Summary
Plant genetic manipulation has led to the development of genetically modified plants (GMPs) expressing various traits. Since their first commercial use in 1996, GMPs have been increasingly used, reaching a global cultivating production area of 114.3 million hectares in 2007. The rapid development of agricultural biotechnology and release of GMPs have provided many agronomic and economic benefits, but has also raised concerns over the potential impact these plants might have on the environment. Among these environmental concerns, the unintentional impact that GMPs might have on soil‐associated microbes, especially rhizosphere‐inhabiting bacteria or rhizobacteria, represents one of the least studied and understood areas. As rhizobacteria are responsible for numerous key functions including nutrient cycling and decomposition, they have been defined as good indicator organisms to assess the general impact that GMPs might have on the soil environment. This minireview summarizes the results of various experiments that have been conducted to date on the impact of GMPs on rhizobacteria. Both biological and technical parameters are discussed and an attempt is made to determine if specific rhizobacterial responses exist for the different categories of GMPs developed to date.
Introduction
In 1996, the first genetically modified seeds were planted in the Unites States for commercial agricultural production. Eleven years later, more than 12 million farmers in 23 countries are cultivating biotech crops, with a global production area of 114.3 million hectares, steadily increasing for the eleventh consecutive year (James, 2007). Genetically modified plants (GMPs) with a wide variety of traits have been developed via genetic engineering. While some genetic modifications seek to directly improve the quality of the plant product, many are designed to aid in the cultivation process. Most GMPs developed to date can be grouped into eight main categories: (i) resistance to herbicides, (ii) resistance to pests, (iii) resistance to pathogens, (iv) resistance to environmental stress, (v) plants with altered root exudates, (vi) plants with altered composition, (vii) ability to produce pharmaceutical or industrial compounds and (viii) elimination of pollutants. Although a diversification in modified traits has been seen, the vast majority of GMPs cultivated commercially in 2007 belonged to only two categories: resistance to herbicides and, to a lesser extent, resistance to pests (James, 2007). Biotech soybean continued to be the main commercially produced GMP in 2007, occupying 58.6 million hectares, followed by maize (35.2 million hectares), cotton (11.5 million hectares) and canola (5.5 million hectares) (James, 2007).
The rapid development of agricultural biotechnology and release of GMPs have provided many agronomic and economic benefits, but have also raised concerns over the potential impact these plants might have on human health and the environment. Among the major environmental concerns are the unintended consequences of potential transgene flow to indigenous plants or organisms through pollen transfer (de Vries et al., 2003) or horizontal gene transfer (Nielsen et al., 2001), plant invasiveness (Dale et al., 2002), the development of resistant pests and pathogens (Dale et al., 2002), and non‐target effects on flora, fauna and microorganisms (Bruinsma et al., 2003; Liu et al., 2005). Among these environmental concerns, the impact of GMPs on soil‐associated microbial communities represents one of the least studied and understood areas. This is possibly due in part to the inherent technical difficulties involved in the study of soil‐borne microorganisms.
Plants are know to have a profound effect on the abundance, diversity and activity of soil microorganisms living in close proximity with their roots in a soil zone defined as the rhizosphere (Pinton et al., 2001). Bacteria inhabiting the rhizosphere, also referred to as rhizobacteria, are responsible for numerous functions including nutrient cycling and decomposition, which can significantly influence vegetation dynamics (Kent and Triplett, 2002). Among these, plant growth‐promoting rhizobacteria represent one of the best‐characterized functional group of rhizobacteria known for playing a significant role in plant health and plant development (Siddiqui, 2006). As it can be assumed that any significant impact of plant genetic transformation might alter these fundamental microbial processes, rhizobacteria have been defined as good indicator organisms and have been studied to assess the general impact that GMPs might have on the soil environment (Lynch et al., 2004).
This minireview seeks to: (i) summarize the work done to date on the impact of GMPs on rhizobacteria, (ii) identify the main variables affecting the impact of GMPs on rhizobacteria, (iii) discuss the resolution of the different technical approaches used to assess such impacts, (iv) determine if specific rhizobacterial responses exist for different categories of GMPs and (v) finally suggest a framework for future assessment of GMP effects on rhizobacteria.
Variables affecting the impact of GMPs on rhizobacteria
Nature of the genetic modification
Many factors come into play when trying to address the impact that GMPs might have on rhizobacteria. The nature of the plant genetic modification certainly represents one of the first key elements that naturally come to mind. Different transgenes, allowing the expression of novel plant proteins, can potentially alter rhizobacteria abundance and diversity in a variety of ways. These novel proteins can, for example, be directly released into the rhizosphere through root exudation or their expression can trigger, through alteration of various metabolic pathways, the production and release of additional non‐proteinaceous compounds (Raubuch et al., 2007). Although the engineered production of some of these compounds is deliberate, targeting, for example, specific pests or pathogens (Mendelsohn et al., 2003), unintentional effects are susceptible to alter non‐targeted rhizobacteria.
The persistence of some of these novel compounds in the rhizosphere, or simply an alteration in the amounts of naturally produced plant compounds due to genetic engineering can also have an impact on rhizobacteria populations (Oger et al., 2000). Even after harvest, decomposition of plant litter can continue releasing novel gene products into the environment (Gebhard and Smalla, 1999), potentially altering rhizobacteria populations over long periods of time. For example, Bt‐recombinant DNA and expressed Cry proteins were released in soils through root‐exudation and plant tissue decomposition where they remained intact and chemically active for extended periods of time, up to 240 days (Palm et al., 1996; Tapp and Stotzky, 1998; Zwahlen et al., 2003). However, while evidence for the persistence of transgenic plant DNA exists (Gebhard and Smalla, 1999), transfer of plant DNA to native soil microorganisms has only been shown under optimized laboratory conditions as a rare event (Nielsen et al., 1997; Gebhard and Smalla, 1998) and has not yet been demonstrated to occur naturally under in situ conditions.
The recent introduction of transgenic crops expressing multiple traits (‘stacked' genes) may further complicate determining the impact of GMPs due to potential interactions among the different transgenes and their respective gene products.
