threat. Public Health Rep. 111: 226–235 [PMC free article] [PubMed] [Google Scholar]
- Felipe M. S., Andrade R. V., Arraes F. B., Nicola A. M., Maranhao A. Q., et al. , 2005. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280: 24706–24714 [DOI] [PubMed] [Google Scholar]
- Fortwendel J. R., Zhao W., Bhabhra R., Park S., Perlin D. S., et al. , 2005. A fungus-specific ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell 4: 1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Henn M. R., Ma L.-J., Cuomo C. A., Birren B., 2005. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 15: 1620–1631 [DOI] [PubMed] [Google Scholar]
- Garaizar J., Brena S., Bikandi J., Rementeria A., Ponton J., 2006. Use of DNA microarray technology and gene expression profiles to investigate the pathogenesis, cell biology, antifungal susceptibility and diagnosis of Candida albicans. FEMS Yeast 6: 987–998 [DOI] [PubMed] [Google Scholar]
- Geber A., Hitchcock C. A., Swartz J. E., Pullen F. S., Marsden K. E., et al. , 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39: 2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N., Brown A., Odds F., 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5: 366–371 [DOI] [PubMed] [Google Scholar]
- Haas H., 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62: 316–330 [DOI] [PubMed] [Google Scholar]
- Hata M., Ishii Y., Watanabe E., Uoto K., Kobayashi S., et al. , 2010. Inhibition of ergosterol synthesis by novel antifungal compounds targeting C-14 reductase. Med. Mycol. 48: 613–621 [DOI] [PubMed] [Google Scholar]
- Holloway A. J., Oshlack A., Diyagama D. S., Bowtell D. D. L., Smyth G. K., 2006. Statistical analysis of an RNA titration series evaluates microarray precision and sensitivity on a whole-array basis. BMC Bioinformatics 7: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L., Hocking-Murray D., Bahrami A. K., Andersson M., Rine J., et al. , 2003. Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol. Biol. Cell 14: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T., Iefuji H., Hiraga Y., Hosomi A., Morita T., et al. , 2008. Multiple functions of ergosterol in the fission yeast Schizosaccharomyces pombe. Microbiol 154: 830–841 [DOI] [PubMed] [Google Scholar]
- Joardar V., Abrams N. F., Hostetler J., Paukstelis P. J., Pakala S., et al. , 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics 13: 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H., Kasuga T., Schaller R. A., Good B., Gardner M. J., et al. , 2006. Phase-specific gene expression underlying morphological adaptations of the dimorphic human pathogenic fungus, Coccidioides posadasii. Fungal Genet. Biol. 43: 545–559 [DOI] [PubMed] [Google Scholar]
- Johnson L., 2008. Iron and siderophores in fungal-host interactions. Mycol. Res. 112: 170–183 [DOI] [PubMed] [Google Scholar]
- Jones D. P., 1986. Intracellular diffusion gradients of O2 and ATP. Am. J. Physiol. 250: 663–675 [DOI] [PubMed] [Google Scholar]
- Jores J., Meens J., Buettner F. F. R., Linz B., Naessens J., et al. , 2009. Analysis of the immunoproteome of Mycoplasma mycoides subsp. mycoides small colony type reveals immunogenic homologues to other known virulence traits in related Mycoplasma species. Vet. Immunol. Immunopathol. 131: 238–245 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D., 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16: 2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana-uchi A., Yamashiro C., Tanabe S., Murayama T., 1997. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. 254: 427–432 [DOI] [PubMed] [Google Scholar]
- Klein B., Tebbets B., 2007. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10: 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D. J., 2003. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47: 1185–1197 [DOI] [PubMed] [Google Scholar]
- Kraus P. R., Hofmann A. F., Harris S. D., 2002. Characterization of the Aspergillus nidulans 14–3-3 homologue, ArtA. FEMS Microbiol. Lett. 210: 61–66 [DOI] [PubMed] [Google Scholar]
- Kupfer D. M., Reece C. A., Clifton S. W., Roe B. A., Prade R. A., 1997. Multicellular ascomycetous fungal genomes contain more than 8000 genes. Fungal Genet. Biol. 21: 364–372 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K., Polacheck I., Popkin T., 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150: 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J., 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66: 279–290 [DOI] [PubMed] [Google Scholar]
