Summary
Acinetobacter baylyi ADP1 was found to tolerate seawater and have a special ability of adhering to an oil–water interface of 10–80 µm emulsified mineral and crude oil droplets. These properties make ADP1 an ideal bacterial chassis for constructing bioreporters that are able to actively search and sense oil spill in water and soils. Acinetobacter baylyi bioreporter ADPWH_alk was developed and applied to the detection of alkanes and alkenes in water, seawater and soils. Bioreporter ADPWH_alk was able to detect a broad range of alkanes and alkenes with carbon chain length from C7 to C36. So far, ADPWH_alk is the only bioreporter that is able to detect alkane with carbon chain length greater than C18. This bioreporter responded to the alkanes in about 30 min and it was independent to the cell growth phase because of two point mutations in alkM promoter recognized by alkane regulatory protein ALKR. ADPWH_alk was applied to detect mineral oil, Brent, Chestnut and Sirri crude oils in water and seawater in the range 0.1–100 mg l−1, showing that the bioreporter oil detection was semi‐quantitative. This study demonstrates that ADPWH_alk is a rapid, sensitive and semi‐quantitative bioreporter that can be useful for environmental monitoring and assessment of oil spills in seawater and soils.
Introduction
Crude oil spill (such as the recent Mexico Gulf oil spill) and contamination associated with crude oil pumping, process and transportation posed a great threat to the environment and also public health. Alkanes and alkenes with various carbon chains are main components of crude oil. Although many microorganisms were able to aerobically or anaerobically degrade alkanes and alkenes (Van Beilen et al., 1994), biodegradation of long chain alkanes and alkenes was limited owing to their low solubility and accessibility to microorganisms, and therefore these compounds resided a relatively long time in the environment. Consequently, a rapid, cheap and sensitive way to detect alkanes or alkenes present in natural habitats was required for environmental monitoring and risk assessment. In regards to this, whole‐cell bioreporters for the detection of alkanes or alkenes are especially useful to environmental management because cell‐based bioreporters could indicate the bioavailability portions of alkanes and alkenes in a complex environment which generally could not be addressed using standard chemical analytical techniques.
Acinetobacter sp. are ubiquitous bacteria in natural aquatic and soil environment (Young et al., 2005) that are frequently found to be capable to degrade a broad range of carbon chain alkenes and alkanes (Lal and Khanna, 1996; DiCello et al., 1997; Ratajczak et al., 1998a,b; Baldi et al., 1999; Choi et al., 1999; Razak et al., 1999; Koma et al., 2001; Pleshakova et al., 2001; Tani et al., 2001; Throne‐Holst et al., 2007; Wentzel et al., 2007; Tanaka et al., 2010). Among these alkane degraders, Acinetobacter baylyi ADP1 is able to utilize alkanes with carbon lengths ranging from 12 up to 36 and the gene regulation for alkane degradation was well characterized (Ratajczak et al., 1998a,b; Throne‐Holst et al., 2007). ADP1 has an AraC/XylS‐like transcriptional regulatory protein ALKR that regulates alkane hydroxylase gene alkM in ADP1 chromosome to initiate alkane oxidization (Ratajczak et al., 1998a,b).
To date, only a few alkane bioreporters have been developed (Sticher et al., 1997; Alkasrawi et al., 1999; Minak‐Bernero et al., 2004), and those bioreporters have been shown to detect short and medium (i.e. C5–C10) carbon chain of alkanes and alkenes. To extend alkane detection spectra, we constructed an alkane/ alkene bioreporter ADPWH_alk that can respond to alkanes and alkenes with various carbon chain lengths ranging from C7 to C36. We found that A. baylyi ADP1 and ADPWH_alk were able to adhere to an interface of oil and water; and to emulsify mineral and crude oils into oil droplets at micrometre level. These special properties enabled ADPWH_alk to overcome alkane's low solubility and accessibility, and to actively search and sense oil spill in water and soils. The ADPWH_alk was used to determine mineral and crude oils in water, seawater and soils.
Results
Genetic structure of alkane bioreporter A. baylyi ADPWH_alk
ADPWH_alk has been constructed by inserting promoterless luxCDABE cassette into alkM in ADP1 and luxCDABE transcription is controlled by ALKR regulation system (Fig. 1). The vector pAlkRM_lux_km was constructed on pGEM‐T backbone which cannot replicate in ADP1, suggesting that the luxCDABE cassette should be inserted in the chromosome of ADP1. Southern blotting confirmed that a single copy of luxCDABE was at ADPWH_alk (data not shown). ADPWH_alk was able to grow on LB agar plate with 300 µg ml−1 ampicillin, indicating that the whole vector pAlkRM_lux_km had been inserted into the chromosome by Campbell‐like integration. The DNA sequences of colony PCR products, which used ADPWH_alk colony as DNA template and ADP1_alk_for/luxC_rev and alk_P_up/ADP1_alk_rev as primer pairs (Table 1), confirmed the genetic structure of ADPWH_alk construct (Fig. 1A). The DNA sequence also indicated that three point mutations at the promoter region of ADPWH_alk, which were introduced by pAlkRM_lux_km. The mutations were within the intergenic region between alkM1‐luxCDABE and alkR (Fig. 1B).
Figure 1.
