Summary
Paenibacillus sp. F6‐B70 was selected from several dozens of isolates with activity against methicillin‐resistant Staphylococcus aureus using a 16S rDNA‐based screening method. F6‐B70 contained polyketide synthase (PKS) and non‐ribosomal peptide synthetase (NRPS) clusters in its genome revealed by PCR amplification of conserved adenylation and ketosynthase (KS) domains. Phylogenetic data suggested that the strain hosts trans‐AT PKSs and their product may be a branched molecule. An antibiotic was subsequently isolated from the methanol extract of F6‐B70 cells. The molecular formula of the antibiotic was deduced to be C33H50NaO6 ([M + Na]+, m/z 565.3505) by analysis of electrospray ionization mass spectral data. Elucidation of the structure by nuclear magnetic resonance and infrared spectroscopy revealed that the active compound, paenimacrolidin (PAM), was a novel 22‐membered macrolide with side‐chains. The new antibiotic, mainly as a bacteriostatic agent, inhibits a couple of multidrug‐resistant Staphylococcus sp. strains. The antibiotic capacity of PAM was compromised by its instability, which can be overcome significantly with addition of an anti‐oxidant. To our knowledge, this is the first report of the isolation of an active macrolide from paenibacilli, which may be a promising source of novel antibiotics.
Introduction
The genus Paenibacillus was created in 1993, originally with 11 species transferred from the members of Bacillus group 3 and was substantially amended in 1997 (Ash et al., 1993; Shida et al., 1997). One of the interesting characteristics of paenibacilli is that some species of the genus are Gram‐negative or Gram‐variable although the Gram‐positive dominate, consistent with their Bacillus originality (Elo et al., 2001; Horn et al., 2005; Rodriguez‐Diaz et al., 2005; Roux et al., 2008). Paenibacillus species, isolated from a variety of ecological niches (Saha et al., 2005), produce a wide range of antibiotics active against different pathogens (Li et al., 2007). Almost half of the genus obtained from the natural environment are estimated to exhibit a broad inhibition spectrum against bacteria and pathogenic fungi (Lorentz et al., 2006). The main antimicrobial agents identified to date are peptide antibiotics, most of which, including polymyxin A to E, paenibacillin, jolipeptin, gavaserin, saltavalin, fusaricidin A to D and gatavalin, are produced by Paenibacillus polymyxa (Pichard et al., 1995; Kajimura and Kaneda, 1997; He et al., 2007). As members of polymyxin‐colistin‐circulin family, polymyxin B and E have been used in clinical practice for many years (Falagas and Kasiakou, 2006).
Many therapeutically useful secondary metabolites in bacteria are synthesized by enzymes encoded by modular biosynthetic gene clusters, such as polyketide synthase (PKS) clusters and non‐ribosomal peptide synthetase (NRPS) clusters (Donadio et al., 2007; Ridley et al., 2008; Scherlach and Hertweck, 2009). Macrolide‐type polyketides are synthesized by the type I polyketide synthases (PKS‐I), which are modularly organized giant enzymes (Weber et al., 2003). The canonical PKS‐I module is composed of at least three domains, acyltransferase (AT), β‐ketoacyl synthase (KS) and acyl carrier protein (ACP), that perform one round of polyketide elongation. Recent studies (Chen et al., 2007; Nguyen et al., 2008; Weber et al., 2008), however, revealed the presence of two different types of PKS‐I systems: the trans‐AT PKS‐Is found in bacilli, myxobacteria and fungi, in which all PKS modules lack an AT domain and are complemented by ATs encoded by isolated genes (Kim and Fuerst, 2006; Simunovic et al., 2006; Nguyen et al., 2008); the hybrid PKS‐Is found in Streptomyces collinus, in which both cis‐ and trans‐AT PKS co‐exist in the genome (Weber et al., 2008). In contrast to the canonical (cis‐AT) KSs that usually cluster tightly into natural product‐specific clades, trans‐KSs generally group according to their substrate types (Nguyen et al., 2008). In paenibacilli, the major antimicrobial secondary metabolites are peptides, predominantly lipopeptides (e.g. polymyxins, fusaricidins), synthesized by NRPS (Li et al., 2007; Choi et al., 2009). While a few macrolide‐type polyketides have been isolated and characterized from the related genus Bacillus (Chen et al., 2006; Schneider et al., 2007), no polyketides produced by Paenibacillus have been reported to date.
It is conceivable that the traditional culture‐based, bioassay‐guided strategies used to screen bioactive secondary metabolites can only discover a small fraction of the biosynthetic capacity encoded in genomes (Bentley et al., 2002; Gross et al., 2007). To facilitate discoveries of novel antimicrobial agents, molecular techniques have been widely employed to determine the presence of target synthesis genes in an organism, which is a good indicator for production of novel active molecules by the organism (McAlpine et al., 2005; Banskota et al., 2006; Komaki et al., 2008; Scherlach and Hertweck, 2009). Since the diversity of biosynthetic pathways can be identified rapidly by polymerase chain reaction (PCR) amplification and sequencing of DNA fragments encoding NRPS/PKS genes, the method is often used to evaluate the genetic potential of newly isolated microorganisms to produce novel compounds (Busti et al., 2006; Kim and Fuerst, 2006).
The increased incidence of bacterial antibiotic resistance has encouraged the search for novel potent antimicrobial agents. Here, we report the discovery of a novel macrocyclic lactone paenimacrolidin (PAM) from Paenibacillus sp. F6‐B70 using antimicrobial screening, 16S rDNA sequencing and in silico analysis of the PKS gene clusters. The subsequent structural and biological characterization demonstrates that PAM possesses the antibiotic activity against methicillin‐resistant Staphylococcus aureus (MRSA), likely through a bacteriostatic mechanism.
Results
Isolation and identification of F6‐B70
Using a competitive inhibition method, a large number of soil bacterial isolates were screened for antimicrobial activity against MRSA ATCC43300 and several dozens showed a distinct area of inhibition on nutrient agar. To rapidly select new strains with a high potential to produce novel antibiotics, 16S rDNA sequences of these obtained isolates were amplified, sequenced and analysed. The 16S rDNA (GenBank Accession No. GQ240305) of one isolate, namely F6‐B70, exhibited a low similarity to that of known bacteria. A consensus phylogenetic tree was constructed (Fig. 1) based on the almost‐complete 16S rDNA gene sequences of F6‐B70 and closely related type strains. F6‐B70 formed a distinct branch within the genus Paenibacillus and was closely related to Paenibacillus ehimensis DSM 11029T (Kim et al., 2004; Lee et al., 2004), with a sequence similarity of 97.1%. The DNA–DNA re‐association between F6‐B70 and P. ehimensis DSM 11029T was 21.8%. All these data indicate that F6‐B70 is a new member of the genus Paenibacillus.
Figure 1.
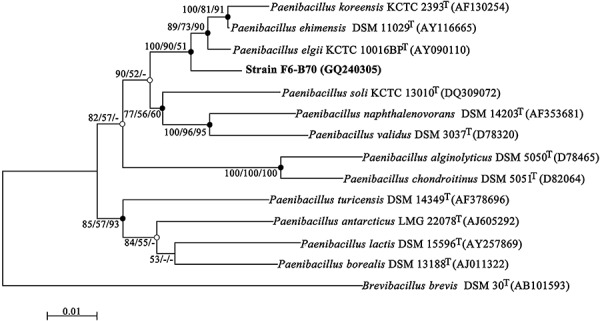
Unrooted neighbour‐joining tree for F6‐B70 within paenibacilli. Closed circles indicate generic branches that were also recovered using the maximum‐parsimony and maximum‐likelihood algorithms; open circles indicate generic branches that were also recovered using the maximum‐parsimony algorithm. Numbers at nodes by the order of neighbour‐joining/maximum‐parsimony/maximum‐likelihood indicate bootstrap values as percentages of 1000, 1000 and 100 replicates. Values lower than 50% are not indicated at the branch points. The 16S rDNA gene sequence of Brevibacillus brevis DSM 30T was included as an outgroup. Bar, 0.01 substitutions per nucleotide position.