- Lees N. D., Lofton S. L., Woods R. A., Bard M., 1980. The effects of varied energy source and detergent on the growth of sterol mutants of Saccharomyces cerevisiae. J. Gen. Microbiol. 118: 209–214 [Google Scholar]
- Lengeler K., Davidson R., D’souza C., Harashima T., 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H., 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R.-m., Cao Y.-b., Fan K.-h., Xu Y., Gao P.-h., et al. , 2009. 2-Amino-nonyl-6-methoxyl-tetralin muriate inhibits sterol C-14 reductase in the ergosterol biosynthetic pathway. Acta Pharmacol. Sin. 12: 1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Ran Y., Gou L., He F., Zhang R., et al. , 2012. Comprehensive transcription analysis of human pathogenic fungus Penicillium marneffei in mycelial and yeast cells. Med. Mycol. 50: 835–842 [DOI] [PubMed] [Google Scholar]
- Lo H. J., Köhler J. R., Didomenico B., Loebenberg D., Cacciapuoti A., et al. , 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949 [DOI] [PubMed] [Google Scholar]
- Maddi A., Fu C., Free S. J., 2012. The Neurospora crassa dfg5 and dcw1 genes encode α-1,6-mannanases that function in the incorporation of glycoproteins into the cell wall. PLoS ONE 7: 38872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah J.H., Yu J.H., 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 5: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M. M., Nash S. L., Hajjeh R. A., Phelan M. A., Conn L. A., et al. , 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. Clin. Infect. Dis. 33: 641–647 [DOI] [PubMed] [Google Scholar]
- Miethke M., Marahiel M. A., 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71: 413–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. P., Clemons K. V., Mirels L. F., Coller J. A., Wu T. D., et al. , 2009. Genomic DNA microarray comparison of gene expression patterns in Paracoccidioides brasiliensis mycelia and yeasts in vitro. Microbiol 155: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H., Netea M., Ferwerda G., Lenardon M., Brown G., et al. , 2011. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun. 79: 1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C., Gow N., 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 39: 41–53 [PubMed] [Google Scholar]
- Munro C., Whitton R., Hughes H., Rella M., Sevaggini S., et al. , 2003. CHS8—a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet. Biol. 40: 146–158 [DOI] [PubMed] [Google Scholar]
- Nes W. D., 2011. Biosynthesis of cholesterol and other sterols. Chem. Rev. 111: 6423–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., Heitman J., 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57: 143–173 [DOI] [PubMed] [Google Scholar]
- O’Connell M. J., Osmani A. H., Morris N. R., Osmani S. A., 1992. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 11: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Tokuoka M., Jin F. J., Takahashi T., Koyama Y., 2010. Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 47: 10–18 [DOI] [PubMed] [Google Scholar]
- Pardo M., Monteoliva L., Vázquez P., Martínez R., Molero G., et al. , 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiol. 150: 4157–4170 [DOI] [PubMed] [Google Scholar]
- Parks L. W., Casey W. M., 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49: 95–116 [DOI] [PubMed] [Google Scholar]
- Pasrija R., Panwar S. L., Prasad R., 2008. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52: 694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C., 2006. Iron uptake in fungi: A system for every source. Biochim. Biophys. Acta. 1763: 636–645 [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Silver J., Oshlack A., Holmes M., Diyagama D., et al. , 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Roosenberg J. M., Lin Y. M., Lu Y., Miller M. J., 2000. Studies and syntheses of siderophores, microbial iron chelators, and analogs as potential drug delivery agents. Curr. Med. Chem. 7: 159–197 [DOI] [PubMed] [Google Scholar]
- Roy J., Zheng B., Rymond B. C., Woolford J. L., 1995. Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol. Cell. Biol. 15: 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan B., Lai P. S., Yeh C. W., Wilson W. K., Pang J., et al. , 2002. Alternative pathways of sterol synthesis in yeast: Use of C27 sterol tracers to study aberrant double-bond migrations and evaluate their relative importance. Steroids 67: 1109–1119 [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., et al. , 2006. TM4 microarray software suite. Methods Enzymol. 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. Fritsch, and T. Maniatis, 1989 Molecular Cloning, Laboratory Manual (Vol. 2nd Edition). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Samonis G., Bafaloukos D., 1992. Fungal infections in cancer patients: an escalating problem. In Vivo 6: 183–193 [PubMed] [Google Scholar]
