Summary
Microorganisms, such as Pseudomonas putida, utilize specific physical properties of cellular membrane constituents, mainly glycerophospholipids, to (re‐)adjust the membrane barrier to environmental stresses. Building a basis for membrane composition/function studies, we inventoried the glycerophospholipids of different Pseudomonas and challenged membranes of growing cells with n‐butanol. Using a new high‐resolution liquid chromatography/mass spectrometry (LC/MS) method, 127 glycerophospholipid species [e.g. phosphatidylethanolamine PE(32:1)] with up to five fatty acid combinations were detected. The glycerophospholipid inventory consists of 305 distinct glycerophospholipids [e.g. PE(16:0/16:1)], thereof 14 lyso‐glycerophospholipids, revealing conserved compositions within the four investigated pseudomonads P. putida KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Furthermore, we addressed the influence of environmental conditions on the glycerophospholipid composition of Pseudomonas via long‐time exposure to the sublethal n‐butanol concentration of 1% (v/v), focusing on: (i) relative amounts of glycerophospholipid species, (ii) glycerophospholipid head group composition, (iii) fatty acid chain length, (iv) degree of saturation and (v) cis/trans isomerization of unsaturated fatty acids. Observed alterations consist of changing head group compositions and for the solvent‐sensitive strain KT2440 diminished fatty acid saturation degrees. Minor changes in the glycerophospholipid composition of the solvent‐tolerant strains P. putida S12 and Pseudomonas sp. VLB120 suggest different strategies of the investigated Pseudomonas to maintain the barrier function of cellular membranes.
Introduction
Driven by technological advances lipidomics and detailed lipid profiling gain currently increasing scientific interest. Lipids play important roles in cell physiology, for example as energy storage, bioactive molecules and main constituents of cellular membranes. The assessment of detailed membrane composition does not equal the extensive characterization of other cell constituents (e.g. proteins) that are accessible by ‘omic’ analyses. Using the now available analytical methods, the response of the membrane composition during changing environmental conditions can be monitored.
Cell membranes consist of a multiplicity of individual protein and lipid species with the main constituents belonging to only few distinct glycerophospholipid classes. The cytoplasmic membrane of many bacteria, including proteobacteria, as well as the inner side of the outer membrane mainly contains phosphatidylethanolamine (PE), phosphatidylglycerol (PG), cardiolipin (CL) and the respective monoacyl‐glycerophospholipids (lyso‐PE, lyso‐PG) (Fig. 1). The latter are part of the de‐acylation/re‐acylation cycle to control the overall lipid species compositions, catalysed by phospholipases, such as phospholipase A2 and lyso‐phospholipases that specifically release fatty acids from the sn2 position of the glycerol backbone (Scheer et al., 2011).
Figure 1.
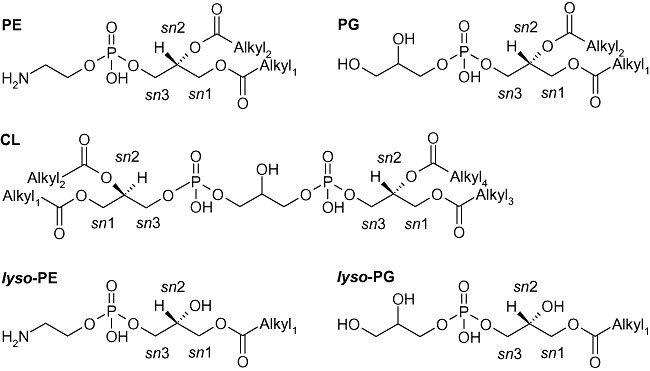
Glycerophospholipid classes of P. putida. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin. lyso‐glycerophospholipid species are representative examples with the acyl moiety in sn1 position. Alkyl1–4 represents alkyl moieties (same or different ones).
In contrast to eukaryotic cells, recent analyses revealed the absence of phosphatidylcholine (PC), phosphatidylinositol (PI) and sphingomyelin in Escherichia coli (Wang et al., 2004) and Pseudomonas putida (Bernal et al., 2007). Indeed, glycerophospholipid biosynthesis in the γ‐proteobacteria prime model organism E. coli (Dowhan, 1997; Cronan, 2003; Karp et al., 2005) misses genes coding for the required biosynthetic reactions. According glycerophospholipid and fatty acid biosynthesis pathways in P. putida can be assembled from genomic information [P. putida strains KT2440 (AE015451.1), GB‐1 (NC_010322.1), F1 (NC_009512.1) and W619 (NC_010501.1) (Kanehisa et al., 2006)] and entries of the KEGG pathway database (Fig. 2). As visualized, prokaryotic membrane lipid diversity originates not from the number of distinct glycerophospholipid classes, but rather from complex fatty acid compositions (Cronan and Gelmann, 1975). Bacterial membranes do not only consist of even‐numbered, straight‐chain fatty acids, which are found in eukaryotic glycerophospholipids, but also odd‐numbered, branched‐chain, as well as cyclic fatty acids (Kaneda, 1991; Fujiwara, 2008).
Figure 2.
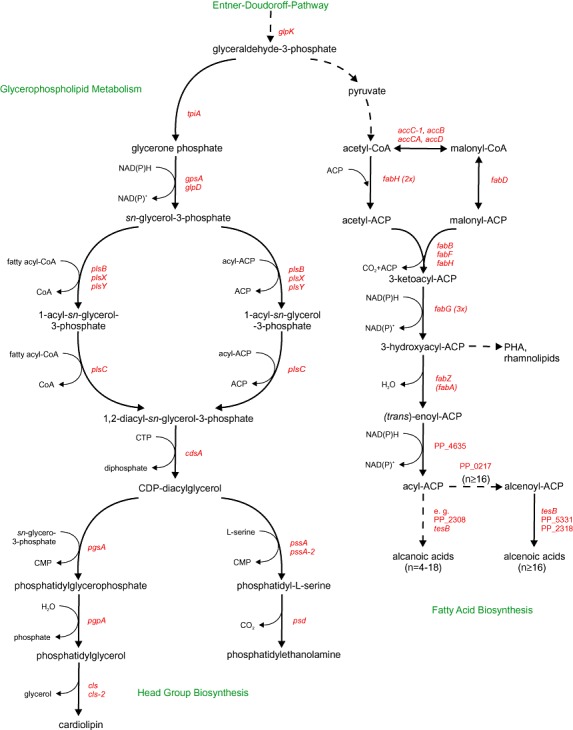
Biosynthesis routes of glycerophospholipids and fatty acids in P. putida. Abbreviations of the enzyme coding genes are: eda– 2‐keto‐3‐deoxy‐6‐phospogluconate aldolase; tpiA– triosephosphate isomerase; gpsA, glpD– glycerol‐3‐phosphate dehydrogenase; plsB, plsX, plsY– glycerol‐3‐phosphate acyltransferase; plsC– 1‐acyl‐sn‐glycerol‐3‐phosphate acyltransferase; cdsA– phosphatidate cytidylyltransferase; pgsA– CDP‐diacylglycerol‐glycerol‐3‐phosphate‐3‐phosphatidyltransferase; pgpA– phosphatidylglycerophosphatase A; cls, cls‐2 – cardiolipin synthase; pssA– phosphatidylserine synthase; pssA‐2 – CDP‐deacylglycerol‐serine O‐phosphatidyltransferase; psd– phosphatidyl serine decarboxylase; accC‐1; accB, accA, accD– acetyl‐CoA carboxylase subunits; fabD– malonyl‐CoA‐[acp] transacylase; fabH– beta‐ketoacyl‐carrier protein synthase I; fabB, fabF– 3‐oxoacyl‐[acp] synthase I/II; fabG– 3‐ketoacyl‐[acp] reductase; fabZ– (3R)‐hydroxymyristol‐[acp] dehydratase; fabA– 3‐hydroxydecanoyl‐[acp] dehydratase/isomerase; tesB– acyl‐CoA thioesterase. Un‐annotated genes are defined as follows: PP_0058 – phospholipid/glycerol acyltransferase; PP_3303 – 3‐oxoacyl‐[acp]‐synthase II; PP_2783 – 3‐oxoacyl‐[cp] reductase; PP_4635 –trans‐2‐enoyl‐CoA reductase; PP_0217 – fatty acid desaturase; PP_2308 – putative acyl‐CoA thioesterase II; PP_5331 – thioesterase superfamily protein; PP_2318 – GDSL family lipase.