The F6‐B70 strain formed pale yellow colonies on nutrient agar, and grew well in nutrient broth under aerobic conditions. Under a transmission electron microscope, the cells of F6‐B70 appeared to be rod‐shaped with peritrichous flagella, suggesting that it is motile. While F6‐B70 was able to form endospores like bacilli, Gram staining showed that it was negative. Aerobic growth of F6‐B70 occurred at 4–37°C and pH 5–10, with optima at 30°C and pH of 7. Strain F6‐B70 tolerated up to 2% NaCl in nutrient broth. The major fatty acid of strain F6‐B70 was anteiso‐C15:0, making up to 58.15% of the total fatty acids, a characteristic of the genus Paenibacillus. The content of iso‐C15:0 was relatively high (17.95%) but the content of iso‐C16:0 (0.93%) was low, which readily distinguished F6‐B70 from P. ehimensis DSM 11029T, possessing 8.1% iso‐C15:0 and 8.6% iso‐C16:0 (Lee et al., 2004). The phenotypic, biochemical and chemotaxonomic characteristics further supported the proposal that F6‐B70 is a novel strain in the genus Paenibacillus.
Secondary metabolite genes
F6‐B70 was able to inhibit growth of MRSA ATCC43300, suggesting that it may contain genes for antimicrobial agents. Given that the analysis of biosynthetic genes for secondary metabolites can provide useful information to determine the nature of the antibiotics in a test organism, attempts to identify PKS and NRPS genes in F6‐B70 were made. Using two primer sets for conserved regions of PKS and NRPS, the isolate yielded distinct bands of the expected size based on sequenced bacterial PKS and NRPS genes. For validation, the PCR products were cloned, sequenced and compared with the available sequences of secondary metabolite genes in GenBank. Among the seven NRPS clones sequenced, four were unique (Table 1). The deduced amino acid sequences of these four NRPS DNA fragments displayed sequence identities of 50.4–62.9% to the known NRPS proteins. On the basis that the NRPS primers were designed to amplify the DNA segment encoding the amino acid specificity pocket of adenylation (A) domains, including 9 of 10 diagnostic residues (Schwarzer et al., 2003; Ehling‐Schulz et al., 2005), the sequences were further analysed for the active‐site residues responsible for amino acid recognition (Rausch et al., 2005). As shown in Table 1, the predicted substrates of the deduced NRPS proteins of F6‐B70 varied. Combined with the low sequenced identities, this result suggests the presence of novel secondary metabolites in the strain.
Table 1.
NRPS/PKS sequences from strain F6‐B70.
| Seq. IDa (accession No.) | Best match of blastx search | Substrateb | ||
|---|---|---|---|---|
| Identity (%) | Genes (accession No., compound) | Origin | ||
| NRPS‐1 (GQ262803) | 62.9 | mycC (AAF08797, mycosubtilin) | Bacillus subtilis | Ser |
| NRPS‐2 (GQ262804) | 62.3 | pmxA (ACA97576, polymyxin) | Paenibacillus polymyxa | Phe/Val |
| NRPS‐3 (GQ262805) | 57.8 | lchA (CAA71582, lichenysin) | Bacillus subtilis | Ile |
| NRPS‐4 (GQ262806) | 50.4 | fusA(ABX38811, fusaricidin) | Paenibacillus polymyxa | Ile |
| PKSI‐2 (GQ262808) | 77.7 | dfnG (ABS74561, difficidin) | Bacillus amyloliquefaciens | Ethylene |
| PKSI‐5 (GQ262809) | 75.2 | mxaN (ABF92489, Myxovirescin) | Myxococcus xanthus | β‐Branch |
| PKSI‐3 (GQ262810) | 73.3 | dfnJ (ABS74558, difficidin) | Bacillus amyloliquefaciens | β‐Branch |
| PKSI‐4 (GQ262811) | 72.6 | dfnJ (ABS74558, difficidin) | Bacillus amyloliquefaciens | β‐Branch |
NRPS (non‐ribosomal peptide synthetase) and PKS (polyketide synthase) prefixes indicate clones deriving from NRPS and PKS PCRs, respectively.
Predicted substrate activated by the corresponding A (adenylation) domain or KS (ketosynthase) domain.
For PKS, 22 independent clones were sequenced and only four different nucleotide sequences were obtained (Table 1). The deduced amino acid sequences of these four unique DNA fragments were highly related (72.6–77.7% identity) to known trans‐AT PKS sequences, suggesting that F6‐B70 hosts trans‐AT PKSs. To verify the structure of PKS in F6‐B70, the primers derived from two independent KS domains were used to amplify DNA fragments in between and a 4209 bp fragment was obtained. Sequence alignment with representative PKS clusters revealed that the fragment contains five domains, including two KS, one dehydratase (DH), one ketoreductase and one ACP (Fig. 2A) (Weber et al., 2008), confirming that strain F6‐B70 contains trans‐PKS. It has been reported that the ∼230‐amino‐acid internal region of KS domains generated by PCR was sufficient to provide structural information (Nguyen et al., 2008). We thus constructed a phylogenetic tree using a neighbour‐joining algorithm based on the unique PKS sequences of F6‐B70 and the closest homologues. As shown in Fig. 2B, all of these four KS sequences were clustered into trans‐AT PKSs clades, indicating that F6‐B70 hosts trans‐AT PKSs. Three KS sequences, PKSI‐3, PKSI‐4 and PKSI‐5, belong to clade I, implicating a branched substrate. On the contrary, PKSI‐2 was grouped into the single‐bond subclade of clade V, suggesting that the substrate may contain a single bond between two specific carbons.
Figure 2.
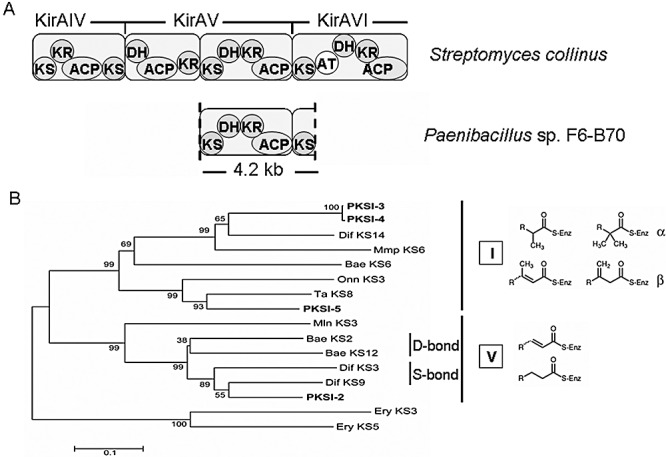
Sequence analyses of KS domains of Paenibacillus sp. F6‐B70. A. A complete module was deduced from a sequenced fragment between two KS domains. The Kirromycin gene cluster was used as the reference as it contains both cis‐ and trans‐AT PKSs (Weber et al., 2008). B. Neighbour‐joining cladogram of KS domains. Bars indicate 10% amino acid sequence divergence. KS numbering refers to the position within the gene cluster starting from the upstream end. The cis‐AT KS3 and KS5 from the erythromycin PKS was used as outgroup. Clade types were shown in roman numbers together with the main substrate type (detailed in Nguyen et al., 2008). The accession numbers of sequences from GenBank database are Bae KS2, ABS74061; Bae KS6, ABS74062; Bae KS12, ABS74064; Dif KS3, ABS74564; Dif KS9, ABS74561; Dif KS14, ABS74558; Ery KS3, AAV51821; Ery KS5, AAV51822; Mln KS3, ABS73797; Mmp KS6, AAM12913; Onn KS3, AAV97870; Ta KS8, ABF92489. Abbreviations: Bae, bacillaene; Dif, difficidin; Mmp, mupirocin; Mln, macrolactin; Onn, onnamide; Ta, myxovirescin.
