Summary
Microbial natural products continue to be an unparalleled resource for pharmaceutical lead discovery, but the rediscovery rate is high. Bacterial and fungal sequencing studies indicate that the biosynthetic potential of many strains is much greater than that observed by fermentation. Prodding the expression of such silent (cryptic) pathways will allow us to maximize the chemical diversity available from microorganisms. Cryptic metabolic pathways can be accessed in the laboratory using molecular or cultivation‐based approaches. A targeted approach related to cultivation‐based methods is the application of small‐molecule elicitors to specifically affect transcription of secondary metabolite gene clusters. With the isolation of the novel secondary metabolites lunalides A and B, oxylipins, cladochromes F and G, nygerone A, chaetoglobosin‐542, ‐540 and ‐510, sphaerolone, dihydrosphaerolone, mutolide and pestalone, and the enhanced production of known secondary metabolites like penicillin and bacitracin, chemical elicitation is proving to be an effective way to augment natural product libraries.
Introduction
Potentially interesting gene clusters, which may encode metabolites that increase competitiveness in natural environments, can remain silent in the unnatural setting of the microbiology laboratory. There are now many examples of bacterial and fungal biosynthetic secondary metabolite gene clusters outnumbering the number of natural products actually synthesized in the laboratory (Gross, 2009) (Fig. 1). These silent (cryptic) gene clusters undoubtedly harbour an enormous reservoir of novel bioactive constituents for drug discovery.
Figure 1.

Selected fully sequenced prokaryotic and eukaryotic secondary metabolite producers (Gross, 2009). Dark bars show the numbers of gene clusters known to be present in the genome, based on the number of isolated secondary metabolites at the time of sequencing. Light bars show the total number of gene clusters deduced from whole genome sequence analysis (Used with permission from Harald Gross.).
Aspergilli are a rich source of secondary metabolites (Schneider et al., 2008); they can harbour 30–50 distinct polyketide synthase (PKS) and non‐ribosomal peptide synthase (NRPS) gene clusters per species (Payne et al., 2006). In Aspergillus niger fewer than 30% of its 31 PKS, 15 NRPS and 9 hybrid PKS‐NRPS (HPN) biosynthetic gene clusters are transcribed under a variety of in vitro culture conditions (Fisch et al., 2009). Sequencing of Streptomyces avermitilis, a known producer of avermectin, revealed at least 25 gene clusters for siderophores, spore pigments and secondary metabolites of polyketide or non‐ribosomal peptide origin (Ömura et al. 2001). More than 6% of the S. avermitilis genome contains genes for secondary metabolite biosynthesis. Salinispora tropica, a marine actinomycete, devotes a very large portion of its genome, approximately 10%, to natural product assembly (Udwary et al., 2007).
One can explore the products of cryptic metabolic pathways using either molecular techniques (Gross, 2009; Hertweck, 2009; Scherlach and Hertweck, 2009) or cultivation‐dependent approaches (Bode et al., 2002). The number of researchers applying cultivation‐dependent approaches appears to be increasing since this decades‐old method was formalized (Schiewe and Zeeck, 1999; Höfs et al., 2000; Bode et al., 2002). Very small changes in cultivation conditions, media composition, pH, temperature or aeration, for example, can completely shift the metabolic profile of various microorganisms (Bode et al., 2002). Bode's group isolated more than 100 compounds from more than 25 structural classes from six different microbes by altering culture conditions (Bode et al., 2002). Cultivation‐dependent approaches can be categorized as biotic or abiotic, and abiotic can entail either physical or chemical means (Radman et al., 2003). A biotic approach using entire organisms (i.e. co‐culture) to stimulate natural product synthesis was recently reviewed (Pettit, 2009).
As an example of a physical method, UV mutagenesis resulted in the synthesis of a new antibiotic, 2,10‐dimethyl 4‐hydroxy‐6‐oxo‐4‐undecen‐7‐yne, from Aspergillus ochra ceus (Awad et al., 2005), and a new 14‐membered macrolide, mutolide 1, from Sphaeropsidales sp. (Bode et al., 2000a). When grown as biofilms in agar lined roller bottles, two Bacillus sp. produced antibiotic activity that was not detected when the strains were grown in shake flasks (Yan et al., 2002). The authors suggest that since these are marine epiphytic bacteria, surface attachment may induce secondary metabolism. Under oxygen limitation, an unidentified red pigment was synthesized by Saccharopolyspora erythraea in either shake flask or batch bioreactor cultures (Clark et al., 1995). In contrast, some Streptomyces natural products are only synthesized under high‐aeration conditions (Bode et al., 2002). The fungus Sphaeropsidales sp., which synthesizes the antifungal spirobisnaphthalene cladospirone bisepoxide, made six new spironaphthalenes when grown under static conditions (Bode et al., 2000b). Other examples of physical conditions that alter secondary metabolite profiles include hydrostatic pressure (Bode et al., 2002).