The molecular basis of bacterial glycerophospholipid diversity has intensively been studied for a number of Gram‐negative and ‐positive bacteria (Langley and Kennedy, 1978; Pinkart and White, 1997; Bukata et al., 2008), foremost E. coli (Dowhan, 1997). Distinguished glycerophospholipid profiles include studies of the actinobacterium Corynebacterium sp. strain 8 (Mazzella et al., 2004), and the methylotrophic bacterium Methyloxiunus trichosporium (Fang and Barcelona, 1998), while a comparative analysis was carried out on five P. putida strains (Fang et al., 2000).
For P. putida extensive evidence for long‐ and short‐term alterations of the inner and outer membrane due to toxic organic solvent exposure exists (Ramos et al., 1997), and growth in the presence of a second phase of toluene (Isken, 2000), styrene (Park et al., 2007) or n‐octanol (Blank et al., 2008) has been shown. Solvent toxicity can be correlated to low logarithms of the partitioning coefficient in an n‐octanol water mixture (logPow < 4) indicating preferred membrane partitioning with disintegration of membrane structure and vital cellular functions (Ramos et al., 2002). Aiming for constant membrane fluidity, described via the transition temperature (Tm), cells try to sustain both proton gradient (ΔpH) and membrane potential (ΔΨ) (Sikkema et al., 1994; Ramos et al., 2002) to enable functional protein embedding (e.g. integral efflux pumps). The transition temperature is determined by the glycerophospholipid composition (Brannigan et al., 2004; Bernal et al., 2007); depends on the glycerophospholipid head group: Tm (CL) > Tm (PE) > Tm (PG); and increases proportional to chain length, saturation degree and cis/trans ratio of the acyl moieties (Weber et al., 1994; Soni et al., 2009).
The latter, i.e. cis/trans isomerization, allows short‐term alterations (in the minute range) of unsaturated fatty acids, the only post‐biosynthetic modification of the acyl‐chain in response to organic solvents (Heipieper and de Bont, 1994). Long‐term alterations embrace modifications of the glycerophospholipid properties including: (i) the saturation degree of fatty acids (Pinkart and White, 1997), (ii) the glycerophospholipid head group composition (Cronan and Gelmann, 1975), (iii) the phospholipid turnover rates, (iv) the fatty acid chain length and (v) the total phospholipid content (Park et al., 2007). Toxic organic solvents entail the inhibition of phospholipid biosynthesis (Sikkema et al., 1994) and alterations of the outer membrane (Blank et al., 2008; Fujiwara, 2008), including denser packing of anionic membrane molecules [e.g. lipopolysaccharides (LPS) and outer membrane proteins (OMP)], to increase hydrophobicity and hamper the accumulation of toxic hydrocarbons (Segura et al., 1999). Notably, both lipid profiling and adaptation studies reported mostly limited analyte numbers (n ≤ 21).
The unambiguous identification of glycerophospholipids is necessary to gain further insights into their complex functionalities. Different mass spectrometric techniques coupled to gas chromatography (GC) or thin‐layer chromatography (TLC) have been established that, however, require additional sample preparations (e.g. derivatization) and potentially introduce undesired artefacts (e.g. analyte oxidation) (Hancock and Meadow, 1969). Full glycerophospholipid profiling at satisfying reproducibility was achieved by high‐performance liquid chromatography coupled online to mass spectrometry by electrospray ionization mass spectrometry (LC/ESI‐MS) (Fang and Barcelona, 1998; Mazzella et al., 2004). Recently, hybrid linear ion trap‐Fourier transform ion cyclotron resonance mass spectrometry (LIT‐FTICR‐MS) enabled the simultaneous profiling of various glycerophospholipid species in complex biological samples, taking advantage of high mass resolution and accuracy. We reported an ESI‐LIT‐FTICR‐MS method coupled to reversed‐phase high‐performance liquid chromatography (RP‐HPLC) (Hein et al., 2009) for the relative quantification of glycerophospholipid species with the advantage of low absolute instrumental detection limits (∼0.05 pmol) and a high linear dynamic range (1.5–2.5 magnitude), and reported one of the few available software solutions for HPLC/ESI‐LIT‐FTICR‐MS data (Hein et al., 2010).
Equipped with this analytical toolbox, we revisited the glycerophospholipid inventory of Pseudomonas in the context of the genetic background and the metabolic pathways of both glycerophospholipid and de novo fatty acid biosynthesis. Based on the current scientific interest in biobutanol production and existing evidence of increased n‐butanol tolerance by P. putida DOT‐T1E and S12, as well as Pseudomonas sp. strain VLB120 (Rühl et al., 2009), we investigated the glycerophospholipid profiles of these strains during n‐butanol exposure with respect to compositional variations of membrane glycerophospholipids at changing environmental conditions.
Results
Glycerophospholipid inventory of P. putida
We revisited the glycerophospholipid inventory of organic solvent‐sensitive and solvent‐tolerant P. putida strains, namely the GRAS classified strain P. putida KT2440, the solvent‐tolerant strains P. putida DOT‐T1E and S12, and the multipurpose [e.g. biocatalysis and solvent tolerance (Park et al., 2007), biofilm producing (Gross et al., 2007)] strain Pseudomonas sp. VLB120. Distinct species of phosphatidylethanolamine, phosphatidylglycerol and cardiolipin were characterized referring to the number of carbon atoms and unsaturated carbon bonds (Fig. 3A), with more detailed information given in Table S1. The novel LIT‐FTICR‐MS technique was thereby complemented with GC/MS analysis of the hydrolysed glycerophospholipids to obtain the relative amounts of the total fatty acid moieties (Fig. 3B).
Figure 3.
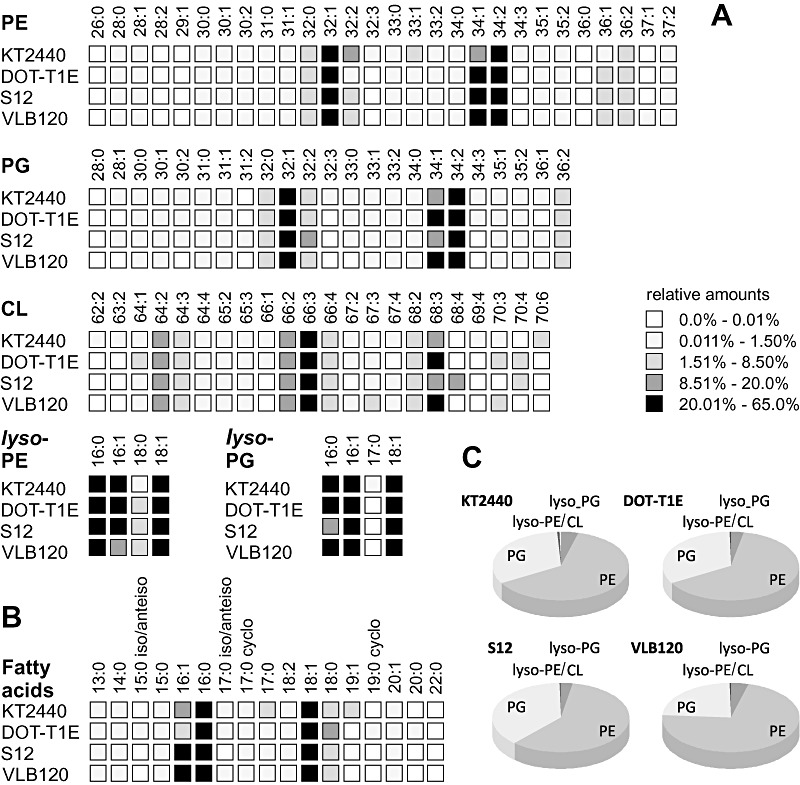
Comparative analysis of the glycerophospholipid inventory of four pseudomonads, P. putida KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Analysis refers to: (A) the relative amounts of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), cardiolipin (CL) and respective lyso‐species; (B) detectable fatty acid moieties within all glycerophospholipid species; (C) the relative distribution of the glycerophospholipid classes. Percentage values are calculated from the ratio of the peak area of a glycerophospholipid (LC/MS) or fatty acid species (GC/MS) to the respective overall areas.
