Abstract
The plant hormone auxin plays a key role in the coordination of many aspects of growth and development. AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) genes encode instable primary auxin responsive regulators of plant development that display a protein structure with four characteristic domains. In the present study, a comprehensive analysis of the 34 members of the maize Aux/IAA gene family was performed. Phylogenetic reconstructions revealed two classes of Aux/IAA proteins that can be distinguished by alterations in their domain III. Seven pairs of paralogous maize Aux/IAA proteins were discovered. Comprehensive root-type and tissue-specific expression profiling revealed unique expression patterns of the diverse members of the gene family. Remarkably, five of seven pairs of paralogous genes displayed highly correlated expression patterns in roots. All but one (ZmIAA23) tested maize Aux/IAA genes were auxin inducible, displaying two types of auxin induction within three hours of treatment. Moreover, 51 of 55 (93%) differential Aux/IAA expression patterns between different root-types followed the expression tendency: crown roots > seminal roots > primary roots > lateral roots. This pattern might imply root-type-specific regulation of Aux/IAA transcript abundance. In summary, the detailed analysis of the maize Aux/IAA gene family provides novel insights in the evolution and developmental regulation and thus the function of these genes in different root-types and tissues.
Introduction
The phytohormone auxin plays an essential role in plant growth and development. Auxin controls many aspects of plant morphology and physiology [1,2] such as apical dominance, tropisms and the differentiation of vascular tissues [3]. Moreover, auxin affects division, elongation, and differentiation of cells [3,4]. On the molecular level, auxin controls gene expression [5,6] and membrane functions [7]. Several auxin-responsive genes have been identified and characterized including the GRETCHEN HAGEN 3 (GH3), SMALL AUXIN-UP RNA (SAUR), and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene families [8]. In etiolated tissue of soybean (Glycine max), Aux/IAA genes were initially identified because of their fast induction by auxin [9]. Subsequently, 29 members of the Arabidopsis thaliana Aux/IAA gene family [10], 26 members of the tomato and sorghum Aux/IAA families [11-13], and 31 Aux/IAA genes were identified in rice [14] and maize [15], respectively. Aux/IAA genes are unique to plants and have not been identified in bacteria, animals or fungi [16]. In Arabidopsis thaliana, not all Aux/IAA genes are inducible by auxin. For instance, AtIAA17 and AtIAA28 display only a minor or no response to exogenous auxin-treatment [17-19]. Members of the Aux/IAA gene family encode short-lived nuclear proteins which consist of four characteristic domains [2,20,21]. Aux/IAA proteins function as transcriptional repressors of downstream auxin-regulated genes [22,23] via a short conserved leucine repeat motif (LxLxLx) in domain I. Domain II, with the conserved degron-sequence GWPPV, is responsible for the stability of Aux/IAA proteins [23]. The interaction of domain II with the F-box protein TIR1 leads to a rapid degradation of Aux/IAA proteins [24]. Point mutations in the degron sequence or deletions of this sequence [25] stabilize Aux/IAA proteins which can result in specific developmental phenotypes [10,16]. Domains III and IV of Aux/IAA proteins homo and heterodimerize with other Aux/IAA proteins or auxin response factors (ARFs) [26,27]. Moreover, interaction of these domains with domain III and IV of ARFs control expression of downstream auxin responsive genes [26,27]. ARF proteins interact with auxin-responsive cis-elements (AuxRE) in the promoter of downstream auxin responsive genes [27-29]. Finally, Aux/IAA proteins contain a nuclear localization signal (NLS) which targets these proteins to the nucleus. Typically, Aux/IAA proteins contain two NLS, one is separated into two parts including the short sequence KR between domain I and II, and a six amino acid sequence in domain II. The second NLS is located at the end of domain IV at the carboxy-terminus [30].
To date only one member of the maize Aux/IAA family has been characterized in detail. RUM1/ZmIAA10 displays characteristics of a canonical Aux/IAA protein including nuclear localization, short half life time and the possibility to interact with ARF proteins [25]. Deletion of 26 amino acids including the degron sequence in the mutant rum1-R resulted in a root-specific phenotype blocking the initiation of embryonic seminal and postembryonic lateral roots in the primary root [31].
In the present study, phylogenetic and syntenic relations of the Aux/IAA gene family members in maize were determined and a comprehensive expression and correlation analysis of different root and shoot tissues of all maize Aux/IAA genes during development was performed, unveiling root-type and tissue-specific expression patterns that might help to understand the diverse functions of these genes in root development.
Materials and Methods
Plant material, growth conditions, and hormone treatment
Seeds of the maize inbred line B73 were sterilized with 6% sodium hypochlorite for 10 min and rinsed in distilled water. Subsequently, seeds were rolled up in germination paper (Anchor paper, www.anchorpaper.com) [32] and transferred to 10 l buckets filled with ~3 l distilled water. Germinating seedlings were incubated at 28 °C with a 16 h light and 8 h dark cycle. Five-day-old maize seedlings were treated with 5 mM α-naphthyl acetic acid (αNAA) working solution for 3 h. The differentiation zone of two to three primary roots per biological replicate was harvested each hour. Coleoptiles were harvested from seedlings grown for four days at 28 °C in the dark. Seedling samples were harvested at different developmental stages and were immediately frozen in liquid nitrogen and stored at -80 °C until RNA isolation.
The Aux/IAA gene family, novel members, phylogeny, and synteny
In the initial version of the maize reference genome sequence B73 RefGen_v1 [33] 31 Aux/IAA genes were predicted among 32,540 protein-encoding genes [15]. To obtain a comprehensive overview of the maize Aux/IAA gene family, the 31 previously identified maize Aux/IAA genes were used as query sequences for blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches of the maize filtered gene set based on genome assembly version AGPv2 (http://www.maizegdb.org/), containing 39,656 high confidence genes.
The protein sequences of the maize Aux/IAA genes were retrieved from MaizeGDB (http://maizegdb.org/). Moreover, the previously identified rice Aux/IAA protein sequences [15] were retrieved from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/index.shtml), and the published sorghum sequences [34] were extracted from Gramene (http://www.gramene.org/). The four conserved domains in the maize Aux/IAA gene family were determined by multiple alignments with ClustalW (http://www.clustal.org/). Synteny of the maize sequences was determined with Comparative Genomics software (CoGe, http://genomevolution.org/CoGe/; [35]) and association with maize subgenomes 1 and 2 were based on [36].
Phylogenetic analyses comparing maize, rice, and sorghum Aux/IAA protein sequences were conducted using the neighbor-joining algorithm in MEGA5 [37] considering 1,000 replications with bootstrap analyses.
RNA isolation and cDNA synthesis
Frozen maize shoot and root tissues were ground and approximately 100 mg per biological replicate were used for total RNA extraction via the RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/). Subsequently, RNA was treated with RNase-free DNAse I (Fermentas, http://www.thermoscientificbio.com/fermentas/). To exclude the possibility of DNA contamination, the RNA samples were tested via PCR with oligonucleotides for maize actin 1 (AY104722) that bind to exon sequences that flank an intron. For cDNA synthesis 500 ng of total RNA was subjected to the qScript cDNA Synthesis Kit protocol (Quanta BioScience, http://www.quantabio.com/). For each root-type and tissue, five biological replicates were analyzed while auxin induction was tested in three biological replicates. Three technical replicates were measured for each sample.