Production and isolation of antibiotics
F6‐B70 grew well and produced active compounds in nutrient broth. While the optimal temperature for bacterial growth was 30°C, production of the antibiotic was maximal at 28°C. The antimicrobial activity reached the detectable level in the nutrient broth approximately 12 h after inoculation, corresponding to the end of the exponential growth phase, and the maximum level at the stationary phase. The antimicrobial activity declined after 24 h of bacterial growth, and disappeared completely after incubation for 84 h. In the fermentation supernatants and the methanol extracts of collected cells, only one antibiotic was detected by high‐pressure liquid chromatography (HPLC) and the antibacterial assay with the same retention time and UV spectra, indicating that the agents in the supernatants and methanol extracts were the same. The maximum UV absorption of the antibiotic was 277 nm, suggesting that it was a triene compound. The antibacterial activity was not detected when the fermentation supernatants were concentrated at 80°C for 10 min. The methanol extracts of F6‐B70 cells therefore were chosen for further studies. The active compound, namely PAM, was purified as described in Experimental procedures with a yield of 136 mg per 48 l of culture medium under the optimal conditions.
Structure elucidation of PAM
PAM was a pale yellow amorphous powder, [α]20D−13.5 (c = 0.57, CH3OH). UV (CH3OH) λmax (logε): 277 (4.57). IR (KBr) νmax: 3338 (OH), 2925, 1724 (C=O), 1669, 1376, 1243, 1071 and 970 cm−1. ESI‐MS (m/z): 565 [M + Na]+. HR‐ESIMS (m/z): 565.3500 [M + Na]+ (calculated for C33H50NaO6, 565.3505). The nuclear magnetic resonance (NMR) spectral data are given in Table 2.
Table 2.
Nuclear magnetic resonance (NMR) data for PAM in CD3OD.
| No. | δHa | δCb | DEPTc |
|---|---|---|---|
| 1 | – | 172.4 | C |
| 2 | 2.44 (2H, m) | 44.4 | CH2 |
| 3 | 4.04 (1H, m) | 67.2 | CH |
| 4 | 1.72 (1H, m); 1.54 (1H, m) | 45.6 | CH2 |
| 5 | 4.22 (1H, m) | 72.0 | CH |
| 6 | 5.38 (1H, dd, J = 15.1, 7.6) | 136.1 | CH |
| 7 | 5.70 (1H, m) | 130.7 | CH |
| 8 | 1.43 (1H, m); 1.50 (1H, m) | 39.2 | CH2 |
| 9 | 3.63 (1H, m) | 69.9 | CH |
| 10 | 1.17 (1H, m); 1.47 (1H, m) | 43.6 | CH2 |
| 11 | 1.85 (1H, m) | 30.4 | CH |
| 12 | 2.07 (1H, m); 2.16 (1H, m) | 38.9 | CH2 |
| 13 | 3.97 (1H, m) | 73.9 | CH |
| 14 | 2.14 (1H, m); 2.28 (1H, m) | 41.7 | CH2 |
| 15 | 5.44 (1H, m) | 134.0 | CH |
| 16 | 5.34 (1H, m) | 127.4 | CH |
| 17 | 2.04 (2H, m); | 30.3 | CH2 |
| 18 | 5.47 (1H, m) | 134.4 | CH |
| 19 | 5.49 (1H, m) | 135.5 | CH |
| 20 | 2.41 (1H, m) | 43.0 | CH |
| 21 | 5.18 (1H, t, J = 7.9) | 80.3 | CH |
| 22 | 5.65 (1H, dd, J = 15.6, 7.9) | 128.7 | CH |
| 23 | 6.90 (1H, d, J = 15.6) | 132.1 | CH |
| 24 | – | 134.2 | C |
| 25 | 6.28 (1H, d, J = 11.6) | 126.8 | CH |
| 26 | 6.52 (1H, t, J = 11.3) | 125.6 | CH |
| 27 | 5.83 (1H, m) | 137.9 | CH |
| 28 | 2.95 (2H, t, 6.4) | 32.9 | CH2 |
| 29 | 5.44 (1H, m) | 130.4 | CH |
| 30 | 4.97 (1H, dd, J = 10.1, 1.6) | 115.7 | CH2 |
| 5.02 (1H, dd, J = 15.4, 1.6) | |||
| 31 | 1.90 (3H, s) | 21.2 | CH3 |
| 32 | 1.00 (3H, d, J = 6.9) | 21.5 | CH3 |
| 33 | 0.96 (3H, d, J = 6.9) | 17.6 | CH3 |
In p.p.m.; multiplicity: singlet (s), doublet (d), triplet (t) and multiplet (m); J in Hz.
In p.p.m. Recorded at 125 MHz.
Multiplicity of Distortionless Enhancement by Polarization Transfer (DEPT) determined by distortionless enhancement by polarization transfer.
PAM had a molecular formula of C33H50O6based on the NMR and HR‐ESIMS data, suggesting the presence of nine double‐bond equivalents. Strong IR absorption bands at 1724 cm−1 were attributable to ester carbonyls, as confirmed by 13C‐NMR signals at δ 172.4 (Table 2). The 1H NMR spectrum of PAM displayed signals for three Me groups [δ 1.90 (s), 1.00 (d, J = 6.9 Hz), and 0.96 (d, J = 6.9 Hz)], a typical AB spin system due to a terminal double bond [δ 4.97 (dd, J = 10.1, 1.6); 5.02 (dd, J = 15.4, 1.6)], 12 olefinic protons, five oxygenated CH groups (δ 5.18, 4.22, 4.04, 3.97 and 3.63), and high‐field proton signals arising from the saturated methylenes (Table 2). Consistent with the molecular formula of PAM, a total of 33 signals were observed in the 13C NMR spectrum, comprising two quaternary, 18 tertiary and 10 secondary C‐atoms, as well as three Me groups. Among them, the 14 downfield carbon signals at δ 137.9–115.7 p.p.m. were due to double‐bond functions. Eight of the nine degrees of unsaturation were due to one carboxyl group and seven double bonds, indicating the monocyclic nature of the new compound. These findings indicated that PAM was a highly unsaturated macrolide with a side‐chain.
The analysis of the 1H NMR, 13C NMR and HMQC spectra of PAM enabled us to assign all the protons to their bonding carbons. Two subunits comprising C‐2 to C‐23 including the methyl group carbons C‐32 and C‐33, and C‐25 to C‐30 indicated by the bold lines (Fig. 3) were mainly established by 1H–1H COSY spectra. The assembly of the two subunits, C‐atoms and hetero‐atoms, was achieved based on clear HMBC findings (Fig. 3). The chemical shift of carbon C‐21 (δH 5.18 p.p.m.; δc 80.3 p.p.m.) and the required ring double‐bond equivalents indicated a ring closure between C‐21 and C‐1 to construct a 22‐membered macrolide, which was confirmed by the cross‐peak of H‐21/C‐1 in the HMBC spectrum. The correlations of the H‐31/C‐23, H‐31/C‐24 and H‐31/C‐25 indicated the presence of a triene moiety, and the absorption maximum at 277 nm (logε 4.57) exhibited in the UV spectrum supported this conclusion. Large coupling constants (Table 2) showed the carbon–carbon double bonds Δ6,7 and Δ22,23 of PAM to have E geometries, as shown in Fig. 3. The geometry of Δ26,27 was determined as Z by the small coupling constant. The E geometry of Δ24,25 was deduced by the NOESY correlation between H‐31 and H‐25. The overlapping proton signals of Δ15,16 and Δ18,19 made it difficult to calculate their coupling constants, so the NOESY spectra were applied to establish the geometry of Δ15,16 and Δ18,19. The proton signal of H‐14 at δ 2.28 corresponded with that of H‐16 (δ 5.34), suggesting the E geometry of Δ15,16. The configuration of Δ18,19 as Z was also solved by the corresponding NOESY correlation of H‐32/H‐17. In summary, the structure of PAM was elucidated as a 22‐membered macrolide with a side‐chain, which was a unique highly unsaturated moiety with a conjugated triene and a terminal double bond.