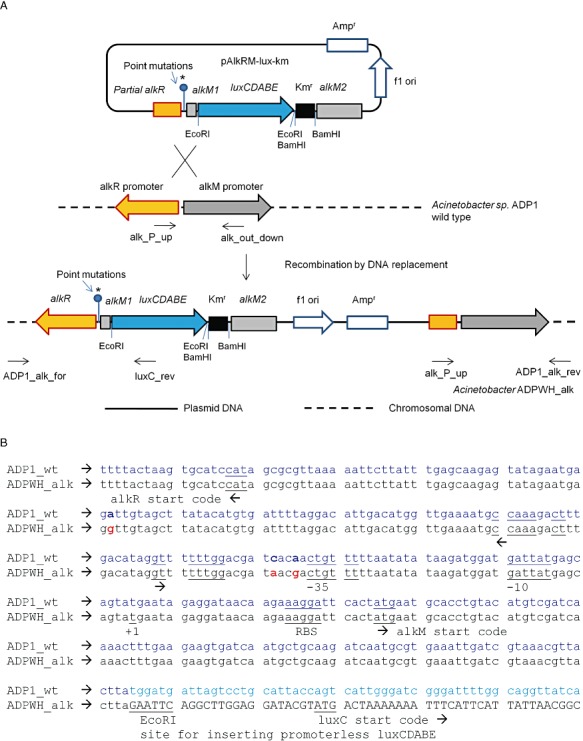
A. Schematic outline of construction of alkane bioreporter Acinetobacter baylyi ADPWH_alk (DNA lengths are not scaled). The three point mutations were marked as  . B. Genetic structure of alkane regulation part of ADPWH_alk. There are three mutation points at promoter region of alkRM. The mutation points were highlighted with bold. The inverted repeat with two mismatches is marked by arrows under the sequence.
. B. Genetic structure of alkane regulation part of ADPWH_alk. There are three mutation points at promoter region of alkRM. The mutation points were highlighted with bold. The inverted repeat with two mismatches is marked by arrows under the sequence.
Table 1.
Primers used in this study.
| Primers | Sequence (5′→3′) |
|---|---|
| alk_P_up | GTCGTACCATCCTGTAATGTGAAATGAG |
| alk_EB_reva | CATCCAGAATTCGGATCCTAAGTAACGTTT |
| alk_EB_fora | AAACGTTACTTAGGATCCGAATTCTGGATG |
| alk_out_down | CGAGCGCCAAATGAGACAGGATATGA |
| ADP1_alk_for | TTGATTAAGCGGATGGCTGGCATATA |
| luxC_rev | GAGAGTCATTCAATATTGGCAGG |
| ADP1_alk_rev | CGAACTATGAAAAGCAGCACTCATCC |
The underlines indicate EcoRI and BamHI restriction sites.
ADPWH_alk actively searching and sensing oils
The solubility of alkane and crude oil in water are usually extremely low; hence alkanes and crude oils are inaccessible to cells, which may hamper oil detections in complex water and soil environments. Acinetobacter baylyi ADP1 and its derivative ADPWH_alk were found to adhere to an oil–water interface and to emulsify oils into small droplets (Fig. 2). In the Escherichia coli DH5α–oil mixture, it was difficult to observe small oil droplets unless vagarious shaking was applied, and E. coli was not associated with oil neither (Fig. 2A). However, in the ADPWH_alk–oil mixture, ADPWH_alk emulsified both mineral and crude oils into 10–80 µm oil droplets and the cells were found attached to the surface of oil droplets but were absent from the water phase (Fig. 2B–D). It indicated that A. baylyi was able to recognize alkanes and twitch their movements to the surface oil droplets. The cell density of ADPWH_alk on the surface of mineral and crude oil droplets was estimated to be 1.4 ± 0.2 × 1010 cells m−2, implying that cells occupied a single layer on the oil surface.
Figure 2.
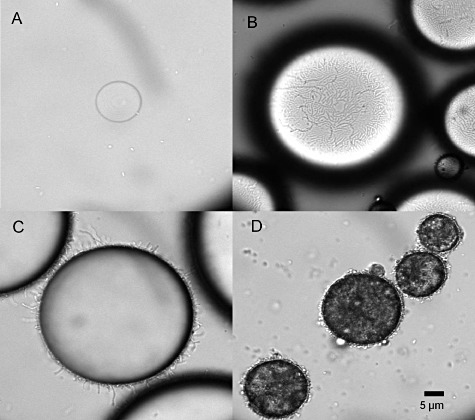
Acinetobacter baylyi ADPWH_alk natural affinity to oil droplets. Unlike E. coli which was unable to emulsify and attach to oil droplets (A), ADPWH_alk, a subclone from ADP1, was able to emulsify and attached to the surface of mineral oil (B, C) and crude oil (D) droplets. The scale bar is 5 µm.
Characterization of alkane bioreporter ADPWH_alk
ADPWH_alk induction was conducted by exposing the bioreporters to 100 µM suspension solution of a series of alkanes including: hexane (C6), dodecane (C12), tetradecane (C14), octadecane (C18), tetracosane (C24), triacontane (C30) and tetracontane (C40). The time‐course of ADPWH_alk activation and the bioluminescence expression were plotted in Fig. 3A. In the presence of dodecane (C12), tetradecane (C14), octadecane (C18), tetracosane (C24) and triacontane (C30), ADPWH_alk was rapidly induced and expressed bioluminescence within 30 min compared with a background level of bioluminescence in the absence of alkanes (Fig. 3A). The maximum induction by octadecane (C18) was approximately two times higher than other alkanes (Fig. 3A), which was consistent with previous report (Ratajczak et al., 1998a). ADPWH_alk was also rapidly induced by Brent crude oil in pure water and seawater and the ADPWH_alk sensing performance to Brent crude oil was not affected by salt and other impurities in seawater (Fig. 3B).
Figure 3.