Quantitative real-time-PCR
Expression of maize Aux/IAA genes was determined by quantitative real-time-PCR in a Bio-Rad CFX 384™ Real-Time System (http://www.bio-rad.de) using gene-specific oligonucleotides (Table S1). The oligonucleotides were designed by Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and checked with NetPrimer (PREMIER Biosoft, http://www.premierbiosoft.com) software. Each reaction contained 4 µl MESA Blue qPCR™ Mastermix Plus for SYBR Assay no ROX (Eurogentec, http://www.eurogentec.com), 1 µl cDNA sample and 250 nM gene-specific oligonucleotide primers to a final volume of 8 µl. The primer efficiency of each oligonucleotide was calculated using the following dilution series: 1, 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, and 1/128. The relative expression levels of the transcripts were calculated with reference to the housekeeping gene myosin (Genbank AC: 486090G09.x1). Differential gene expression was determined by a two sided Student´s t-test.
Identification of expression patterns for auxin inducible Aux/IAA genes
Two types of expression patterns, A and B, were identified for auxin-inducible Aux/IAA genes in maize within 3 hours of auxin induction. For all genes auxin induction at three time points t1, t2, t3 was compared to the control at t0. Pattern A is characterized by a significantly increased expression at one time point compared to the control to. In pattern A increased expression remained at later time points significantly above the control. In pattern B, expression at t1 or t2 was significantly increased compared to t0, while expression at t2 or t3 was significantly decreased compared to t1 or t2, respectively.
Results
The maize Aux/IAA family consists of 34 members
To obtain a comprehensive overview of the maize Aux/IAA gene family, the previously identified 31 maize Aux/IAA sequences [15] were used as query to screen version 2 of the maize filtered gene set comprising 39,656 high confidence genes (ZmB73_5b_FGS; http://www.maizegdb.org/). As a result, three novel maize Aux/IAA genes which display the canonical four domain structure were discovered including ZmIAA32 (GRMZM2G366373), ZmIAA33 (GRMZM2G359924), and ZmIAA34 (GRMZM2G031615) increasing the total number of maize Aux/IAA genes to 34 (Table 1). Aux/IAA domains were identified via a SMART (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/smart/set_mode.cgi) search. Proteins encoded by 29 Aux/IAA genes displayed all four domains. By contrast, in ZmIAA25 domains I and II are lacking and domains III and IV are incomplete. Moreover, domain II is absent in ZmIAA24 and ZmIAA26 whereas domain IV is missing in ZmIAA22 and ZmIAA31. Furthermore, in ZmIAA28 domain IV is split in two parts by the insertion of nine amino acids (Figure S1). A summary of the characteristics of the 34 maize Aux/IAA genes and the proteins encoded by them is provided in Table 1. The four Aux/IAA domains are highlighted in an alignment of the Aux/IAA protein sequences by ClustalW (Figure S1). The five ZmIAA proteins ZmIAA3, ZmIAA9, ZmIAA13, ZMIAA24, and ZmIAA26 displayed a modified LxLxPP instead of the predominant LxLxLP motif in domain I, whereas the 13 Aux/IAA protein sequences ZmIAA4, ZmIAA6, ZmIAA9, ZmIAA11, ZmIAA12, ZmIAA16, ZmIAA17, ZmIAA18, ZmIAA20, ZmIAA23, ZmIAA30, ZmIAA33, and ZmIAA34 displayed a variation in the conserved motif of domain III.
Table 1. Characteristics of the maize Aux/IAA gene family.
| Name | AC maizeGDB | Maize chromo-some | Genome location | Strand | Protein length (aa) | Sub-genome 1 | Sub-genome 2 | Maize chromo-some with syntenic region | ZmIAA gene in syntenic region | Rice chromo-some with syntenic region | Sorghum chromo-some with syntenic region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZmIAA1 | GRMZM2G079957_T2 | 1 | 170.535.129- 170.536.982 | 1 | 227 | X | 3 | ZmIAA8 | 12 | 8 | |
| ZmIAA2 | GRMZM2G159285_T1 | 1 | 275.031.309- 275.034.076 | -1 | 237 | X | 5 | ZmIAA14 | 3 | 1 | |
| ZmIAA3 | GRMZM5G809195_T1 | 1 | 288.394.822- 288.396.335 | -1 | 202 | X | 5 | ZmIAA13 | 3 | 1 | |
| ZmIAA4 | GRMZM2G104176_T1 | 3 | 7.117.449- 7.120.087 | -1 | 229 | X | 1 | 3 | |||
| ZmIAA5 | GRMZM2G004696_T1 | 3 | 10.073.330- 10.076.684 | 1 | 220 | X | 8 | ZmIAA27 | 1 | 3 | |
| ZmIAA6 | GRMZM2G074742_T1 | 3 | 48.981.283- 48.983.063 | 1 | 198 | X | 1 | 3 | |||
| ZmIAA7 | GRMZM2G138268_T1 | 3 | 117.766.041- 117.770.646 | -1 | 271 | X | 12 | 8 | |||
| ZmIAA8 | GRMZM2G167794_T1 | 3 | 118.064.610- 118.066.326 | -1 | 230 | X | 1 | ZmIAA1 | 12 | 8 | |
| ZmIAA9 | GRMZM2G057067_T1 | 3 | 199.305.498- 199.307.986 | -1 | 357 | X | 1 | 3 | |||