The evaluation of these detailed profiles resulted in relative distributions of the glycerophospholipid species (head group composition) (Fig. 3C). All here investigated Pseudomonas strains revealed phosphatidylethanolamine as the main membrane component, contributing on average two‐thirds of the total glycerophospholipid fraction. At low amounts of cardiolipin (< 5%), phosphatidylglycerol was the second most abundant class accounting for approximately one‐third. The lyso‐forms of phosphatidylethanolamine and phosphatidylglycerol were detected in traces (< 1% of the total glycerophospholipid pool) (Fig. 3C). Notably, the four strains revealed similar glycerophospholipid composition, only Pseudomonas sp. strain VLB120 exhibited increased phosphatidylethanolamine (72% compared with 62%) at the expense of phosphatidylglycerol (23% compared with 33%). The determined glycerophospholipid compositions are comparable to literature: the membrane fractions of phosphatidylethanolamine in P. putida S12 and P. putida DOT‐T1E were reported to be 60% (Heipieper et al., 1996) and 75% (Ramos et al., 1997) respectively.
Existing variations between our data and previous reports, for example the detection of the phosphatidylethanolamine derivates dimethyl‐ and monomethylphosphatidyl ethanolamine (DMPE and MMPE) (Martinez‐Morales et al., 2003), might result from different analytical methods and/or culture conditions, as shown for growth rate‐dependent phosphatidylethanolamine and phosphatidylglycerol synthesis in E. coli (Ballesta and Schaechter, 1972).
The relative amount of the different glycerophospholipid species varied only slightly between the four strains (Fig. 3A). Each major glycerophospholipid class contained more than 20 individual species that were characterized by their total number of carbon atoms within both fatty acid moieties and degree of saturation, for example PE(32:1) (Table S2). Additionally, MSn experiments provided information on the individual fatty acid moieties. Hence, PE(32:1) comprises not only (16:0/16:1) but also contains the combination (15:0/17:1). Saturated and (mono‐)unsaturated fatty acids with a chain length of 13–22 carbon atoms were detected, with (16:0/16:1) and (18:0/18:1) as the major fatty acid moieties (Table S3), accounting for 93% up to 99% of the total acyl moieties (Table 1). In agreement with reports from the groups of Heipieper and de Bont (Heipieper et al., 1992; Weber et al., 1994), only small relative amounts of the cyclic fatty acids 17:0‐cyclo and 19:0‐cyclo and the methylated (branched‐chain) fatty acids 15:0‐iso/anteiso and 17:0‐iso/anteiso were present in P. putida during exponential growth (Fig. 3B). Notably, about 77% of the total glycerophospholipids consisted of the most abundant phosphatidylethanolamine PE(32:1) and PE(34:1)/PE(34:2) and phosphatidylglycerol species PG(32:1) and PG(34:1)/PG(34:2) (Table 1).
Table 1.
Relative amounts of the major glycerophospholipid species and fatty acid moieties.
| Relative amount of 32:1/34:1/34:2 (%) | Relative amount of 16:0/16:1/18:0/18:1 (%) | |||
|---|---|---|---|---|
| PE | PG | Total glycerophospholipids | ||
| P. putida KT2440 | ||||
| 0% n‐butanol non‐treated | 81 | 85 | 78 | 93 ± 6 |
| 0% n‐butanol treated | 81 | 84 | 78 | 82 ± 6 |
| 1% n‐butanol non‐treated | 81 | 75 | 75 | 99 ± 0 |
| 1% n‐butanol treated | 84 | 14 | 52 | 99 ± 0 |
| P. putida DOT‐T1E | ||||
| 0% n‐butanol non‐treated | 78 | 84 | 78 | 99 ± 0 |
| 0% n‐butanol treated | 79 | 84 | 78 | 99 ± 0 |
| 1% n‐butanol non‐treated | 12 | 6 | 9 | 54 ± 1 |
| 1% n‐butanol treated | 83 | 88 | 81 | 98 ± 2 |
| P. putida S12 | ||||
| 0% n‐butanol non‐treated | 81 | 78 | 77 | 99 ± 0 |
| 0% n‐butanol treated | 81 | 78 | 78 | 99 ± 0 |
| 1% n‐butanol non‐treated | 77 | 57 | 61 | 95 ± n.d. |
| 1% n‐butanol treated | 82 | 66 | 73 | 99 ± 0 |
| Pseudomonas sp. VLB120 | ||||
| 0% n‐butanol non‐treated | 79 | 82 | 76 | 99 ± 0 |
| 0% n‐butanol treated | 78 | 82 | 76 | 99 ± 0 |
| 1% n‐butanol non‐treated | 80 | 75 | 72 | 98 ± 0 |
| 1% n‐butanol treated | 79 | 81 | 76 | 99 ± 0 |
The relative amounts of 32:1/34:1/34:2 phosphatidylethanolamine and phosphatidylglycerol were calculated from their abundance within the respective glycerophospholipid class multiplied with the relative amount of these classes. The relative amount of the major fatty acid moieties 16:0/16:1/18:0/18:1 was calculated for the total glycerophospholipid pool. Free fatty acids were not considered. Errors represent the standard deviation of independent experiments (n = 2–8). n.d.: not determined because of missing replicates.
Besides glycerophospholipid profiling with respect to acyl moieties and lipid species, we determined features of the fatty acid moieties like the degree of saturation and the ratio between the cis‐ and trans‐isomers of unsaturated fatty acids. Please note that this was only determined for 18:1 and not for 16:1 unsaturated fatty acids. The latter fatty acid isomers were not resolved by the applied GC separation. Still cis/trans isomerization could be addressed as isomerization of both 16:1 and 18:1 fatty acid moieties respond qualitatively comparable in response to organic solvents (Guckert et al., 1986; Weber et al., 1994; Huertas et al., 2000; Heipieper et al., 2001). These modifications are important for membrane function as they define the membrane structure and hence the transition temperature (Tm), which directly influences membrane fluidity and the movement (e.g. free rotation and lateral movements) of membrane lipids and proteins (Heipieper et al., 2003; Castellanos et al., 2007). We defined the degree of saturation as the number of double bonds over the total number of detected glycerophospholipids. LC/MS analyses indicated saturation degrees of about 70% (P. putida DOT‐T1E 67 ± 4%, KT2440 71 ± 6%, S12 70 ± 11%, Pseudomonas sp. strain VLB120 67 ± 15%). These values are in agreement with previous studies, for example with a saturation degree of 67% for P. putida S12 grown on glucose or LB medium (Weber et al., 1994).
Membrane density changes in accordance with the existence and conformation of unsaturated carbon bonds. Saturated fatty acids have straight acyl chains and cis‐unsaturated isomers introduce a kink in the acyl chain (Isken and de Bont, 1998), whereas trans‐isomers almost resemble saturated fatty acids. The cis/trans ratio of 18:1 unsaturated fatty acids is therefore an interesting characteristic, whose rapid alteration (in the minute range) enables solvent‐tolerant pseudomonads to survive environmental stresses (Ramos et al., 2002). Calculated for 18:1 fatty acids a three‐ to fivefold excess of the cis‐isomer was detected for the P. putida strains KT2440, DOT‐T1E and S12 (Table 2). In contrast Pseudomonas sp. strain VLB120 exhibited an 18:1‐cis/trans ratio of 1.6 ± 0.2, which depicts the main difference within the glycerophospholipid profiles of the investigated strains.
Table 2.
Glycerophospholipid profiles of non‐treated and treated P. putida strains: the relative amount of the glycerophospholipid classes, the degree of saturation and the cis/trans ratio.
| Glycerophospholipid classes (%) | Degree of saturationa | Cis/trans ratiob | |||
|---|---|---|---|---|---|
| PE | PG | CL | |||
| P. putida KT2440 | |||||
| 0% n‐butanol non‐treated | 62 ± 2 | 33 ± 3 | 4± 1 | 71 ± 6 | 3.4 ± 0.8 |
| 0% n‐butanol treated | 2.3 ± 0.7 | ||||
| 1% n‐butanol non‐treated | 59 ± 1 | 36 ± 1 | 4 ± 0 | 71 ± 4 | 5.2 ± 0.3 |
| 1% n‐butanol treated | 56 ± 8 | 41 ± 6 | 3 ± 1 | 46 ± 13 | 6.4 ± 0.3 |
| P. putida DOT‐T1E | |||||
| 0% n‐butanol non‐treated | 64 ± 2 | 33 ± 2 | 3± 0 | 67 ± 4 | 4.9 ± 0.5 |
| 0% n‐butanol treated | 4.8 ± 0.5 | ||||
| 1% n‐butanol non‐treated | 53 ± 0 | 45 ± 0 | 2 ± 0 | 18 ± 0 | n.a. |
| 1% n‐butanol treated | 66 ± 1 | 30 ± 0 | 3 ± 0 | 67 ± 0 | 4.6 ± n.d. |
| P. putida S12 | |||||
| 0% n‐butanol non‐treated | 59 ± 2 | 38 ± 2 | 3± 0 | 70 ± 11 | 3.2 ± 1.0 |
| 0% n‐butanol treated | 2.4 ± 0.3 | ||||
| 1% n‐butanol non‐treated | 71 ± 0 | 12 ± 0 | 15 ± 0 | 65 ± 1 | 1.2 ± n.d. |
| 1% n‐butanol treated | 62 ± 0 | 34 ± 0 | 4 ± 0 | 74 ± 5 | 2.3 ± 0.1 |
| Pseudomonas sp. VLB120 | |||||
| 0% n‐butanol non‐treated | 72 ± 2 | 24 ± 1 | 4± 1 | 67 ± 0 | 1.6 ± 0.2 |
| 0% n‐butanol treated | 2.7 ± 0.6 | ||||
| 1% n‐butanol non‐treated | 74 ± 6 | 18 ± 9 | 7 ± 3 | 69 ± 0 | 1.1 ± 0.8 |
| 1% n‐butanol treated | 72 ± 2 | 23 ± 2 | 5 ± 0 | 67 ± 0 | 1.7 ± n.d. |
The degree of saturation was calculated as the number of saturated double bonds over the total number of fatty acids.