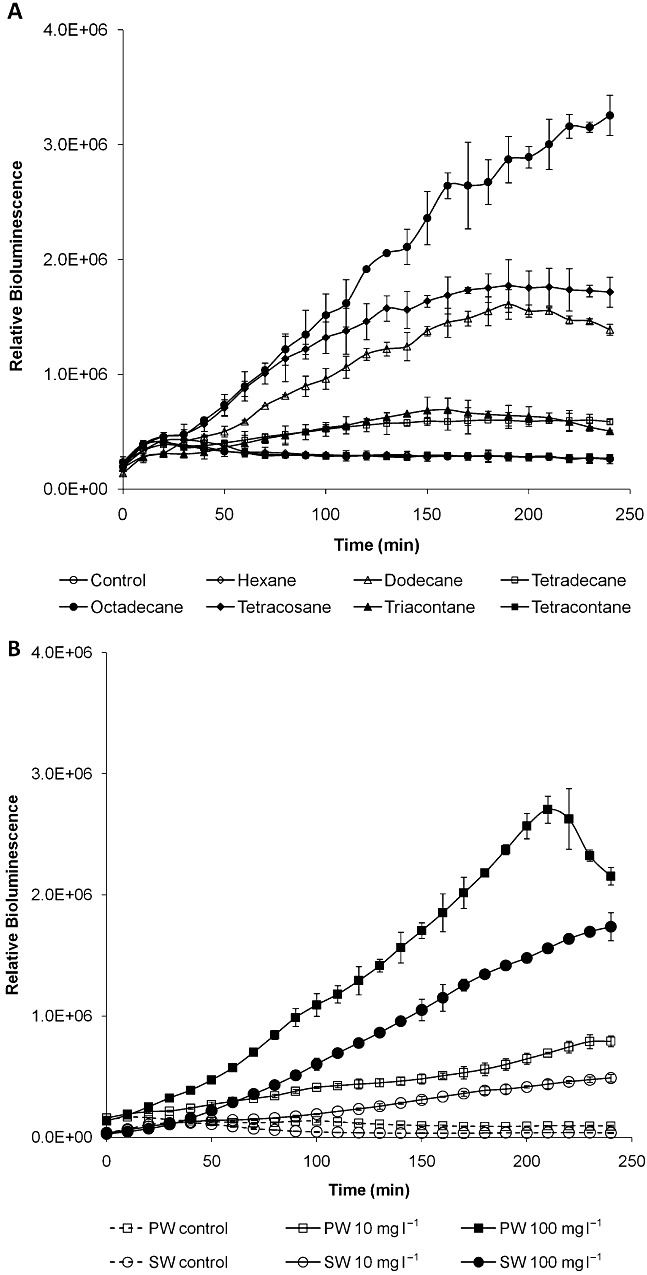
Dynamic ADPWH_alk response to alkanes with 100 µM of each compound (A); and 100 mg l−1 Brent crude oil in pure water (PW) and seawater (SW) (B).
Detection of a broad range of carbon chain alkenes and alkanes (C7–C36)
Figure 4 represented that ADPWH_alk was sensitive to alkanes and alkenes with carbon chain length ranging from C7 to C36. Although A. baylyi could only utilize alkanes with carbon chain longer than C12 (Ratajczak et al., 1998a; Wentzel et al., 2007), ADPWH_alk was able to respond to alkanes with low carbon chain length down to C7, meaning that this bioreporter could cover a broad range of alkenes and alkanes (i.e. C7–C36). Alkanes with shorter (e.g. hexane – C6) or longer carbon chains (e.g. tetracontane – C40) did not activate ADPWH_alk (Figs 3A and 4). ADPWH_alk induction ratio varied to alkanes or alkenes with different lengths of carbon chain, while alkanes or alkenes with the same carbon chain length, such as C12 dodecane/dodecene and C18 octadecane/octadecene, exhibited similar induction ratios (Fig. 4). The induction ratios of C12, C18 and C24 alkanes or alkenes were significantly higher than alkanes/alkenes with other carbon chain length (Fig. 3), suggesting that alkanes or alkenes with specific carbon chain lengths had effects on the complex of ALKR–promoter–RNAP and favoured alkM transcription initiation.
Figure 4.
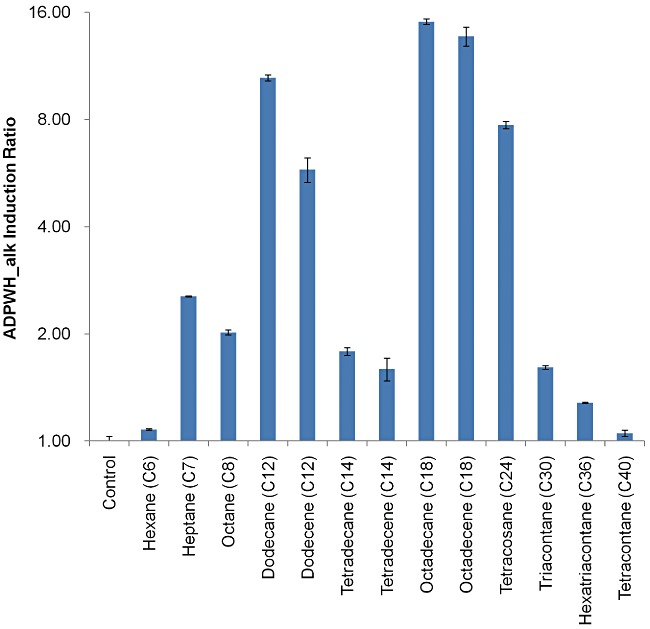
ADPWH_alk response to alkanes and alkenes with various carbon chain length from C6 to C40. The alkanes or alkenes were emulsified by ultrasound and the final concentration of each alkane or alkene was 100 µM. The P‐values of induction ratios to C7–C36 < 0.001 (against control).