| ZmIAA10 | GRMZM2G037368_T1 | 3 | 209.094.740- 209.098.102 | -1 | 269 | X | 8 | ZmIAA29 | 1 | 3 | |
| ZmIAA11 | GRMZM2G059544_T2 | 4 | 39.557.305- 39.558.529 | -1 | 251 | X | 8 | 7 | |||
| ZmIAA12 | GRMZM2G142768_T1 | 4 | 171.373.109- 171.375.531 | 1 | 293 | X | 2 | 4 | |||
| ZmIAA13 | GRMZM2G152796_T1 | 5 | 4.200.507- 4.201.934 | -1 | 181 | X | 1 | ZmIAA3 | 3 | 1 | |
| ZmIAA14 | GRMZM2G077356_T1 | 5 | 7.772.505- 7.775.451 | 1 | 228 | X | 1 | ZmIAA2 | 3 | 1 | |
| ZmIAA15 | GRMZM2G128421_T1 | 5 | 17.343.149- 17.344.582 | 1 | 224 | ||||||
| ZmIAA16 | GRMZM2G121309_T1 | 5 | 151.712.395- 151.716.302 | 1 | 289 | X | 2 | 4 | |||
| ZmIAA17 | GRMZM2G030465_T1 | 5 | 214.411.995- 214.413.229 | 1 | 206 | X | 2 | 4 | |||
| ZmIAA18 | GRMZM2G000158_T4 | 6 | 79.914.748- 79.916.495 | -1 | 197 | X | 9 | ZmIAA30 | 6 | 10 | |
| ZmIAA19 | GRMZM2G079200_T1 | 6 | 103.139.630- 103.141.327 | -1 | 198 | X | 6 | 10 | |||
| ZmIAA20 | GRMZM5G864847_T1 | 6 | 130.004.758- 130.006.167 | -1 | 234 | ||||||
| ZmIAA21 | GRMZM2G147243_T2 | 6 | 133.196.856- 133.200.588 | 1 | 244 | X | 8 | ZmIAA28 | 5 | 9 | |
| ZmIAA22 | GRMZM2G141205_T1 | 6 | 146.505.719- 146.506.414 | -1 | 231 | ||||||
| ZmIAA23 | GRMZM2G074427_T2 | 6 | 160.166.708- 160.170.208 | 1 | 346 | X | 5 | 9 | |||
| ZmIAA24 | GRMZM2G149449_T1 | 7 | 9.392.913- 9.393.634 | 1 | 115 | ||||||
| ZmIAA25 | GRMZM2G115357_T2 | 7 | 10.970.395- 10.971.721 | -1 | 66 | X | 7 | 2 | |||
| ZmIAA26 | GRMZM2G048131_T1 | 7 | 142.773.908- 142.775.365 | 1 | 139 | X | 9 | 2 | |||
| ZmIAA27 | GRMZM2G130953_T2 | 8 | 18.318.828- 18.320.919 | -1 | 186 | X | 3 | ZmIAA5 | 1 | 3 | |
| ZmIAA28 | GRMZM2G035465_T3 | 8 | 110.812.538- 110.816.166 | -1 | 256 | X | 6 | ZmIAA21 | 1 | 9 | |
| ZmIAA29 | GRMZM2G163848_T5 | 8 | 150.560.417- 150.563.479 | 1 | 272 | X | 3 | ZmIAA10 | 5 | 3 | |
| ZmIAA30 | GRMZM2G001799_T1 | 9 | 16.249.556- 16.251.444 | 1 | 216 | X | 6 | ZmIAA18 | 6 | 10 | |
| ZmIAA31 | GRMZM2G134517_T1 | 10 | 134.255.159- 134.255.926 | -1 | 255 | ||||||
| ZmIAA32 | GRMZM2G366373_T2 | 1 | 253.302.210- 253.303.545 | -1 | 226 | ||||||
| ZmIAA33 | GRMZM2G359924_T1 | 8 | 108,568,427- 108,570,670 | 1 | 228 | ||||||
| ZmIAA34 | GRMZM2G031615_T2 | 4 | 16,418,192- 16,426,336 | 1 | 355 | X | 11 | 5 |
Phylogeny and synteny of the maize Aux/IAA family
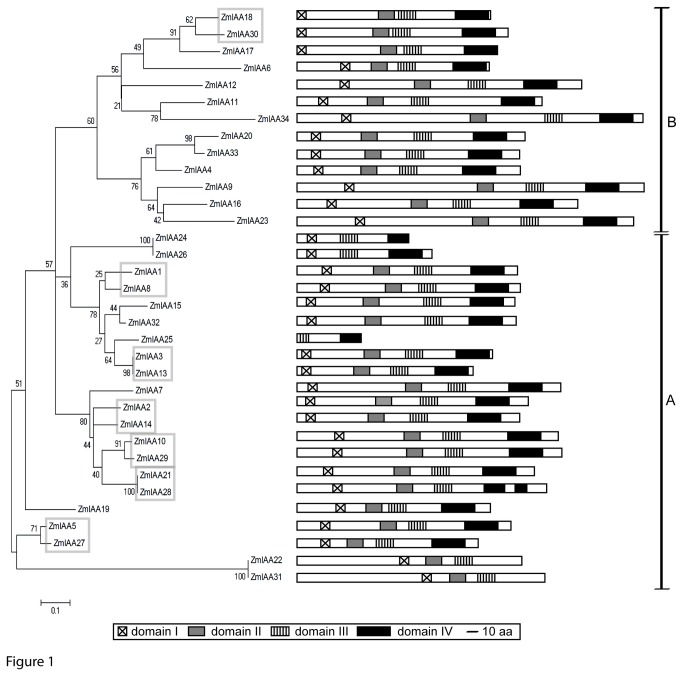
Phylogenetic reconstructions were based on the full-length sequences of the 34 maize Aux/IAA proteins. Two major groups of Aux/IAA proteins (class A and class B) were observed (Figure 1), which coincided with the alteration in the conserved motif in domain III (Figure S1).
Figure 1. Unrooted phylogenetic tree and distribution of conserved domains in maize Aux/IAA proteins.
The phylogenetic tree reveals two classes (A and B) of Aux/IAA proteins that differ in the sequence of domain III. The structure of the Aux/IAA proteins and the distribution of their domains are displayed to the right. The scale bar corresponds to 10 amino acids (aa). Paralogous Aux/IAA genes are encircled. The values associated to each branch are bootstrap percentages. The size bar indicates sequence divergence : 0.1 = 10%.
Evolution and synteny of the maize Aux/IAA gene family was studied via CoGe (http://genomevolution.org/CoGe/; Table 1). In total, seven pairs of paralogous maize Aux/IAA genes were identified (Table 1, Figure 1). Moreover, for seven Aux/IAA genes (ZmIAA15, ZmIAA20, ZmIAA22, ZmIAA24, ZmIAA31, ZmIAA32, and ZmIAA33) no orthologs were found in rice and sorghum suggesting that these genes are the result of gene duplications after the separation of maize from rice and sorghum.
Functional diversification of monocot maize, rice, and sorghum Aux/IAA genes was studied by a phylogenetic reconstruction of the Aux/IAA proteins of these species (Figure S2).
Expression of the Aux/IAA gene family during development
Aux/IAA gene expression was determined in embryonic primary and seminal roots and post-embryonic lateral and shoot-borne roots. Moreover, primary roots were surveyed at different developmental stages. Finally, different root tissues of the primary root including the meristematic and elongation zones and cortex and stele tissues of the differentiation zone were analyzed. Expression levels were determined by quantitative RT-PCR for 30 of 34 Aux/IAA genes. For the closely related genes ZmIAA22/ZmIAA31 and ZmIAA24/ZmIAA26 (Figure 1) no specific oligonucleotides were available that allowed to distinguish between them. Moreover, these four genes encode for Aux/IAA proteins that do not display the canonical four domain structure.