Figure 3.

Structure of paenimacrolidin (PAM) isolated from strain F6‐B70 (A) and selected 2D‐NMR correlations of the new compound (B).
Antimicrobial activities
The antimicrobial activities of purified PAM and relevant antibiotics in clinical use were compared against reference strains and clinical isolates using the agar diffusion method (Table 3). The results showed that PAM had an inhibitory effect on growth of MRSA and ampicillin‐resistant Staphylococcus epidermidis with a similar or even more potent antimicrobial activity than vancomycin (VAN). Quantitative experiments were subsequently performed to determine the minimum inhibitory concentration (MIC), the concentration that totally prevented microbial growth in Mueller–Hinton broth at 37°C for 18 h (Table 4). The MICs of PAM (16–32 µg ml−1) were unexpectedly higher than those of VAN (1–2 µg ml−1) (Table 4), which was not consistent with the qualitative experiments.
Table 3.
Antibacterial activity of PAM in comparison with reference compounds.
| Strain | Inhibition zone size (mm) | |||
|---|---|---|---|---|
| PAM (10 µg) | PAM (30 µg) | AMP (10 µg) | VAN (30 µg) | |
| S. aureus ATCC25923 | 19.8 ± 2.1 | 20.2 ± 1.2 | 28.6 ± 4.6 | 19.2 ± 0.8 |
| S. aureus ATCC29213 | 22.0 ± 2.6 | 22.8 ± 1.8 | 24.0 ± 3.3 | 22.1 ± 1.2 |
| MRSA ATCC43300 | 16.7 ± 2.0 | 17.0 ± 1.4 | 19.5 ± 2.7 | 16.7 ± 0.5 |
| MRSA B6172 | 20.1 ± 3.1 | 20.5 ± 1.1 | 0 | 18.4 ± 0.4 |
| MRSA B4524 | 18.7 ± 2.2 | 19.4 ± 1.5 | 0 | 17.9 ± 0.9 |
| S. epidermidis CMCC26069 | 21.9 ± 2.9 | 24.1 ± 1.4 | 0 | 22.1 ± 3.2 |
| B. subtilis CGMCC1.1470 | 0 | 0 | 30.5 ± 4.1 | 24.3 ± 2.6 |
| E. faecalis ATCC29212 | 0 | 0 | 22.0 ± 2.0 | 17.9 ± 1.0 |
| E. coli ATCC35218 | 0 | 0 | 20.7 ± 3.5 | 0 |
Table 4.
The MICs of PAM and reference compounds.
| Strain | MIC (µg ml−1)a | ||
|---|---|---|---|
| PAM | VAN | AMP | |
| S. aureus ATCC25923 | 32 | 2 | 0.125 |
| S. aureus ATCC29213 | 16 | 1 | 1 |
| MRSA ATCC43300 | 32 | 1 | 4 |
The MIC was defined as the antibiotic concentrations that totally prevent microbial growth in Mueller–Hinton broth at 37°C for 18 h.
To further evaluate the antimicrobial activity against S. aureus strains, time‐kill experiments were conducted. As shown in Fig. 4, PAM under 32 µg ml−1 was effective in inhibiting growth of either methicillin‐sensitive S. aureus ATCC25923 or MRSA ATCC43300 for up to 6 h but lost the activity completely after 24 h. When the concentration of the chemical increased to128 µg ml−1 it restrained growth of either strain but hardly reduced log10cfu ml−1 values through the 24 h test period. Evidently, these data indicate that PAM is a bacteriostatic rather than bactericidal agent.
Figure 4.
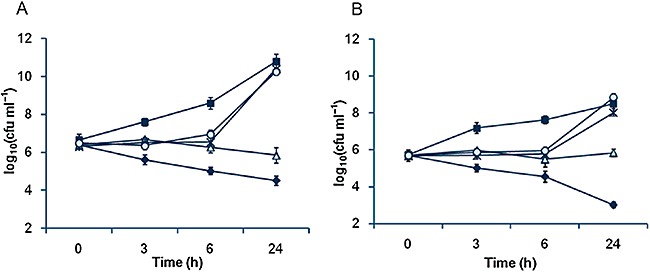
Antibacterial activity of PAM against S. aureus by time‐kill curve methods against (A) MRSA ATCC43300 and (B) S. aureus ATCC25923 strains. In both panels, non‐PAM control, closed square; 1 µg ml−1 PAM, open circle; 32 µg ml−1 PAM, asterisk; 128 µg ml−1 PAM, open triangle; 4 µg ml−1 vancomycin (VAN), closed diamond.
Considering that PAM is a member of the polyene family, which is sensitive to pH, temperature, oxygen and light, we speculate that the unusual high MICs of PAM may be due to its instability. To this end, the stability of PAM in Mueller–Hinton broth at 37°C was accessed. As shown in Fig. 5A, the remaining PAM was rapidly reduced to 20.6%, 6%, 3.8%, 0% of the initial concentration (128 µg ml−1) at 6, 12, 18 and 24 h after inoculation, respectively. A similar phenomenon was observed in LB medium at 37°C or 28°C (data not shown). These data indicate that the decay of PAM appears to be exponential with a half‐life of approximately 4 h, suggesting that the chemical is unstable.
Figure 5.
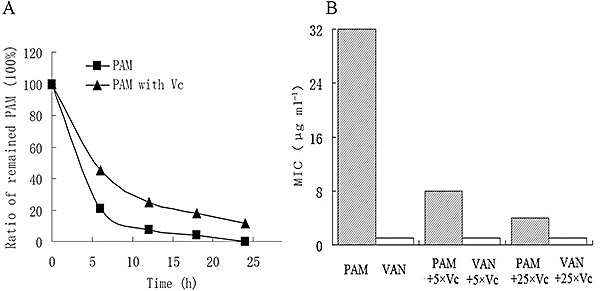
The relationship of the antibiotic activity of PAM and its stability. MIC was defined as the lowest antibiotic concentration that completely prevented visible growth of MRSA ATCC43300 after incubation at 37°C for 18 h. A. PAM (128 µg ml−1) was prepared in sterile Mueller–Hinton broth without ( ) or with VC (208 µg ml−1) (▴), and incubated at 37°C. Compared with the initial concentration of PAM, the ratio of the remaining PAM was determined after incubation at 37°C for 6, 12, 18 and 24 h, by HPLC analysis on a Diamonsil C18 reversed‐phase column. B. The MICs of PAM or VAN without or with VC in the molar ratio of 1:25 and 1:5, respectively. Abbreviations: PAM, paenimacrolidin; VAN, vancomycin; VC, l‐ascorbic acid.