Calibration of ADPWH_alk responding to mineral and crude oils in water, seawater and soil
ADPWH_alk induction ratios increased in response to higher concentrations of mineral, Brent, Chestnut and Sirri crude oils in the range of 0.1–100 mg l−1 in a semi‐quantitative manner (Fig. 5), and the detection limit (0.1 mg l−1) was equivalent to the US EPA crude oil contamination limit (US EPA water standard and gold book). Optimal time of ADPWH_alk for oil quantitative estimation was 200–240 min. The three crudes tested were typical oils, and Brent crude was a standard crude oil used by world oil trade. Although the mineral oil and crude oils are consist of a mixture of alkanes and alkenes with different compositions, ADPWH_alk induction patterns to different oils were similar (Fig. 5), which suggested that a single general calibration curve was sufficient to estimate the oil concentration in water in practice. ADPWH_alk induction in seawater and in pure water was similar, indicating that seawater had little impact on the bioreporter's performance and that ADPWH_alk could be applied to detect oil spill in seawater. A curve of ADPWH_alk induction ratio against crude oil contents in a standard soil was plotted in Fig. 6, which shows that the induction ratio increased with higher contents of crude oil in soils. On the one hand, the more soil added into the bioreporter, the more accuracy of the estimation. On the other hand, less amount of soil would be desirable since soil would interfere with the bioreporter induction measurement by blocking the bioluminescence light. Hence, in Fig. 6, each measurement point was present at its optimal condition which was identified as the maximum bioreporter induction ratio with the minimal soil content. This problem can be overcome by applying magnetic functionalized nanoparticles to the bioreporter (Zhang et al., 2011).
Figure 5.
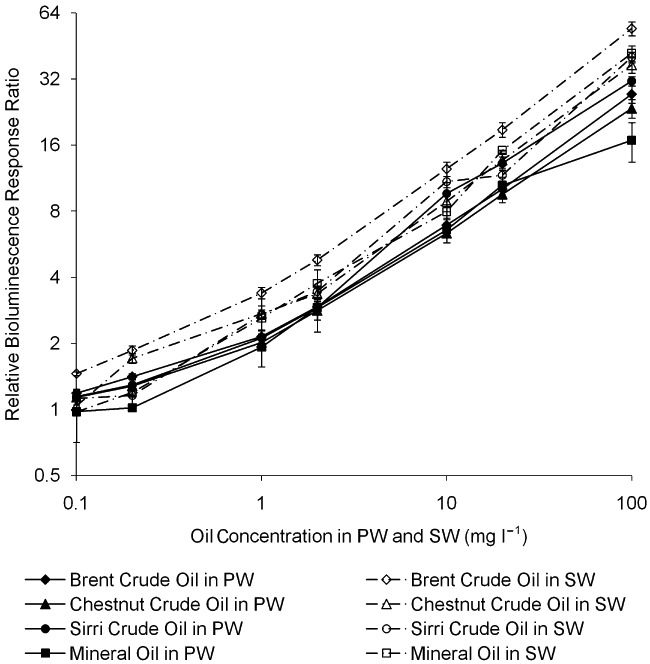
ADPWH_alk response to different concentrations of mineral oil and Brent, Chestnut and Sirri crude oils in pure water and seawater.
Figure 6.
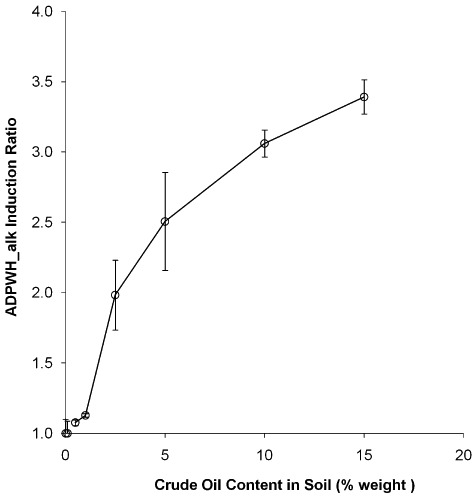
ADPWH_alk response to crude oil in soils.
Discussion
Actively searching and sensing alkanes and oils
It is crucial that bacteria should access to insoluble alkanes or oils before they can detect them. Oil emulsification would significantly increase the oil surface area and provide a much larger interface for biochemical reactions. Alkane chemotaxis provides another advantage of oil‐degrading bacteria. Acinetobacter baylyi is able to carry out both alkane emulsification and chemotaxis, which make ADPWH_alk a desirable oil‐detecting bioreporter as it actively searches and senses oils in water and soils. We observed that it took ADPWH_alk ∼20 min to emulsify and bind alkane droplets. Given that the cells take ∼10 min to carry out signal transport, regulation process and gene expression, the minimal response time for the alkane bioreporter would be about 30 min. Additionally, it was observed that ADPWH_alk could also be activated by alkanes within 40 min after stored in water at 4°C for more than 1 month. Although the alkM–lux fusion and the wild‐type alkM gene in ADPWH_alk may suffer a second cross‐over event to remove luxCDABE cassette, ADPWH_alk unstability was not observed in our lab even after 200 continuous generations. In practice, negative and positive controls are always required to rule out possible false‐negative results.
The transformant ADPWH_alk was able to respond to alkanes with various lengths of carbon chains. To rule out the possibility that activation of ADPWH_alk was due to alkanes being served as substrates of luciferase LuxAB instead of ALKR‐based transcriptional regulation, a series of controls and tests were carried out. The results showed that ADPWH_alk was induced by octadecane while other bioreporters such as ADPWH_lux (salicylate) (Huang et al., 2005), ADPWH_Tol (toluene) (Huang et al., 2008) and ADPWH_recA (toxicity) (Song et al., 2009), which also contained luxCDABE as reporter as well, could not be activated by octadecane and the addition of octadecane did not increase bioluminescence expression of those bioreporters (Fig. S2).