Alterations in the chemical composition of growth media can have profound effects on secondary metabolite profiles. Changing the water used to make the media from tap to distilled resulted in the isolation of six new secondary metabolites from Paraphaeosphaeria quadriseptata, cytosporones F‐I, quadriseptin A and 5′‐hydroxymonocillin III (Paranagama et al., 2007). When grown in a modified malt extract medium containing soluble starch instead of glucose, Gymnascella dankaliensis produces the unusual steroids dankasterone A and B (Amagata et al., 2007). Aspergillus nidulans synthesizes novel prenylated quinolin‐2‐one alkaloids when grown on rice medium, but not on a variety of other media (Scherlach and Hertweck, 2006).
An offshoot of the cultivation‐dependent approach is the rational use of small‐molecule modifiers that target control of cryptic natural product pathways. Studies from almost 20 years ago in which such non‐nutrient additives, or elicitors, were shown to affect microbial growth rates or ultrastructure (e.g. Akiyama et al., 1992; Benhamou, 1992) contributed to the current and rapidly expanding interest in elicitation of biologically active secondary metabolites. This review summarizes the current state of microbial small‐molecule‐directed elicitation, including, where known, mechanisms of elicitation at the transcription, translation or enzyme level. Examples are provided of enhanced production of known metabolites, and completely new metabolites.
DNA methyltransferase and histone deacetylase inhibitors
Fungal biosynthetic gene clusters are often located in the distal regions of chromosomes (Nierman et al., 2005; Shwab et al., 2007; Fisch et al., 2009). These regions exist in a heterochromatin state whose genes are often transcriptionally controlled by epigenetic regulation such as histone deacetylation and DNA methylation (Jenuwein and Allis, 2001). Chromatin remodelling by DNA methyltransferase and histone deacetylase inhibitors can result in activation of cryptic biosynthetic gene clusters. Selective manipulation of epigenetic targets using small‐molecule inhibitors of histone deacetylase and DNA methyltransferase activities leads to enhanced expression of PKS, NRPS and HPN biosynthetic pathways, and production of new secondary metabolites (Shwab et al., 2007; Fox and Howlett, 2008; Williams et al., 2008; Fisch et al., 2009; Henrikson et al., 2009).
Silencing of secondary metabolite gene clusters can be reversed by removing genes important to the establishment of a repressive chromatin configuration (Bok et al., 2009). In A. nidulans, loss of function CcIA (involved in histone H3 lysine 4 methylation) strains allowed for expression of at least two silent gene clusters, one yielding active anthraquinone constituents, and another active polyketides (Bok et al., 2009). Deletion of A. nidulans hdA, which encodes a histone deacetylase, causes transcriptional activation of two telomere‐proximal gene clusters, and increased levels of their gene products sterigmatocystin and penicillin (Shwab et al., 2007).
Transcriptional activation also occurs with small‐molecule epigenetic modifiers. Twelve diverse fungi, including Aspergillus, Cladosporium, Clonostachys, Diatrype and Penicillium, were treated with DNA methyltransferase and histone deacetylase inhibitors and metabolic profiles evaluated by HPLC, MS, 1H‐NMR and TLC (Williams et al., 2008). Eleven of the 12 fungi were responsive to one or more epigenetic treatments based on the production of new natural products and/or enhanced accumulation of constitutive secondary metabolites. Stimulation with amphotericin C, cycloheximide, 5‐fluorouracil or multiple media types did not result in new or enhanced production of secondary metabolites. Treatment of Alternaria alternata and Penicillium expansum with the histone deacetylase inhibitor trichostatin A resulted in a statistically significant increase in numerous unidentified secondary metabolites in both species (Shwab et al., 2007).
Treatment of a Diatrype sp. with the DNA methyltransferase inhibitor 5‐azacytidine led to the production of two new polyketides, lunalides A and B (Williams et al., 2008) (Fig. 2). Exposure of Cladosporium cladosporioides to 5‐azacytidine or the histone deacetylase inhibitor suberoylanilide hydroxamic acid led to dramatic restructuring of its metabolome (Williams et al., 2008). 5‐azacytidine elicited the de novo production of several oxylipins in C. cladosporioides (Fig. 2). Suberoylanilide hydroxyamic acid induced two new perylenequinones, cladochromes F and G (Fig. 2), four known cladochromes, A, B, D and E, and calphostin B in C. cladosporioides. According to the authors, this was the first reported co‐occurrence of such an extensive range of cladochrome‐calphostin metabolites from a single source. Elicitation of cladochromes A and B is especially noteworthy because these compounds were first reported as the unique products of Cladosporium infection of Cucumis sativus seedlings, which were not obtained from a large number of monoculture fermentations (Overeem et al., 1967; Arnone et al., 1988). The small‐molecule elicitation results eliminated the possibility that the compounds are the products of mixed Cladosporium–Cucumis biosynthesis.