Genetic markers
As it is essential to confirm that a genetic transformation event is successful, selectable genetic markers, usually conferring the ability to survive in the presence of a normally toxic compound, are commonly used to co‐transform plants with transgenes of interest. The most widely used selectable genetic marker in plants is the nptII gene, which was originally derived from Escherichia coli transposon Tn5, and encodes a neomycin phosphotransferase that confers resistance to the aminoglycoside antibiotics neomycin and kanamycin (Gasson, 2000). Such antibiotics are added to growth media when transgenic plants are regenerated from transgenic cells, maintaining selection (Dyer, 1996). It is surprising that the impact of such genetic markers on rhizobacteria has almost been ignored to date. Among the very few studies performed to address this question, Burris and colleagues (2008) showed that overexpression of Atwbc in Arabidopsis thaliana, which confers kanamycin resistance, had no effect on E. coli populations, and Demanèche and colleagues (2008) showed that no significant differences were observed in antibiotic‐resistance levels when comparing transgenic and non‐transgenic corn fields. However, LeBlanc and colleagues (2007) clearly showed that white spruce transformed with either nptII, cry1Ab or both genes had a significant impact on the diversity of rhizobacteria, but this impact was different and specific for each transgene. The use of such important antibiotic‐resistance gene products in GMPs could also represent additional concerns as they may, for example, inactivate oral doses of antibiotics if the plant is ingested by humans and/or favour antibiotic resistance build‐up in microorganisms (Gasson, 2000). According to a study performed in 2003, a DNA construct composed of the nptII transgene remained detectable by polymerase chain reaction (PCR) in potato field soil after 4 years of laboratory incubation (de Vries et al., 2003). New generations of transgenic plants that do not harbour antibiotic‐resistance genes of bacterial origin are however in development, which could help address this concern.
Crops
Different crops, and in some cases even different cultivars of the same plant species, are known to favour specific rhizobacteria populations (Mazzola et al., 2004). It has been demonstrated that this impact of different cultivars on rhizobacteria populations can even be more pronounced than plant genetic transformation. For example, Milling and colleagues (2005) have shown that the non‐genetically transformed potato cultivar SOLANA displayed a different rhizobacteria population when compared with the non‐genetically modified potato cultivar (SIBU) or a transgenic version altered in starch composition (SIBU SI), while rhizobacteria populations associated with SIBU and SIBU SI were not significantly different. This is why appropriate controls, not only including the non‐GM parental cultivar, but also additional cultivars of the same plant species, should be included in studies addressing the impact of GMPs on rhizobacteria. Different crop‐ or cultivar‐related factors such as the architecture of the root system, differences in soil nutrient utilization, as well as amounts and composition of root exudates, can be determinant in specific plant–rhizobacteria interactions (Brimecombe et al., 2001).
Production management systems
In general, agricultural, horticultural and forestry practices create a huge perturbation in soil systems, and changes in production management practices can have serious and long‐lasting effects on rhizobacteria (Höflich et al., 1999). Soil properties and soil management practices, including tillage, fertilizer use, pesticide applications, herbicide treatments and crop rotation, are of significant importance (Marschner et al., 2004). A ‘baseline of normal variation’ should be established for each crop under study to assess the effect caused by these practices, allowing to gauge the impact that plant genetic transformation might have as compared with this ‘normal variation’. Experimental design for risk assessment studies on GMPs should also take into consideration the changes in management practices associated with cultivation of GMPs. If, for example, the use of a Bt‐crop allows avoiding usage of insecticides as compared with the conventional farming approach which relies on their uses, it appears relevant to determine if the effect of genetic transformation on rhizobacteria is still significant as compared with the impact of insecticide application.
Ecosystems parameters
Other biotic and abiotic sources of variation, such as season, weather, plant developmental stage and field location, have been identified as important drivers of microbial community structure alteration in the rhizosphere (Marschner et al., 2004). Proper experimental design is crucial for assessing GMP‐induced effects, and should include controls for natural variation within plant–soil systems. Monitoring schemes must relate to ecologically relevant levels of environmental variation, and sampling procedures should take spatial and temporal changes into account. Although time‐ and resource‐consuming, the evaluation of parameters, such as seasonal and year‐to‐year variations, should be identified and the impact of plant genetic transformation be gauged against them.
Methods used to assess the impact of GMPs on rhizobacteria abundance and diversity
Soil ecosystems are complex, heterogeneous and represent a challenge when it comes to isolate, culture or identify most microorganisms inhabiting this environment. There is strong evidence that most soil bacteria seen under the microscope are viable but fail to be detected by classical culture‐dependent microbiological techniques (Ward et al., 1990). Methodological advances, especially the introduction of culture‐independent molecular biological techniques (Torsvik, 1980), now enable a thorough examination of GMP‐induced effect on rhizobacteria. It is important to keep in mind that different methodological approaches present different resolutions and limitations, which will inevitably affect the conclusions drawn from risk assessment studies investigating the impact of GMPs on rhizobacteria. Although a trend towards molecular and polyphasic approaches is developing, which offer more complete pictures of microbial community dynamics, a significant portion of studies performed to date on the risk assessment of GMPs on rhizobacteria mainly relied on classical culture‐dependent methods (Lynch et al., 2004).
Low‐resolution techniques
General measures, such as bacterial counts (usually expressed as ‘colony‐forming units’ or ‘most probable numbers’) or total microbial biomass, can detect changes in terms of microbial abundance only at a rudimentary level. Bacterial counts mainly rely on plating methods which are the most widely used classical culture‐dependent methods in microbiology. As counts are usually quite variable, strongly dependent on the incubation conditions used and don't allow the detection of the great majority of soil bacteria because they are not culturable using standard methods, the information provided is very limited (Liu et al., 2005). Microbial biomass measurements, usually determined using chemical approaches, such as the chloroform fumigation–extraction method, are also quite variable and often not precise (Olfs and Scherer, 1996).
Intermediate‐resolution techniques
Community‐level physiological profiling (CLPP) of microbial populations (e.g. the BiOLOG assay) has been used to characterize microbial diversity based on a pattern of substrate utilization in 96‐well microtiter plates. Although these assays can depict community changes between treatments, mainly providing functional ability information, they rely on culture of microorganisms and usually fail at identifying the taxonomical groups associated with functional changes (Garland and Mills, 1991).
Microbial lipid analysis, such as phospholipid fatty acid analysis, fatty acid methyl ester analysis (FAME), ester‐link fatty acid analysis and neutral lipid fatty acid analysis, rely on the detection and/or quantification of microbial membrane fatty acid profiles specific to different taxonomical groups of microorganisms. They can provide information on a variety of microbial characteristics, such as abundance, taxonomic and functional identity, and overall community diversity. However, it has been shown that membrane fatty acid components of bacteria may change in response to the availability of nutrients and environmental stress, thereby altering lipid signatures (Kieft et al., 1994). Although the use of lipid‐based techniques for microbial community analysis in soil is advantageous because they do not require culturing and can provide quantitative data on entire communities, it has been shown that combined effects of physiological and phylogenetic changes on the lipid composition of a community can confound interpretation of the data (Liu et al., 2005).