Mutation in alkM shortens response time to alkanes and oils
Those point mutations affected at alkM promoter alkR regulatory binding and RNA transcription in response to alkanes. ADPWH_alk responded to alkanes in about 30 min and was independent of the growth phases (Fig. 3A and Fig. S3), while previous report showed that strain WH405 (alkM::lacZ) was silenced for about 600 min (10 h) until the strain reached a transition period between exponential and stationary phases (Ratajczak et al., 1998a). Such differences might be associated with the point mutations in alkM promoter within alkR–alkM intergenic region which controls the reporter genes luxCDABE. ALKR is a transcriptional regulator belonging to AraC/XylS family and is constitutively transcribed at low level in different growth phases (Ratajczak et al., 1998a), which was observed as constant bioluminescent background of the controls in Fig. 3. ALKR regulatory protein includes C‐terminal DNA‐binding domain (CTD) for promoter binding and N‐terminal domain (NTD) for inducer recognition (Gallegos et al., 1997; Tropel and van der Meer, 2004; Dominguez‐Cuevas et al., 2010). Alkanes were presumably recognized by ALKR regulatory protein and their interaction triggered a conformation change of ALKR dimers which isomerizes the promoter–RNAP complex and led to the activation of alkM gene. The inverted repeat region shown in Fig. 1B was presumptively the binding site of ALKR dimmers. There were two point mutations between −35 region and the inverted repeat in ADPWH_alk (Fig. 1A), which might affect binding and interaction between ALKR dimers and alkM promoter. It was assumed that this change enabled cells to overcome some limiting factors related to growth phases.
In summary, we developed a bioreporter ADPWH_alk which is able to detect alkanes and oils in seawater and soils. It provides a rapid, sensitive and quantitative tool for environmental monitoring and assessment for oil spills in seawater and soils. The bioreporter was constructed in the chromosome of an environmental bacterium –A. baylyi, which is able to actively search and sense oils. This bioreporter is more robust in terms of sensing performance (e.g. responding time, tolerance of seawater) and long‐term storage. In comparison with previously reported alkane bioreporters, ADPWH_alk detects a broader carbon chain of alkanes/alkenes, responds in shorter time and its detection limit can be extremely low to some alkanes (e.g. octadecane) (Table 2). To our knowledge, so far ADPWH_alk is the only bioreporter that is able to detect alkane with carbon chain length greater than C18.
Table 2.
Comparison of different alkane bioreporters.
| Host | Regulation system | Reporter | Response time | Detection limit | Alkane detection range | Reference |
|---|---|---|---|---|---|---|
| E. coli DH5α | alkB‐alkS | gfp | 2.5 h | 0.011 mg l−1a | C6–C10 | Jaspers et al. (2001) |
| E. coli DH5α | alkB‐alkS | luxAB | 1.0−2.0 h | 0.011 mg l−1a | C6–C10 | Sticher et al. (1997), Tecon et al. (2010) |
| E. coli DH5α | alkBFGHJT | luxAB | – | 0.78 mg l−1 | C6–C10 | Minak‐Bernero et al. (2004) |
| Y. lipolytica | – | OD600 | 24 h | 0.85 mg l−1 | C9–C12 | Alkasrawi et al. (1999) |
| A. baylyi ADP1 | alkM‐alkR | lacZ | 10 h | – | C7–C18 | Ratajczak et al. (1998a) |
| A. baylyi ADP1 | alkM‐alkR | luxCDABE | 0.5 h | 0.1 mg l−1b | C7–C36 | Zhang et al. (2011) and this study |
The detection limit for octane only.
The detection limit for octadecane is 8.6 × 10−4 mg l−1.
Bioreporters employ live cells (usually genetically engineered bacteria) as detection elements and they can be a complementary tool to chemical analysis. Bioreporters have a few advantages: rapid, simple, cheap, in situ and importantly providing information of bioavailability. Bioavailability is one of main concerns of environmental risk management since these two factors directly link environmental contamination to human health risk. Most environmental samples are mixtures of complex contaminants. The additive, antagonistic and synergistic effects caused by complex physical or chemical interactions would make the risk assessment unpredictable if only chemical analysis (e.g. GC‐MS or HPLC) was applied to the estimation of bioavailability of contaminated samples. Chemical analysis of contaminated soil and water samples usually requires sample pre‐treatment and extraction, which makes inert or active portions of contaminants indistinguishable, while risk assessment is concerned to the active portion (bioavailability). In contrast, bioreporters detect contaminants' active or bioavailable portions, and it directly links contamination to biological effects and make bioavailability assessment more relevant to human health risk. It is a challenge to construct bioreporters with high reliability, robustness and reproducibility. With recent advances in molecular cell biology, synthetic biology and nanotechnology, bioreporters can be endowed with new functions which enhanced the sensing performance (Zhang et al., 2011). It could reduce the gap between laboratory work and real‐world application.
Experimental procedures
Culture media and strains
The bacterial strains and plasmids used in this study are listed in Table 3. Acinetobacter baylyi ADP1 and its mutants were incubated at 30°C, and E. coli at 37°C. All chemicals were obtained from Sigma‐Aldrich and were analytical grade reagents. Luria–Bertani (LB) medium or minimal medium (MM) (Huang et al., 2005) was used for the cultivation of bacteria as appropriate. One litre of MM contains 2.5 g of Na2HPO4, 2.5 g of KH2PO4, 1.0 g of NH4Cl, 0.1 g of MgSO4·7H2O, 10 µl of saturated CaCl2 solution, 10 µl of saturated FeSO4 solution and 1 ml of Bauchop & Elsden solution. MM succinate (MMS) was prepared by adding 20 mM succinate to MM. Ampicillin (Amp) at 100 µg ml−1 and 50 µg ml−1 kanamycin (Km) was used for E. coli, and 300 µg ml−1 Amp and 10 µg ml−1 Km for A. baylyi ADP1 or its mutants when required.
Table 3.