- Samson R.A., Yilmaz N., Houbraken J., Spierenburg H., Seifert K.A., et al. , 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 70: 159–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Blas G., Travassos L., Fries B., Goldman D., 2000. Fungal morphogenesis and virulence. Med. Mycol. 38: 79–86 [PubMed] [Google Scholar]
- Saville S., Thomas D., López-Ribot J., 2005. Use of genome information for the study of the pathogenesis of fungal infections and the development of diagnostic tools. Rev. Iberoam. Micol. 22: 238–241 [DOI] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., et al. , 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. C., 2006. Implications of sterol structure for membrane lipid composition, fluidity and phospholipid asymmetry in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 1047–1051 [DOI] [PubMed] [Google Scholar]
- Si H., Rittenour W. R., Xu K., Nicksarlian M., Calvo A. M., et al. , 2012. Morphogenetic and developmental functions of the Aspergillus nidulans homologues of the yeast bud site selection proteins Bud4 and Axl2. Mol. Microbiol. 85: 252–270 [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Speed T. P., 2003. Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Smyth G. K., 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3. [DOI] [PubMed] [Google Scholar]
- Smyth, G. K., 2005a Limma linear models for microarray data, pp. 397–420 in Gentleman, R., V. Carey, S. Dut, R. Irizarry, and W. Huber (Editors), Bioinformatics and Computational Biology Solutions using R and Bioconductor, Springer, New York. [Google Scholar]
- Smyth G. K., Michaud J., Scott H., 2005b Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sosinska G., De Groot P., Brul S., 2009. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 9: 1013–1028 [DOI] [PubMed] [Google Scholar]
- Steinbach W. J., Cramer R. A., Perfect B. Z., Asfaw Y. G., Sauer T. C., et al. , 2006. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., 2011. Motors in fungal morphogenesis: cooperation vs. competition. Curr. Opin. Microbiol. 14: 660–667 [DOI] [PubMed] [Google Scholar]
- Timberlake W., Boylan M., Mirabito P., Willett C., Zimmerman C., 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 3: 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman M. M., Woods J. P., 1999. Ferric reduction is a potential iron acquisition mechanism for Histoplasma capsulatum. Infect. Immun. 67: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd R., Fraser J., Wong K., Davis M., Hynes M., 2005. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot. Cell 4: 1646–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H., Chang Y., Washburn R., Wheeler M. H., Kwon-Chung K. J., 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180: 3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. F., Washburn R. G., Chang Y. C., Kwon-Chung K. J., 1997. Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol. Microbiol. 26: 175–183 [DOI] [PubMed] [Google Scholar]
- Vanittanakom N., Cooper C. R., Fisher M. C., Sirisanthana T., 2006. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev. 19: 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Aisen P., Casadevall A., 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63: 3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. D., 1986. Iron, infection, and neoplasia. Clin. Physiol. Biochem. 4: 50–60 [PubMed] [Google Scholar]
- Weinberg E. D., 2009. Iron availability and infection. Biochim. Biophys. Acta. 1790: 600–605 [DOI] [PubMed] [Google Scholar]
- Wheeler M., Bell A., 1988. Melanins and their importance in pathogenic fungi. Curr. Top. Med. Mycol. 2: 338–387 [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., et al. , 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277: 31079–31088 [DOI] [PubMed] [Google Scholar]
- Yu, J.-H., 2010 Regulation of development in Aspergillus nidulans and, Aspergillus fumigatus. Mycobiol. 38: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.-Y., Pascal G., Wong S. S. Y., Glaser P., Woo P. C. Y., et al. , 2003. Exploring the Penicillium marneffei genome. Arch. Microbiol. 179: 339–353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.