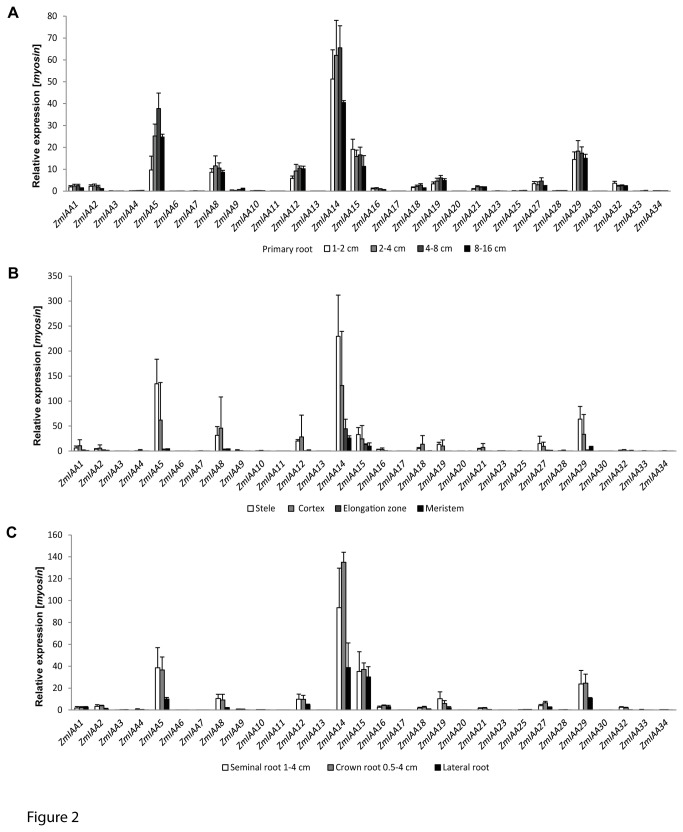
Expression patterns largely differed between the different maize Aux/IAA genes (Figure 2). ZmIAA5, ZmIAA8, ZmIAA12, ZmIAA14, ZmIAA15, and ZmIAA29 displayed the highest expression levels in primary roots (Figure 2A). ZmIAA14 contributes ~40% of all Aux/IAA transcripts in 1-2 cm primary roots and ~35% in later developmental stages of the primary root. While ZmIAA14 levels in young primary roots (up to the 4-8 cm class) are not significantly different, expression of this gene significantly decreases during the later developmental stages 4-8 cm and 8-16 cm (Figures S3 and S4). ZmIAA5 displayed the second highest Aux/IAA expression in primary roots and provides 8% of the Aux/IAA transcripts in 1-2 cm primary roots and 20% in 8-16 cm primary roots. ZmIAA5 displayed a significant increase in gene expression between 1-2 cm primary roots and 4-8 cm primary roots (Figures S3 and S4). As in ZmIAA14, expression of ZmIAA5 decreases between 4-8 cm and 8-16 cm primary roots.
Figure 2. Expression of Aux/IAA genes during development in different root-types and tissues.
Relative expression of 30 Aux/IAA genes was surveyed via qRT-PCR relative to myosin during primary root development (A), in different primary root tissues (B), and in seminal, crown and lateral roots (C).
These six Aux/IAA genes also displayed the highest expression in primary root tissues three days after germination (Figure 2B). Preferential expression in cortex and stele tissues compared to the meristematic and elongation zones was detected for 27 of 30 tested Aux/IAA genes (Figure S3). Only ZmIAA11 and ZmIAA13 displayed the highest expression in the root apical meristem while in ZmIAA34 expression in the meristematic and elongation zones were slightly higher than in the stele. The six genes that displayed the highest expression in primary roots also displayed the highest expression in seminal, crown and lateral roots (Figure 2C).
When analyzing differential gene expression of maize Aux/IAA genes between different root-types, a total 19 of genes (ZmIAA1, ZmIAA2, ZmIAA4, ZmIAA5, ZmIAA8, ZmIAA13, ZmIAA14, ZmIAA15, ZmIAA16, ZmIAA18, ZmIAA20, ZmIAA21, ZmIAA23, ZmIAA25, ZmIAA27, ZmIAA28, ZmIAA30, ZmIAA33, ZmIAA34) displayed significantly higher expression in crown roots compared to at least one stage of primary root development (Figures S3 and S4). In contrast, only ZmIAA6 was significantly lower expressed in crown roots than in the three stages of primary root development (Figure S3 and S4). Moreover, four genes were preferentially expressed in crown versus lateral roots (ZmIAA2, ZmIAA13, ZmIAA28, ZmIAA30) and another four genes were preferentially expressed in crown versus seminal roots (ZmIAA16, ZmIAA27, ZmIAA28, ZmIAA30). Hence, among 27 genes differentially expressed between crown roots and other root-types 26 display a higher expression in crown roots. When comparing differential gene expression between primary, seminal and lateral roots eleven genes were differentially expressed between primary and lateral roots (ZmIAA5, ZmIAA7, ZmIAA8, ZmIAA9, ZmIAA12, ZmIAA13, ZmIAA18, ZmIAA21, ZmIAA27, ZmIAA32, ZmIAA34). All of these genes except ZmIAA27 displayed a higher expression in primary versus lateral roots. Moreover, nine genes were differentially expressed between at least one stage of primary root development and seminal roots (ZmIAA3, ZmIAA6, ZmIAA9, ZmIAA13, ZmIAA16, ZmIAA21, ZmIAA25, ZmIAA28, ZmIAA30). Only ZmIAA6 and ZmIAA9 were preferentially expressed in primary roots, while the remaining seven genes were preferentially expressed in seminal roots. Finally, seven genes were preferentially expressed in seminal versus lateral roots (ZmIAA7, ZmIAA8, ZmIAA10, ZmIAA13, ZmIAA20, ZmIAA21, ZmIAA34). These differential expression patterns display a strong bias for expression levels between the different root-types, which applies to 51 of 55 (93%) of the pairwise differential expression patterns between different root-types observed in the present study. This pattern suggests that differentially expressed Aux/IAA genes follow in most instances the following hierarchy of expression: crown roots > seminal roots > primary roots > lateral roots.
To complement the expression profile of the Aux/IAA gene family, different shoot tissues of young maize seedlings were analyzed (Figure S3). The genes ZmIAA6, ZmIAA9, and ZmIAA20 displayed a significantly higher expression in shoot tissues than in any root tissue. In all three instances the genes were highly expressed in the mesocotyl. Moreover, for some Aux/IAA genes expression in the coleoptile was higher than in some of the root tissues (Figure S3). Such a preferential expression was however not observed for leaves and coleoptilar node.
Correlation of gene expression in different root-types and tissues of paralogous Aux/IAA gene pairs
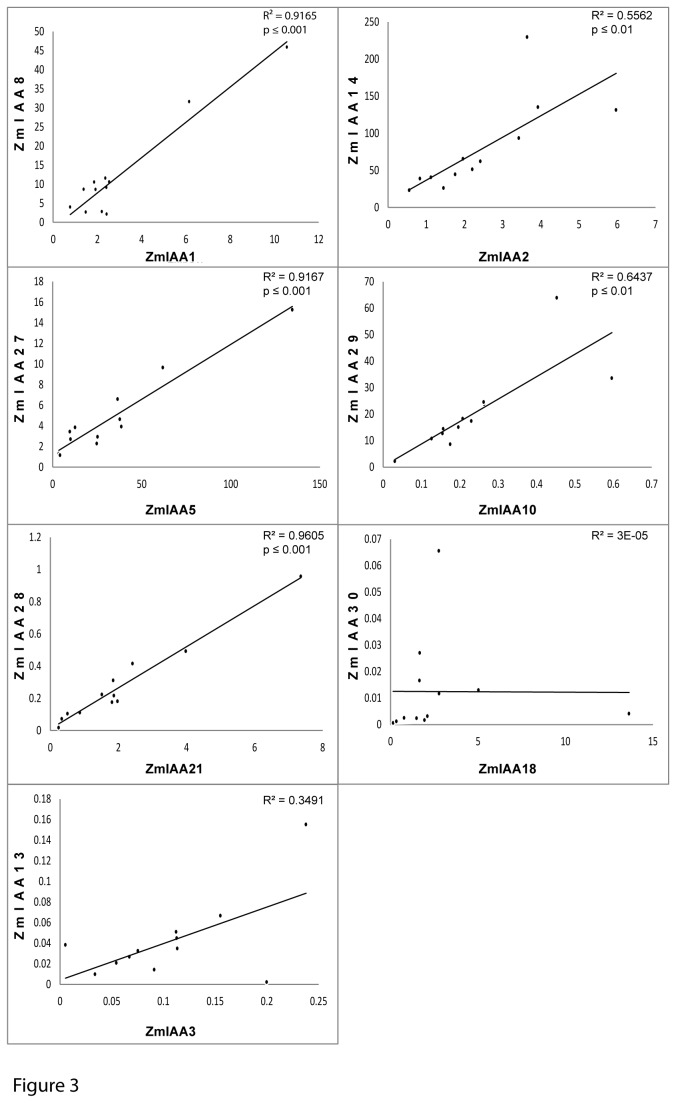
To compare expression patterns between the seven paralogous maize gene pairs in roots, coefficients of determination were calculated based on the gene expression data (Figure 3). This analysis revealed a strong linear correlation (p ≤0.01) for five of seven gene pairs up to R2 = 0.96 (ZmIAA21/ZmIAA28). Nevertheless, for all five pairs of paralogs one of the genes displayed on average a significantly higher expression level than the other one. The paralogous pairs ZmIAA3/ZmIAA13 and ZmIAA18/ZmIAA30 did not display any significant correlation with respect to their expression patterns in roots.