Figure 2.
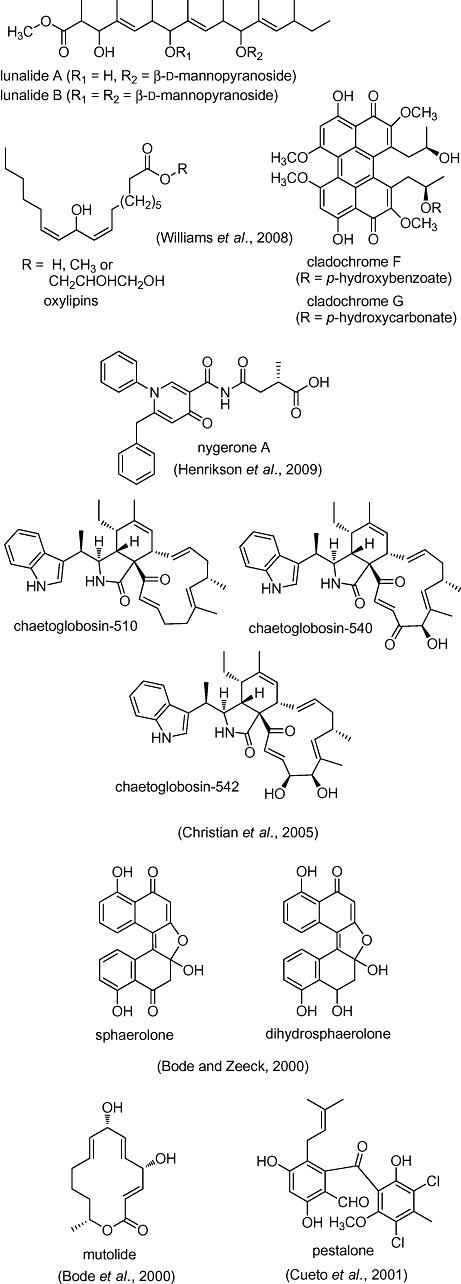
Novel microbial secondary metabolites elicited with small molecules.
More than 70% of the 31 PKS‐, 15 NRPS‐ and 9 HPN‐encoding gene clusters in A. niger are transcriptionally suppressed under standard laboratory culture conditions; all but seven of these are transcriptionally upregulated when the fungus is exposed to suberoylanilide hydroxamic acid or 5‐azacytidine (Fisch et al., 2009). Small‐molecule toxins targeting different cellular processes, cycloheximide, amphotericin B and 5‐fluorocytosine, did not induce similar changes in natural product production in this organism. In a related study, the addition of suberoylanilide hydroxamic acid to semi‐solid cultures of A. niger yielded a new metabolite containing a unique 1‐phenylpyridin‐4 (1H)‐one core, nygerone A (Fig. 2) (Henrikson et al., 2009). Nygerone A was not detected when A. niger was grown under varied fermentation conditions (not reported).
Histone deacetylase inhibitors include peptides. The cyclodepsipeptide jasplakinolide elicited synthesis of three new secondary metabolites in Phomospis asparagi (Christian et al., 2005). Chaetoglobosin‐542 and ‐540 (Fig. 2) exhibited cancer cell line cytotoxicity and disrupted actin filaments, while chaetoglobosin‐510 (Fig. 2) was inactive in these screens. The polypeptide antibiotic goadsporin induces antibiotic activity (not isolated) in a wide variety of streptomycetes (Onaka et al., 2001). Whether jasplakinolide and goadsporin specifically inhibit histone deacetylase or have another target that results in an altered metabolome remains to be seen. One target of jasplakinolide is known; it stabilizes actin filaments in vitro, but can disrupt actin filaments in vivo.
Oligosaccharides
In Penicillium chrysogenum, mannan oligosaccharides (MO) enhance penicillin G levels and penicillin G intermediates, increase germination rates, hyphal tip numbers, clump area and spore counts, and decrease levels of reactive oxygen species (Ariyo et al., 1998; Tamerler et al., 2001; Radman et al., 2004a,b, 2006). Early evidence that increased production of penicillin in the presence of alginate or alginate‐derived oligomannuronate (OM) is due to higher transcript levels of the three penicillin biosynthetic genes pcbAB, pcbC and penDE was provided by Gang and colleagues (2001), using Northern analysis and reporter gene studies. The stimulatory effect was observed in a defined medium and to a lesser extent in a complex medium containing corn steep liquor. The authors postulate that the difference could be due to the presence of elicitors in corn steep.