Stable isotope probing is a culture‐independent method used for linking function with taxonomic identity, which involves using stable isotope‐enriched substrates for the identification of metabolically active members within a complex natural environment (Radajewski et al., 2000). This technique is particularly interesting when several uncultivated microorganisms are involved in a process. However, it fails at depicting a portrait of the total community structure present in a sample.
Several other broad‐targeted culture‐independent molecular methods are now also available for the detection of shifts in total microbial community structure without the need for culturing microorganisms. Most of these methods rely on a three step procedure, an initial DNA extraction step from samples under study (i.e. rhizosphere soil samples), a PCR‐amplification step targeting either taxonomical (e.g. 16S rDNA) or functional relevant genomic DNA sequences, then followed by a third step allowing comparison of the fragments obtained based on either their sizes or their sequences. Depending on the various specificities of the primers used for the PCR amplification step to zoom in on specific microbial lineages, and the nature of the third step to separate PCR amplicons, the resolution of these approaches can vary from intermediate to high.
At the intermediate resolution level, denaturing/temperature gradient gel electrophoresis (DGGE/TGGE), single‐stranded conformational polymorphism (SSCP) and terminal restriction fragment length polymorphism (T‐RFLP) approaches are the most widely used. In DGGE/TGGE analysis, PCR amplicons with the same length but different nucleotide sequences are separated in polyacrylamide gels by a denaturing gradient or a temperature gradient for DGGE and TGGE respectively. DGGE/TGGE bands can then be excised from the gel and sequenced, allowing the identification of the dominant bands. The SSCP, like DGGE/TGGE, detects sequence variations between different PCR amplicons. However, this technical approach relies on using one PCR primer phosphorylated at the 5′ end, leading to the degradation of one DNA strand following PCR amplification by selective digestion using a lambda exonuclease. Resulting single strands are separated by electrophoresis under non‐denaturing conditions in a polyacrylamide gel, allowing intra‐molecular folding of the DNA strand dependent upon DNA variations. The T‐RFLP analysis is based on the restriction endonuclease digestion of fluorescent end‐labelled PCR amplicons, often 16S rDNA sequences. The amplicons are then separated by gel or capillary electrophoresis. Usually quick to perform as compared with high‐resolution techniques, DGGE/TGGE, SSCP and T‐RFLP analyses provide a good overview of the microbial diversity present in a sample but generally fail at detecting microorganisms present in low quantities (Lynch et al., 2004). Finally, although being used less frequently since the development of more powerful approaches, culture‐independent hybridization techniques, such as slot/dot blots or FISH (fluorescence in situ hybridization) rely on the use of oligonucleotidic probes homologous to phylogenetic relevant coding or non‐coding regions. In the FISH method, probes are labelled with fluorescent dyes and used for in situ detection of cells in environmental samples by whole‐cell hybridization (Amann et al., 1990). In slot/dot blots, DNA is directly extracted from the samples under study and hybridized with labelled probes of interest. Probes can target phylogenetic groups at different taxonomical levels, ranging from domain to subspecies and show the relative abundance of major phylotypes in soil microbial communities (Lynch et al., 2004). These approaches are however time‐consuming and mainly used to target specific microbial groups of interest instead of whole community analysis.
High‐resolution techniques
Culture‐independent molecular based high‐resolution techniques available to study microbial diversity mainly include ribosomal intergenic spacer analysis (RISA and ARISA), amplified ribosomal DNA restriction analysis (ARDRA and T‐ARDRA) and PCR‐cloning sequencing (Lynch et al., 2004).
The RISA or the automated version of RISA called ARISA are based on analysis of length polymorphism of PCR‐amplified cloned ribosomal intergenic spacer regions located between 16S and 23S rDNA. This region varies both in length and nucleotide sequence, even between closely related microbial strains. The digested PCR amplicons obtained are then separated by gel electrophoresis (RISA) or first labelled using a forward fluorescent PCR primer and then separated by capillary electrophoresis systems (ARISA). The ARDRA follows basically the same technical approach as RISA with the exception that 16S rDNA is targeted instead of the intergenic spacer region and that this gene only significantly varies in nucleotide composition and not in length. Again, the digested amplicons can either be separated by gel electrophoresis or the approach can be automated (called terminal ARDRA or T‐ARDRA) which first involves labelling the amplicons using a forward fluorescent PCR primer and then separation by capillary electrophoresis systems. Both RISA and ARDRA amplicons can be sequenced to identify fragments of interest. If no DNA digestion step is used, the amplicons obtained (either from 16S rDNA or ribosomal intergenic spacer regions) can be cloned, then directly sequenced, an approach generally identified as PCR‐cloning‐sequencing or referred to as 16S rDNA analysis if the 16S rDNA gene is specifically targeted (Lynch et al., 2004).
Among emerging culture‐independent techniques allowing to characterize microbial community dynamics with intermediate to high resolution are metagenomics and taxonomic microarrays. In metagenomics, total genomic DNA (metagenome) derived from microbial communities can be cloned into large vectors, such as bacterial artificial chromosomes, to study microbial diversity and/or functional aspects, as well as the interaction between both (Rondon et al., 2000). In taxonomic microarrays, mainly 16S rRNA‐based microarrays, short oligonucleotides targeting phylogenetic groups at different taxonomical levels are hybridized in a high‐throughput format with DNA extracted from samples of interest (Sanguin et al., 2006). This allows quick identification of the whole microbial community present, requiring less time and reducing costs as compared with library methods.
Case studies: microbial communities affected by GMPs
Resistance to herbicides
The GMP‐resisting or ‐tolerating herbicides have consistently been predominant since 1996 in biotech agriculture. Mainly two transgenes, the 5‐enolpyruvylshikimate‐3‐phosphate synthase gene and the phosphinothricin‐acetyltransferase gene, conferring resistance to the herbicide glyphosate and glufosinate respectively, have been commonly used for plant genetic transformation.