Figure 3. Correlation of gene expression of the seven paralogous Aux/IAA gene pairs in maize roots.
Five of seven paralogous Aux/IAA pairs showed a significant correlation in their gene expression patterns in roots (coefficient of determination R2 >0.5; p ≤0.01). Only ZmIAA30/ZmIAA18 and ZmIAA13/ZmIAA3 did not display significant expression correlation in the tested root-types and tissues.
The paralogous Aux/IAA pairs ZmIAA1/ZmIAA8, ZmIAA5/ZmIAA27, ZmIAA21/ZmIAA28, and ZmIAA3/ZmIAA13 revealed a higher expression level of the gene located in maize subgenome 1 compared to the paralog in subgenome 2. In the other three pairs ZmIAA2/ZmIAA14, ZmIAA10/ZmIAA29, and ZmIAA18/ZmIAA30 the gene of subgenome 2 was preferentially expressed.
Two types of auxin-induced Aux/IAA expression kinetics in maize
Screening for cis–elements in the regulatory region 3 kb upstream of ATG revealed that 28 of 34 analyzed maize Aux/IAA promoters contain canonical auxin response elements (AuxRE) 5´ TGTCTC 3´ or its inverse complement sequence 5´ GAGACA 3´ (Table S2). Moreover, all 34 maize Aux/IAA promoters contain several multiple tandem copies of the AuxRE core sequence 5´ TGTC 3´ or 5´GACA 3´. The presence of AuxRE motifs and its derivatives suggests that these genes are regulated by auxin. Therefore, auxin induction of the maize Aux/IAA gene family was tested in the differentiation zone of young maize primary roots over a time period of 3 h by quantitative real-time PCR (Figure S5). Two distinct expression patterns were observed after auxin treatment (Figure 4). In pattern A at least one time point displayed significantly increased expression compared to t0 and expression at t3 was still significantly higher than at t0 (Figure 4A and Figure S4). Moreover, in pattern B expression at t1 or t2 was significantly increased compared to t0, whereas expression at t2 or t3 was significantly decreased compared to t2 or t1, respectively (Figure 4B and Figure S4). The majority (22 of 27) of maize Aux/IAA genes displayed pattern A (Figure 4A and Figure S4). Pattern B was observed for ZmIAA8, ZmIAA13, ZmIAA20, ZmIAA25, and ZmIAA33 (Figure 4B and Figure S4). Moreover, ZmIAA30 and ZmIAA34 were below the detection limit to observe any expression. ZmIAA23 was the only maize gene that was not induced by auxin (Figure S4).
Figure 4. Auxin (αNAA) induction of maize Aux/IAA genes.
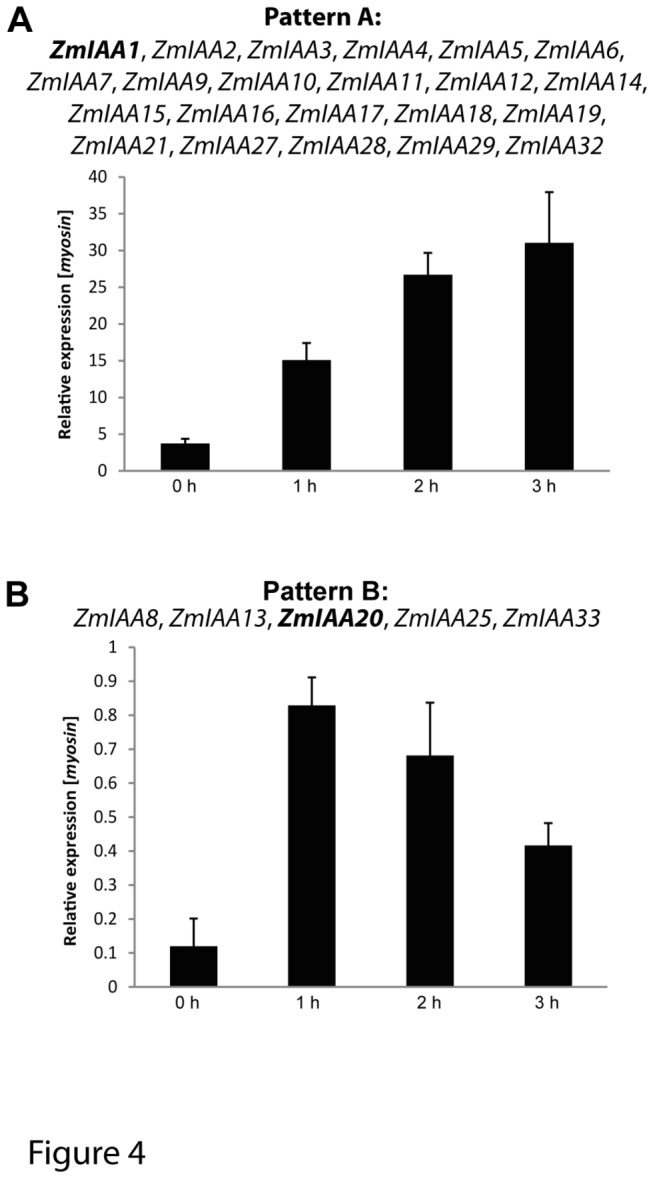
Two patterns of auxin induction were identified by qRT-PCR in the differentiation zone of 5-day-old maize primary roots after 5 µM αNAA (α-Naphthalene Acetic Acid) treatment over three hours. Genes were either constitutively induced (A) or expression decreased relative to the initial induction (B). Each pattern is illustrated by one example (in bold) while other Aux/IAA genes that followed these induction patterns are listed. All tested Aux/IAA genes were αNAA inducible except ZmIAA23. A detailed account of the Aux/IAA induction results is provided in Figure S5.
Discussion
Novel Aux/IAA genes and structural analyses of maize Aux/IAA family
Aux/IAA genes are plant-specific transcriptional regulators [16]. Initially, 31 maize Aux/IAA genes were discovered in the maize genome [15]. Improved annotation allowed for the identification of three novel Aux/IAA genes (ZmIAA32: GRMZM2G366373, ZmIAA33: GRMZM2G359924, ZmIAA34: GRMZM2G031615) in the present study, increasing the total number in maize to 34. Similarly, the rice genome contains 31 Aux/IAA genes [14] while sorghum harbors 26 Aux/IAA genes in its genome.