The absolute quantification of penicillin biosynthetic gene copy number in the presence of MO and OM was recently reported using quantitative PCR (Nair et al., 2009). Increased production was noted in both shaken flasks and bioreactors (where pH, oxygen concentration and temperature were maintained) containing P. chrysogenum. In shaken flasks, penicillin production was 133% higher in single elicitor‐added cultures (MO added 48 h post‐inoculation) compared with control cultures. Production increased another 71% when two different elicitors were used (MO at 48 h followed by OM at 96 h). Increased transcription levels were also similar in shaken flasks and bioreactors, with an increase of 216%, 61% and 200% in the pcbAB, pcbC and penDE transcript levels for single elicitor‐added cultures over control cultures. Addition of a second elicitor resulted in increases of 116% (pcbAB), 39% (pcbC) and 33% (penDE) transcript levels compared with single elicitor cultures. Strains of P. chrysogenum synthesize the pigment chrysogenin. Alginate and pectin oligosaccharides enhance chrysogenin production up to 40% in shake flasks and bioreactors (Asilonu et al., 2000). Adding the oligosaccharide elicitors at the beginning of fermentation did not have a detectable effect on secondary metabolite production. The optimal time for elicitor addition was 24–48 h, but chrysogenin yields improved through 72 h.
Oligosaccharides also elicit increased synthesis of antibiotics in bacteria. Bacitracin yield improvements of 29%, 27% and 16% over control Bacillus licheniformis cultures were achieved in a defined medium with oligoguluronate (OG), MO and OM respectively (Murphy et al., 2007a). When OG was added at 0 h and MO at 24 h, bacitracin production was enhanced a further 13.2%, while pH profiles and biomass were unaffected (Murphy et al., 2007b). Evidence for transcriptional level control was obtained using real‐time PCR, where a direct correlation between enhancement of bacitracin A production and the transcription of its biosynthetic genes was made. Elicitor supplementation (OG and MO as above) resulted in increased transcription of the bacitracin biosynthetic genes bacABC (Murphy et al., 2007b), and overexpression of bcrABC genes encoding the bacitracin ABC transporter system (Murphy et al., 2008). Polysaccharides have also been used to enhance production of secondary metabolites in bacteria and fungi (Patterson and Bolis, 1997; Zhu et al., 2008).
Enzyme inhibitors
Enzyme inhibitors can influence secondary metabolite production. Liang and colleagues (2010) tested the hypothesis that P450‐type enzymes, involved in the regulation of biosynthesis of secondary metabolites such as triterpenoids, could potentially increase production of anti‐tumor ganoderic acids. The addition of 100 µM phenobarbital (P450 inducer) to a two‐stage cultivation involving shake culture followed by static culture resulted in an increase in the levels of ganoderic acid‐Mk, ‐T, ‐S and ‐Me by 47%, 28%, 36% and 64% respectively. Transcription of three key genes in the triterpene biosynthetic pathway, 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase, squalene synthase and lanosterol synthase, was upregulated under phenobarbital induction.
Lycopene is a precursor of cyclic carotenoids and undergoes a number of metabolic reactions (e.g. cyclization) to form carotenoids (Bhosale, 2004). The chemical inhibitors pyridine and imidazole stimulate lycopene formation in Blakeslea trispora by inhibiting the enzymes responsible for cyclization of lycopene.
Two new bisnaphthalenes, sphaerolone and dihydrosphaerolone (Fig. 2), from the accumulated 1,3,8‐trihydroxynaphthalene, were isolated when Sphaeropsidales sp. was grown in the presence of enzyme inhibitors such as tricyclazole (Bode and Zeeck, 2000), an antifungal that inhibits 1,3,8‐trihydroxynaphthalene reductase. A new macrolide mutolide (Fig. 2) was also elicited by tricyclazole in this strain (Bode et al., 2000a). Mutolide had weak antibacterial activity. Interestingly, both UV mutagenesis and tricyclazole apparently activate a silent gene cluster responsible for mutolide biosynthesis (Bode et al., 2000a).
Solvents and heavy metals
Organic compounds such as ethanol or dimethylsulfoxide (DMSO) have also been used to elicit secondary metabolite biosynthesis. Ethanol and DMSO cause mistranslation (So and Davie, 1964, 1965), and presumably have other mechanisms such as induction of the stress response (Chen et al., 2000). Although their precise mechanism (more likely multiple mechanisms) of inducing secondary metabolite synthesis is unknown, they are included due to their ease of use, inexpensive nature and dramatic results.
In the presence of 3% DMSO, chloramphenicol and tetracenomycin C production increased approximately threefold in Streptomyces venezuelae and S. glaucescens respectively (Chen et al., 2000). Thiostrepton production increased about twofold in S. azureus with 3% DMSO. When treated and untreated cultures were compared, no significant differences in biomass were seen. These elicitation results are remarkable given that these antibiotics are from three different biosynthetic families. Substantial differences in HPLC patterns and increased pigment production were noted when cultures of three other species of Streptomyces were supplemented with DMSO. Dimethyl sulfone, a metabolite of DMSO, had similar but less pronounced stimulatory effects on antibiotic production. Ethanol (1–3%) stimulated tetracenomycin C production, but inhibited chloramphenicol production.