The cis/trans ratio represents the ration of 18:1‐cis to 18:1‐trans fatty acids. Errors represent the standard deviation of independent experiments (n = 2–8). n.d.: not determined because of missing replicates; n.a.: not analysed.
Application: n‐butanol exposition
Equipped with the comprehensive inventory of glycerophospholipid species, we wanted to understand the effect of the short‐chain alcohol n‐butanol [logPow 0.8, maximum membrane accumulation of 1.59 M (Neumann et al., 2005)], a promising synthon for chemical industry, on the glycerophospholipid composition of P. putida. We previously suggested P. putida DOT‐T1E, S12 and Pseudomonas sp. strain VLB120 as possible candidates for biobutanol production as treated cells, sequentially transferred between LB agar plates in an airtight desiccator with an n‐butanol saturated gas phase, had lower energy requirements for cell maintenance than non‐treated cells of the same strains when grown in the presence of 1% (v/v) n‐butanol (Rühl et al., 2009).
Hereafter, we refer to treated cells when strains underwent the sequential transfer procedure, while exposed cells were grown in shake flask where 1% (v/v) n‐butanol was added at the time of inoculation. At reference conditions (w/o n‐butanol exposure) the glycerophospholipid profiles of non‐treated and treated cells revealed few differences. Only minor changes (below 2.5% relative difference) were observed for the cardiolipin species CL(70:3) and CL(70:4) in P. putida DOT‐T1E and KT2440. In contrast, the relative amounts of the major fatty acids (16:1/16:0) and (18:1/18:0) changed significantly towards the unsaturated species (Fig. 4B), while the total relative amount of these fatty acids (about 99%) was not affected in the solvent‐tolerant strains P. putida DOT‐T1E, S12 and Pseudomonas sp. VLB120 (Table 1). Pseudomonas putida strain KT2440 slightly shifted the composition of fatty acid moieties during n‐butanol treatment from 16:0 and 18:1 towards the fatty acids 17:0/17:1 and 20:1 (see Table 1).
Figure 4.
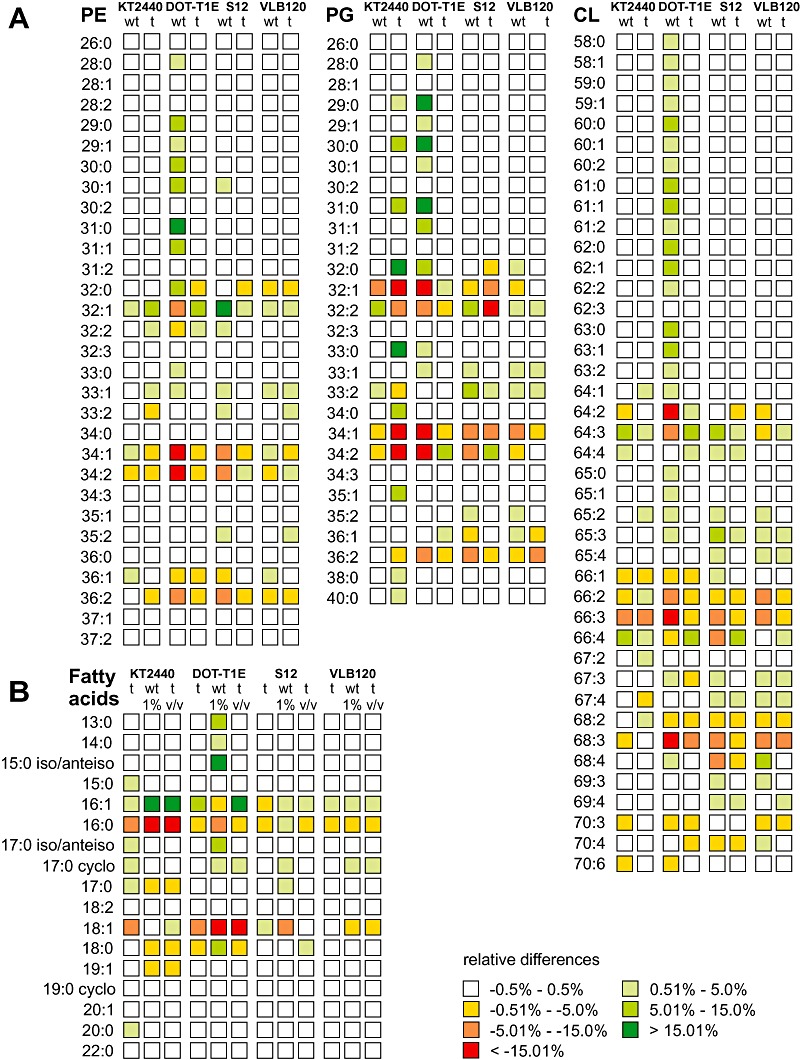
Relative differences of the glycerophospholipid species and fatty acid moieties of P. putida following n‐butanol exposure. A. Relative differences in the relative amounts of glycerophospholipid species were calculated compared with the average of non‐treated and treated cells grown at reference conditions. B. Relative differences of the relative amounts of fatty acid moieties compared with the average abundance of fatty acids of non‐treated cells grown at reference conditions. Abbreviations: wt, non‐treated; t, treated cells.
Exposed to the non‐lethal n‐butanol concentration of 1% (v/v), the four Pseudomonas strains specifically responded by both short‐ and long‐term adaptation mechanisms. We first analysed general modifications of the glycerophospholipid compositions including: (i) the glycerophospholipid species, (ii) the glycerophospholipid head group composition and (iii) the chain length of fatty acid moieties.
Major alterations of the glycerophospholipid composition were only detected for the main species within each class (Fig. 4A). Generally, PE(32:1) levels were enhanced, while most experimental conditions showed decreased fractions of PE(34:1)/PE(34:2) and PG(32:1)/PG(34:1)/PG(34:2). As expected from the consecutive biochemical pathway (Fig. 2), the cardiolipins resembled the distribution of the phosphatidylglycerol species. Significantly altered relative amounts and overall contributions of the main phosphatidylglycerol species were observed for only two experimental conditions: (i) treated cells of P. putida KT2440 exposed to n‐butanol had a considerably diminished relative amount of the major phosphatidylglycerol species PG(32:1), PG(34:1) and PG(34:2), and (ii) non‐treated cells of the solvent‐tolerant P. putida strain DOT‐T1E showed the relative amounts of PG(32:1), PG(34:1) and PG(34:2) and the corresponding phosphatidylethanolamine species, PE(32:1), PE(34:1) and PE(34:2), declining in the same manner (Table 1).
The relative amounts of the three glycerophospholipid classes, phosphatidylethanolamine, phosphatidylglycerol and cardiolipin, were also affected by n‐butanol exposure. Both non‐treated P. putida DOT‐T1E and KT2440 exhibited a shift from phosphatidylethanolamine to phosphatidylglycerol at slightly decreasing cardiolipin amounts (e.g. 64% compared with 53% for phosphatidylethanolamine and 33% compared with 45% for phosphatidylglycerol in P. putida DOT‐T1E). A different reaction was observed for non‐treated cells of P. putida S12 and Pseudomonas sp. strain VLB120, which reacted to n‐butanol by enhanced phosphatidylethanolamine and cardiolipin levels at contemporaneously diminished relative amounts of phosphatidylglycerol (Table 2).