Sequence analysis of the maize Aux/IAA protein family revealed that five maize Aux/IAA proteins do not contain all four domains characteristic of this protein family. Similarly, domain II which is required for the degradation of the Aux/IAA proteins is partially or totally missing in OsIAA4, OsIAA8, OsIAA27, OsIAA28, and OsIAA29 in rice [14] and in AtIAA20, AtIAA30, AtIAA31, AtIAA32, AtIAA33, and AtIAA34 in Arabidopsis [38]. Moreover, tomato Sl-IAA32 does not have domain II and Sl-IAA33 is lacking domains I and II [12].
Furthermore, in several genes modifications of characteristic amino acid sequences were observed. The LxLxLx motif in domain I is known to function as a repressor and is characteristic of flowering plants. In contrast, LxLxPP is typically found in mosses or in one of the three Aux/IAA genes of the vascular non-seed plant S. moellendorffii [39]. Nevertheless, the sequence LxLxPP was also observed in five maize Aux/IAA proteins (ZmIAA3, ZmIAA9, ZmIAA13, ZmIAA24, and ZmIAA26), while in Arabidopsis and in rice no LxLxPP motives were found. The vascular non-seed plant S. moellendorffii and flowering plants do not form monophyletic groups, suggesting that the motif was established independently in each lineage. In Arabidopsis, it was reported that specific point mutations in codons that encode the leucine residue in the domain I motif lead to a weaker repression of ARF-mediated transcription [23]. Although there is no evidence that LxLxPP functions as a repressor domain, it was proposed to be functionally important due to its wide distribution outside the flowering plants [39].
Synteny and correlation in gene expression
The diversity and complexity of the Aux/IAA gene family in modern maize can be explained by gene and genome duplications and by gene loss because of partial fractionation. The maize genome was duplicated ~5-12 million years ago [40]. As a consequence, pairs of paralogous Aux/IAA genes emerged in maize. Today, seven pairs of paralogous Aux/IAA genes are retained in the maize genome. This implies that the maize Aux/IAA gene family was already diversified by gene duplications before this whole genome duplication event. Based on their synteny with sorghum, a species whose genome has not been duplicated, maize genes can be allocated to the maize subgenomes 1 or 2. The chromosomal regions with less gene loss were designated subgenome maize 1 [41]. A classical model of gene duplication assumes that one duplicated gene maintains the original function, while a second copy is lost, silenced or evolves a new function [42]. Hence, some of these gene pairs might have diversified by subfunctionalization or neofunctionalization [43]. Five of the seven paralogous Aux/IAA gene pairs displayed a significant correlation of their gene expression patterns in roots. Among those, only one pair (ZmIAA21/ZmIAA28) also showed similar expression patterns in shoot tissues. The correlation in gene expression for five of seven Aux/IAA gene pairs might also imply functional redundancy of these gene pairs. Nevertheless, even for these gene pairs functional diversification cannot be excluded because the average expression levels of these gene pairs are significantly different. Similarly, functional redundancy has been reported for Arabidopsis and rice Aux/IAA genes [17,39,44].
A subset of the duplicated genes has been lost since the ancient genome duplication by intrachromosomal recombination, silencing or null mutations [40], an ongoing process called fractionation [36]. In the maize Aux/IAA gene family, thirteen genes which can be assigned to either subgenome 1 or 2 but for which no paralog was retained are the result of fractionation. Finally, seven Aux/IAA genes were not assigned to any of the maize subgenomes. These genes likely emerged after the ancient maize genome duplication. Therefore, the Aux/IAA gene family in modern maize is the result of ancient gene duplications, a more recent whole genome duplication, partial fractionation, and modern gene duplications.
Expression profiling of Aux/IAA during development
In the present study, a systematic expression analysis of different maize root-types, tissues and developmental stages revealed root and tissue-specific expression patterns. The six maize Aux/IAA genes ZmIAA5, ZmIAA8, ZmIAA12, ZmIAA14, ZmIAA15, and ZmIAA29 displayed an overall high expression in all tested embryonic and postembryonic roots. This might suggest that these Aux/IAA genes play a constitutive role during maize root development. Similarly, the rice genes OsIAA5, OsIAA6, and OsIAA23 displayed the highest expression levels in roots [14]. Among those, OsIAA5 is the ortholog of the maize paralogs ZmIAA10/rum1 and ZmIAA29. Hence, both of the closely related genes OsIAA5 and ZmIAA29 display a very high expression in roots [14]. Similarly, OsIAA23 and ZmIAA5 which both display high expression levels in roots map to the same phylogenetic clade. However, while ZmIAA14 displays the highest transcript level of all tested maize Aux/IAA genes, its rice ortholog OsIAA13 displays only low expression in roots. Hence, while some rice and maize genes might have conserved their function in root development during evolution other members of the gene family might have not. Interestingly, none of ten cotton Aux/IAA genes displayed a major expression peak in roots [29].
Among the 30 maize Aux/IAA genes tested in the present study, only three genes displayed significantly higher expression in non-root tissue than in any of the tested root samples. In contrast, among the 31 rice Aux/IAA genes only OsIAA6 and OsIAA23 revealed their expression maximum in six day-old roots compared to shoot tissues. The maize ortholog of OsIAA6 is ZmIAA9, which displayed its expression maximum in the mesocotyl. In contrast, ZmIAA19, which is the ortholog of the rice gene OsIAA23, displays higher expression levels in all root-types than in any of the analyzed shoot tissues. This supports the notion that even among the closely related maize and rice Aux/IAA gene families a functional diversification of the gene family members has occurred during evolution [14]. However, very different tissues and developmental stages were analyzed in these maize and rice studies which makes them difficult to compare.
Thus far, mutant analyses of maize Aux/IAA genes revealed a developmental phenotype only for ZmIAA10 which corresponds to rootless with undetectable meristem 1 (rum1). The mutant does not initiate seminal roots and lacks lateral roots at the primary root, while primary and shoot-borne roots were not affected [25]. In the present study, ZmIAA10 displays the highest expression in stele and cortex tissues of three-day old primary roots. This is consistent with the observed phenotype because lateral roots are initiated from pericycle cells of the stele and from endodermis cells of the cortex [45]. Moreover, ZmIAA10/rum1 displays significantly lower expression in the elongation zone than in any root-type or tissue.
When Aux/IAA gene expression was compared pairwise between the four major maize root-types primary, seminal, lateral, and crown roots, 55 differential gene expression patterns were observed among the 30 maize Aux/IAA genes. Remarkably, among these differential expression patterns there was a strong bias with respect to expression levels between the different roots types with the tendency: crown roots > seminal roots > primary roots > lateral roots. Since this is a very general trend this might reflect differential control of auxin signal transduction in the molecular context of different root-types. Root-type-specific expression levels of Aux/IAA genes are controlled by upstream factors that bind to AuxRE in the promoter of Aux/IAA genes. Abundance of Aux/IAA transcripts and their proteins also affects the activity of downstream genes and thus contributes to the specific forms and functions of the different root-types of maize.