) or with VC (208 µg ml−1) (▴), and incubated at 37°C. Compared with the initial concentration of PAM, the ratio of the remaining PAM was determined after incubation at 37°C for 6, 12, 18 and 24 h, by HPLC analysis on a Diamonsil C18 reversed‐phase column. B. The MICs of PAM or VAN without or with VC in the molar ratio of 1:25 and 1:5, respectively. Abbreviations: PAM, paenimacrolidin; VAN, vancomycin; VC, l‐ascorbic acid.
Attempts of reducing the decay rate of PAM were made with the supplementation of l‐ascorbic acid (Vitamin C, VC). VC is a well‐known anti‐oxidant and its MIC against MRSA ATCC43300 is too high (> 4 mg ml−1) to interfere the results. The addition of VC to the solution of PAM with the molar ratio of 25:1 or 5:1, significantly inhibited the rapid increase of PAM's MICs (Fig. 5B). In the presence of VC at 25:1 and 5:1, the MICs of PAM were reduced roughly eight and four times, respectively. In addition, the decay assay revealed that the half‐life of PAM was about 5.5 h when VC (208 µg ml−1, the molar ratio of VC to PAM was 5:1) was added (Fig. 5A). Similar results were obtained when VC was substituted with vitamin E (data not shown). All these results indicate that PAM is a unique antimicrobial agent active against MRSA ATCC43300 and ampicillin‐resistant S. epidermidis although it is relatively unstable compared with the commercialized counterparts. Moreover, the chemical can be substantially stabilized by the addition of antioxidants, suggesting great possibilities for further improvement.
Discussion
For many years actinomycetes have been prolific sources of antibiotics (Nguyen et al., 2008; Ridley et al., 2008). Only recently, bacteria in other taxonomic groups loom as a practical source for new bioactive compounds and several successful attempts have been made (Zhang et al., 2006; Wright and Sutherland, 2007; Blunt et al., 2009). In this study, new environmental isolates displaying antimicrobial activity were subjected to 16S rDNA sequence screening to exclude the well‐documented antibiotic producers. The method proved to be effective and efficient, generating a number of candidates for further exploration. Among them, F6‐B70 warrants further investigation because (i) the strain belongs to the genus Paenibacillus, whose type species P. polymyxa is renowned for its ability to produce antimicrobial chemicals (Beatty and Jensen, 2002; Raza et al., 2008), (ii) the genus used to be classified into Bacillus, which has emerged as a promising antibiotic source too (Mannanov and Sattarova, 2001; Butcher et al., 2007; Nguyen et al., 2008), (iii) the only sequenced strain Paenibacillus sp. JDR‐2 has an unusually large genome of 7.2 MB, comparable to those of actinomycetes (http://img.jgi.doe.gov), and (iv) little has been done to identify active metabolites in other paenibacilli.
We initially speculated that the antimicrobial chemical(s) produced by Paenibacillus sp. F6‐B70 would be peptides on the basis that it is the case of the closely related strain P. polymyxa (Raza et al., 2008). It is serendipitous that PCR with primers for KS domain sequences generated the products with expected size. Both KS sequence analysis and complete module sequencing indicated that F6‐B70 hosts trans‐AT PKSs. Furthermore, all sequenced A domains of NRPS and KS domains of PKS showed less than 80% sequence similarity to the known secondary metabolic genes, strongly suggesting that the bacterium may host novel enzymes for secondary metabolite synthesis. Interestingly, no PKS analogy was identified in the genome of Paenibacillus sp. JDR‐2 despite of its enormous size. Unlike cis‐AT PKSs whose evolution is dominated by vertical KS acquisition, horizontal gene transfer (HGT) is a major driving force in evolution of trans‐AT PKSs (Nguyen et al., 2008; Ridley et al., 2008). In fact, HGT is particularly favoured in Bacillus and its close relatives, from which exclusively trans‐AT clusters have been identified, by natural transformation (Stewart and Carlson, 1986; Albertini et al., 1995; Chen et al., 2006). By the same token, the genus Paenibacillus appears to be a promising frontier for new antimicrobial chemicals.
Paenimacrolidin, displaying inhibition activity against drug‐resistant staphylococcal strains, was isolated and purified from Paenibacillus sp. F6‐B70. Although the presence of multiple NRPS A and KS domains in strain F6‐B70 implicates more than one metabolite, PAM is the only antimicrobial chemical obtained from supernatants and methanol extracts of cells. This may not be surprising given that the number of genes encoding biosynthetic enzymes typically outnumbers the isolated secondary metabolites in most microorganisms (Scherlach and Hertweck, 2009). Nevertheless, it is equally likely that the culture conditions adopted in the present study may not have allowed expression of all NRPS and PKS genes. Therefore, a great effort will be required to explore the fermentation conditions in order to gain full access to the untapped reservoir of potentially bioactive agents.
PAM is a novel macrocyclic lactone antibiotic containing three separate monoene structure elements in a 22‐membered lactone ring. To our knowledge, a macrolide produced by paenibacilli has never been reported before. Macrolides are widespread secondary metabolites and have been isolated from the related genus Bacillus, such as macrolactins, difficidin and oxydifficidin (Schneider et al., 2007), all of which demonstrate potent antibacterial or antiviral, and cytotoxic activities (Zimmerman et al., 1987; Romero‐Tabarez et al., 2006). It is worth noting that one of macrolactins, 7‐O‐malonylmacrolactin A (MMA), was effective against many multidrug‐resistant Gram‐positive bacterial pathogens, including MRSA and VAN‐resistant enterococci (Romero‐Tabarez et al., 2006). The antibiotic capacity of PAM was compromised by its instability. This is not surprising because PAM may share many of the drawbacks of erythromycin and other macrolide antibiotics, including acid intolerance and a short serum half‐life. In addition, the instability of PAM may be explained in part by its highly unsaturated moiety with a conjugated triene and a terminal double bond, which is likely sensitive to pH, temperature, oxygen and light. Although the instability can be rescued significantly by addition of anti‐oxidant agents such as VC and VE, structural changes by chemical modification may introduce more profound improvements (Zuckerman, 2004). Substitutions of hydroxyl group(s) with a methoxy group may be worth trying on the basis that such a substitution drastically increases stability of erythromycin (Watanabe et al., 1993).
Given that multiple NRPS/PKS gene clusters are present in a genome, to assign the genes responsible for synthesis of certain metabolites remains challenging. Recently, macrolactin A and difficidin/oxydifficidin were found to be synthesized by enzymes encoded by gene clusters pks2 and pks3, respectively, in Bacillus amyloliquefaciens FZB42 using cassette mutagenesis in combination with advanced mass spectrometric techniques (Chen et al., 2006; Schneider et al., 2007). However, this strategy is not readily applicable to Paenibacillus sp. F6‐B70 because the strain is found to be hardly amicable to genetic manipulation after numerous attempts. Instead, in silico analyses appear to be feasible to correlate KS domains with the identified chemical PAM given that KSs of trans‐AT PKSs generally form clusters according to their substrate specificity (Nguyen et al., 2008). In this study, four partial KS domains were obtained by PCR and the analysis of these KSs revealed two lines of evidence in support of the notion that most, if not all, of them are related with the biosynthesis of PAM although direct evidence correlating any of the genes with PAM was lacking. First, the deduced amino acid sequences of three KS domains are found to share high levels of similarity to those of PKSs for difficidin/oxydifficidin synthesis, consistent with the finding that the structure of PAM is similar to that of difficidin/oxydifficidin (Zimmerman et al., 1987; Zweerink and Edison, 1987). Second, three KS domains belong to clade I of the KS‐based phylogenetic tree, agreeing well with the finding that PAM is a β‐branched molecule (Fig. 2) (Nguyen et al., 2008).