So far, all studies performed on the impact of glyphosate‐resistant GMPs on rhizobacteria have shown a clear significant impact on rhizobacterial diversity (Siciliano et al., 1998; Siciliano and Germida, 1999; Dunfield and Germida, 2001; 2003) (Table 1). Different approaches, mainly CLPP and FAME, have revealed clear microbial shifts in the rhizosphere and/or the root interior of different oilseed rape (canola) cultivars, the only glyphosate‐resistant plant species tested so far for its impact on rhizobacteria. Interestingly, two studies have addressed this impact under field conditions over a 2 year period (Dunfield and Germida, 2001; 2003). Both showed through PCA analysis that the rhizobacteria population inhabiting the rhizosphere of the genetically modified Quest cultivar had different CLPP and FAME profiles as compared with the population inhabiting the rhizosphere of the closely non‐modified Excel cultivar (Dunfield and Germida, 2001; 2003) or the more distantly related non‐genetically modified Fairview cultivar (Dunfield and Germida, 2001). This significant effect, although detected at most sampling times, was not persistent the next field season following winter at the pre‐seeding stage. This clearly suggests that changes in the rhizobacteria community observed in the rhizosphere of glyphosate‐resistant GMPs are temporary and dependent on the presence of the transgenic plant. As only one study has addressed the impact of glyphosate‐resistant GMPs on the abundance of rhizobacteria without showing any significant effect (Dunfield and Germida, 2001) (Table 1), it is difficult at this point to clearly conclude on the potential impact that glyphosate‐resistant GMPs may have on rhizobacteria abundance.
Table 1.
Studies addressing the impact of GMPs on the abundance and/or diversity of rhizobacteria.
| Modified plant | Modified trait | Technique used | Impact on abundance of rhizobacteriaa | Impact on diversity of rhizobacteriaa | Reference |
|---|---|---|---|---|---|
| Resistance to herbicides | |||||
| Canola | Glyphosate resistance | CFU, CLPP, FAME, | No | Yes | Dunfield and Germida (2001) |
| Canola | Glyphosate resistance | CLPP, FAME, T‐ARDRA | – | Yes | Dunfield and Germida (2003) |
| Canola | Glyphosate resistance | FAME | – | Yes | Siciliano and Germida (1999) |
| Canola | Glyphosate resistance | CLPP, FAME | – | Yes | Siciliano et al. (1998) |
| Canola | Glufosinate resistance | DGGE, microbial biomass | Minor effect | Minor effect | Sessitsch et al. (2004) |
| Canola | Glufosinate resistance | DGGE | – | Minor effect | Gyamfi et al. (2002) |
| Maize | Glufosinate resistance | SSCP | – | No | Schmalenberger and Tebbe (2002) |
| Maize | Glufosinate resistance | CLPP, ELFA, PLFA | Minor effect | Minor effect | Griffiths et al. (2007) |
| Sugar beet | Glufosinate resistance | SSCP | – | No | Schmalenberger and Tebbe (2003) |
| Resistance to pests | |||||
| Cotton | Bt (Cry1Ac) | CFU | Yes | – | Rui et al. (2005) |
| Cotton | Bt (Cry1Ab) | CLPP | – | No | Shen et al. (2006) |
| Maize | Bt (Cry1Ab) | CLPP, PLFA | No | No | Griffiths et al. (2006) |
| Maize | Bt (Cry1Ab) | CLPP, PLFA | No | No | Griffiths et al. (2005) |
| Maize | Bt (Cry1Ab) | SSCP | – | Minor effect | Baumgarte and Tebbe (2005) |
| Maize | Bt (Cry1Ab) | CFU | No | – | Saxena and Stotzky (2001) |
| Maize | Bt (Cry1Ab) | CLPP, PLFA | No | Minor effect | Blackwood and Buyer (2004) |
| Maize | Bt (Cry1Ab) | DGGE | – | Yes | Castaldini et al. (2005) |
| Maize | Bt (Cry1Ab) | ARISA, CFU, CLPP | Minor effect | Yes | Brusetti et al. (2004) |
| Maize | Bt (Cry1F) | CLPP, PLFA | No | Minor effect | Blackwood and Buyer (2004) |
| Maize | Bt (Cry3Bb) | Microbial biomass, T‐RFLP | No | No | Devare et al. (2004) |
| Maize | Bt (Cry3Bb) | Microbial biomass | Yes | – | Devare et al. (2007) |
| Potato | Cystatin production | PLFA | Yes | – | Cowgill et al. (2002) |
| Potato | Concavalin A production | CLPP | – | No | Griffiths et al. (2000) |
| Potato | Galanthus nivalis agglutinin production | CLPP | – | Minor effect | Griffiths et al. (2000) |
| Rice | Bt (Cry1Ab) | DGGE, T‐RFLP | – | Minor effect | Liu et al. (2008) |
| Spruce | Bt (Cry1Ab) | ARDRA | – | Yes | LeBlanc et al. (2007) |
| Resistance to pathogens | |||||
| Potato | Cecropin production | ARDRA, ARISA | – | Yes | Sessitsch et al. (2003) |
| Potato | Attacin and cecropin prod. | T‐RFLP | – | Minor effect | Rasche et al. (2006) |
| Potato | T4 lysozyme | CFU, CLPP, DGGE | Minor effect | Minor effect | Heuer et al. (2002) |
| Potato | T4 lysozyme | T‐RFLP | – | Minor effect | Rasche et al. (2006) |
| Potato | T4 lysozyme | CFU, CLPP, FAME | No | No | Lottmann et al. (1999) |
| Potato | T4 lysozyme | CLPP, FAME, rep‐PCR | – | No | Lottmann and Berg (2001) |
| Potato | Magainin II production | CFU | Yes | – | O'Callaghan et al. (2004) |
| Potato | Barnase/Barstar | T‐RFLP | – | Minor effect | Lukow et al. (2000) |
| Papaya | RP mutant | CFU | Yes | – | Wei et al. (2006) |
| Plants with altered root exudates | |||||
| Alfalfa | neMDH overexpression | CFU, CLPP, 16S rDNA analysis | No | Yes | Tesfaye et al. (2003) |
| Birdsfoot trefoil | Opine production | CFU | Yes | – | Oger et al. (1997) |
| Birdsfoot trefoil | Opine production | ARDRA, CFU | Yes | Yes | Oger et al. (2004) |
| Black nightshade | Opine production | CFU | Yes | – | Mansouri et al. (2002) |
| Tobacco | Ferritin overexpression | CFU | Yes | – | Robin et al. (2006b) |
| Tobacco | Ferritin overexpression | ARISA, PCR‐RFLP | – | Yes | Robin et al. (2006a) |
| Tobacco | Ferritin overexpression | CFU, RAPD | No | Yes | Robin et al. (2007) |
| Tobacco | N‐AHLS | CFU, DGGE, PLFA | No | No | d'Angelo‐Picard et al. (2004) |
| Plants with altered composition | |||||
| Aspen | Altered stem lignin | NLFA, PLFA | Minor effect | Minor effect | Bradley et al. (2007) |
| Alfalfa | Ovalbumin production | CFU | Yes | Yes | Faragova et al. (2005) |
| Potato | Altered starch composition | DGGE | – | Minor effect | Milling et al. (2005) |
| Troyer citrange | Phytohormone balance | CFU, rep‐PCR | Minor effect | Minor effect | Cirvilleri et al. (2005) |
| Troyer citrange | Phytohormone balance | ARDRA, CLPP, DGGE | – | Minor effect | Abbate et al. (2005) |
| Ability to produce pharmaceutical or industrial compounds | |||||
| Alfalfa | Lignin peroxy. production | CLPP, rep‐PCR | – | Yes | Di Giovanni et al. (1999) |
| Alfalfa | Lignin peroxy. production | ARDRA, CFU, CLPP | Yes | Yes | Donegan et al. (1999) |
| Alfalfa | Alpha‐amylase production | CLPP, rep‐PCR | – | Yes | Di Giovanni et al. (1999) |
| Alfalfa | Alpha‐amylase production | ARDRA, CFU, CLPP | Yes | Minor effect | (Donegan et al., 1999) |
| Tobacco | HSA production | CFU, MPN | No | – | Sabharwal et al. (2007) |
| Elimination of pollutants | |||||
| Tobacco | Nitroreductase overexpression | CFU, CLPP, DGGE | Yes | Yes | Travis et al. (2007) |
A minor effect indicates that an effect is reported or suggested but that it is either not clearly supported by statistical analyses or that results of statistical analyses are not clearly significant.
