Abstract
Genome-wide association studies are a powerful method to dissect the genetic basis of traits, although in practice the effects of complex genetic architecture and population structure remain poorly understood. To compare mapping strategies we dissected the genetic control of flavonoid pigmentation traits in the cereal grass sorghum by using high-resolution genotyping-by-sequencing single-nucleotide polymorphism markers. Studying the grain tannin trait, we find that general linear models (GLMs) are not able to precisely map tan1-a, a known loss-of-function allele of the Tannin1 gene, with either a small panel (n = 142) or large association panel (n = 336), and that indirect associations limit the mapping of the Tannin1 locus to Mb-resolution. A GLM that accounts for population structure (Q) or standard mixed linear model that accounts for kinship (K) can identify tan1-a, whereas a compressed mixed linear model performs worse than the naive GLM. Interestingly, a simple loss-of-function genome scan, for genotype-phenotype covariation only in the putative loss-of-function allele, is able to precisely identify the Tannin1 gene without considering relatedness. We also find that the tan1-a allele can be mapped with gene resolution in a biparental recombinant inbred line family (n = 263) using genotyping-by-sequencing markers but lower precision in the mapping of vegetative pigmentation traits suggest that consistent gene-level resolution will likely require larger families or multiple recombinant inbred lines. These findings highlight that complex association signals can emerge from even the simplest traits given epistasis and structured alleles, but that gene-resolution mapping of these traits is possible with high marker density and appropriate models.
Keywords: quantitative trait loci, null alleles, structured populations, genome scan, grain pigmentation
Genome-wide association studies (GWAS) have been used to dissect the genetic basis of complex traits in humans (Rosenberg et al. 2010; Visscher et al. 2012), model systems (Atwell et al. 2010; Flint and Eskin 2012), and crop species (Beló et al. 2008; Cockram et al. 2010; Huang et al. 2010, 2012; Morris et al. 2013). By the use of natural populations instead of biparental families traditionally used in linkage mapping, GWAS offers a number of potential advantages: avoiding the need for crosses; capturing greater genetic diversity for the trait of interest; and providing greater mapping resolution through the use of historical recombination events. However, complex genetic architectures and structured traits can create spurious signals and indirect associations in GWAS, and the interactions between genetic architecture and population structure remain poorly understood. Complex genetic architecture can arise from allelic heterogeneity (multiple independent alleles at the same gene) or genetic heterogeneity (multiple genes controlling the trait), and epistasis (nonadditive interactions among multiple genes; Platt et al. 2010; Segura et al. 2012). A simple, common form of epistasis is complementary dominance, in which functional alleles of two or more genes are required for the expression of the dominant phenotype, so loss-of-function alleles interact epistatically. When multiple functional alleles are present on the same haplotype block, synthetic associations with shared ancestral alleles may result in positively misleading GWAS signals (Dickson et al. 2010; Platt et al. 2010). Complex genetic interactions are thought to underlie cases in which known causative alleles do not emerge as the most significant associations in GWAS, such as the case of FRIGIDA expression in Arabidopsis (Atwell et al. 2010; Segura et al. 2012).
Given the complexity of mapping traits in diverse populations, much effort has gone to developing and characterizing various statistical approaches to GWAS. It is well-known that the simplest GWAS methods that ignore population structure, such as Wilcoxon rank sum tests or general linear models (GLMs), will yield inflated association signals when used for structured traits (Atwell et al. 2010; Huang et al. 2010; Zhao et al. 2011). Conversely, approaches that control for inflated signals by accounting for population structure [including the structured association (Pritchard et al. 2000) and mixed linear models (Yu et al. 2006)] can yield false-negative results when causal variants are structured (Bergelson and Roux 2010). Given the trade-offs in the use of simple vs. structured models, further empirical studies of validated functional variants are needed to inform investigations of novel trait loci. Moreover, it has been argued that the impact and prevalence of synthetic associations will only be determined empirically (Goldstein 2011).
To better characterize the interaction of genetic architecture and population structure, and compare methods for GWAS in structured populations, we investigated the genome-wide association signals of flavonoid pigmentation traits. Flavonoid pigmentation shows abundant natural variation in many plant species and therefore has been a classic empirical model in genetics (Darwin 1859; Nilsson-Ehle 1909; Sax 1923; McClintock 1950; Huang et al. 2010). As such, the flavonoid pathway has been almost completely elucidated in Arabidopsis and maize, and many of the enzymes, regulators, and transporters underlying flavonoid traits are widely conserved across plant families (Petroni and Tonelli 2011; Supporting Information, Table S1). Although the core components of the flavonoid network are relatively well-understood, there are a number of areas that remain to be elucidated, including the polymerization and transport of tannins (Zhao et al. 2010) and pathways responsible for lineage-specific end products involved in defense and environmental adaptation (Ibraheem et al. 2010). Moreover, even when the genes underlying a trait are known, the characterization of functional allelic variation remains an important and challenging goal (Tian et al. 2009).
In sorghum [Sorghum bicolor (L.) Moench] there is abundant natural variation for flavonoid pigmentation, which underlies a number of agronomic traits, such as grain mold (Esele et al. 1993) and anthracnose resistance (Ibraheem et al. 2010), and nutritional traits, such as digestibility (Kaufman et al. 2012) and anti-inflammatory properties (Moraes et al. 2012). The role of pigmentation in crop diversification and improvement is complex (Gross and Olsen 2010), as exemplified by grain tannins, which provide defense against molding and bird predation but also impart bitterness and astringency (Doggett 1988). Classical inheritance and linkage studies have mapped loci controlling pigmentation of several sorghum tissues including the testa (inner seed coat; B1 and B2), pericarp (outer seed coat; R and Y), coleoptile (seedling leaf sheath; Rs1 and Rs2), and adult vegetative leaf and stem (P and Q) (Vinall and Cron 1921; Stephens 1946; Doggett 1988; Mace and Jordan 2010). In addition, two flavonoid genes have been cloned in sorghum, which can be used to validate mapping approaches: a MYB transcription factor (Y1; Yellow seed1), which controls pericarp pigmentation and phytoalexin production (Ibraheem et al. 2010), and a WD40 regulator (Tannin1), which controls the presence of tannins in the testa (Wu et al. 2012). Although testa pigmentation segregates as a simple Mendelian trait, from the standpoint of GWAS it may present a complex genetic architecture because of multiple tan1 loss-of-function alleles (tan1-a and tan1-b) and complementary dominance between at least two loss-of-function loci (Wu et al. 2012). Moreover, sorghum has complex population structure because of extensive ancient crop diffusion and a propensity for inbreeding, which presents a challenge for GWAS of agroclimatic traits (Morris et al. 2013). Here, we take advantage of high-resolution genotyping-by-sequencing single-nucleotide polymorphism (SNP) maps to compare the ability of several genome-wide mapping approaches to identify known flavonoid pigmentation loci, contrast linear model GWAS to a simple loss-of-function genome scan, and use GWAS to identify additional loci that may underlie flavonoid pigmentation in sorghum.