Ethanol (1%) elicited synthesis of a new chlorinated benzophenone antibiotic, pestalone, by the marine fungus Pestalotia (Cueto et al., 2001) (Fig. 2). Pestalone has potent activity against methicillin‐resistant Staphylococcus aureus and vancomycin‐resistant Enterococcus faecium, and marginal activity against the National Cancer Institute's 60 human tumour cell line screen. The antibiotic is undetectable in non‐elicited Pestalotia cultures. Pestalone is also synthesized when Pestalotia is co‐cultured with a marine bacterium (Cueto et al., 2001).
Ethanol (6%) dramatically increases synthesis of the benzoxazolophenanthridine antibiotic jadomycin B in S. venezuelae (Doull et al., 1994). Addition of ethanol (0.2%) to cultures of Phaffia rhodozyma resulted in increased carotenoid production, apparently due to activation of oxidative metabolism and induction of HMG‐CoA reductase (Gu et al., 1997). Polyphenol production (and biomass) by the medicinal fungus Inonotus obliquus is enhanced by the continuous addition of a low concentration of hydrogen peroxide (1 mM) to batch cultures (Zheng et al., 2009). Superoxide dismutase and catalase activities were also enhanced.
In a review of the effect of heavy metals on secondary metabolite production/cellular differentiation, Weinberg (1990) summarizes evidence that regulation primarily occurs at the level of transcription. Furthermore, three heavy metals, Mn2+, Fe3+ and Zn2+, are identified as having a key role in the secondary metabolism of a wide variety of microorganisms. In a more recent study, non‐toxic concentrations of the heavy metal ions Cu2+, Cd2+ and Cr3+ significantly increased production of the polyketide monocillin I by the plant‐associated fungus P. quadriseptata (Paranagama et al., 2007). Extracts of metal tolerant actinobacteria grown in the presence of Ni2+ (in either complex or minimal media) inhibited the growth of a variety of bacteria and fungi (Haferburg et al., 2009). Because many antibiotics and other secondary metabolites can scavenge heavy metals (Demain and Fang, 2000), the authors propose that synthesis of such antimicrobial metabolites is increased when metals are added to the fermentation medium of heavy metal tolerant strains (Haferburg et al., 2009). Certain enzymes require metals as cofactors. Mn2+, for example, is a cofactor for enzymes involved in carotenoid synthesis, and this heavy metal has been shown to enhance carotenoid production in various microbes (Bhosale, 2004). Other heavy metal ions, La3+, Ce3+ and Nd3+, had a stimulatory effect on carotenoid synthesis in Phaffia rhodozyma, while slightly inhibiting growth (Wang et al., 1999).
Conclusions and prospects for the future
Results of genome analyses reveal why manipulation of microbial growth can be such a fertile avenue for secondary metabolite discovery. Certain groups of bacteria and fungi have dozens of secondary metabolite pathways that are not expressed under standard laboratory growth conditions. Microbial small‐molecule secondary metabolites resulting from several billion years of biosynthetic reactions have been a tremendous resource for natural product drug discovery, and their usefulness will continue if the substantial proportion of cryptic biosynthetic pathways can be accessed.
Transcriptional suppression of secondary metabolites may be a mechanism to protect microbes from autotoxicity. Biosynthesis of metabolically expensive and potentially autotoxic secondary metabolites is presumably only elicited when an appropriate environmental trigger is encountered (Patterson and Bolis, 1997; Fisch et al., 2009). The use of small‐molecule elicitors, in some cases targeted, non‐random elicitors such as methyltransferase and histone deacetylase inhibitors, allows researchers to overcome transcriptional suppression of potentially important gene clusters in the laboratory. To date, more than a dozen new compounds from microbes have been identified with small‐molecule elicitation. Enhanced production of rare metabolites is another important application of this method, and there are numerous reports of increased production of well‐known metabolites using elicitors. Interestingly, the de novo production of some secondary metabolites can be elicited with both a small molecule or UV (mutolide), or a small molecule or co‐culture (pestalone).
Treatment of microbes with one of the many small‐molecule elicitors is a simple, inexpensive method to more thoroughly exploit the metabolic potential of microbes. Small‐molecule elicitation allows identification of new secondary metabolites and enhanced synthesis of low yield secondary metabolites in a broad spectrum of microorganisms. These types of experiments can be performed in almost any lab, and are much less cumbersome than varying myriad cultivation parameters. Furthermore, this technique uses native hosts as opposed to heterologous hosts, which can be fraught with complications.
Acknowledgments
Financial support was provided by grants R01 CA90441‐01‐05, 2R56 CA090441‐06A1, and 5R01 CA090441‐07 from the Division of Cancer Treatment, Diagnosis and Centers, National Cancer Institute, DHHS, and the Arizona Biomedical Research Commission. Many thanks to F. Hogan for preparing Figure 2.