In 27 of 30 maize Aux/IAA genes surveyed in the present study, preferential expression in cortex and stele tissues of the differentiation zone compared to the meristematic and elongation zones was observed. This expression pattern correlates with auxin response in maize roots as visualized by DR5:RFP [46]. DR5:RFP reports sites where strong Aux/IAA protein degradation occurs [46]. These sites typically correlate with auxin maxima which enhance the transcription of Aux/IAA genes. Hence, DR5:RFP is also an indirect sensor for Aux/IAA transcriptional activity. In maize roots DR5:RFP maxima were observed in metaxylem elements and phloem poles in the stele [46]. Moreover, at early stages of lateral root development auxin response maxima were detected in pericycle and endodermis cells [46]. Remarkably, DR5:RFP also displays a strong signal in the meristematic zone of maize roots [46]. In this zone, only moderate Aux/IAA transcription was observed for most members of the gene family. However, while the DR5:RFP peak was mainly localized in the root cap, expression was surveyed in root tips that went beyond the root cap and also included the meristematic zone.
Auxin induced gene expression
Auxin-responsive (AuxRE) cis-elements are characteristic of the promoters of auxin-responsive genes [21]. Promoter analyses illustrated that all maize Aux/IAA genes contain canonical auxin-response elements or their core sequence. In contrast, in the promoter region of Aux/IAA genes of Vitis vinifera no AuxRE motifs were identified [47]. Similarly, in Arabidopsis only the promoters of AtIAA26 and AtIAA29 contain an AuxRE motif [48]. Consequently, auxin-induced gene expression may be directed by tissue-specific factors different than Aux/IAA proteins in these plants [47,48]. Other promoter elements like MYB and bZIP related binding sites play a role in auxin-mediated transcription [49,50]. Similar results were presented for a putative ocs element in Arabidopsis [51]. Consistent with the presence of AuxRE or their core elements, 27 of 28 expressed maize Aux/IAA genes were auxin inducible in qRT-PCR experiments. Similarly, in rice 24 of 29 expressed Aux/IAA genes were auxin inducible [44]. The kinetics of Aux/IAA gene expression is unique and depends on the variability of the regulation of free auxin, tissue-specific auxin receptors and different regulation of transcriptional and posttranscriptional events [14]. In maize, two auxin-dependent expression patterns were observed after αNAA treatment. While 22 Aux/IAA genes were constantly induced over time, five Aux/IAA genes showed a significant decrease after an initial increase in expression. Similarly, in rice 12 of 24 Aux/IAA genes displayed continuously increased expression during the time course while 12 genes displayed decreased expression at later time points [44]. In tomato, up-regulation of transcript levels of 17 of 19 Sl-Aux/IAA genes was detected in seedlings upon auxin treatment [12].
In summary, the detailed analysis of the maize Aux/IAA genes provides novel insights into the organization and expression of this large gene family that plays a crucial role in auxin signal transduction and thus the regulation of maize development. Moreover, tissue and root-type-specific expression profiles and induction studies provide interesting starting points for genetic analyses of candidate genes that might be involved in the initiation, emergence or specification of specific root-types in the complex maize root stock.
Supporting Information
Alignment of the maize Aux/IAA protein sequences. Aux/IAA protein sequences were compared by multiple alignments of the four conserved domains with ClustalW. Differences in the amino acid sequences of domain III, which distinguish class A and class B Aux/IAA proteins (see Figure 1) are boxed. The four domains are highlighted.
(PDF)
Phylogenetic reconstruction of the Aux/IAA protein families in different monocot species. Phylogenetic reconstruction of maize (Zea mays, Zm) sorghum (Sorghum bicolor, Sb), and rice (Oryza sativa, Os) Aux/IAA protein families in an unrooted tree with the neighbor-joining algorithm of MEGA5. Monocot specific clades are encircled. The values associated to each branch are bootstrap percentages. The size bar indicates sequence divergence: 0.05 = 5%.
(PDF)
Summary of Aux/IAA gene expression patterns in maize. Gene expression patterns obtained by qRT-PCR experiments in root and shoot tissues. Expression values in whole roots are highlighted in black, expression in primary root tissues in dark grey, and expression in shoot organs in light grey. ZmIAA10, ZmIAA13, and ZmIAA30 did not display any expression in shoot tissues. l: light, d:dark, N.D.: no expression detected.
(PDF)
Summary of pairwise Student´s t-tests of Aux/IAA gene expression comparisons in root and shoot tissues. Pairwise comparison of differential gene expression patterns between the various roots and shoot tissues by a two-sided Student´s t-test. Different significance levels are highlighted in color. Red: p ≤0.05; yellow: p ≤0.01; green: p ≤0.001. N. D. Expression was not detected in one of these tissues.
(PDF)
Summary of maize Aux/IAA gene induction by αNAA. Auxin induction patterns of the maize Aux/IAA genes determined by qRT-PCR in the differentiation zone of 5-day-old maize primary roots after 5 µM αNAA (α-Naphthalene Acetic Acid) treatment over three hours. A summary of these results in provided in Figure 4.
(PDF)
Oligonucleotide primers used in the present study.
(XLSX)
Promoter analyses of 3 kb upstream of the ATG start codon of maize Aux/IAA genes.
(XLSX)
Funding Statement
This work was supported by a DFG (Deutsche Forschungsgemeinschaft) grant to F.H. and funding by the University of Bonn. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9-17. doi: 10.1104/pp.111.1.9. PubMed: 8685277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana . J Mol Biol 251: 533-549. doi: 10.1006/jmbi.1995.0454. PubMed: 7658471. [DOI] [PubMed] [Google Scholar]
- 3. Went FW, Thimann KV (1937) Phytohormones. New York: Macmillan. [Google Scholar]
- 4. Estelle M (1992) The plant hormone auxin - insight in sight. BioEssays 14: 439-444. doi: 10.1002/bies.950140703. PubMed: 1332699. [DOI] [PubMed] [Google Scholar]
- 5. Guilfoyle TJ, Key JL (1986) Auxin regulated gene expression in higher plants. Crit Rev Plant Sci 4: 247-276. doi: 10.1080/07352688609382226. [DOI] [Google Scholar]
- 6. Theologis A (1986) Rapid gene regulation by auxin. Annu Rev Plant Physiol: 407-438. [Google Scholar]
- 7. Blatt MR, Thiel G (1993) Hormonal control of ion channel gating. Annu Rev Plant Physiol Plant Mol Biol 44: 543-567. doi: 10.1146/annurev.pp.44.060193.002551. [DOI] [Google Scholar]
- 8. Guilfoyle T (1999) Auxin-regulated genes and promoters. In: Hooykaas PJJ, Hall MA, Libbenga KR. Biochemistry and molecular biology of plant hormones Elservier, Amsterdam, The Netherlands [Google Scholar]
- 9. Walker JC, Key JL (1982) Isolation of cloned cDNAs to auxin-responsive poly(A) RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci U_S_A 79: 7185-7189. doi: 10.1073/pnas.79.23.7185. PubMed: 16593257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387-400. doi: 10.1023/A:1015255030047. PubMed: 12036262. [DOI] [PubMed] [Google Scholar]
- 11. Wu J, Peng Z, Liu S, He Y, Cheng L et al. (2012) Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol Genet Genomics 287: 295-311. doi: 10.1007/s00438-012-0675-y. PubMed: 22314799. [DOI] [PubMed] [Google Scholar]
- 12. Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M et al. (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53: 659-672. doi: 10.1093/pcp/pcs022. PubMed: 22368074. [DOI] [PubMed] [Google Scholar]
- 13. Wang SK, Bai YH, Shen CJ, Wu YR, Zhang SN et al. (2010) Auxin-related gene families in abiotic stress response in Sorghum bicolor . Funct Integr Genomics 10: 533-546. doi: 10.1007/s10142-010-0174-3. PubMed: 20499123. [DOI] [PubMed] [Google Scholar]
- 14. Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK et al. (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6: 47-59. doi: 10.1007/s10142-005-0005-0. PubMed: 16200395. [DOI] [PubMed] [Google Scholar]