Materials and Methods
Flavonoid-related candidate genes
To identify potential components of the flavonoid network in sorghum and to define an a priori candidate gene set for comparison with mapping results, we conducted a systematic survey of flavonoid-related gene families in the reference sorghum genome (Table S1 and File S1; Paterson et al. 2009). Because Arabidopsis is by far the best-understood model for the flavonoid network (Winkel-Shirley 2001; Zhao et al. 2010; Petroni and Tonelli 2011), we defined the candidate gene set primarily based on the Arabidopsis flavonoid-related genes in TAIR (www.arabidopsis.org; Lamesch et al. 2011). Sorghum homologs of the reference genes were obtained from Phytozome (www.phytozome.org; Goodstein et al. 2011). Note that because most flavonoid-related genes are conserved across diverse plant species, an Arabidopsis-based homology search captures the sorghum orthologs of many flavonoid-related genes in maize, rice, and other plant species (Schnable and Freeling 2011; Petroni and Tonelli 2011), as well as the two cloned flavonoid genes in sorghum, Yellow seed1 and Tannin1 (Ibraheem et al. 2010; Wu et al. 2012).
Genotyping-by-sequencing (GBS)
Genotypes for this study were generated with genotyping-by-sequencing (Elshire et al. 2011) using the GBS pipeline 3.0 in the TASSEL software package (Bradbury et al. 2007) and the BTx623 genome as a reference (Paterson et al. 2009). Genotypes for the association panels at 265,487 SNPs were previously obtained (Morris et al. 2013). For this study we also genotyped the same 265,487 SNPs in 263 F6-7 recombinant inbred lines (RILs; ICSV 745 × PB 15520) that were developed as a stem borer resistance mapping population but also segregate flavonoid pigmentation phenotypes (Vinayan 2010). Note, SNP positions in the GBS data may differ slightly (several base pairs) from the reference genome because of small indel polymorphisms. Missing genotype calls were imputed using the FastImputationBitFixedWindow plugin in TASSEL 4.0 (http://sourceforge.net/projects/tassel/).
Pigmentation phenotypes
Tannin phenotypes for the small association panel (n = 142) were previously published (Wu et al. 2012). This panel represents a subset of early-maturing, semi-dwarf accessions from the U.S. Sorghum Association Panel (Casa et al. 2008). For the large association panel, seeds for the full Sorghum Association Panel were obtained from the U.S. National Plant Germplasm System via the Germplasm Resource Information Network (http://www.ars-grin.gov). The presence or absence of a pigmented testa on the grains was visually assessed (three seeds per accession) after removal of the pericarp on the dorsal side and scored as 1 and 0, respectively (File S2). Eleven accessions that showed a segregating phenotype were dropped from the analysis. Pericarp pigmentation was visually assessed for three seeds per accession, scored as 0 for white and 1 for red or yellow (File S2). Brown pericarp accessions were dropped from the analysis because this phenotype is known to be caused by the spread of tannin from the testa and masks the expression of the R and Y genes (Doggett 1988). For analysis of structuring of tannin phenotypes, we obtained testa pigmentation data (n = 14,785) in world sorghum collections from the Germplasm Resource Information Network. For the RIL family, phenotyping was performed in the 2007 and 2008 kharif (rainy) season in Patancheru, India, with presence/absence of pigmentation scored for 10-wk-old seedlings (coleoptile color) or physiologically mature plants (testa and adult plant color; Vinayan 2010).
Genomic analysis
Genome-wide mapping in RILs and the association panels was carried out using a GLM, mixed linear model (MLM), or compressed mixed linear model (CMLM) with population parameters previously determined (Zhang et al. 2010) as implemented in the Genomic Association and Prediction Integrated Tool (Lipka et al. 2012). When a population structure (Q) term was included, the model selection feature of the Genomic Association and Prediction Integrated Tool was used to determine the optimal number of principal components (Zhu and Yu 2009). Genetic structure of the world sorghum populations was estimated by the use of principal components analysis implemented by cmdscale in R (R Core Team 2012).
Loss-of-function genome scan P-value for a given SNP (loss of function, i.e., LOF) is calculated with binomial tests in R (R Core Team 2012) as follows:
where L1 and L2 are the counts of the loss-of-function phenotype for alleles 1 and 2 (“successes”), respectively, W1 and W2 are the counts of the wild-type phenotype for alleles 1 and 2 (“failures”), respectively, and P is the overall proportion of loss-of-function phenotypes given by P = (L1+ L2)/(L1+ L2+ W1+ W2).
Results
Grain tannin GWAS in a small association panel
To identify loci underlying natural variation in grain tannin pigmentation, we first characterized associations between published tannin (presence/absence) phenotypes from a small (n = 142) global diversity panel (Wu et al. 2012) and genotypes from a 265,487 SNP genotyping-by-sequencing data set (Morris et al. 2013). Included in the GBS SNP map is a G-to-T transition in the Tannin1 coding region (S4_61667908) that is 218 bp upstream of, and in perfect linkage disequilibrium with, the G-deletion that is causative for the tan1-a null allele (n = 161) (Wu et al. 2012). Therefore, we can use this SNP (hereafter referred to as the tan1-a SNP) as a positive control to compare mapping approaches. Using a simple model without statistical control for population structure (General Linear Model; GLM) we find that, as expected, the most significant association signals lies at ~61 Mb on chromosome 4 colocalized with the Tannin1 locus (Figure S1A-B and Table 1). However, the tan1-a SNP is not the most significant association in this region (rank = 230; P < 10−8). Instead, the most significant association peaks are found in the ~1-Mb region surrounding Tannin1, at 61.1 Mb (S4_61121403; P < 10−12), 61.2 Mb (S4_61233495; P < 10−10), and 61.8 Mb (S4_61862778; P < 10−12). Including the Tannin1 locus, we observe at least 13 peaks of association that are significant at a nominal Bonferroni-corrected P-value of 0.01 (P < 2 × 10−7), among which are additional candidate loci for genetic control of tannin pigmentation (File S3).