References
- Akiyama H., Endo T., Nakakita R., Murata K., Yonemoto Y., Okayama K. Effect of depolymerized alginates on the growth of bifidobacteria. Biosci Biotechnol Biochem. 1992;56:355–356. doi: 10.1271/bbb.56.355. [DOI] [PubMed] [Google Scholar]
- Amagata T., Tanaka M., Yamada T., Doi M., Minoura K., Ohishi H. Variation in cytostatic constituents of a sponge‐derived Gymnascella dankaliensis by manipulating the carbon source. J Nat Prod. 2007;70:1731–1740. doi: 10.1021/np070165m. et al. [DOI] [PubMed] [Google Scholar]
- Ariyo B., Tamerler C., Bucke C., Keshavarz T. Enhanced penicillin production by oligosaccharides from batch cultures of Penicillium chrysogenum in stirred‐tank reactors. FEMS Microbiol Lett. 1998;166:165–170. doi: 10.1111/j.1574-6968.1998.tb13198.x. [DOI] [PubMed] [Google Scholar]
- Arnone A., Assante G., Di Modugno V., Merlini L., Nasini G. Perylenequinones from cucumber seedlings infected with Cladosporium cucumerinum. Phytochemistry. 1988;27:1675–1678. [Google Scholar]
- Asilonu E., Bucke C., Keshavarz T. Enhancement of chrysogenin production in cultures of Pencillium chrysogenum by uronic acid oligosaccharides. Biotechnol Lett. 2000;22:931–936. [Google Scholar]
- Awad G., Mathieu F., Coppel Y., Lebrihi A. Characterization and regulation of new secondary metabolites from Aspergillus ochraceus M18 obtained by UV mutagenesis. Can J Microbiol. 2005;51:59–67. doi: 10.1139/w04-117. [DOI] [PubMed] [Google Scholar]
- Benhamou N. Ultrastructural and cytochemical aspects of chitosan on Fusarium oxysporum f. sp. radicis‐lycopersici agent of tomato crown and root rot. Phytopathology. 1992;82:1185–1193. [Google Scholar]
- Bhosale P. Environmental and cultural stimulants in the production of carotenoids from microorganisms. Appl Microbiol Biotechnol. 2004;63:351–361. doi: 10.1007/s00253-003-1441-1. [DOI] [PubMed] [Google Scholar]
- Bode H.B., Zeeck A. Sphaerolone and dihydrosphaerolone, two bisnaphthyl‐pigments from the fungus Sphaeropsidales sp. F‐24'707. Phytochemistry. 2000;54:597–601. doi: 10.1016/s0031-9422(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Bode H.B., Walker M., Zeeck A. Secondary metabolites by chemical screening, 41+ Structure and biosynthesis of mutolide, a novel macrolide from a UV mutant of the fungus F‐24'707. Eur J Org Chem. 2000a;8:1451–1456. [Google Scholar]
- Bode H.B., Walker M., Zeeck A. Secondary metabolites by chemical screening, 42+‡+ cladospirones B to I from Sphaeropsidales sp. F‐24'707 by variation of culture conditions. Eur J Org Chem. 2000b;18:3185–3193. [Google Scholar]
- Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bok J.W., Chiang Y.‐M., Szewczyk E., Reyes‐Dominguez Y., Davidson A.D., Sanchez J.F. Chromatin‐level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wang G.‐Y.‐S., Li X., Waters B., Davies J. Enhanced production of microbial metabolites in the presence of dimethyl sulfoxide. J Antibiot. 2000;53:1145–1153. doi: 10.7164/antibiotics.53.1145. [DOI] [PubMed] [Google Scholar]
- Christian O.E., Compton J., Christian K.R., Mooberry S.L., Valeriote F.A., Crews P. Using jasplakinolide to turn on pathways that enable the isolation of new chaetoglobosins from Phomospis asparagi. J Nat Prod. 2005;68:1592–1597. doi: 10.1021/np050293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G.J., Langley D., Bushell M.E. Oxygen limitation can induce microbial secondary metabolite formation: investigations with miniature electrodes in shaker and bioreactor culture. Microbiology. 1995;141:663–669. [Google Scholar]
- Cueto M., Jensen P.R., Kauffman C., Fenical W., Lobkovsky E., Clardy J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod. 2001;64:1444–1446. doi: 10.1021/np0102713. [DOI] [PubMed] [Google Scholar]
- Demain A.L., Fang A. The natural functions of secondary metabolites. Adv Biochem Engin. 2000;69:1–39. doi: 10.1007/3-540-44964-7_1. [DOI] [PubMed] [Google Scholar]
- Doull J.L., Singh A.K., Hoare M., Ayer S.W. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- Fisch K.M., Gillaspy A.F., Gipson M., Henrikson J.C., Hoover A.R., Jackson L. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J Ind Microbiol Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. et al. [DOI] [PubMed] [Google Scholar]