The treatment procedure resulted in membrane alterations that differed from that of non‐treated cells. Treated cells of the three solvent‐tolerant strains P. putida DOT‐T1E, S12 and Pseudomonas sp. strain VLB120 almost maintained their original membrane lipid composition with the relative differences of the major glycerophospholipid species mostly below ± 5% (Fig. 4A). Differing results were obtained from the application of treated P. putida KT2440 where n‐butanol treatment enforced the extent of glycerophospholipid alterations after n‐butanol exposure.
Considering environmental stresses, like n‐butanol exposure, the structural composition of the fatty acid moieties has to be addressed in addition to the glycerophospholipid inventory. The acyl moieties at sn1 position, which are introduced by the sn‐glycerol‐3‐phosphate acyltransferase (PlsB), are one possible regulation site within glycerophospholipid synthesis (Wilkison and Bell, 1997). Exposed to n‐butanol, the relative amounts of the major fatty acid moieties, 16:0, 16:1, 18:0 and 18:1, of both non‐treated and treated cells were maintained at the constant level of approximately 99% (Table 1). Only non‐treated cells of P. putida DOT‐T1E revealed significant relative amounts of the fatty acids 13:0, 14:0 and 15:0 iso/anteiso (Fig. 4). As a result, the contribution of 16:1/16:0 and 18:1/18:0 fatty acids to the total acyl moieties decreased from 99 ± 0% to 54 ± 1%.
Besides the above depicted possibilities of compositional membrane changes, pseudomonads can alter their fatty acids in response to organic solvents by: (i) changing the degree of saturation and (ii) changing the cis/trans isomeric ratio of the unsaturated fatty acids. In this study, diverse changes in the fatty acid composition were recorded (Table 2). While the overall degree of saturation in non‐treated cells did not change in the presence of n‐butanol, a lower content of saturated fatty acid moieties within the phosphatidylglycerol species was observed for treated P. putida KT2440 [(Fig. 4), degree of saturation approximately 15%], which resulted in a significant decrease of the overall saturation (71 ± 4% to 46 ± 13%). In this strain no cis/trans isomerization of 18:1 unsaturated fatty acid moieties was observed, but an increase in the relative amount of cis‐unsaturated 18:1 fatty acid moieties (Table 2). A behaviour contrary to that of P. putida KT2440 was depicted in solvent‐tolerant P. putida DOT‐T1E. Non‐treated cells of this strain revealed a more than threefold lower degree of saturation, 18 ± 0% compared with 67 ± 4%, after n‐butanol exposure, while treated cells were able to maintain both saturation degree and cis/trans ratio (Table 2).
Notably, only slight changes of the saturation degree were observed for the solvent‐tolerant strains P. putida S12 and Pseudomonas sp. strain VLB120. These strains reacted to both the treatment procedure and n‐butanol exposure by cis/trans isomerization of 18:1 unsaturated fatty acid moieties. For example, non‐treated P. putida S12 grown in the presence of n‐butanol had a higher content of trans‐18:1. Interestingly, the cis/trans ratio of treated Pseudomonas sp. strain VLB120 exposed to n‐butanol equalled that of non‐treated cells under normal growth conditions (1.6 ± 0.2 and 1.7 ± n.d. respectively), thereby indicating a higher content of cis‐unsaturated fatty acids in treated Pseudomonas sp. VLB120 cells without n‐butanol exposure (2.7 ± 0.6).
Differences in the change of the glycerophospholipid composition between the solvent‐tolerant strains and P. putida KT2440 were observed. These differences in the response to n‐butanol exposure, irrespective of the conserved genetic inventory, imply differences in the (transcriptional) regulation of both fatty acid and glycerophospholipid biosynthesis.
Discussion
Glycerophospholipid inventory of P. putida
The analytical possibilities in life sciences are rapidly expanding, including the determination and quantification of lipid species. We used these new possibilities to revisit in depth the glycerophospholipid composition of the Gram‐negative γ‐proteobacterium P. putida, as distinct strains can alter their membrane composition to allow growth in the presence of highly toxic organic solvents (Weber et al., 1993; Sikkema et al., 1994; Chen et al., 1995; Heipieper et al., 2001), including octanol and styrene. The Pseudomonas genus is characterized by the presence of the common bacterial phospholipids phosphatidylethanolamine, phosphatidylglycerol and cardiolipin (Diedrich and Cota‐Robles, 1974; Ramos et al., 2002). We observed congruent compositions of the three major phospholipid classes and their respective lyso‐forms in four strains; in total 305 distinct glycerophospholipids. Notably, unlike Fang and colleagues (2000), no dimethyl‐phosphatidylethanolamine or monomethyl‐phosphatidylethanolamine were detected. These differences might be explained by glycerophospholipid composition changes that occur in dependence of growth conditions (Ohta et al., 1974; Pierucci, 1979), the exposure to cyclic hydrocarbons [e.g. BTEX and phenol (Heipieper and de Bont, 1994; Weber et al., 1994; Pinkart and White, 1997)], aliphatic alcohols [e.g. ethanol (Dombek and Ingram, 1984)] and other organic solvents (Ingram, 1977; Gustafson and Tagesson, 1985).
Application: n‐butanol exposition
Since P. putida was suggested as host for n‐butanol production (Nielsen et al., 2009), we investigated the response of the glycerophospholipid composition in the presence of this aliphatic alcohol rather than to the traditionally investigated aromatic hydrocarbons (Isken and de Bont, 1998) that are important during bioremediation. Exposed to a non‐lethal n‐butanol concentration of 1% (v/v), the investigated strains showed changes in the cis/trans ratio and modified head group compositions; responses previously reported during adaptation of P. putida to toluene and other aromatic carbohydrates (Ramos et al., 1997). An increase of trans‐unsaturated fatty acids compensates for fluidizing effects due to organic solvents and suggests high cis/trans isomerase (Cti) activity. The compositional alteration of the glycerophospholipid inventory of Pseudomonas was strain specific. Notably, the glycerophospholipid composition of cells, which were exposed to n‐butanol before the actual experiment, differed significantly from phosphatidylglycerol compositions of non‐treated cells, suggesting some kind of long‐term adaptation.
Cis/trans isomerases exist in all tested Pseudomonas strains [P. putida DOT‐T1E and S12 (Heipieper et al., 1996; Bernal et al., 2007) and Pseudomonas sp. strain VLB120 (J. Kalinowski, L.M. Blank and A. Schmid, unpubl. result)] and are not limited to solvent‐tolerant strains [P. putida KT2440 (AE015451.1, geneID 1045408) (Benson et al., 2005)]. Notably, our results suggest that the Cti of P. putida KT2440 is non‐functional in the presence of n‐butanol.
High accumulation of extracellular n‐butanol in the cytoplasmic membrane subsequently disintegrates the lipid bilayer. The here applied n‐butanol concentration (1% v/v) at the measured n‐butanol decrease of 24 ± 5 mmol l−1 h−1 (loss due to evaporation and consumption) fully induced adaptation mechanisms over the experimental time‐course. Full activity of the cis/trans isomerase can be assumed, as the half maximum trans/cis ratio (trans/cis50) in P. putida S12 was assigned at 41.2 mM [0.38% (v/v)] of n‐butanol (Heipieper et al., 1995). With n‐butanol degradation in P. putida KT2440 equal to that of tolerant strains, n‐butanol consumption can be neglected as reason for strain‐specific effects on the glycerophospholipid composition. Nevertheless, the slight changes in the glycerophospholipid profiles, relating to mostly growth and energy independent mechanisms [e.g. cis/trans isomerization (Heipieper et al., 1995)], support our hypothesis that the reduced n‐butanol effect on treated P. putida[i.e. long‐term exposed to n‐butanol (Rühl et al., 2009)] originated from membrane alterations.