Table 1. Summary of testa pigmentation GWAS.
| SNP (Relevance) | S4_61667908 (tan1-a allele) | S4_61862778 (tags Tan1, -0.2 Mb) | S4_61121403 (tags Tan1, -0.5 Mb) | S4_62353772 (tags Tan1, +0.7 Mb) |
|---|---|---|---|---|
| Small association panel (n = 142) | ||||
| GLM | 230 (3 × 10−7) | 1 (4 × 10−11) | 2 (5 × 10−11) | 38 (8 × 10−8) |
| CMLM (K) | 82 (2 × 10−5) | 3 (6 × 10-8) | 1 (2 × 10−8) | 159,802 (0.76) |
| CMLM (Q + K) | 82 (2 × 10−5) | 3 (6 × 10−8) | 1 (2 × 10−8) | 159,802 (0.76) |
| MLM (K) | 176 (1 × 10−6) | 2 (7 × 10−10) | 1 (7 × 10−10) | 12 (8 × 10−9) |
| Large association panel (n = 336) | ||||
| GLM | 9 (1 × 10−14) | 3 (2 × 10−17) | 14 (4 × 10−15) | 1 (2 × 10−22) |
| GLM (Q) | 1 (9 × 10−15) | 3 (5 × 10−12) | 5 (2 × 10−10) | 14 (2 × 10−8) |
| CMLM (K) | 69,491 (0.76) | 77,062 (0.86) | 1 (1 × 10−11) | 61,782 (0.64) |
| CMLM (Q + K) | 14355 (0.09) | 39,506 (0.33) | 1 (9 × 10−11) | 3381 (0.01) |
| MLM (K) | 1 (2 × 10−13) | 115,088 (0.83) | 2 (1 × 10−11) | 87,702 (0.57) |
| Loss-of-function | 1 (3 × 10−19) | 4 (1 × 10−13) | 22 (1 × 10−9) | 26 (2 × 10−9) |
Mapping results for various GWAS models giving the rank of the given SNP of 265,487 SNPs (P values in parenthesis). GWAS, genome-wide association studies; GLM, general linear model; CMLM, compressed mixed linear model; MLM, mixed linear model; SNP, single-nucleotide polymorphism.
Given the extensive population structure in sorghum, as well as kinship among elite sorghum lines, we also carried out GWAS using MLMs that take into account population structure (Q) and/or kinship (K) (Table 1). We fit both standard MLMs (Yu et al. 2006), which treat each individual as an independent group when fitting the random effect of kinship, and CMLMs, which have been shown to outperform GLMs and standard MLMs in some cases (Zhang et al. 2010). In addition, we estimated the optimal number of principal components to include as fixed effects (Q) on the basis of model selection and Bayesian information criteria (Zhu and Yu 2009). The genomic locations of GWAS peaks from the GLM are largely unchanged with the CMLM (Figure S1C-D and Table 1), although the nominal significance of the peaks is reduced. All the nominally significant association peaks identified with the GLM remain in the CMLM, although most fall below the threshold for genome-wide significance. The top associations remain near Tannin1 on chromosome 4 but controlling for population structure and kinship with a MLM (MLM and CMLM) does not improve the mapping resolution at the Tannin1 locus, as the tan1-a SNP remains less significant (P < 10−4) than several other SNPs in the ~1 Mb region surrounding Tannin1 (P < 10−6).
Grain tannin GWAS in a large association panel
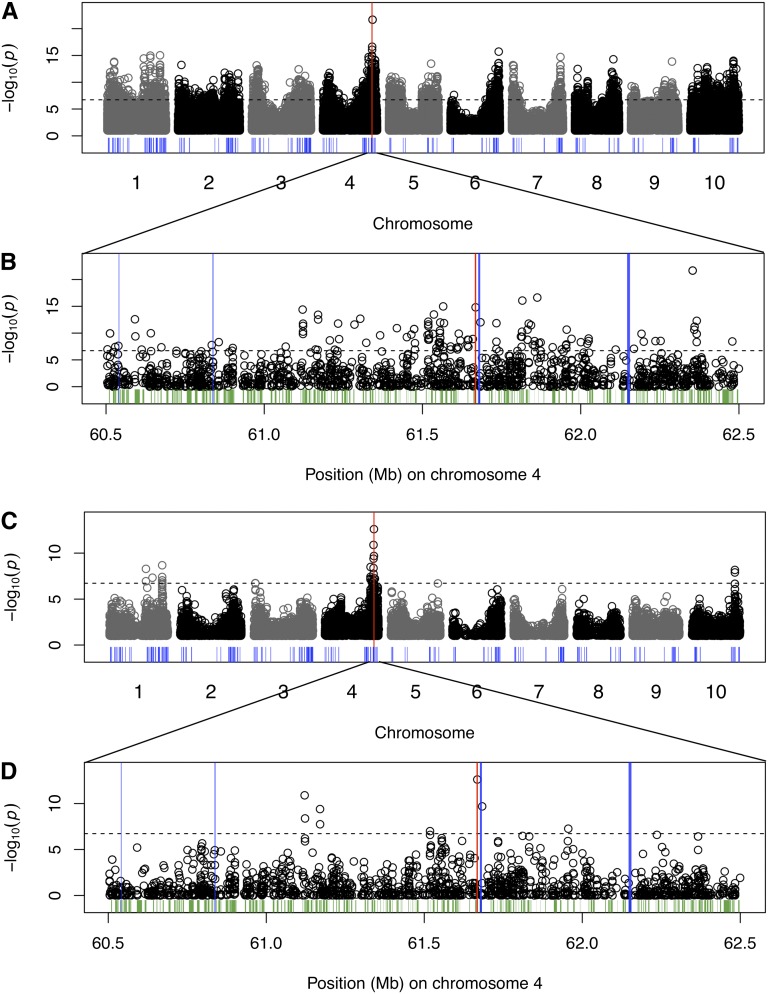
If the inability to precisely map Tannin1 in the small association panel is caused by a small sample size, then increasing the size of the association panel should improve the mapping resolution. Therefore, we phenotyped testa presence/absence in the sorghum association panel (Casa et al. 2008) and performed GWAS by using a variety of linear models (n = 336; Table 1). Fitting a GLM with this larger panel, the association for the tan1-a SNP is increased in rank and significance (Figure 1A-B; rank = 9; P < 10−14), but is still not the top association. Adding control for population structure and kinship using CMLM (K or Q+K), we find the rank and significance of the tan1-a SNP is actually reduced compared with the results using a GLM (Table 1; rank = 14355; P = 0.09). Finally, a GLM with population term (Q) or a standard MLM (K only; Figure 1C-D) that treats each individual as a separate group for estimating kinship does identify the tan1-a SNP precisely (rank = 1; P < 10−15 and P < 10−12, respectively).