- Fox E.M., Howlett B.J. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 2008;11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Gang L., Casqueiro J., Gutiérrez S., Kosalkova K., Castillo N.‐I., Martín J.F. Elicitation of penicillin biosynthesis by alginate in Penicillium chrysogenum, exerted on pcbAB, pcbC, and penDE genes at the transcriptional level. J Microbiol Biotechnol. 2001;11:812–818. [Google Scholar]
- Gross H. Genomic mining – a concept for the discovery of new bioactive natural products. Curr Opin Drug Discov Devel. 2009;2:207–219. [PubMed] [Google Scholar]
- Gu W.L., An G.H., Johnson E.A. Ethanol increases carotenoid production in Phaffia rhodozyma. J Ind Microbiol. 1997;19:114–117. doi: 10.1038/sj.jim.2900425. [DOI] [PubMed] [Google Scholar]
- Haferburg G., Groth I., Möllmann U., Kothe E., Sattler I. Arousing sleeping genes: shifts in secondary metabolism of metal tolerant actinobacteria under conditions of heavy metal stress. Biometals. 2009;22:225–234. doi: 10.1007/s10534-008-9157-4. [DOI] [PubMed] [Google Scholar]
- Henrikson J.C., Hoover A.R., Joyner P.M., Cichewicz R.H. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- Hertweck C. Hidden biosynthetic treasures brought to light. Nat Chem Biol. 2009;5:450–452. doi: 10.1038/nchembio0709-450. [DOI] [PubMed] [Google Scholar]
- Höfs R., Walker M., Zeeck A. Hexacyclinic acid, a polyketide from Streptomyces with a novel carbon skeleton. Angew Chem Int Ed. 2000;39:3258–3261. doi: 10.1002/1521-3773(20000915)39:18<3258::aid-anie3258>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Liang C.‐X., Li Y.‐B., Xu J.‐W., Wang J.‐L., Miao X.‐L., Tang Y.‐J. Enhanced biosynthetic gene expressions and production of ganoderic acids in static liquid culture of Ganoderma lucidum under phenobarbital induction. Appl Microbiol Biotechnol. 2010;86:1367–1374. doi: 10.1007/s00253-009-2415-8. et al. [DOI] [PubMed] [Google Scholar]
- Murphy T., Parra R., Radman R., Roy I., Harrop A., Dixon K., Keshavarz T. Novel application of oligosaccharides as elicitors for the enhancement of bacitracin A production in cultures of Bacillus licheniformis. Enzyme Microb Technol. 2007a;40:1518–1523. [Google Scholar]
- Murphy T., Roy I., Harrop A., Dixon K., Keshavarz T. Effect of oligosaccharide elicitors on bacitracin A production and evidence of transcriptional level control. J Biotechnol. 2007b;131:397–403. doi: 10.1016/j.jbiotec.2007.07.943. [DOI] [PubMed] [Google Scholar]
- Murphy T., Roy I., Harrop A., Dixon K., Keshavarz T. Elicitation effects of oligosaccharides on the transcriptional level of bacitracin ABC transporter genes in Bacillus licheniformis. Biotechnol Lett. 2008;30:1665–1670. doi: 10.1007/s10529-008-9743-0. [DOI] [PubMed] [Google Scholar]
- Nair R., Roy I., Bucke C., Keshavarz T. Quantitative PCR study on the mode of action of oligosaccharide elicitors on penicillin G production by Penicillium chrysogenum. J Appl Microbiol. 2009;107:1131–1139. doi: 10.1111/j.1365-2672.2009.04293.x. [DOI] [PubMed] [Google Scholar]
- Nierman W.C., Pain A., Anderson M.J., Wortman J.R., Kim H.S., Arroyo J. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. et al. [DOI] [PubMed] [Google Scholar]
- Ömura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Tabata H., Igarashi Y., Sato Y., Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes I. Purification and characterization. J Antibiot. 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- Overeem J.C., Sijpesteijn A., Fuchs A. Formation of perylenequinones in etiolated cucumber seedlings infected with Cladosporium cucumerinum. Phytochemistry. 1967;6:99–105. [Google Scholar]
- Paranagama P.A., Wijeratne E.M.K., Gunatilaka A.A.L. Uncovering biosynthetic potential of plant‐associated fungi: effect of culture conditions on metabolite production by Paraphaeosphaeria quadriseptata and Chaetomium chiversii. J Nat Prod. 2007;70:1939–1945. doi: 10.1021/np070504b. [DOI] [PubMed] [Google Scholar]
- Patterson G.M.L., Bolis C.M. Fungal cell‐wall polysaccharides elicit an antifungal secondary metabolite (phytoalexin) in the cyanobacterium Scytonema ocellatum. J Phycol. 1997;33:54–60. [Google Scholar]