CFU, colony‐forming unit; CLPP, phospholipid fatty acid analysis; ELFA, ester‐link fatty acid analysis; MPN, most probable numbers; NLFA, neutral lipid fatty acid analysis; neMDH, nodule‐enhanced malate dehydrogenase; PLFA, phospholipid fatty acid analysis.
Contrary to the results indicating that glyphosate‐resistant GMPs can alter the diversity of rhizobacteria, studies addressing the impact of glufosinate‐resistant GMPs have only shown minor effects (Gyamfi et al., 2002; Sessitsch et al., 2004; Griffiths et al., 2007) or no effects (Schmalenberger and Tebbe, 2002; 2003) on rhizobacterial diversity or abundance (Table 1). Although studies addressing the impact of glufosinate‐resistant GMPs have usually addressed impacts under short periods of time and in some cases were conducted under controlled soil conditions (i.e. in pots), at least two studies reported results obtained under field conditions during two consecutive growth seasons, and their conclusions were not different (Schmalenberger and Tebbe, 2003; Griffiths et al., 2007). Different plant species also did not significantly affected the results as minor effects of glufosinate‐resistant canola and maize on rhizobacteria were reported (Gyamfi et al., 2002; Griffiths et al., 2007), while studies reporting no effects used either glufosinate‐resistant maize or sugar beet (Schmalenberger and Tebbe, 2002; 2003). The impact of agricultural practices, soil type and seasons was considered more determinant than the incorporation of a glufosinate‐resistant gene in the plant's genome for altering rhizobacterial diversity and/or abundance (Griffiths et al., 2007).
Resistance to pests
The GMP engineered for improved resistance against pests, especially insects, have been mainly obtained by the incorporation of cry genes, encoding the production of insecticidal crystal proteins (Cry), also known as Bt toxins. Different cry genes, encoding various forms of Cry proteins with specific insecticidal effects, were originally isolated from the bacteria Bacillus subtilis. Crops engineered with such genes, commonly referred to as Bt‐crops, have proved to be effective at controlling a wide variety of insect pests. The GMP producing cystein proteinase inhibitors (cystatins) conferring resistance to nematodes, as well as GMP producing lectins such as concavalin A and Galanthus nivalis agglutinin, improving resistance against invertebrate pests, have also been developed.
Numerous studies have investigated the impact of Bt‐crops on rhizobacteria. Plants, such as cotton, maize, rice and even ligneous species such as spruce, have been engineered to produce various Cry proteins, mainly Cry1Ab, Cry1Ac, Cry1Af and Cry3Bb, each active against specific insect groups (Mendelsohn et al., 2003). The impact of such GMPs on rhizobacteria ranged from non‐significant to significant effects on rhizobacterial diversity and abundance (Table 1). Neither plant species nor cry transgene type used can yet be correlated with specific responses. However, most studies performed under longer periods of time, ranging from 2 (Castaldini et al., 2005; Liu et al., 2008) to 3 years (Baumgarte and Tebbe, 2005; Devare et al., 2007), have shown at least some sort of effects on either rhizobacterial diversity or abundance. This effect was generally reported as being less important than field site, soil and plant growth stage in terms of impact on rhizobacteria. Interestingly, many studies have incorporated non‐GMPs as well as non‐GMPs treated with insecticides as controls in their experimental set‐up (Devare et al., 2004; 2007; Liu et al., 2008). The effect of insecticides application on rhizobacteria was in all cases either minor or absent, again suggesting that other production management system or ecosystem parameters were more critical for the alteration of rhizobacterial communities.
Potato plants engineered to produce cystatins (Cowgill et al., 2002) altered rhizobacterial abundance under field conditions, while potato plants producing lectins (Griffiths et al., 2000) had either minor or no effect on rhizobacterial diversity under controlled soil conditions (Table 1). As only very few studies were performed on the impact of cystatin‐ and lectin‐producing plants on rhizobacteria, it is difficult at this point to clearly conclude on the real impact these plants might have.
Resistance to pathogens
Genetic engineering of plants with the aim of achieving better resistance against pathogens has relied on the use of transgenes encoding various lytic peptides and enzymes, mainly with bactericidal and/or fungicidal activities. Such peptides include cercopins, attacins and megainin, while T4 lysozyme is the main enzyme that has been produced so far in GMPs to control pathogens. Risk assessment studies investigating the impact of pathogen‐resistant GMPs on rhizobacteria have all used potato, with one exception using papaya engineered with a replicase (RP) mutant gene conferring resistance to the papaya ringspot virus (Wei et al., 2006).