Strains and plasmids used in this study.
| Bacteria and plasmids | Description | Reference |
|---|---|---|
| Acinetobacter baylyi strains | ||
| ADP1(BD413) | Wild type | Juni and Janik (1969) |
| ADPWH_lux | Bioreporter of salicylate. Promoterless luxCDABE from pSB417 were inserted between salA and salR genes in the chromosome of ADP1 | Huang et al. (2005) |
| ADPWH_Tol (ADPWH‐Pu‐lux‐xylR) | Bioreporter of toluene. Promoterless luxCDABE from pSB417 were fused with Pu promoter and inserted between salA and salR genes, and xylR gene was inserted at salA in the chromosome of ADP1 | Huang et al. (2008) |
| ADPWH_recA | Bioreporter of genotoxicity. Promoterless luxCDABE from pSB417 were inserted in recA gene in the chromosome of ADP1 | Song et al. (2009) |
| ADPWH_alk | Bioreporter of alkanes. Promoterless luxCDABE were inserted in alkM gene in the chromosome of ADP1 | This study |
| Escherichia coli | ||
| JM109 | High efficiency competent cells | Promega |
| DH5α | F‐endA1 glnV44 thi‐1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA‐argF)U169, hsdR17(rK‐mK+), λ‐ | Grant et al. (1990) |
| Plasmids | ||
| pGEM‐T | Ampr, T7 and SP6 promoters, lacZ, vector | Promega |
| pSB417 | luxCDABE source plasmid, luxCDABE from Photorhabdus (Xenorhabdus) luminescens ATCC2999 (Hb strain) | Winson et al. (1998) |
| pUTKm1 | Source of kanamycin resistance gene | Delorenzo et al. (1990) |
| pAlkRM_EB | alkRM with EcoRI/BamHI (878 bp) inserted into pGEM‐T | This study |
| pAlkRM_lux_km | luxCDABE (5846 bp) from pSB417 and Km (1708 bp) from pUTKm1 were inserted at EcoRI and BamHI site of pAlKRM_EB | This study |
DNA manipulation and cloning alkane bioreporter ADPWH_alk
First, a DNA fragment alkRM was amplified by PCR from wild‐type ADP1, as shown in Fig. S1. Briefly, PCR reactions were made by using a colony of wild‐type ADP1 as DNA template and primer pairs of alk_P_up/alk_EB_rev and alk_EB_for/alk_out_down (Table 1). The two PCR products were fused by overlap extension PCR (Huang et al., 2005) using a primer pair of alk_P_up and alk_out_down to create EcoRI and BamHI restriction sites between alkR and alkM fragments. The PCR product alkRM with EcoRI/BamHI (878 bp) was gel purified and cloned into pGEM‐T (Promega, UK) vector to create pAlkRM_EB (Table 3). Second, the Km gene from mini‐Tn5 plasmid pUTKm1 (Delorenzo et al., 1990) and a promoterless luxCDABE cassette from pSalAR_lux (Winson et al., 1998; Huang et al., 2005) were separately cloned into BamHI and EcoRI site of pAlkRM‐EB (Fig. 1). The luxCDABE‐km cassette was flanked by 452 and 415 bp homologous DNA fragments. The resulting pAlkRM‐lux‐km plasmid was used as transformation carrier to introduce luxCDABE and Km genes into the alkM in the chromosome of ADP1 (Fig. 1). The transformation was carried out as previous description (Huang et al., 2005; 2008). The transformants were selected in LB agar with 10 µg ml−1 kanamycin and 1 µl ml−1 liquid paraffin. Colonies with strong bioluminescence were selected and designated as ADPWH_alk.
To confirm the integration of luxCDABE in the chromosome, PCR was performed using a primer pairs ADP1_alk_for/luxC_rev and alk_P_up/ADP1_alk_rev (Table 1), in which the primers ADP1_alk_for and ADP1_alk_rev were excluded in the vector pAlkRM_lux_km (Fig. 1A). The resultant PCR products were purified, cloned into pGEM‐T vector and sequenced.
Quantifying oil adhesiveness to ADPWH_alk
Bacterial cells were harvested by 3000 r.p.m. centrifugation after grown in LB liquid medium overnight at 30°C (ADPWH_alk) or 37°C (E. coli DH5α). The cells were then resuspended in mineral medium with 20 mM sodium succinate as sole carbon source. Ten microlitres of mineral oil or Brent crude oil was added into 1 ml of bacteria solution (∼108 cells ml−1), gently mixed and incubated at 30°C for 30 min. The emulsified bacteria/oil mixture was applied for microscopy examination. The cells found on 30 oil droplets were counted and the diameters of oil droplets were measured for oil surface calculation. The cell density on oil surface was then calculated as number of cells per m2 surface.
Preparation of alkane bioreporter ADPWH_alk
Luria–Bertani and MMS were used for the cultivation of bioreporters. After grown in LB medium at 30°C overnight, ADPWH_alk cells were harvested by centrifugation at 3000 r.p.m. for 10 min at 4°C, and subsequently washed twice and resuspended in MM medium with the same volume. Subsequently, ADPWH_alk bioreporters were cultured at 30°C for 2 h, followed by centrifugation at 3000 r.p.m. for 10 min at 4°C, and resuspended in water with the same volume as bioreporter stock solution. ADPWH_alk bioreporters were stored at 4°C and were ready for use, which be maintained for more than 1 month without changing performance and sensitivity. This is consistent with a previous report of A. baylyi‐based toxicity biosensor (Song et al., 2009). The other three bioreporter strains including ADPWH_lux, ADPWH_Tol and ADPWH_recA (Table 3) were prepared using the same method as described above.
Induction of bioreporter ADPWH_alk
Before induction experiments, all the bioreporter stock solutions, including ADPWH_alk, ADPWH_lux, ADPWH_Tol and ADPWH_recA, were centrifuged at 3000 r.p.m. for 5 min and then resuspended in the same volume of MMS medium. This treatment would optimize the bioreporter's response.