- 15. Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays . p. L.). Mol Biol Rep 37: 3991-4001 [DOI] [PubMed]
- 16. Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420-425. doi: 10.1016/S1360-1385(01)02042-8. PubMed: 11544131. [DOI] [PubMed] [Google Scholar]
- 17. Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana . Plant Cell 17: 3282-3300. doi: 10.1105/tpc.105.036723. PubMed: 16284307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301-319. doi: 10.1105/tpc.010283. PubMed: 11884676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465-480. doi: 10.2307/3871400. PubMed: 11251090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ainley WM, Walker JC, Nagao RT, Key JL (1988) Sequence and characterization of two auxin-regulated genes from soybean. J Biol Chem 263: 10658-10666. PubMed: 2899079. [PubMed] [Google Scholar]
- 21. Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373-385. doi: 10.1023/A:1015207114117. PubMed: 12036261. [DOI] [PubMed] [Google Scholar]
- 22. Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809-2822. doi: 10.2307/3871536. PubMed: 11752389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533-543. doi: 10.1105/tpc.017384. PubMed: 14742873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441-445. doi: 10.1038/nature03543. PubMed: 15917797. [DOI] [PubMed] [Google Scholar]
- 25. von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X et al. (2011) Rootlesswithundetectablemeristem1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341-353 [DOI] [PubMed]
- 26. Kim J, Harter K, Theologis A (1997) Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U_S_A 94: 11786-11791. doi: 10.1073/pnas.94.22.11786. PubMed: 9342315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963-1971. doi: 10.1105/tpc.9.11.1963. PubMed: 9401121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533-543. doi: 10.1105/tpc.008417. PubMed: 12566590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han X, Xu X, Fang DD, Zhang T, Guo W (2012) Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum . Gene 503: 83-91. doi: 10.1016/j.gene.2012.03.069. PubMed: 22575728. [DOI] [PubMed] [Google Scholar]
- 30. Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci U_S_A 91: 326-330. doi: 10.1073/pnas.91.1.326. PubMed: 8278386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS et al. (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1 . Plant Physiol 139: 1255-1267. doi: 10.1104/pp.105.067330. PubMed: 16215225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845-857. doi: 10.1046/j.1365-313X.1996.10050845.x. [DOI] [Google Scholar]
- 33. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112-1115. doi: 10.1126/science.1178534. PubMed: 19965430. [DOI] [PubMed] [Google Scholar]
- 34. Wang YJ, Lü Y-P, Xie Q, Deng D-X, Bian Y-L (2010) Whole-genome sequence characterization of primary auxin-responsive Aux/IAA gene family in sorghum (Sorghum bicolor . p. L.). Acta Agron Sin 36: 688-694
- 35. Lyons E, Freeling M (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J 53: 661-673. doi: 10.1111/j.1365-313X.2007.03326.x. PubMed: 18269575. [DOI] [PubMed] [Google Scholar]
- 36. Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U_S_A 108: 4069-4074. doi: 10.1073/pnas.1101368108. PubMed: 21368132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al. (2011) MEGA5: molecular evolutionary geneticsa analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731-2739. doi: 10.1093/molbev/msr121. PubMed: 21546353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699-714. doi: 10.1105/tpc.105.039172. PubMed: 16489122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paponov IA, Teale W, Lang D, Paponov M, Reski R et al. (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9: 126. doi: 10.1186/1471-2148-9-126. PubMed: 19493348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D et al. (2010) Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLOS Biol 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schnable JC, Freeling M (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLOS ONE 6: e17855-e17855. doi: 10.1371/journal.pone.0017855. PubMed: 21423772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X et al. (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23: 469-478. PubMed: 16280546. [DOI] [PubMed] [Google Scholar]
- 43. Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459-473. PubMed: 10629003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Y, Wang L, Xiong L (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229: 577-591. doi: 10.1007/s00425-008-0853-7. PubMed: 19034497. [DOI] [PubMed] [Google Scholar]
- 45. Fahn A (1990) Plant Anatomy. Oxford: Pergamon Press. [Google Scholar]
- 46. Jansen L, Roberts I, De Rycke R, Beeckman T (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci 367: 1525-1533. doi: 10.1098/rstb.2011.0239. PubMed: 22527395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cakir B, Kilickaya O, Olcay AC (2013) Genome-wide analysis of Aux/IAA genes in Vitis vinifera: cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta Physiol Plants 35: 365-377. [Google Scholar]
- 48. Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738-1752. doi: 10.1104/pp.104.039669. PubMed: 15247399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berendzen KW, Weiste C, Wanke D, Kilian J, Harter K et al. (2012) Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. BMC Plant Biol 12: 125. doi: 10.1186/1471-2229-12-125. PubMed: 22852874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS et al. (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440-2453. doi: 10.1105/tpc.107.050963. PubMed: 17675404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen W, Singh KB (1999) The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J 19: 667-677. doi: 10.1046/j.1365-313x.1999.00560.x. PubMed: 10571852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the maize Aux/IAA protein sequences. Aux/IAA protein sequences were compared by multiple alignments of the four conserved domains with ClustalW. Differences in the amino acid sequences of domain III, which distinguish class A and class B Aux/IAA proteins (see Figure 1) are boxed. The four domains are highlighted.
(PDF)
Phylogenetic reconstruction of the Aux/IAA protein families in different monocot species. Phylogenetic reconstruction of maize (Zea mays, Zm) sorghum (Sorghum bicolor, Sb), and rice (Oryza sativa, Os) Aux/IAA protein families in an unrooted tree with the neighbor-joining algorithm of MEGA5. Monocot specific clades are encircled. The values associated to each branch are bootstrap percentages. The size bar indicates sequence divergence: 0.05 = 5%.
(PDF)
Summary of Aux/IAA gene expression patterns in maize. Gene expression patterns obtained by qRT-PCR experiments in root and shoot tissues. Expression values in whole roots are highlighted in black, expression in primary root tissues in dark grey, and expression in shoot organs in light grey. ZmIAA10, ZmIAA13, and ZmIAA30 did not display any expression in shoot tissues. l: light, d:dark, N.D.: no expression detected.
(PDF)
Summary of pairwise Student´s t-tests of Aux/IAA gene expression comparisons in root and shoot tissues. Pairwise comparison of differential gene expression patterns between the various roots and shoot tissues by a two-sided Student´s t-test. Different significance levels are highlighted in color. Red: p ≤0.05; yellow: p ≤0.01; green: p ≤0.001. N. D. Expression was not detected in one of these tissues.
(PDF)
Summary of maize Aux/IAA gene induction by αNAA. Auxin induction patterns of the maize Aux/IAA genes determined by qRT-PCR in the differentiation zone of 5-day-old maize primary roots after 5 µM αNAA (α-Naphthalene Acetic Acid) treatment over three hours. A summary of these results in provided in Figure 4.
(PDF)
Oligonucleotide primers used in the present study.
(XLSX)
Promoter analyses of 3 kb upstream of the ATG start codon of maize Aux/IAA genes.
(XLSX)