Our findings from both metabolic pathway analysis (Rühl et al., 2009) and glycerophospholipid profiling hint for the regulation of environmental effects on the transcriptional level. For example, significantly increased fractions of shorter‐chain‐length fatty acids and reduced fractions of saturated fatty acids in n‐butanol exposed non‐treated P. putida DOT‐T1E might result from n‐butanol inhibition of de novo fatty acids biosynthesis by the FasII system, as previously reported for ethanol (Heipieper and de Bont, 1994). Furthermore, regulation of fatty acid and glycerophospholipid biosynthesis on the transcriptional level can be correlated to: (i) expression of the fabA and fabB genes, which are regulated by the transcription factors of fatty acid biosynthesis (FabR) and degradation (FadR) (Feng and Cronan, 2009; Zhu et al., 2009), (ii) the sn‐glycerol‐3‐phosphate acyltransferase PlsB and the availability of acyl donors (acyl‐ACP or acyl‐CoA), and (iii) the CDP‐diacylglycerol‐glycerol‐3‐phosphate‐3‐phosphatidyltransferase PgsA playing a role in maintaining head group composition. Indeed, we observed in experiments with P. putida S12 and Pseudomonas sp. strain VLB120 slight changes in head group composition after n‐butanol exposure (Table 2). Different reactions of non‐treated and treated cells of these strains with respect to the glycerophospholipid classes could be correlated to the respective sensitivity towards n‐butanol accumulation in the phospholipid bilayer and required stabilizing effects by higher phosphatidylethanolamine or cardiolipin amounts. Here, the role of glycerophospholipids via feedback inhibition onto the FasII enzymes (Fujita et al., 2007; Zhang et al., 2008) enables the coordination of glycerophospholipids with cell growth in dependence of the encountered environment, which is a basic function for organic solvent (n‐butanol) resistance. With a high number of analytes, investigation of their origin from either directed regulation or enzymatic side‐activity has to be investigated. Based on our results biological consequences of minor changes in the glycerophospholipid profiles could be further addressed in more detail.
Conclusion
The glycerophospholipid compositions of cellular membranes of Pseudomonas were comprehensively determined. The relative abundance of molecular lipid species is the basis for biophysical models that describe and predict structure–function relationships of cellular membranes. Such detailed understanding is necessary to comprehend the different adaptation strategies, which we observed for the investigated strains when exposed to n‐butanol. More generally, we expect major findings from detailed glycerophospholipid analysis, which is mainly driven by the availability of new high‐resolution analytical techniques, as the one used here, for the so‐called lipidome analysis.
Experimental procedures
Chemicals
Acetonitrile, methanol and water were of LC/MS grade, chloroform and n‐propanol were of HPLC grade. All solvents were purchased from Carl Roth GmbH & Co.KG (Karlsruhe, Germany) or Sigma‐Aldrich Chemie GmbH. Ammonium acetate and acetic acid of analytical grade were obtained from Merck KGaA (Darmstadt, Germany). n‐Butanol and media components were purchased from Sigma Aldrich/Fluka Chemie AG (Buchs, Switzerland) and Difco Laboratories (Detroit, USA) at the highest grade available. Trimethylsulfoniumhydroxid (0.25 M in methanol) for derivatization of fatty acids was obtained from Macherey‐Nagel (Düren, Germany).
Bacterial strains, culture media, treatment and growth conditions
Pseudomonas putida DOT‐T1E (Ramos et al., 1998), KT2440 (Ramos‐Diaz and Ramos, 1998), S12 (Weber et al., 1993) and Pseudomonas sp. strain VLB120 (Park et al., 2007) were investigated in this study. Glucose‐supplemented LB medium was used for cultivation, containing (per litre) 10 g of peptone/tryptone, 5 g of yeast extract and 5 g of sodium chloride. All strains were incubated at 30°C in a horizontal shaker (200 r.p.m.) using 50 ml of medium in 500 ml baffled shake flasks. Growth was monitored by measuring the optical density at a wavelength of 600 nm (OD600) using a plate reader (Infinite 200 Pro series, Tecan GmbH, Crailsheim, Germany). An OD600 value of 1.0 correlated to a cell dry weight (CDW) of 1.18 gCDW l−1.
Adaptation to n‐butanol was carried out as published previously (Rühl et al., 2009). Briefly, cells were exposed to n‐butanol during growth on LB agar plates using an airtight desiccator with an n‐butanol saturated gas phase at 30°C. Colonies were repeatedly transferred every 2 days to new plates for at least 15 times, before harvesting and storage at −80°C prior to shake‐flask experiments. Cells that underwent this procedure are referred to as treated cells.
Shake‐flask experiments were started from P. putida overnight cultures with an inoculation volume of 1% (v/v). Cells harvested from cultures with addition of 1% (v/v) of n‐butanol are referred to as exposed cells. Bacteria were harvested at a biomass concentration of 1 gCDW l−1 (OD600 = 0.8). For glycerophospholipid extraction the method of Bligh and Dyer was modified (Bligh and Dyer, 1959), omitting the use of an aqueous phase to increase the recovery of acidic glycerophospholipids. Lipid extraction was started with 15 mgCDW using the appropriate volume of culture medium. For high analyte recovery, cell suspensions were transferred to Teflon centrifuge tubes and cells were harvested by centrifugation (5 min, 4633 g, −6°C; Heraeus Multifuge 1 R‐S). The cell pellet was gently washed with 3 ml of deionized water (0°C) and centrifuged again (5 min, 4633 g, −6°C) before resuspension in 3 ml of methanol (0°C) to quench all metabolic processes. For lipid extraction 6 ml of chloroform was added. The extraction was carried out by sonication (10 min), shaking (30 min), sonication (10 min) and shaking (60 min). Cell residues were separated by centrifugation (10 min, 4000 g, 0°C), the extracts transferred to silylated 1.5 ml glass vials, dried under nitrogen stream at 30°C and stored at −20°C. For analysis, the samples were reconstituted in a mixture of acetonitrile/methanol/chloroform (49:49:2, by volume).
High‐performance liquid chromatography/mass spectrometry
For HPLC/MS analysis the reconstituted samples were adjusted to a final concentration of 5 mM ammonium acetate/50 mM acetic acid (pH 3.75). Chromatographic separations were performed with a Surveyor MS pump and Surveyor autosampler (Thermo Scientific, San Jose, CA, USA) with a 5 µl full loop injection using a microbore C4‐Nucleodur Gravity column (150 mm × 1 mm i.d., particle size 5 µm) from Macherey‐Nagel (Düren, Germany). The following binary gradient at a flow rate of 115 µl min−1 was applied: 50% solvent B isocratic for 2 min, followed by 50–75% in 33 min, 75–100% in 10 min, 100% isocratic for 20 min, from 100% to 50% in 1 min, and then 50% solvent B isocratic for 14 min. Mobile phase A consisted of 2.5% acetonitrile, 2.5% methanol and 95% H2O (v/v) with 5 mM ammonium acetate/50 mM acetic acid (pH 3.75). Mobile phase B contained 5% H2O, 25% n‐propanol, 35% acetonitrile and 35% methanol (v/v) with 5 mM ammonium acetate/50 mM acetic acid (pH 3.75).
ESI‐LIT‐FTICR‐MS experiments were carried out using a LTQ‐FT‐Fourier transform ion cyclotron resonance hybrid‐mass spectrometer (Thermo Scientific, Bremen, Germany) with a 7.0 Tesla actively shielded superconducting magnet and electrospray ionization source operated in the data‐dependent mode. Survey centroid MS spectra in the mass range m/z 185–1850 were acquired in the FTICR with a resolution R = 25 000 at m/z 400 (target accumulation value 5 000 000, maximal ion accumulation time 750 ms). The two most intensive ions were sequentially isolated for accurate mass measurements by a FTICR ‘SIM scan’ in a narrow mass window (± 5 Da, R = 50 000, target accumulation value 100 000, maximal ion accumulation time 750 ms) in the profile mode. Subsequent fragmentations (MS2, MS3) were performed in the linear ion trap by collision‐induced dissociation (CID) (target accumulation value 10 000, maximal ion accumulation time 150 ms). Former target ions selected for MS/MS were dynamically excluded for 60 s with a total cycle time of approximately 4.6 s. General MS conditions were: −3.5 kV spray voltage, 30 arbitrary units sheath gas flow, 5 arbitrary units auxiliary gas flow and 2 arbitrary units sweep gas flow. The temperature of the ion transfer tube was set to 225°C. Parameters for CID MS2 and MS3 experiments: 30% normalized collision energy, activation at q = 0.25 for 30 ms. Ion selection thresholds were 500 counts for SIM scans, 500 counts for MS2 and 100 counts for MS3 experiments.