In conclusion, a polyunsaturated macrolactone, PAM, was discovered from Paenibacillus sp. F6‐B70 by a combination of antimicrobial screening, 16S rDNA sequencing and analysis of its secondary metabolite genes. PAM is a unique bacteriostatic antibiotic that inhibits a couple of tested Staphylococcus sp. strains, including MRSA. Although PAM is unpleasantly unstable, addition of anti‐oxidants extends its activity substantially. On the basis of its bioactivity and physicochemical properties, PAM is considered a unique antibiotic among the few reported triene macrolides, which has potential for use on clinical isolates of MRSA. In addition, this study not only demonstrates that paenibacilli are a new group of bacteria promising for novel antibiotics but also provides an effective way to discover them.
Experimental procedures
Strain isolation and screening
Soil samples were collected from Tianmu Mountain National Natural Reserve, Zhejiang, China. Samples were air‐dried at 37°C for 3–5 days and screened for microorganisms that produce antimicrobial agents. Briefly, soil samples were suspended in sterile water and homogenized using a blender. The suspensions were serially diluted and plated on nutrient agar, potato dextrose (potato extracts 0.5%, glucose 2.0%) agar, Gause's synthetic agar (soluble starch 2.0%, KNO3 0.1%, K2HPO4 0.05%, MgSO4 0.05%, NaCl 0.05%, FeSO4 0.001%, agar 2.0%, pH 7.4) and YG (yeast extracts 1.0%, glucose 1.0%, pH 7.2) agar. The plates were incubated at 28°C for 14 days. The bacterial colonies were subsequently isolated and the isolates were maintained on nutrient agar. Each isolate pre‐cultured for 24–72 h at 28°C was inoculated on nutrient agar freshly seeded with MRSA ATCC43300. The plates were incubated at 28°C for 1–3 days to manifest inhibition areas. The isolates with significant inhibitory activity against MRSA were selected for further study.
16S rDNA analysis and DNA–DNA hybridization
To determine the 16S rDNA gene sequences of the isolates with obvious inhibitory activity against ATCC43300, genomic DNA was extracted and purified using a genomic DNA extraction kit (Axygen Scientific, Union City, CA) according to the instructions of the manufacturer. The 16S rDNA genes were amplified using PCR with the universal primers 27F (5′‐AGAGTTTGATCCTGGCTCAG‐3′) and 1541R (5′‐AAGGAGGTGATCCAGCCGCA‐3′) and sequenced (Weisburg et al., 1991). The 16S rDNA sequence of each isolate was aligned with related reference sequences retrieved from NCBI GenBank databases using blast. Isolate F6‐B70, showing a low similarity to that of known bacteria, was selected for further study. Phylogenetic analyses were constructed using neighbour‐joining and maximum‐parsimony algorithm within MEGA4 (Tamura et al., 2007), and using maximum‐likelihood algorithm within PHYLIP (Felsenstein, 2005) based on the 16S rDNA gene sequences of F6‐B70 and related organisms. The DNA–DNA hybridization between F6‐B70 and its closest related strain P. ehimensis DSM 11029T was performed by thermal denaturation method (De Ley et al., 1970).
Chemotaxonomic analyses, morphology and physiological characterizations of F6‐B70
Fatty acid methyl esters were extracted and determined by the Sherlock Microbial Identification system (MIDI, Newark, DE) according to the manufacturer's instructions. The isomer type of the diaminopimelic acid within the peptidoglycan was analysed as described previously (Hasegawa et al., 1983). The morphology of F6‐B70 was examined by Gram staining, spore staining and transmission electron microscope. Physiological characterization was performed to determine the temperature and pH ranges for growth and requirement for and tolerance to NaCl.
Analysis of secondary metabolite genes
To amplify ∼700 bp segments covering the adenylation (A) domain of NRPSs from F6‐B70 and the ketosynthase (KS) domain of PKSs, the primers NRPSA‐F/R (5′‐TSGCSATGGACCCSCAGCAG)/(5′‐CCSGTSCCGTGSGCCTCSAC) and PKSF/R (5′‐GGWCDACHGGHMANCCHAARGG)/(5′‐GGCAKCCATYTYGCCARGTCNCCKGT) were used with F6‐B70 genomic DNA as a template (Izumikawa et al., 2003; Ehling‐Schulz et al., 2005). The PCR products of expected size were cloned in the pMD19‐T vector and the inserted segments were sequenced. Sequences were blasted against published sequences in GenBank for homologous counterparts and a phylogenetic tree was constructed using a neighbour‐joining algorithm based on obtained KS sequences. To acquire the complete sequence of a PKS module, primers KSW‐F/R (5′‐ATTGAAGCGCACGGTACAGG)/(5′‐GCAGCGTCCGTCTTCAGATAGCAT), based on the two identified KS domains, were used.
Optimization of fermentation media
Strain F6‐B70 from the stock was streaked onto nutrient agar and incubated at 30°C for 72 h to generate starter cultures. The following five media were tested for antagonistic factor production: nutrient broth (peptone 0.5%, beef extract 3.0%, NaCl 0.5%, agar 1.5%), LB medium, potato dextrose broth, YS medium [2.6% yeast extracts, 4.0% starch, 0.05% K2HPO4, 0.25% MgSO4·7H2O, 0.4% (NH4)2SO4 and 0.3% CaCO3, pH 7.0] and YG broth. The liquid media were inoculated with starter cultures and incubated at 28°C, 200 r.p.m. for 72 h. The activity of compounds produced by the strain against MRSA ATCC43300, Escherichia coli ATCC35218 and Bacillus subtilis CGMCC 1.1470 was accessed every 24 h by the paper disc method. Among the five media tested, nutrient broth turned out to be the best medium to optimize the production of active compounds.
Production and purification of active compounds
Strain F6‐B70 was cultivated in 4 l of nutrient broth at 28°C for 24 h in a 7.5 l NBS Bioflo 110 Fermentor (New Brunswick Scientific, Edison, NJ). A total of 48 l of culture was used for isolation of active compounds. The cells pelleted by centrifugation were extracted twice with 2 l of methanol. The extract was concentrated with a Rotavapor at 37°C and the aqueous residual was removed by centrifugation at 10 000 g. The precipitation was resuspended in methanol and purified by preparative HPLC using a Hypersil BDS C8 column (250 by 20 mm; Dalian Elite, Dalian, China). The HPLC was performed on an Elite system (Dalian Elite, Dalian, China) with a linear gradient from 30% to 70% acetonitrile at a flow rate of 10 ml min−1 in 60 min and UV detection at 210 nm. Each peak was collected and tested for antimicrobial activity. Active fractions were evaporated to remove the acetonitrile and then freeze‐dried with a lyophilizer. The resulting purified antibiotic was subjected to structure elucidation, To prevent the rapid degradation of the active compound during preparation, samples at every step were stored at −20°C.
Spectroscopic measurements
The UV‐visible absorption spectra and optical rotation of the active compound were determined in methanol on a SHIMADZU UV‐2550 UV‐Vis scanning spectrometer (Shimadzu, Kyoto, Japan) and a JASCO P1030 Polarimeter (Tokyo, Japan). The infrared spectra were obtained with a NEXUS 870 PT‐IR spectrometer. High‐ and low‐resolution mass spectra were recorded on FTICR‐mass spectrometer (APEX III, Bruker, Bremen, Germany) and Bruker Esquire‐LC mass spectrometers (Bruker, Karlsruhe, Germany), respectively, equipped with electrospray ion source (ESI, positive ion mode). For nuclear magnetic resonance spectroscopy, the sample was dissolved in deuterated methanol‐d4, or chloroform‐d1, and the data were obtained with an AVANCE DMX‐600 spectrometer (Bruker) at 27°C.