Figure 1.
Genome-wide association study of testa presence using linear models. Manhattan plots using a GLM (A, B) and standard MLM kinship (K) (C, D) with 265,487 SNPs and 336 accessions, with (B, D) detailed view at Tannin1 locus on chromosome 4 for GLM and MLM (K), respectively. Although the Tannin1 locus is identified as the major effect locus for tannin presence with GLM, the Tannin1 gene (red bar) is precisely identified with a MLM. Other flavonoid-related genes are indicated by the blue bars, whereas all other annotated genes in the detailed view are indicated in green.
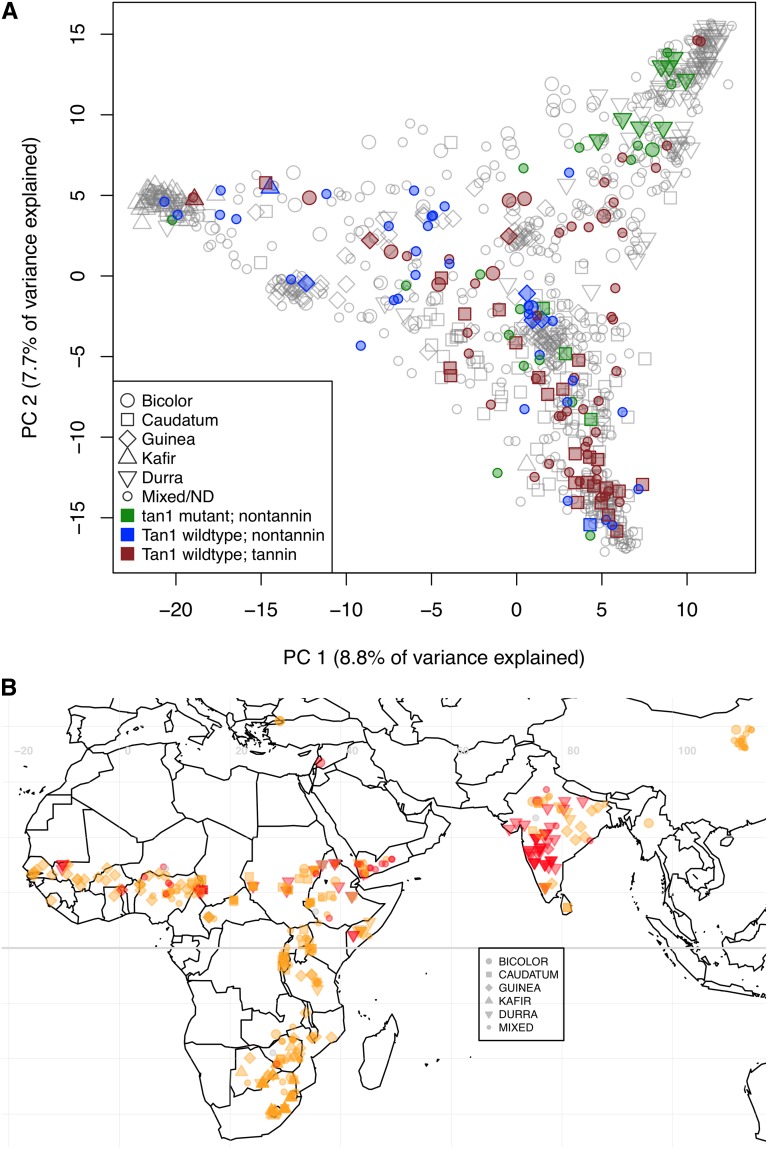
Why do some models that account for population structure (CMLM) perform worse than a naive model (GLM), generating a false-negative result for the tan1-a SNP? To better understand the population structure of natural variation in tannins, we characterized the distribution of pigmented testa phenotype in worldwide sorghum collections. The tannin trait segregates in all the botanical races of sorghum but shows modest population structure, with durra and guinea types having the lowest proportion of tannin accessions (15%) and caudatum and guinea-caudatum accessions having the greatest proportion (76% and 83%, respectively). However, with respect to model selection in GWAS, the structuring of the trait itself may be less important than structuring of the alleles underlying the trait. The tan1-a allele is found at high frequency in African and Indian durra accessions and at low frequency in Chinese and southern African accessions (Figure 2). The tan1-a allele explains the tannin phenotypes in all durra-derived accessions studied, but only partially accounts for the phenotype in caudatum accessions and not at all in guinea accessions. Among caudatum, guinea, and kafir types there are numerous accessions that have wild-type Tannin1 coding regions yet have nontannin phenotypes, which suggests that the population structuring of heterogeneous alleles may account for the overcorrection by the CMLM and the effective correction by the MLM.
Figure 2.
Distribution of tannin phenotype and loss-of-function alleles in a worldwide sorghum diversity panel. (A) Accessions plotted according to the first two principal components of sorghum population structure (small panel; n = 142), with phenotyped accessions color-coded by Tannin1 allele and tannin phenotype (nonphenotyped accessions in gray). Although some nontannin accessions are explained by tan1-a or tan1-b (green), others must be due to additional loss-of-function alleles (blue). (ND: Not Determined) (B) Distribution of wild-type allele (G; orange) and loss-of-function allele (T; red) for the tan1-a SNP (S4_61667908) in source-identified sorghum accessions.