Different types of inducers: hexane (13.1 µl), heptanes (14.7 µl), octane (16.2 µl), dodecane (22.7 µl), tetradecane (26.0 µl), octadecane (25.5 mg), tetracosane (33.9 mg), triacontane (42.3 mg), hexatriacontane (50.7 mg) and tetracontane (56.3 mg), were dissolved in 100 ml of deionized water respectively. After homogenization using a 40 kHz ultrasound for 30 s, the n‐alkanes emulsion stock solutions were obtained with final concentrations of 1.0 mM. The emulsion treatment enabled alkanes to homologically distribute in water although they were insoluble.
For oil and crude oils pre‐treatment, 12.5 µl of mineral oil (Sigma) or crude oils including Brent crude oil (from Brent Reservoir, North Sea, UK), Chestnuts crude oil (from Chestnuts Reservoir, North Sea, UK) and Sirri crude oil (from Sirri Island, Iran) were dissolved in 100 ml of deionized water respectively. The mixtures were then homogenized using a 40 kHz ultrasound for 30 s to make up approximately 100 mg l−1 oil stock solution. The stock solutions were diluted to the final series concentrations of 0.1, 0.2, 1.0, 2.0, 10, 20 and 100 mg l−1 with deionized water or seawater.
Applying ADPWH_alk for the detection of oils in soils
Standard crude oil was added into chloroform to make 0, 1, 2, 5, 10, 25, 50, 100 and 150 mg ml−1 stock solutions. Ten millilitres of oil–chloroform mixtures were added into 10 g of soils respectively and volatilized at 30°C for 24 h to remove any trace of chloroform. The final crude oil contents in the soil were 0.0%, 0.1%, 0.25%, 0.5%, 1.0%, 2.5%, 5.0%, 10% and 15% (% weight). For soil sample pre‐treatment and detection, 5–200 mg of soil samples with different oil contents were transferred into tubes with eight replicates for each sample. Each tube was added into 5 ml of deionized water and exposed to 40 kHz ultrasound for 300 s for homogenization. After static settlement for 10 min, the supernatants of soil/water mixtures were carefully taken out for ADPWH_alk detection.
Bioluminescence detection
Application of bioreporters for various detections were all carried out in MMS medium. For all water sample detection, 180 µl of bioreporters in MMS medium were added into each well of a black clear‐bottom 96‐well microplate (Corning Costa, USA). Then, 20 µl of each water sample was added into relative wells. For all soil sample detection, 180 µl of supernatants of soil/water mixtures were mixed with 20 µl of ADPWH_alk in MMS medium. In total 200 µl of sample–bioreporter mixture was added into a well of 96‐well microplate for measurement. At least three biological replicates and three measurement replicates were carried out for each sample.
The microplates were loaded into Synergy 2 Multi‐mode Microplate Reader (BioTek Instruments, UK) equipped with Gen5 analysis software. The microplates were incubated at 30°C during the measurements. The bioluminescence and OD600 were measured every 10 min. Before each measurement, 30 s of vertical shaking was used for oil inducer dispersion. Relative bioluminescence was calculated by dividing induced bioluminescence by OD600. Bioluminescence induction ratio was evaluated by dividing relative bioluminescence of samples by relative bioluminescence of controls (non‐induced samples).
Acknowledgments
We thank Royal Society Research Grant, NERC Innovative Fund and China National Natural Science Foundation (Project 40730738). We also thank Huada Genomics Institute for providing travelling fund for Y.H.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Overlap extension PCR to create EcoRI/BamHI sites at alkR–alkM DNA fragment.
Fig. S2. The effect of alkanes on bioluminescence expression. ADPWH_alk was activated by octadecane while the bioreporters ADPWH_lux (salicylate), ADPWH_Tol (toluene), ADPWH_recA (toxicity) cannot be activated by octadecane and the addition of octadecane did not increase bioluminescence expression.
Fig. S3. Biosensor ADPWH_lux growth curve in LB supplemented with 100 μM alkane with different carbon chain lengths.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alkasrawi M., Nandakumar R., Margesin R., Schinner F., Mattiasson B. A microbial biosensor based on Yarrowia lipolytica for the off‐line determination of middle‐chain alkanes. Biosens Bioelectron. 1999;14:723–727. doi: 10.1016/s0956-5663(99)00046-9. [DOI] [PubMed] [Google Scholar]
- Baldi F., Ivosevic N., Minacci A., Pepi M., Fani R., Svetlicic V., Zutic V. Adhesion of Acinetobacter venetianus to diesel fuel droplets studied with in situ electrochemical and molecular probes. Appl Environ Microbiol. 1999;65:2041–2048. doi: 10.1128/aem.65.5.2041-2048.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.H., Hori K., Tanji Y., Unno H. Microbial degradation kinetics of solid alkane dissolved in nondegradable oil phase. Biochem Eng J. 1999;3:71–78. [Google Scholar]
- Delorenzo V., Herrero M., Jakubzik U., Timmis K.N. Mini‐Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram‐negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCello F., Pepi M., Baldi F., Fani R. Molecular characterization of an n‐alkane‐degrading bacterial community and identification of a new species, Acinetobacter venetianus. Res Microbiol. 1997;148:237–249. doi: 10.1016/S0923-2508(97)85244-8. [DOI] [PubMed] [Google Scholar]