Most studies conducted so far on the impact of pathogen‐resistant GMPs have shown at least some sort of effects on rhizobacterial diversity or abundance (Table 1). While all studies investigating the impact of antimicrobial peptide‐producing GMPs have shown some effects either on rhizobacterial diversity or abundance (Sessitsch et al., 2003; O'Callaghan et al., 2004; Rasche et al., 2006), studies investigating the impact of T4 lysozyme‐producing GMPs have either shown minor or no effects (Lottmann et al., 1999; Lottmann and Berg, 2001; Heuer et al., 2002; Rasche et al., 2006). Only two studies looked at the impact of T4 lysozyme‐producing GMPs over more than one growth season under field conditions and showed no effect on rhizobacteria (Lottmann et al., 1999; Lottmann and Berg, 2001), clearly suggesting that T4 lysozyme‐producing GMPs have at most only a very minor effect of rhizobacteria. As only one study for each transgene used is available in this category, except for T4 lysozyme‐producing GMPs, it is difficult at this point to determine trends or to generalize about the results obtained. The only study performed on GMPs other than potato showed an alteration of rhizobacteria abundance in the rhizosphere of RP papaya.
Plants with altered root exudates
As rhizobacteria populations and microbial gene expression are known for being rapidly altered by root exudation changes (Matilla et al., 2007; Attila et al., 2008), GMPs have been engineered to produce altered exudation patterns. These GMPs have mainly been developed for risk assessment studies and not for commercial production. The rationale being that if plants with altered root exudation patterns are incapable of altering rhizobacteria populations, other genetic changes, not specifically designed to alter rhizobacteria, should probably cause no significant changes. The GMPs have been developed to produce opines, which are small molecules found in the crown gall tumours induced by the phytopathogenic bacterium Agrobacterium tumefaciens, and being used as a growth substrate by the bacteria (Oger et al., 1997; 2004; Mansouri et al., 2002). The GMPs have also been engineered to produce N‐acyl homoserine lactones, which regulates quorum sensing in Gram‐negative bacteria (d'Angelo‐Picard et al., 2004). Finally, plants overexpressing ferritins, which are multimeric proteins that store iron in a soluble and biologically available form, thereby contributing to plant iron homeostasis (Robin et al., 2006a,b), or plants overexpressing a nodule‐enhanced malate dehydrogenase, which catalyses the reversible reduction of oxaloacetate to malate, leading to an increased exudation of organic acids in the rhizosphere (Tesfaye et al., 2003), have also been developed.
All studies performed on GMPs with altered root exudates showed a clear impact on rhizobacterial diversity and abundance, with one exception, N‐acyl homoserine lactone‐producing tobacco (d'Angelo‐Picard et al., 2004) (Table 1). These results confirm that plant genetic transformation specifically engineered to alter rhizobacteria can produce the expected effect. This effect appears highly dependent on the transgene used. Clearly, as it is now possible to develop GMPs that will have a significant targeted effect on rhizobacteria, it appears justified to confirm through risk assessment studies that GMP expressing other traits do not exert unintentional impacts on rhizobacteria.
Plants with altered composition
The GMPs with altered stem lignin or starch composition, production of a high‐methionine protein ovalbumin or modifications in phytohormone balance have been developed. As traditional breeding has been intensively used to improve plant composition over the years, it appears likely that improving specific plant composition traits through genetic engineering will become very important. This technology has however not yet been significantly used under large‐scale commercial field conditions.
All studies performed to date on the impact of GMPs with altered composition have shown at least minor effects on rhizobacterial diversity and abundance (Table 1). As only very few studies have been performed in this category, usually only one study per transgene, it is difficult to generalize about the results obtained. When looking at the results gathered over more than one growth season, only the impact of Troyer citrange [Poncirus trifoliate (L.) Raf. x Citrus sinensis (L.) Osbeck] was looked at, showing only minor effects on rhizobacterial diversity and abundance (Abbate et al., 2005; Cirvilleri et al., 2005). Other parameters, such as soil and plant growth stage, were considered more determinant than genetic alteration for their impact on rhizobacteria communities.
Ability to produce pharmaceutical or industrial compounds
The GMPs have been engineered to produce pharmaceutical compounds, such as human serum albumin (HSA) used as a plasma expander, stabilizing agent, drug delivery protein and an adjuvant for novel drugs (Sabharwal et al., 2007). Plants genetically modified to produce industrial relevant compounds, such as alpha‐amylase used in starch processing for the production of juice, beer, wine and bread, or lignin peroxydase used for large‐scale lignin degradation and as a bleaching agent in biopulping processes, have also been developed (Di Giovanni et al., 1999; Donegan et al., 1999).
All risk assessment studies performed in this category revealed effects on rhizobacterial diversity and abundance, except one study on HSA‐producing tobacco (Table 1). Very few studies have however been performed on the impact of GMP producing pharmaceutical or industrial compounds on rhizobacteria. Also, the studies performed to date have tested only two different plant species: alfalfa and tobacco. There is no doubt that plants engineered to produce such commercially important compounds will likely become a significant part of tomorrow's biotech agribusiness.
Elimination of pollutants
Only one study reported on the impact of GMPs engineered to eliminate pollutants on rhizobacteria populations. Transgenic tobacco plants overexpressing a bacterial nitroreductase gene were capable of detoxifying soil contaminated with the highly explosive 2,4,6‐trinitrotoluene (TNT). These plants significantly altered rhizobacterial diversity and abundance. The effect was however dependent on the concentration of TNT present in soil (Travis et al., 2007).
Concluding remarks
One of the most striking conclusions concerning risk assessment studies performed on the impact of GMPs on rhizobacteria is that so far a wide variety of techniques have been used to examine numerous parameters and confusion still exists as to which information is really relevant in this context. To date, most studies investigating the impact of GMPs have shown at least minor effects on the abundance and/or the diversity of rhizobacteria (Fig. 1). Despite the limited number of studies performed on this topic, we can now conclude with certainty that the effects observed are not artefacts. Clear rhizobacterial community changes have been shown in the rhizosphere of a variety of plants using different transgenes. The results obtained were reproducible and, in many cases, rhizobacterial responses to different categories of GMPs appeared more dependent on the nature of the transgene used than any other considerations. However, based on the limited information presently available in the literature, we can suspect that these effects, although not negligible, can be considered ‘minor’ when compared with other ‘normal’ sources of variation, such as field site, soil type, seasonal variation, plant growth stage, etc. Agricultural practices, such as irrigation, crop rotation, tillage and use of herbicides and pesticides, have been shown to influence rhizobacteria populations with similar impact and in many cases more importantly than plant genetic transformation. So far, no study has been able to clearly show a long‐term negative effect of plant genetic transformation on rhizobacterial populations. Regardless of the plant species, transgenes and growth conditions used, all rhizobacteria community changes reported during active growth seasons have diminished to undetectable levels in a few weeks or months following plant harvesting.