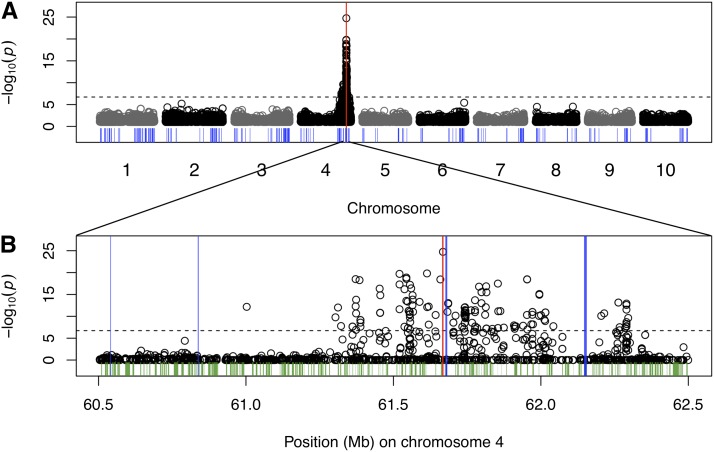
Mapping of flavonoid traits in a RIL family
Given that complex association signals made the precise mapping of tan1-a in an association population difficult, we considered whether the reduced genetic and allelic heterogeneity of a biparental family would allow precise mapping of Tannin1 using linear models. We phenotyped testa pigmentation (tannin presence) in a population of 263 RILs that we genotyped at 265,487 SNPs. There is a single locus associated with the pigmented testa in this family that is precisely colocalized with Tannin1 on chromosome 4, with the most significant SNP being the tan1-a SNP (Figure 3). To determine whether this gene resolution mapping in RILs is likely to be a typical result, we also mapped two other flavonoid pigmentation traits (coleoptile color and adult plant color) that are segregating in this RIL family. We mapped coleoptile color to a region around 54 Mb on chromosome 6 (Figure S2). This peak colocalizes with the classical Rs1 locus and a priori candidate gene Sb06g025060, a putative basic helix-loop-helix (bHLH) transcription factor, and a sorghum co-ortholog of Arabidopsis TRANSPARENT TESTA8 and maize B1/R1 anthocyanin regulators (File S3). However, the most significant SNP (S6_53849573) is 220 kb upstream of Sb06g025060, and no promising a posteriori candidate genes are found at S6_53849573, suggesting gene resolution was not achieved in this case. Adult plant color maps to 58 Mb on chromosome 6, colocalized with the classical P locus (Doggett 1988; Mace and Jordan 2010) and a large cluster of putative reductase genes that are homologous to Arabidopsis TRANSPARENT TESTA3 and BANYULS and maize ANTHOCYANINLESS1 (File S1). Here again, the most significant SNP (S6_57865283) is not colocalized precisely with the a priori candidates (TT3/BANYULS cluster) but 260 kb upstream (Figure S3).
Figure 3.
Genome-wide mapping of testa presence in a RIL biparental family. Manhattan plots for a linear model using 263 recombinant inbred lines genotyped at 265,487 SNPs, (A) scanning genome-wide and (B) with a detailed view at Tannin1 locus on chromosome 4, with Tannin1 indicated by the red bar. Other flavonoid-related genes are indicated by the blue bars, whereas all other annotated genes in the detailed view are indicated in green.
Loss-of-function genome scan on grain tannin
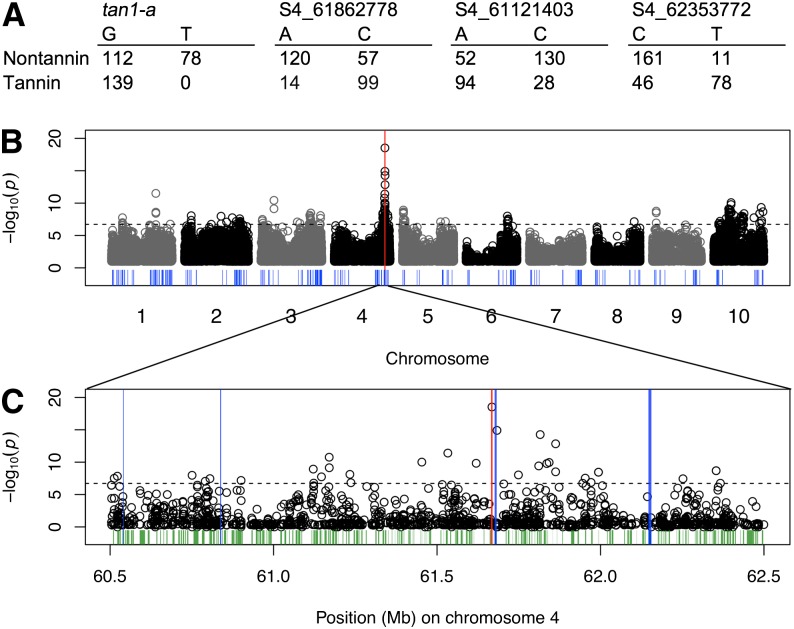
Why did several of the linear models we tested fail to precisely identify the tan1-a allele in association panels, even though it is common and highly penetrant? A comparison of the 2 × 2 contingency tables for the tan1-a SNP vs. the more significant SNPs provides some insight here (Figure 4A). The loss-of-function tan1-a allele shows striking covariation with the testa phenotype (T allele: nontannin = 78 vs. tannin = 0) but little signal of covariation for the wild-type allele (G allele: nontannin = 112 vs. tannin = 139). Although this lack of covariation for the wild-type allele is to be expected (because there is no reason that accessions carrying the wild-type allele at Tannin1 cannot carry loss-of-function alleles at other loci), it reduces the significance of a linear model fitting the genotype-phenotype association. In contrast, the other more significant SNPs near Tannin1 show covariation for both alleles (in opposite directions), with the wild-type allele more often found with the wild-type phenotype (Figure 4A). This pattern of covariation increases the significance of the fit of a linear model or contingency test, even though the genotype-phenotype covariation for the wild-type allele is irrelevant when considering a loss-of-function polymorphism.
Figure 4.
Genome-wide mapping of testa presence using a loss-of-function genome scan. (A) Allele counts for the tan1-a SNP and other SNPs near Tannin1 that are associated with a nontannin phenotype. The nominal P-value is from a binomial test on the putative loss-of-function allele at each SNP, (B) scanning genome-wide, and (C) a detailed view of the Tannin1 locus on chromosome 4, with Tannin1 indicated by the red bar. Other flavonoid-related genes are indicated by the blue bars, whereas all other annotated genes in the detailed view are indicated in green.