Gas chromatography/mass spectrometry
Aliquots (100 µl) of the reconstituted samples were transferred to silylated 200 µl glass inserts for 1.8 ml autosampler vials and dried under a nitrogen stream at 30°C. Samples were supplemented with 30 µl of chloroform and 70 µl of trimethyl‐sulfoniumhydroxid (0.25 M in methanol), mixed thoroughly, incubated for 60 min at 60°C and cooled down. A Focus GC coupled to a Polaris Q quadrupole ion trap mass spectrometer (both Thermo Scientific, Dreieich, Germany) equipped with a HP‐5 MS column (30 m; 0.25 mm i.d.; 0.25 µm film thickness; GGA, Moers, Germany) with the following temperature profile was used for analysis: 150°C (4 min), 2°C min−1, 250°C. One microlitre of sample was injected in splitless mode at 250°C injector temperature and a transfer capillary temperature of 280°C. The following mass spectrometric parameters were used: acquisition delay 3 min, ion source temperature 200°C, full scan range m/z 35–500, 70 eV electron impact ionization in the positive mode.
Profiler‐Merger‐Viewer software
For conversion of the raw files into text files the implemented file converter of Xcalibur (Thermo Scientific) was used. Text files were further processed by the Profiler‐Merger‐Viewer tool written in Java (Hein et al., 2010).
Acknowledgments
This work was supported by the German Ministry of Science and Education (BMBF, Project ERA‐NET SysMO, No. 0313980A) (VAPMdS). The authors acknowledge financial support from Deutsche Bundesstiftung Umwelt (DBU), the PAKT Project of the Leibniz Association ‘Integrated Cell Analysis’, and financial support by the Ministerium für Innovation, Wissenschaft, Forschung und Technologie des Landes Nordrhein‐Westfalen (Düsseldorf, Germany) and the Bundesministerium für Bildung und Forschung (Bonn, Germany) is gratefully acknowledged.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Average distribution of the relative amounts of glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Average relative amounts are calculated from both experimental and analytical replicates for each experimental set‐up. Data from replicates are provided in Tables S1a–S1d. Errors refer to the standard deviation of the data from replicates.
Table S2. Identified distinct glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Distinct glycerophospholipid species are characterized by the respective fatty acid moieties in sn1/sn2 position. Fatty acid moieties written in bold letters refer to the prominent combinations that can be found in almost all samples.
Table S3. Average distribution of the relative amounts of fatty acid moieties of the distinct glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Average relative amounts are calculated from experimental replicates for each experimental set‐up. Data from replicates are provided in Table S3a. Errors refer to the standard deviation of the data from replicates.
Table S1a. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida KT2440.
Table S1b. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida DOT‐T1E.
Table S1c. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida S12.
Table S1d. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of Pseudomonas sp. strain VLB120.
Table S3a. Distribution of the relative amounts of fatty acid moieties of the distinct glycerophospholipid species for the single measurements (experimental replicates) of the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ballesta J.P., Schaechter M. Dependence of the rate of synthesis of phosphatidylethanolamine and phosphatidylglycerol on the rate of growth of Escherichia coli. J Bacteriol. 1972;110:452–453. doi: 10.1128/jb.110.1.452-453.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch‐Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D33–D38. [Google Scholar]
- Bernal P., Segura A., Ramos J.L. Compensatory role of the cis‐trans‐isomerase and cardiolipin synthase in the membrane fluidity of Pseudomonas putida DOT‐T1E. Environ Microbiol. 2007;9:1658–1664. doi: 10.1111/j.1462-2920.2007.01283.x. [DOI] [PubMed] [Google Scholar]
- Blank L.M., Ionidis G., Ebert B., Bühler B., Schmid A. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 2008;275:5173–5190. doi: 10.1111/j.1742-4658.2008.06648.x. [DOI] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brannigan G., Tamboli A.C., Brown F.L.H. The role of molecular shape in bilayer elasticity and phase behavior. J Chem Phys. 2004;121:3259–3271. doi: 10.1063/1.1770569. [DOI] [PubMed] [Google Scholar]
- Bukata L., Altabe S., de Mendoza D., Ugalde R.A., Comerci D.J. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J Bacteriol. 2008;190:8197–8203. doi: 10.1128/JB.01069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos M., Kushiro K., Lai S.K., Shuler M.L. A genomically/chemically complete module for synthesis of lipid membrane in a minimal cell. Biotechnol Bioeng. 2007;97:397–409. doi: 10.1002/bit.21251. [DOI] [PubMed] [Google Scholar]
- Chen Q., Janssen D.B., Witholt B. Growth on octane alters the membrane lipid fatty acids of Pseudomonas oleovorans due to the induction of alkB and synthesis of octanol. J Bacteriol. 1995;177:6894–6901. doi: 10.1128/jb.177.23.6894-6901.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J.E. Bacterial membrane lipids: where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- Cronan J.E., Gelmann E.P. Physical properties of membrane lipids: biological relevance and regulation. Bacteriol Rev. 1975;39:232–256. doi: 10.1128/br.39.3.232-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrich D.L., Cota‐Robles E.H. Heterogeneity in lipid‐composition of outer membrane and cytoplasmic membrane of Pseudomonas Bal‐31. J Bacteriol. 1974;119:1006–1018. doi: 10.1128/jb.119.3.1006-1018.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek K.M., Ingram L.O. Effects of ethanol on the Escherichia coli plasma membrane. J Bacteriol. 1984;157:233–239. doi: 10.1128/jb.157.1.233-239.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Fang J.S., Barcelona M.J. Structural determination and quantitative analysis of bacterial phospholipids using liquid chromatography electrospray ionization mass spectrometry. J Microbiol Methods. 1998;33:23–35. [Google Scholar]
- Fang J., Barcelona M.J., Alvarez P.J.J. Phospholipid compositional changes of five pseudomonad archetypes grown with and without toluene. Appl Microbiol Biotechnol. 2000;54:382–389. doi: 10.1007/s002530000389. [DOI] [PubMed] [Google Scholar]
- Feng Y., Cronan J.E. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Matsuoka H., Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y. Cyclic phosphatidic acid – a unique bioactive phospholipid. Biochim Biophys Acta. 2008;1781:519–524. doi: 10.1016/j.bbalip.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R., Hauer B., Otto K., Schmid A. Microbial biofilms: new catalysts for maximizing productivity of long‐term biotransformations. Biotechnol Bioeng. 2007;98:1123–1134. doi: 10.1002/bit.21547. [DOI] [PubMed] [Google Scholar]
- Guckert J.B., Hood M.A., White D.C. Phospholipid ester‐linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl Environ Microbiol. 1986;52:794–801. doi: 10.1128/aem.52.4.794-801.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson C., Tagesson C. Influence of organic solvent mixtures on biological membranes. Br J Ind Med. 1985;42:591–595. doi: 10.1136/oem.42.9.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock I.C., Meadow P.M. Extractable lipids of Pseudomonas aeruginosa. Biochim Biophys Acta. 1969;187:366–379. doi: 10.1016/0005-2760(69)90010-1. [DOI] [PubMed] [Google Scholar]
- Hein E.M., Blank L.M., Heyland J., Baumbach J.I., Schmid A., Hayen H. Glycerophospholipid profiling by high‐performance liquid chromatography/mass spectrometry using exact mass measurements and multi‐stage mass spectrometric fragmentation experiments in parallel. Rapid Commun Mass Spec. 2009;23:1636–1646. doi: 10.1002/rcm.4042. [DOI] [PubMed] [Google Scholar]
- Hein E.M., Bodeker B., Nolte J., Hayen H. Software tool for mining liquid chromatography/multi‐stage mass spectrometry data for comprehensive glycerophospholipid profiling. Rapid Commun Mass Spec. 2010;24:2083–2092. doi: 10.1002/rcm.4614. [DOI] [PubMed] [Google Scholar]
- Heipieper H.J., de Bont J.A.M. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty‐acid composition of membranes. Appl Environ Microbiol. 1994;60:4440–4444. doi: 10.1128/aem.60.12.4440-4444.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper H.J., Diefenbach R., Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol‐degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper H.J., Loffeld B., Keweloh H., de Bont J.A.M. The cis/trans isomerization of unsaturated fatty‐acids in Pseudomonas putida S12 – an indicator for environmental‐stress due to organic‐compounds. Chemosphere. 1995;30:1041–1051. [Google Scholar]