Figure 1.
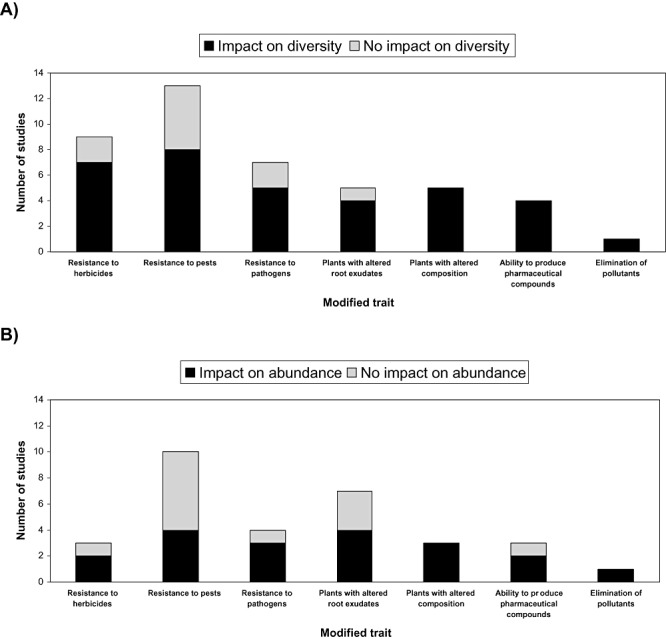
Studies assessing impacts induced by GMPs on rhizobacterial (A) diversity and (B) abundance. Number of studies showing impact or no impact on rhizobacterial diversity or abundance is presented by categories of plant modification.
Framework for future assessment of GMP effects on rhizobacteria
The use of GMPs in a commercial context is in continuous climb. Although it has so far been mainly restricted to agriculture, we can reasonably assume that GMPs will, in the near future, be increasingly used in horticulture and in forestry. Even if no significant long‐term negative impact on rhizobacteria has been clearly identified to date, it is imperative that more studies addressing potential non‐target effects of GMPs be conducted. One of the main reasons for this is that the technology is recent and only a very limited amount of studies have so far been conducted in this area, limiting the conclusions that can be drawn.
In order for future studies to be as informative as possible, the following framework is proposed. Risk assessment studies on the impact of GMPs on rhizobacteria and microorganisms in general should be conducted under field or ‘natural’ conditions over long‐term periods (i.e. over at least 2 years). These studies should incorporate as many treatments as possible (i.e. using different cultivars, field sites, soil types, sampling at different growth stages etc.) to clearly define a baseline representative of ‘natural variation’, and should as well incorporate non‐genetically transformed control plants and control plants transformed using genetic markers only. The experimental approaches could also follow changes in a rotational cropping context to determine if any effects could still be detected in non‐genetically modified intercrops. Finally, from a technical point of view, polyphasic detection approaches should be used to assess microbial abundance and diversity in this context. In order for these recommendations to be realistic in terms of time, work and financial resources, here is a proposed streamlined set of guidelines for risk assessment studies on the impact of GMPs on rhizobacteria, which could also be applied to study the impact of other perturbations (environmental, chemical of physical) on rhizobacteria populations:
At least two different cultivars of the same plant species should be used.
At least two field sites (each with a different soil type) should be studied.
Sampling should be performed at least three times per growth season (at planting, mid season and at harvesting).
GMPs, non‐genetically transformed control plants and control plants transformed using genetic markers only should be used.
One culture‐dependent and one culture‐independent technical approach should be used. BIOLOG assays combined with PCR‐cloning sequencing assays represent a powerful combination. As sequencing costs have gone down, it is recommended that 96 clones should be analysed per sample. Alternatively, laboratories on a tight budget could consider other high‐resolution techniques, such as RISA, ARISA or ARDRA to cut down on costs.
At least three replicates per treatment should be used. Based on the experimental set‐up proposed here (2 cultivars × 2 field sites × 3 harvesting periods × 3 categories of plant transformation × 3 replicates), this represents 108 samples to be analysed using two different technical approaches (i.e. BIOLOG and PCR‐cloning‐sequencing assays).
The conclusion and suggestions on risk assessment mentioned here should not be viewed as static or absolute. They are based on the knowledge presently available. Some key issues concerning the impact of GMPs on rhizobacteria and soil microorganisms might develop or change in light of novel findings.