To investigate an approach that may be appropriate for loss-of-function alleles we used a simple heuristic genome scan based on a binomial test (see the section Materials and Methods). We identified SNPs with alleles that are often found in individuals with the loss-of-function phenotype (testa absent) and rarely or never found in individuals with the wild-type phenotype (testa present). In effect, the phenotypes at the putative wild-type allele at each SNP is ignored. With this loss-of-function genome scan approach, the tan1-a SNP is precisely identified whereas other SNPs near the Tannin1 locus that had strong indirect associations with linear modeling have reduced association signals (Figure 4, B and C, Table 1, and Figure S4). Thus, in this case we find that a simple heuristic scan that considers the underlying genetics of the trait outperforms more sophisticated models.
Pericarp pigmentation GWAS
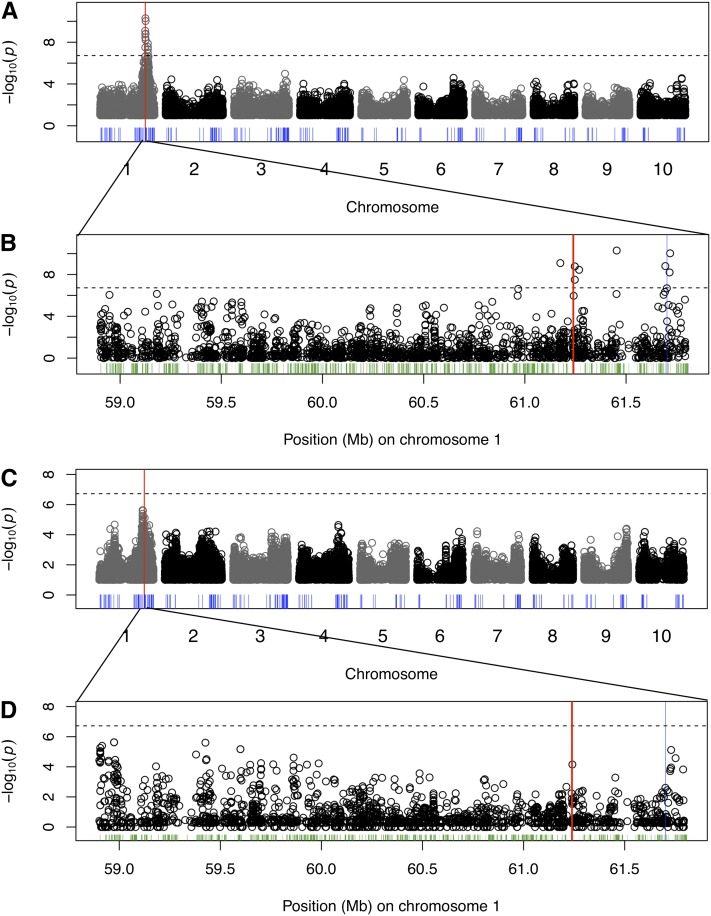
Although we found that gene-resolution mapping was possible with Tannin1, we wondered whether this would be true for other flavonoid pigmentation traits. To assess the mapping resolution in this panel with another trait, we used the white pericarp phenotype, which reflects a lack of flavonoid pigmentation in the outer seed coat (Doggett 1988; Ibraheem et al. 2010). It is known that white pericarp phenotype can be caused by loss-of-function mutations in the Yellow seed1 gene (Y1, Chr1: 61,237,360–61,241,520; Ibraheem et al. 2010). Note, the Y1 gene was cloned based on the excision of a transposable element, not genetic analysis of natural variation, so we do not have a validated genetic variant of Y1 in the SNP data as we did for the Tannin1 case. However, it is known from the classical inheritance literature that natural variation at the Y locus exists (Rooney 2000). As expected, the most significant associations were found around 61 Mb on chromosome 1 (Figure 5). As was the case for Tannin1, though, the Y1 gene was broadly but not precisely identified: among the top associations are SNPs that are flanking Y1 (5 kb from Y1, S1_61246791; P < 10−8) but the most significant association are ~0.5 Mb from Y1 (S1_61717652). Because white pericarp represents loss-of-function phenotype, we also used the loss-of-function genome scan approach for the pericarp pigmentation trait. In this case, the loss-of-function genome scan does not improve the mapping resolution as compared to the linear models.
Figure 5.
GWAS of white pericarp phenotype. Manhattan plots for GWAS using CMLM kinship (K) (A, B) and a loss-of-function genome scan (C, D) and with 265,000 SNPs and 347 accessions, showing a (A, C) genome-wide and (B, D) a detailed view around Y1 on chromosome 1, with the Y1 gene indicated by the red bar. Other flavonoid-related genes are indicated by the blue bars, whereas all other annotated genes in the detailed view are indicated in green. ×
Discussion
Genetics of flavonoid pigmentation
GWAS have been useful to characterize the contribution of known and novel flavonoid pigmentation genes in several plant species (Atwell et al. 2010; Huang et al. 2010; Cockram et al. 2010). Given that control of flavonoids by a WD40-bHLH-MYB regulatory system is broadly conserved (Petroni and Tonelli 2011), it seems likely that these regulators underlie some natural variation in sorghum pigmentation. Is there evidence of natural variation in each of the three types of transcription factors that control tannins in plants (WD40, bHLH, MYB)? From classical inheritance studies, it is known that at least two loci control the presence of brown coloration in grain subcoat (B1 and B2) that have been mapped to chromosome 2 and chromosome 4, and one of these was cloned as WD40 gene Tannin1 (Rami et al. 1998; Mace and Jordan 2010; Wu et al. 2012). On the basis of a comparison of B1/B2 genotypes from classical inheritance studies (Doggett 1988; Rooney 2000) and Tannin1 genotypes (Wu et al. 2012), it can be inferred that Tannin1 corresponds to B2. Multiple studies have identified a second major effect locus controlling tannin presence at around 8 Mb on chromosome 2, and from the Tannin1-controlled GWAS (File S3), there is evidence that the gene underlying B1 is the putative bHLH transcription factor Sb02g006390. This study is based on qualitative tannin phenotypes (presence/absence) so quantitative phenotyping may reveal additional loci underlying the observed variation among intermediate and high tannin varieties (Gu et al. 2004).