- Heipieper H.J., Meulenbeld G., van Oirschot Q., de Bont J.A.M. Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl Environ Microbiol. 1996;62:2773–2777. doi: 10.1128/aem.62.8.2773-2777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper H.J., de Waard P., van der Meer P., Killian J.A., Isken S., de Bont J.A.M. Regiospecific effect of 1‐octanol on cis‐trans isomerization of unsaturated fatty acids in the solvent‐tolerant strain Pseudomonas putida S12. Appl Microbiol Biotechnol. 2001;57:541–547. doi: 10.1007/s002530100808. et al. [DOI] [PubMed] [Google Scholar]
- Heipieper H.J., Meinhardt F., Segura A. The cis‐trans isomerase of unsaturated fatty acids in Pseudomonas and Vibrio: biochemistry, molecular biology and physiological function of a unique stress adaptive mechanism. FEMS Microbiol Lett. 2003;229:1–7. doi: 10.1016/S0378-1097(03)00792-4. [DOI] [PubMed] [Google Scholar]
- Huertas M.J., Duque E., Molina L., Rossello‐Mora R., Mosqueda G., Goddy P. Tolerance to sudden organic solvent shocks by soil bacteria and characterization of Pseudomonas putida strains isolated from toluene polluted sites. Environ Sci Technol. 2000;34:3395–3400. et al. [Google Scholar]
- Ingram L.O. Changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food additives. Appl Environ Microbiol. 1977;33:1233–1236. doi: 10.1128/aem.33.5.1233-1236.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken S. WU; 2000. ) Physiology of Solvent Tolerance in Pseudomonas putida S12. Wageningen, the Netherlands: [Google Scholar]
- Isken S., de Bont J.A.M. Bacteria tolerant to organic solvents. Extremophiles. 1998;2:229–238. doi: 10.1007/s007920050065. [DOI] [PubMed] [Google Scholar]
- Kaneda T. Iso and anteiso‐fatty acids in bacteria – biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki‐Kinoshita K.F., Itoh M., Kawashima S. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P.D., Ouzounis C.A., Moore‐Kochlacs C., Goldovsky L., Kaipa P., Ahren D. Expansion of the Biocyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;19:6083–6089. doi: 10.1093/nar/gki892. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley K.E., Kennedy E.P. Origin of membrane asymmetry – pathway of synthesis of membrane lipids of Bacillus megaterium Km in vivo. Fed Proc. 1978;37:1691. [Google Scholar]
- Martinez‐Morales F., Schobert M., Lopez‐Lara I.M., Geiger O. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology. 2003;149:3461–3471. doi: 10.1099/mic.0.26522-0. [DOI] [PubMed] [Google Scholar]
- Mazzella N., Molinet J., Syakti A.D., Dodi A., Doumenq P., Artaud J., Bertrand J.C. Bacterial phospholipid molecular species analysis by ion‐pair reversed‐phase HPLC/ESI/MS. J Lipid Res. 2004;45:1355–1363. doi: 10.1194/jlr.D300040-JLR200. [DOI] [PubMed] [Google Scholar]
- Neumann G., Kabelitz N., Zehnsdorf A., Miltner A., Lippold H., Meyer D. Prediction of the adaptability of Pseudomonas putida DOT‐T1E to a second phase of a solvent for economically sound two‐phase biotransformations. Appl Environ Microbiol. 2005;71:6606–6612. doi: 10.1128/AEM.71.11.6606-6612.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D.R., Leonard E., Yoon S.H., Tseng H.C., Yuan C., Prather K.L.J. Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng. 2009;11:262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Ohta A., Shibuya I., Maruo B., Ishinaga M., Kito M. Extremely labile phosphatidylserine synthetase of an Escherichia coli mutant with temperature‐sensitive formation of phosphatidylethanolamine. Biochim Biophys Acta. 1974;348:449–454. [PubMed] [Google Scholar]
- Park J.B., Bühler B., Panke S., Witholt B., Schmid A. Carbon metabolism and product inhibition determine the epoxidation efficiency of solvent‐tolerant Pseudomonas sp. strain VLB120 Delta C. Biotechnol Bioeng. 2007;98:1219–1229. doi: 10.1002/bit.21496. [DOI] [PubMed] [Google Scholar]
- Pierucci O. Phospholipid synthesis during the cell‐division cycle of Escherichia coli. J Bacteriol. 1979;138:453–460. doi: 10.1128/jb.138.2.453-460.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkart H.C., White D.C. Phospholipid biosynthesis and solvent tolerance in Pseudomonas putida strains. J Bacteriol. 1997;179:4219–4226. doi: 10.1128/jb.179.13.4219-4226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Rodriguez‐Herva J.‐J., Godoy P., Haidour A., Reyes F., Fernàndez‐Barrero A. Mechanisms for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Godoy P., Segura A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT‐T1E. J Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Gallegos M.T., Godoy P., Ramos‐Gonzalez M.I., Rojas A. Mechanisms of solvent tolerance in Gram‐negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. et al. [DOI] [PubMed] [Google Scholar]
- Ramos‐Diaz M.A., Ramos J.L. Combined physical and genetic map of the Pseudomonas putida KT2440 chromosome. J Bacteriol. 1998;180:6352–6363. doi: 10.1128/jb.180.23.6352-6363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl J., Schmid A., Blank L.M. Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations. Appl Environ Microbiol. 2009;75:4653–4656. doi: 10.1128/AEM.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer M., Grote A., Chang A., Schomburg I., Munaretto C., Rother M. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Duque E., Mosqueda G., Ramos J.L., Junker F. Multiple responses of Gram‐negative bacteria to organic solvents. Environ Microbiol. 1999;1:191–198. doi: 10.1046/j.1462-2920.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- Sikkema J., de Bont J.A.M., Poolman B. Interactions of cyclic hydrocarbons with biological‐membranes. J Biol Chem. 1994;269:8022–8028. [PubMed] [Google Scholar]
- Soni S.P., Ward J.A., Sen S.E., Feller S.E., Wassall S.R. Effect of trans unsaturation on molecular organization in a phospholipid membrane. Biochemistry. 2009;48:11097–11107. doi: 10.1021/bi901179r. [DOI] [PubMed] [Google Scholar]
- Wang X.G., Scagliotti J.P., Hu L.T. Phospholipid synthesis in Borrelia burgdorferi: BB0249 and BB0721 encode functional phosphatidylcholine synthase and phosphatidylglycerolphosphate synthase proteins. Microbiology. 2004;150:391–397. doi: 10.1099/mic.0.26752-0. [DOI] [PubMed] [Google Scholar]
- Weber F.J., Ooijkaas L.P., Schemen R.M.W., Hartmans S., de Bont J.A.M. Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl Environ Microbiol. 1993;59:3502–3504. doi: 10.1128/aem.59.10.3502-3504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F.J., Isken S., de Bont J.A.M. Cis/trans isomerization of fatty acids as a defense mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology. 1994;140:2013–2017. doi: 10.1099/13500872-140-8-2013. [DOI] [PubMed] [Google Scholar]
- Wilkison W.O., Bell R.M. sn‐Glycerol‐3‐phosphate acyltransferase from Escherichia coli. Biochim Biophys Acta. 1997;1348:3–9. doi: 10.1016/s0005-2760(97)00099-4. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu W.Z., Hu T.C., Du L., Luo C., Chen K.X. Structural basis for catalytic and inhibitory mechanisms of beta‐hydroxyacyl‐acyl carrier protein dehydratase (FabZ) J Biol Chem. 2008;283:5370–5379. doi: 10.1074/jbc.M705566200. et al. [DOI] [PubMed] [Google Scholar]
- Zhu K., Zhang Y.M., Rock C.O. Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J Biol Chem. 2009;284:34880–34888. doi: 10.1074/jbc.M109.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Average distribution of the relative amounts of glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Average relative amounts are calculated from both experimental and analytical replicates for each experimental set‐up. Data from replicates are provided in Tables S1a–S1d. Errors refer to the standard deviation of the data from replicates.
Table S2. Identified distinct glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Distinct glycerophospholipid species are characterized by the respective fatty acid moieties in sn1/sn2 position. Fatty acid moieties written in bold letters refer to the prominent combinations that can be found in almost all samples.
Table S3. Average distribution of the relative amounts of fatty acid moieties of the distinct glycerophospholipid species for the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120. Average relative amounts are calculated from experimental replicates for each experimental set‐up. Data from replicates are provided in Table S3a. Errors refer to the standard deviation of the data from replicates.
Table S1a. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida KT2440.
Table S1b. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida DOT‐T1E.
Table S1c. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of P. putida S12.
Table S1d. Distribution of the relative amounts of glycerophospholipid species for the single measurements (experimental and analytical replicates) of Pseudomonas sp. strain VLB120.
Table S3a. Distribution of the relative amounts of fatty acid moieties of the distinct glycerophospholipid species for the single measurements (experimental replicates) of the investigated P. putida strains KT2440, DOT‐T1E, S12 and Pseudomonas sp. strain VLB120.