- Payne G.A., Nierman W.C., Wortman J.R., Pritchard B.L., Brown D., Dean R.A. Whole genome comparison of Aspergillus flavus and A. oryzae. Med Mycol. 2006;44:S9–S11. doi: 10.1080/13693780600835716. et al. [DOI] [PubMed] [Google Scholar]
- Pettit R.K. Mixed fermentation for natural product drug discovery. Appl Microbiol Biotech. 2009;83:19–25. doi: 10.1007/s00253-009-1916-9. [DOI] [PubMed] [Google Scholar]
- Radman R., Saez T., Bucke C., Keshavarz T. Elicitation of plants and microbial cell systems. Biotechnol Appl Biochem. 2003;37:91–102. doi: 10.1042/ba20020118. [DOI] [PubMed] [Google Scholar]
- Radman R., Bucke C., Keshavarz T. Elicitor effects on reactive oxygen species in liquid cultures of Penicillium chrysogenum. Biotechnol Lett. 2004a;26:147–152. doi: 10.1023/b:bile.0000012897.28356.cb. [DOI] [PubMed] [Google Scholar]
- Radman R., Bucke C., Keshavarz T. Elicitor effects on Penicillium chrysogenum morphology in submerged cultures. Biotechnol Appl Biochem. 2004b;40:229–233. doi: 10.1042/BA20040062. [DOI] [PubMed] [Google Scholar]
- Radman R., Bland E.J., Sangworachat N., Bucke C., Keshavarz T. Effects of oligosaccharides and polysaccharides on the generation of reactive oxygen species in different biological systems. Biotechnol Appl Biochem. 2006;44:129–133. doi: 10.1042/BA20050217. [DOI] [PubMed] [Google Scholar]
- Scherlach K., Hertweck C. Discovery of aspoquinolones A‐D, prenylated quinoline‐2‐one alkaloids from Aspergillus nidulans, motivated by genome mining. Org Biomol Chem. 2006;4:3517–3520. doi: 10.1039/b607011f. [DOI] [PubMed] [Google Scholar]
- Scherlach K., Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- Schiewe H.‐J., Zeeck A. Cineromycins, γ‐butyrolactones and ansamycins by analysis of the secondary metabolite pattern created by a single strain of Streptomyces. J Antibiot. 1999;52:635–642. doi: 10.7164/antibiotics.52.635. [DOI] [PubMed] [Google Scholar]
- Schneider P., Misiek M., Hoffmeister D. In vivo and in vitro production options for fungal secondary metabolites. Mol Pharm. 2008;5:234–242. doi: 10.1021/mp7001544. [DOI] [PubMed] [Google Scholar]
- Shwab E.K., Bok J.W., Tribus M., Galehr J., Graessle S., Keller N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A.G., Davie E.W. The effects of organic solvents on protein biosynthesis and their influence on the amino acid code. Biochemistry. 1964;3:1165–1169. doi: 10.1021/bi00896a027. [DOI] [PubMed] [Google Scholar]
- So A.G., Davie E.W. Effects of amino acids, s‐RNA, and ethanol on coding ambiguity with polyuridylic acid. Biochemistry. 1965;4:1973–1979. [Google Scholar]
- Tamerler C., Ariyo B., Bucke C., Keshavarz T. Effect of mannan and alginate oligosaccharides on production in bioreactors of penicillin G and its biosynthetic intermediates. Ann Microbiol. 2001;51:53–60. [Google Scholar]
- Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiao Y., Ding Y., Wang Z. Effects of La3+, Ce3+, Nd3+ on growth and carotenoids content of Phaffia rhodozyma. Weishengwuxue Tongbao. 1999;26:117–119. [Google Scholar]
- Weinberg E.D. Roles of trace metals in transcriptional control of microbial secondary metabolism. Biol Metals. 1990;2:191–196. doi: 10.1007/BF01141358. [DOI] [PubMed] [Google Scholar]
- Williams R.B., Henrikson J.C., Hoover A.R., Lee A.E., Cichewicz R.H. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Yan L., Boyd K.G., Burgess J.G. Surface attachment induced production of antimicrobial compounds by marine epiphytic bacteria using modified roller bottle cultivation. Mar Biotechnol. 2002;4:356–366. doi: 10.1007/s10126-002-0041-x. [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhao Y., Zhang M., Wei Z., Miao K., Sun W. Oxidative stress response of Inonotus obliquus induced by hydrogen peroxide. Med Mycol. 2009;47:814–823. doi: 10.3109/13693780802653933. [DOI] [PubMed] [Google Scholar]
- Zhu L.‐W., Zhong J.‐J., Tang Y.‐J. Significance of fungal elicitors on the production of ganoderic acid and Ganoderma polysaccharides by the submerged culture of medicinal mushroom Ganoderma lucidum. Process Biochem. 2008;43:1359–1370. [Google Scholar]