Given that flavonoid-related gene families are well-characterized and well-conserved across distant plant lineages, we would expect that most association peaks would colocalize to a priori candidate genes. Although a number of peaks do colocalize precisely with candidate genes, many more of them do not, despite the large candidate gene set and the liberal inclusion criteria. It is possible that some unexpected signals represent true associations at novel genes, though it is unlikely for most peaks. In some cases, these unexpected signals may represent stochastic noise, spurious associations that do not reflect any underlying genetic heritability. However, most of these peaks increased in significance with a larger sample size so they are unlikely to represent stochastic noise and more likely to represent indirect associations (Platt et al. 2010). In particular, the significant SNPs near Tannin1 are likely to represent synthetic associations (Goldstein 2011), instances in which a phenotype caused by multiple rare alleles is spuriously assigned to a common allele that is linked.
Mapping loss-of-function alleles
The development of experimental design strategies and statistical methods that account for complex genetic architecture and population structure in genome-wide mapping studies is an active area of research (Zhang et al. 2010; Zhao et al. 2011; Huang et al. 2012; Lipka et al. 2012; Segura et al. 2012). Here we were able to compare several mapping approaches empirically by using the testa presence/absence phenotype in sorghum and a GBS SNP map that includes a validated SNP in the Tannin1 major effect gene. All the approaches we tested here identified the Tannin1 locus, broadly defined (+/− 1 Mb of the Tannin1 gene), as the major locus underlying testa presence/absence, but the precise identification of the Tannin1 gene was not consistently obtained. Increasing the panel size increased the significance of the tan1-a SNP relative to the indirect associations. Given that minor allele frequencies (MAFs) of indirect associations (i.e., the SNPs near Tannin1 that have greater association signals than the tan1-a SNP) are greater (MAF = 0.3 – 0.45) than the tan1-a SNP (MAF = 0.2), these indirect associations fit the expectation for synthetic associations: older, more widespread SNPs that tag a haplotype block on which multiple loss-of-function alleles of Tannin1 have arisen (Dickson et al. 2010; Orozco et al. 2010). Although methods have been developed to account for epistasis by stepwise model fitting (Segura et al. 2012), it will be difficult to derive benefits from these models if the most significant association in the first model step is itself an indirect association. Given that allelic heterogeneity is abundant among the handful of well-characterized sorghum genes [Tannin1 (Wu et al. 2012), Maturity1 (Murphy et al. 2011), and Shattering1 (Lin et al. 2012)] synthetic associations could be a common feature of major effect loci in sorghum and other species of similar population structure.
Loss-of-function variants are a common source of natural variation, and recently methods have been developed to identify multiple, low-frequency loss-of-function alleles at the same gene in GWAS (Liu et al. 2011). The loss-of-function genome scan approach we used here allowed us to precisely map a high frequency loss-of-function allele to the exclusion of nearby indirect associations. Note, that although we applied this approach to binary phenotypes the principle does not depend on binary phenotypes. Rather, it depends on the epistasis that exists because the loss-of-function allele cannot be rescued (i.e., wild-type phenotype is never found with loss-of-function allele) while the wild-type allele can easily be found in an individual that harbors an independent loss-of-function allele at a different locus (i.e., loss-of-function phenotype is often found with wild-type allele).
Improving resolution with genomic data
We found that mapping in a biparental family using high-density SNP markers can achieve gene resolution. Traditionally, high-resolution SNP maps have not been used for biparental families due to technical limitations and the expectation that the small number of recombination events limits the utility of greater marker density. The gene-resolution mapping of Tannin1 in a modest-sized (n = 263) RIL family with simple phenotyping (field-based scoring of testa presence/absence) demonstrates that the combination of advanced mapping populations with high-resolution genotyping is an effective strategy for trait dissection (Bergelson and Roux 2010; Brachi et al. 2011). Still, the lower-resolution mapping results for the adult plant color and coleoptile color traits in the RILs suggest that consistent gene-level mapping will require larger families and/or advanced multi-parent mapping approaches (Jordan et al. 2011). Given the substantial investment that has already been made in RIL development, the cost-effectiveness of GBS, and the suitability of the system for “genotype once, phenotype many times” approach, a broader effort to make high-density genotypes available for existing advanced mapping populations seems warranted.
It is worth noting that if the tan1-a SNP had not, by chance, been represented in the GBS SNP map, the results of the loss-of-function genome scan would have been qualitatively equivalent to the linear models. This highlights that to derive a benefit from the loss-of-function genome scan approach over linear models the density of genotyping must be high enough that causative SNPs, or SNPs in perfect LD with the causative polymorphism (e.g., tan1-a), are represented in the genotyping data. A lack of a causative or perfectly linked SNP for pericarp pigmentation would explain why both the linear model and loss-of-function genome scan approaches achieved just Mb resolution for this trait. Similarly in the RIL mapping, had the tan1-a SNP not been represented the other SNPs in the data set would not have precisely tagged the Tannin1 gene. Although the identification of a priori candidates through comparative genomics provides some guidance, there are typically many reasonable candidates within Mb-scale mapping intervals; for example, around 60−63 Mb on chromosome 4, there are four other homologs of tannin genes that would have been equally promising candidates. As genomic coverage increases, imputation methods improve, and mapping populations are refined and expanded, genome-wide mapping approaches will increasingly need to be optimized to identify causative variants as opposed to tagging SNPs (Huang et al. 2012). Given the ever-lowering costs of sequencing vs. the high cost of candidate gene validation efforts, the use of whole-genome resequencing to increase the resolution of mapping studies is likely to be cost effective. Although this study of relatively simple pigmentation traits highlights the trade-offs between population- and family-based mapping approaches because of the complexity of association signals, in either approach high density genotyping can facilitate gene resolution mapping of traits.
Supplementary Material
Acknowledgments
We thank National Science Foundation for providing funds (ID: IOS-0965342) to carry out this research under Basic Research to Enable Agricultural Development project. This work was also supported by a grant from United States Department of Agriculture-National Institute of Food and Agriculture Plant Feedstock Genomics for Bioenergy Program (#2011-03502) to S. K. This work has been undertaken as part of the CGIAR Research Program on Dryland Cereals. We thank the editors and two anonymous reviewers for helpful suggestions.