Biological assay of in vitro activities
In vitro antimicrobial activities of products were tested using the paper disc method. Mueller–Hinton agar plates were inoculated with 0.1 ml of tested strain cells that had been pre‐cultured and diluted to the proper concentration (108 cells ml−1 ultimately). A paper disc (6 mm in diameter) impregnated with 10 µl of active compound was placed on the surface of the agar. The diameter of the inhibition zone was measured after incubation at 37°C for 18 h. The tested strains included S. aureus ATCC29213, S. aureus ATCC25923, S. epidermidis CMCC 26069, B. subtilis CGMCC 1.1470, Enterococcus faecalis ATCC29212, E. coli ATCC35218, MRSA ATCC43300, and several clinically isolated MRSA strains (B6172 and B4524).
The MIC was determined in a 96‐well microtitre plate using a method reported previously (Romero‐Tabarez et al., 2006). MeOH (10 µl) and Mueller–Hinton (meat extracts 0.2%, acid digest of casein 1.75%, starch 0.15%) broth alone were used as controls. The plate was incubated for 24 h and visible growth and optical density (OD) at 630 nm were recorded after 6, 12, 18 and 24 h of incubation and read with a Multiscan MK3 UV microtitre reader (THERMO ELECTRON, Shanghai, China). MIC was defined as the lowest antibiotic concentration that completely prevented visible growth after incubation at 37°C for 18 h. Vancomycin and ampicillin, freshly prepared in sterile‐distilled water, were used as positive controls. In addition, time‐kill experiments were performed to evaluate the antibacterial activity of PAM against S. aureus strains according to the procedure described previously (Jacqueline et al., 2003).
Stability studies
Purified active compound (128 µg ml−1) was prepared in sterile Mueller–Hinton broth and LB medium, and incubated at 28°C and 37°C for 24 h. The residual concentration of the antibiotic was determined every 6 h on a Diamonsil C18 reversed‐phase column (250 × 4.6 mm, 5 µm) of a SHIMADZU LC‐6A system (Shimadzu, Kyoto, Japan) with a linear gradient from 10% to 90% acetonitrile at a flow rate of 1 ml min−1 in 60 min, and UV detection at 280 nm by the HPLC analysis. The peak current exhibited a linear relationship with PAM concentration in the range of 1–150 µg ml−1.
Nucleotide accession numbers
The nucleotide sequence of 16S rDNA of strain F6‐B70 has been deposited in the GenBank under the Accession No. GQ240305. The sequences of secondary metabolite genes obtained in this study were also deposited in GenBank, and the corresponding accession numbers are reported in Table 1.
Acknowledgments
We gratefully acknowledge L.P. Shi, the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, for providing the NMR spectral data for PAM. We thank X.H. Jiang, College of Life Sciences, Zhejiang University, for providing the MS measurements. We also thank Sir Run Run Shaw Hospital for providing the MRSA test strains. This work was supported by Major State Basic Research Development Program (973 Program: 2010CB833803) to H.G. and X‐C.W.
References
- Albertini A.M., Caramori T., Scoffone F., Scotti C., Galizzi A. Sequence around the 159o region of the Bacillus subtilis genome: the pksX locus spans 33.6 kb. Microbiology. 1995;141:299–309. doi: 10.1099/13500872-141-2-299. [DOI] [PubMed] [Google Scholar]
- Ash C., Priest F.G., Collins M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Van Leeuwenhoek. 1993;64:253–260. doi: 10.1007/BF00873085. [DOI] [PubMed] [Google Scholar]
- Banskota A.H., Mcalpine J.B., Sørensen D., Ibrahim A., Aouidate M., Piraee M. Genomic analyses lead to novel secondary metabolites. Part 3. ECO‐0501, a novel antibacterial of a new class. J Antibiot (Tokyo) 2006;59:533–542. doi: 10.1038/ja.2006.74. et al. [DOI] [PubMed] [Google Scholar]
- Beatty P.H., Jensen S.E. Paenibacillus polymyxa produces fusaricidin‐type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can J Microbiol. 2002;48:159–169. doi: 10.1139/w02-002. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Chater K.F., Cerdeno‐Tarraga A.M., Challis G.L., Thomson N.R., James K.D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. et al. [DOI] [PubMed] [Google Scholar]
- Blunt J.W., Copp B.R., Hu W.‐P., Munro M.H.G., Northcote P.T., Prinsep M.R. Marine natural products. Nat Prod Rep. 2009;26:170–244. doi: 10.1039/b805113p. [DOI] [PubMed] [Google Scholar]
- Busti E., Monciardini P., Cavaletti L., Bamonte R., Lazzarini A., Sosio M., Donadio S. Antibiotic‐producing ability by representatives of a newly discovered lineage of actinomycetes. Microbiology. 2006;152:675–683. doi: 10.1099/mic.0.28335-0. [DOI] [PubMed] [Google Scholar]
- Butcher R.A., Schroeder F.C., Fischbach M.A., Straight P.D., Kolter R., Walsh C.T., Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.‐H., Vater J., Piel J., Franke P., Scholz R., Schneider K. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 2006;188:4024–4036. doi: 10.1128/JB.00052-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I. Comparative analysis of the complete genome sequence of the plant growth‐promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. et al. [DOI] [PubMed] [Google Scholar]
- Choi S.‐K., Park S.‐Y., Kim R., Kim S.‐B., Lee C.‐H., Kim J.F., Park S.‐H. Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J Bacteriol. 2009;191:3350–3358. doi: 10.1128/JB.01728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley J., Cattoir H., Reynaert A. Quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Donadio S., Monciardini P., Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- Ehling‐Schulz M., Vukov N., Schulz A., Shaheen R., Andersson M., Martlbauer E., Scherer S. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl Environ Microbiol. 2005;71:105–113. doi: 10.1128/AEM.71.1.105-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo S., Suominen I., Kampfer P., Juhanoja J., Salkinoja‐Salonen M., Haahtela K. Paenibacillus borealis sp. nov., a nitrogen‐fixing species isolated from spruce forest humus in Finland. Int J Syst Evol Microbiol. 2001;51:535–545. doi: 10.1099/00207713-51-2-535. [DOI] [PubMed] [Google Scholar]
- Falagas M.E., Kasiakou S.K. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care. 2006;10:R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Gross H., Stockwell V.O., Henkels M.D., Nowak‐Thompson B., Loper J.E., Gerwick W.H. The Genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Takizawa M., Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. [Google Scholar]
- He Z., Kisla D., Zhang L., Yuan C., Green‐Church K.B., Yousef A.E. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl Environ Microbiol. 2007;73:168–178. doi: 10.1128/AEM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M.A., Ihssen J., Matthies C., Schramm A., Acker G., Drake H.L. Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov. and Paenibacillus terrae strain MH72, N2O‐producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int J Syst Evol Microbiol. 2005;55:1255–1265. doi: 10.1099/ijs.0.63484-0. [DOI] [PubMed] [Google Scholar]
- Izumikawa M., Murata M., Tachibana K., Ebizuka Y., Fujii I. Cloning of modular type I polyketide synthase genes from salinomycin producing strain of Streptomyces albus. Bioorg Med Chem. 2003;11:3401–3405. doi: 10.1016/s0968-0896(03)00337-7. [DOI] [PubMed] [Google Scholar]