- Dominguez‐Cuevas P., Ramos J.L., Marques S. Sequential XylS‐CTD binding to the Pm promoter induces DNA bending prior to activation. J Bacteriol. 2010;192:2682–2690. doi: 10.1128/JB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos M.T., Schleif R., Bairoch A., Hofmann K., Ramos J.L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.G.N., Jessee J., Bloom F.R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation‐restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Wang H., Huang L.F., Zheng H.J., Singer A.C., Thompson I.P., Whiteley A.S. Chromosomally located gene fusions constructed in Acinetobacter sp ADP1 for the detection of salicylate. Environ Microbiol. 2005;7:1339–1348. doi: 10.1111/j.1462-5822.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Singer A.C., Spiers A.J., Preston G.M., Whiteley A.S. Characterizing the regulation of the Pu promoter in Acinetobacter baylyi ADP1. Environ Microbiol. 2008;10:1668–1680. doi: 10.1111/j.1462-2920.2008.01583.x. [DOI] [PubMed] [Google Scholar]
- Jaspers M.C.M., Meier C., Zehnder A.J.B., Harms H., van der Meer J.R. Measuring mass transfer processes of octane with the help of an alkS‐alkB : gfp‐tagged Escherichia coli. Environ Microbiol. 2001;3:512–524. doi: 10.1046/j.1462-2920.2001.00218.x. [DOI] [PubMed] [Google Scholar]
- Juni E., Janik A. Transformation of Acinetobacter calcoaceticusBacterium anitratum. J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koma D., Hasumi F., Yamamoto E., Ohta T., Chung S.Y., Kubo M. Biodegradation of long‐chain n‐paraffins from waste oil of car engine by Acinetobacter sp. J Biosci Bioeng. 2001;91:94–96. doi: 10.1263/jbb.91.94. [DOI] [PubMed] [Google Scholar]
- Lal B., Khanna S. Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J Appl Bacteriol. 1996;81:355–362. doi: 10.1111/j.1365-2672.1996.tb03519.x. [DOI] [PubMed] [Google Scholar]
- Minak‐Bernero V., Bare R.E., Haith C.E., Grossman M.J. Detection of alkanes, alcohols, and aldehydes using bioluminescence. Biotechnol Bioeng. 2004;87:170–177. doi: 10.1002/bit.20089. [DOI] [PubMed] [Google Scholar]
- Pleshakova E.V., Muratova A.Y., Turkovskaya O.V. Degradation of mineral oil with a strain of Acinetobacter calcoaceticus. Appl Biochem Microbiol. 2001;37:342–347. [PubMed] [Google Scholar]
- Ratajczak A., Geissdorfer W., Hillen W. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n‐alkanes and requires the transcriptional activator AlkR. J Bacteriol. 1998a;180:5822–5827. doi: 10.1128/jb.180.22.5822-5827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak A., Geissdorfer W., Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral‐membrane hydrocarbon hydroxylases. Appl Environ Microbiol. 1998b;64:1175–1179. doi: 10.1128/aem.64.4.1175-1179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razak C.N.A., Wang W.F., Rahman S., Basri M., Salleh A.B. Isolation of the crude oil degrading marine Acinetobacter sp E11. Acta Biotechnol. 1999;19:213–223. [Google Scholar]
- Song Y.Z., Li G.H., Thornton S.F., Thompson I.P., Banwart S.A., Lerner D.N., Huang W.E. Optimization of bacterial whole cell bioreporters for toxicity assay of environmental samples. Environ Sci Technol. 2009;43:7931–7938. doi: 10.1021/es901349r. [DOI] [PubMed] [Google Scholar]
- Sticher P., Jaspers M.C.M., Stemmler K., Harms H., Zehnder A.J.B., vanderMeer J.R. Development and characterization of a whole‐cell bioluminescent sensor for bioavailable middle‐chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D., Takashima M., Mizuta A., Tanaka S., Sakatoku A., Nishikawa A. Acinetobacter sp Ud‐4 efficiently degrades both edible and mineral oils: isolation and characterization. Curr Microbiol. 2010;60:203–209. doi: 10.1007/s00284-009-9525-5. et al. [DOI] [PubMed] [Google Scholar]
- Tani A., Ishige T., Sakai Y., Kato N. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp strain M‐1. J Bacteriol. 2001;183:1819–1823. doi: 10.1128/JB.183.5.1819-1823.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R., Beggah S., Czechowska K., Sentchilo V., Chronopoulou P.M., McGenity T.J., van der Meer J.R. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ Sci Technol. 2010;44:1049–1055. doi: 10.1021/es902849w. [DOI] [PubMed] [Google Scholar]
- Throne‐Holst M., Wentzel A., Ellingsen T.E., Kotlar H.K., Zotchev S.B. Identification of novel genes involved in long‐chain n‐alkane degradation by Acinetobacter sp strain DSM 17874. Appl Environ Microbiol. 2007;73:3327–3332. doi: 10.1128/AEM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropel D., van der Meer J.R. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev. 2004;68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beilen J.B., Wubbolts M.G., Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- Wentzel A., Ellingsen T.E., Kotlar H.K., Zotchev S.B., Throne‐Holst M. Bacterial metabolism of long‐chain n‐alkanes. Appl Microbiol Biotechnol. 2007;76:1209–1221. doi: 10.1007/s00253-007-1119-1. [DOI] [PubMed] [Google Scholar]
- Winson M.K., Swift S., Hill P.J., Sims C.M., Griesmayr G., Bycroft B.W. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini‐Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. et al. [DOI] [PubMed] [Google Scholar]
- Young D.M., Parke D., Ornston L.N. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu Rev Microbiol. 2005;59:519–551. doi: 10.1146/annurev.micro.59.051905.105823. [DOI] [PubMed] [Google Scholar]
- Zhang D., Fakhrullin R.F., Özmen M., Wang H., Wang J., Paunov V.N. Functionalization of whole‐cell bacterial reporters with magnetic nanoparticles. Microb Biotechnol. 2011;4:89–97. doi: 10.1111/j.1751-7915.2010.00228.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Overlap extension PCR to create EcoRI/BamHI sites at alkR–alkM DNA fragment.
Fig. S2. The effect of alkanes on bioluminescence expression. ADPWH_alk was activated by octadecane while the bioreporters ADPWH_lux (salicylate), ADPWH_Tol (toluene), ADPWH_recA (toxicity) cannot be activated by octadecane and the addition of octadecane did not increase bioluminescence expression.
Fig. S3. Biosensor ADPWH_lux growth curve in LB supplemented with 100 μM alkane with different carbon chain lengths.