Footnotes
Communicating editor: D.-J. De Koning
Literature Cited
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beló A., Zheng P., Luck S., Shen B., Meyer D., et al. , 2008. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol. Genet. Genomics 279: 1–10 [DOI] [PubMed] [Google Scholar]
- Bergelson J., Roux F., 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11: 867–879 [DOI] [PubMed] [Google Scholar]
- Brachi B., Morris G. P., Borevitz J. O., 2011. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Casa A. M., Pressoir G., Brown P. J., Mitchell S. E., Rooney W. L., et al. , 2008. Community resources and strategies for association mapping in sorghum. Crop Sci. 48: 30–40 [Google Scholar]
- Cockram J., White J., Zuluaga D. L., Smith D., Comadran J., et al. , 2010. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc. Natl. Acad. Sci. USA 107: 21611–21616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C., 1859. On the Origin of the Species by Means of Natural Selection: Or, The Preservation of Favoured Races in the Struggle for Life. John Murray, London: [PMC free article] [PubMed] [Google Scholar]
- Dickson S. P., Wang K., Krantz I., Hakonarson H., Goldstein D. B., 2010. Rare variants create synthetic genome-wide associations. PLoS Biol. 8: e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett H., 1988. Sorghum. Longman Scientific & Technical, Essex, UK [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esele J. P., Frederiksen R. A., Miller F. R., 1993. The association of genes controlling caryopsis traits with grain mold resistance in sorghum. Phytopathology 83: 490–495 [Google Scholar]
- Flint J., Eskin E., 2012. Genome-wide association studies in mice. Nat. Rev. Genet. 13: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. B., 2011. The importance of synthetic associations will only be resolved empirically. PLoS Biol. 9: e1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., et al. , 2011. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Olsen K. M., 2010. Genetic perspectives on crop domestication. Trends Plant Sci. 15: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Kelm M. A., Hammerstone J. F., Beecher G., Holden J., et al. , 2004. Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J. Nutr. 134: 613–617 [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., et al. , 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Wei X., Li C., Wang A., et al. , 2012. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44: 32–39 [DOI] [PubMed] [Google Scholar]
- Ibraheem F., Gaffoor I., Chopra S., 2010. Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor. Genetics 184: 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan D. R., Mace E. S., Cruickshank A. W., Hunt C. H., Henzell R. G., 2011. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 51: 1444–1457 [Google Scholar]
- Kaufman R. C., Herald T. J., Bean S. R., Wilson J. D., Tuinstra M. R., 2013. Variability in tannin content, chemistry and activity in a diverse group of tannin containing sorghum cultivars. J. Sci. Food Agric. 93: 1233–1241 [DOI] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., et al. , 2011. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Li X., Shannon L. M., Yeh C.-T., Wang M. L., et al. , 2012. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., et al. , 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399 [DOI] [PubMed] [Google Scholar]
- Liu F., Struchalin M. V., van Duijn K., Hofman A., Uitterlinden A. G., et al. , 2011. Detecting low frequent loss-of-function alleles in genome wide association studies with red hair color as example. PLoS ONE 6: e28145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace E. S., Jordan D. R., 2010. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 121: 1339–1356 [DOI] [PubMed] [Google Scholar]
- McClintock B., 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36: 344–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes É. A., Natal D. I. G., Queiroz V. A. V., Schaffert R. E., Cecon P. R., et al. , 2012. Sorghum genotype may reduce low-grade inflammatory response and oxidative stress and maintains jejunum morphology of rats fed a hyperlipidic diet. Food Res. Int. 49: 553–559 [Google Scholar]
- Morris G. P., Ramu P., Deshpande S. P., Hash C. T., Shah T., et al. , 2013. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 110: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. L., Klein R. R., Morishige D. T., Brady J. A., Rooney W. L., et al. , 2011. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson-Ehle H., 1909. Kreuzunguntersuchungen an Häfer und Weizen. Lunds Universitets Arsskrift 5: 1–122 [Google Scholar]
- Orozco G., Barrett J. C., Zeggini E., 2010. Synthetic associations in the context of genome-wide association scan signals. Hum. Mol. Genet. 19: R137–R144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Petroni K., Tonelli C., 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181: 219–229 [DOI] [PubMed] [Google Scholar]
- Platt A., Vilhjálmsson B. J., Nordborg M., 2010. Conditions under which genome-wide association studies will be positively misleading. Genetics 186: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2012. R: A Language and Environment for Statistical Computing. R Core Team, Vienna, Austria [Google Scholar]
- Rami J.-F., Dufour P., Trouche G., Fliedel G., Mestres C., et al. , 1998. Quantitative trait loci for grain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor L. Moench). TAG Theoret. Appl. Genet .97: 605–616 [Google Scholar]
- Rooney W. L., 2000. Genetics and cytogenetics, pp. 261–307 in Sorghum: Origin, History, Technology, and Production, edited by Smith C. W., Frederiksen R. A. John Wiley and Sons, New York [Google Scholar]
- Rosenberg N. A., Huang L., Jewett E. M., Szpiech Z. A., Jankovic I., et al. , 2010. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K., 1923. The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8: 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J. C., Freeling M., 2011. Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V., Vilhjálmsson B. J., Platt A., Korte A., Seren Ü., et al. , 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens J. C., 1946. A second factor for subcoat in sorghum seed. Agron. J. 38: 340–342 [Google Scholar]
- Tian Z., Qian Q., Liu Q., Yan M., Liu X., et al. , 2009. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 106: 21760–21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinall H. N., Cron A. B., 1921. Improvement of sorghums by hybridization. J. Hered. 12: 435–443 [Google Scholar]
- Vinayan, M. T., 2010 Genetic Architecture of Spotted Stem Borer Resistance in Sorghum as Inferred From QTL Mapping and Synteny with the Maize Genome, Ph.D. Thesis, Tamil Nadu Agricultural University, Tamil Nadu, India. [Google Scholar]
- Visscher P. M., Brown M. A., McCarthy M. I., Yang J., 2012. Five years of GWAS discovery. Am. J. Hum. Genet. 90: 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B., 2001. Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li X., Xiang W., Zhu C., Lin Z., et al. , 2012. Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proc. Natl. Acad. Sci. USA. 109: 10281–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Pang Y., Dixon R. A., 2010. The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 153: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C.-W., Eizenga G. C., Wright M. H., Ali M. L., et al. , 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Yu J., 2009. Nonmetric multidimensional scaling corrects for population structure in association mapping with different sample types. Genetics 182: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.