- Jacqueline C., Caillon J., Le Mabecque V., Miègeville A., Donnio P., Bugnon D., Potel R. In vitro activity of linezolid alone and in combination with gentamicin, vancomycin or rifampicin against methicillin‐resistant Staphylococcus aureus by time‐kill curve methods. J Antimicrob Chemother. 2003;51:857–864. doi: 10.1093/jac/dkg160. [DOI] [PubMed] [Google Scholar]
- Kajimura Y., Kaneda M. Fusaricidins B, C and D, new depsipeptide antibiotics produced by Bacillus polymyxa KT‐8: isolation, structure elucidation and biological activity. J Antibiot (Tokyo) 1997;50:220–228. [PubMed] [Google Scholar]
- Kim D.‐S., Bae C.‐Y., Jeon J.‐J., Chun S.‐J., Oh H.W., Hong S.G. Paenibacillus elgii sp. nov., with broad antimicrobial activity. Int J Syst Evol Microbiol. 2004;54:2031–2035. doi: 10.1099/ijs.0.02414-0. et al. [DOI] [PubMed] [Google Scholar]
- Kim T.K., Fuerst J.A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture‐dependent and culture‐independent approaches. Environ Microbiol. 2006;8:1460–1470. doi: 10.1111/j.1462-2920.2006.01040.x. [DOI] [PubMed] [Google Scholar]
- Komaki H., Fudou R., Iizuka T., Nakajima D., Okazaki K., Shibata D. PCR detection of type I polyketide synthase genes in myxobacteria. Appl Environ Microbiol. 2008;74:5571–5574. doi: 10.1128/AEM.00224-08. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.‐S., Pyun Y.‐R., Bae K.S. Transfer of Bacillus ehimensis and Bacillus chitinolyticus to the genus Paenibacillus with emended descriptions of Paenibacillus ehimensis comb. nov. and Paenibacillus chitinolyticus comb. nov. Int J Syst Evol Microbiol. 2004;54:929–933. doi: 10.1099/ijs.0.02765-0. [DOI] [PubMed] [Google Scholar]
- Li J., Beatty P.K., Shah S., Jensen S.E. Use of PCR‐targeted mutagenesis to disrupt production of fusaricidin‐type antifungal antibiotics in Paenibacillus polymyxa. Appl Environ Microbiol. 2007;73:3480–3489. doi: 10.1128/AEM.02662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz R.H., Artico S., Silveira A.B., Einsfeld A., Corcao G. Evaluation of antimicrobial activity in Paenibacillus spp. strains isolated from natural environment. Lett Appl Microbiol. 2006;43:541–547. doi: 10.1111/j.1472-765X.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- McAlpine J.B., Bachmann B.O., Piraee M., Tremblay S., Alarco A.‐M., Zazopoulos E., Farnet C.M. Microbial genomics as a guide to drug discovery and structural elucidation: eco‐02301, a novel antifungal agent, as an example. J Nat Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- Mannanov R.N., Sattarova R.K. Antibiotics produced by Bacillus bacteria. Chem Nat Comp. 2001;37:117–123. [Google Scholar]
- Nguyen T., Ishida K., Jenke‐Kodama H., Dittmann E., Gurgui C., Hochmuth T. Exploiting the mosaic structure of trans‐acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. et al. [DOI] [PubMed] [Google Scholar]
- Pichard B., Larue J.‐P., Thouvenot D. Gavaserin and saltavalin, new peptide antibiotics produced by Bacillus polymyxa. FEMS Microbiol Lett. 1995;133:215–218. doi: 10.1111/j.1574-6968.1995.tb07887.x. [DOI] [PubMed] [Google Scholar]
- Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D.H. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza W., Yang W., Shen Q.‐R. Paenibacillus polymyxa: antibiotics, hydrolytic enzymes and hazard assessment. J Plant Pathol. 2008;90:403–414. [Google Scholar]
- Ridley C.P., Lee H.Y., Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Diaz M., Lebbe L., Rodelas B., Heyrman J., De Vos P., Logan N.A. Paenibacillus wynnii sp. nov., a novel species harbouring the nifH gene, isolated from Alexander Island, Antarctica. Int J Syst Evol Microbiol. 2005;55:2093–2099. doi: 10.1099/ijs.0.63395-0. [DOI] [PubMed] [Google Scholar]
- Romero‐Tabarez M., Jansen R., Sylla M., Lunsdorf H., Haussler S., Santosa D.A. 7‐O‐malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin‐resistant Staphylococcus aureus, vancomycin‐resistant enterococci, and a small‐colony variant of Burkholderia cepacia. Antimicrob Agents Chemother. 2006;50:1701–1709. doi: 10.1128/AAC.50.5.1701-1709.2006. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux V., Fenner L., Raoult D. Paenibacillus provencensis sp. nov., isolated from human cerebrospinal fluid, and Paenibacillus urinalis sp. nov., isolated from human urine. Int J Syst Evol Microbiol. 2008;58:682–687. doi: 10.1099/ijs.0.65228-0. [DOI] [PubMed] [Google Scholar]
- Saha P., Mondal A.K., Mayilraj S., Krishnamurthi S., Bhattacharya A., Chakrabarti T. Paenibacillus assamensis sp. nov., a novel bacterium isolated from a warm spring in Assam, India. Int J Syst Evol Microbiol. 2005;55:2577–2581. doi: 10.1099/ijs.0.63846-0. [DOI] [PubMed] [Google Scholar]
- Scherlach K., Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- Schneider K., Chen X.‐H., Vater J., Franke P., Nicholson G., Borriss R., Süssmuth R.D. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod. 2007;70:1417–1423. doi: 10.1021/np070070k. [DOI] [PubMed] [Google Scholar]
- Schwarzer D., Finking R., Marahiel M.A. Nonribosomal peptides: from genes to products. Nat Prod Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- Shida O., Takagi H., Kadowaki K., Nakamura L.K., Komagata K. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol. 1997;47:289–298. doi: 10.1099/00207713-47-2-289. [DOI] [PubMed] [Google Scholar]
- Simunovic V., Zapp J., Rachid S., Krug D., Meiser P., Müller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3‐hydroxy‐3‐methylglutaryl‐coa synthases, and trans‐acting acyltransferases. Chembiochem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- Stewart G.J., Carlson C.A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–231. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Morimoto S., Adachi T., Kashimura M., Asaka T. Chemical modification of erythromycins. IX. Selective methylation at the C‐6 hydroxyl group of erythromycin A oxime derivatives and preparation of clarithromycin. J Antibiot (Tokyo) 1993;46:647–660. doi: 10.7164/antibiotics.46.647. [DOI] [PubMed] [Google Scholar]
- Weber T., Welzel K., Pelzer S., Vente A., Wohlleben W. Exploiting the genetic potential of polyketide producing streptomycetes. J Biotechnol. 2003;106:221–232. doi: 10.1016/j.jbiotec.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Weber T., Laiple K.J., Pross E.K., Textor A., Grond S., Welzel K. Molecular analysis of the kirromycin biosynthetic gene cluster revealed β‐alanine as precursor of the pyridone moiety. Chem Biol. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. et al. [DOI] [PubMed] [Google Scholar]
- Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.D., Sutherland A.D. New strategies for combating multidrug‐resistant bacteria. Trends Mol Med. 2007;13:260–267. doi: 10.1016/j.molmed.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang H.W., Song Y.C., Tan R.X. Biology and chemistry of endophytes. Nat Prod Rep. 2006;23:753–771. doi: 10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- Zimmerman S.B., Schwartz C.D., Monaghan R.L., Pelak B.A., Weissberger B., Gilfillan E.C. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. I. Production, taxonomy and antibacterial activity. J Antibiot (Tokyo) 1987;40:1677–1681. doi: 10.7164/antibiotics.40.1677. et al. [DOI] [PubMed] [Google Scholar]
- Zuckerman J.M. Macrolides and ketolides: azithromycin, clarithromycin, telithromycin. Infect Dis Clin North Am. 2004;18:621–649. doi: 10.1016/j.idc.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Zweerink M.M., Edison A. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J Antibiot (Tokyo) 1987;40:1692–1697. doi: 10.7164/antibiotics.40.1692. [DOI] [PubMed] [Google Scholar]