References
- Abbate C., Ascher J., Pietramellara G., Ambrosoli R., Gennari M. Analysis of bacterial communities in the rhizosphere of transgenic rolABC citrange Troyer: preliminary studies. Fresenius Environ Bull. 2005;14:867–872. [Google Scholar]
- Amann R.I., Krumholz L., Stahl D.A. Fluorescent‐oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo‐Picard C., Faure D., Carlier A., Uroz S., Raffoux A., Fray R., Dessaux Y. Bacterial populations in the rhizosphere of tobacco plants producing the quorum‐sensing signals hexanoyl‐homoserine lactone and 3‐oxo‐hexanoyl‐homoserine lactone. FEMS Microbiol Ecol. 2004;51:19–29. doi: 10.1016/j.femsec.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Attila C., Ueda A., Cirillo S.L.G., Cirillo J.D., Chen W., Wood T.K. Pseudomonas aeruginosa PA01 virulence factors and poplar tree response in the rhizosphere. Microb Biotechnol. 2008;1:17–29. doi: 10.1111/j.1751-7915.2007.00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarte S., Tebbe C.C. Field studies on the environmental fate of the Cry1Ab Bt‐toxin produced by transgenic maize (MON810) and its effect on bacterial communities in the maize rhizosphere. Mol Ecol. 2005;14:2539–2551. doi: 10.1111/j.1365-294X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- Blackwood C.B., Buyer J.S. Soil microbial communities associated with Bt and non‐Bt corn in three soils. J Environ Qual. 2004;33:832–836. doi: 10.2134/jeq2004.0832. [DOI] [PubMed] [Google Scholar]
- Bradley K.L., Hancock J.E., Giardina C.P., Pregitzer K.S. Soil microbial community responses to altered lignin biosynthesis in Populus tremuloides vary among three distinct soils. Plant Soil. 2007;294:185–201. [Google Scholar]
- Brimecombe M.J., De Leij F.A., Lynch J.M. The effect of root exudates on rhizosphere microbial populations. In: Pinton R., Varanini Z., Nannipieri P., editors. Marcel Dekker, Inc; 2001. pp. 95–140. [Google Scholar]
- Bruinsma M., Kowalchuk G.A., Van Veen J.A. Effects of genetically modified plants on microbial communities and processes in soil. Biol Fertil Soils. 2003;37:329–337. [Google Scholar]
- Brusetti L., Francia P., Bertolini C., Pagliuca A., Borin S., Sorlini C. Bacterial communities associated with the rhizosphere of transgenic Bt 176 maize (Zea mays) and its non transgenic counterpart. Plant Soil. 2004;266:11–21. et al. [Google Scholar]
- Burris K., Mentewab A., Ripp S., Stewart C.N., Jr. An Arabidopsis thaliana ABC transporter that confers kanamycin resistance in transgenic plants does not endow resistance to Escherichia coli. Microb Biotechnol. 2008;1:191–195. doi: 10.1111/j.1751-7915.2007.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldini M., Turrini A., Sbrana C., Benedetti A., Marchionni M., Mocali S. Impact of Bt corn on rhizospheric and on beneficial mycorrhizal symbiosis and soil eubacterial communities in experimental microcosms. Appl Environ Microbiol. 2005;71:6719–6729. doi: 10.1128/AEM.71.11.6719-6729.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirvilleri G., Spina S., Scuderi G., Gentile A., Catara A. Characterization of antagonistic root‐associated fluorescent pseudomonads of transgenic and non‐transgenic citrange Troyer plants. J Plant Pathol. 2005;87:179–186. [Google Scholar]
- Cowgill S.E., Bardgett R.D., Kiezebrink D.T., Atkinson H.J. The effect of transgenic nematode resistance on non‐target organisms in the potato rhizosphere. J Appl Ecol. 2002;39:915–923. [Google Scholar]
- Dale P.J., Clarke B., Fontes E.M.G. Potential for the environmental impact of transgenic crops. Nat Biotechnol. 2002;20:567–574. doi: 10.1038/nbt0602-567. [DOI] [PubMed] [Google Scholar]
- Demanèche S., Sanguin H., Poté J., Navarro E., Bernillon D., Mavingui P. Antibiotic‐resistant soil bacteria in transgenic plant fields. Proc Nat Acad Sci USA. 2008;105:3957–3962. doi: 10.1073/pnas.0800072105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devare M., Londono R.L., Thies J.E. Neither transgenic Bt maize (MON863) nor tefluthrin insecticide adversely affect soil microbial activity or biomass: a 3‐year field analysis. Soil Biol Biochem. 2007;39:2038–2047. [Google Scholar]
- Devare M.H., Jones C.M., Thies J.E. Effect of Cry3Bb transgenic corn and tefluthrin on the soil microbial community: biomass, activity, and diversity. J Environ Qual. 2004;33:837–843. doi: 10.2134/jeq2004.0837. [DOI] [PubMed] [Google Scholar]
- Di Giovanni G.D., Watrud L.S., Seidler R.J., Widmer F. Comparison of parental and transgenic alfalfa rhizosphere bacterial communities using Biolog GN metabolic fingerprinting and enterobacterial repetitive intergenic consensus sequence‐PCR (ERIC‐PCR) Microb Ecol. 1999;37:129–139. doi: 10.1007/s002489900137. [DOI] [PubMed] [Google Scholar]
- Donegan K.K., Seidler R.J., Doyle J.D., Porteous L.A., Di Giovanni G., Widmer F., Watrud L.S. A field study with genetically engineered alfalfa inoculated with recombinant Sinorhizobium meliloti: effects on the soil ecosystem. J Appl Ecol. 1999;36:920–936. [Google Scholar]
- Dunfield K.E., Germida J.J. Diversity of bacterial communities in the rhizosphere and root interior of field‐grown genetically modified Brassica napus. FEMS Microbiol Ecol. 2001;38:1–9. [Google Scholar]
- Dunfield K.E., Germida J.J. Seasonal changes in the rhizosphere microbial communities associated with field‐grown genetically modified canola (Brassica napus. Appl Environ Microbiol. 2003;69:7310–7318. doi: 10.1128/AEM.69.12.7310-7318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer W.E. Techniques for producing herbicide‐resistant crops. In: Duke S.O., editor. Lewis Publishers; 1996. pp. 37–52. [Google Scholar]
- Faragova N., Farago J., Drabekova J. Evaluation of abundance of aerobic bacteria in the rhizosphere of transgenic and non‐transgenic alfalfa lines. Folia Microbiol. 2005;50:509–514. doi: 10.1007/BF02931439. [DOI] [PubMed] [Google Scholar]
- Garland J.L., Mills A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community‐level sole‐carbon‐source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M.J. Gene transfer from genetically modified‐food. Curr Opin Biotechnol. 2000;11:505–508. doi: 10.1016/s0958-1669(00)00136-1. [DOI] [PubMed] [Google Scholar]
- Gebhard F., Smalla K. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl Environ Microbiol. 1998;64:1550–1554. doi: 10.1128/aem.64.4.1550-1554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard F., Smalla K. Monitoring field releases of genetically modified sugar beets for persistence of transgenic plant DNA and horizontal gene transfer. FEMS Microbiol Ecol. 1999;28:261–272. [Google Scholar]
- Griffiths B.S., Geoghegan I.E., Robertson W.M. Testing genetically engineered potato, producing the lectins GNA and Con A, on non‐target soil organisms and processes. J Appl Ecol. 2000;37:159–170. [Google Scholar]
- Griffiths B.S., Caul S., Thompson J., Birch A.N.E., Scrimgeour C., Andersen M.N. A comparison of soil microbial community structure, protozoa and nematodes in field plots of conventional and genetically modified maize expressing the Bacillus thuringiens CryIAb toxin. Plant Soil. 2005;275:135–146. et al. [Google Scholar]
- Griffiths B.S., Caul S., Thompson J., Birch A.N.E., Scrimgeour C., Cortet J. Soil microbial and faunal community responses to Bt maize and insecticide in two soils. J Environ Qual. 2006;35:734–741. doi: 10.2134/jeq2005.0344. et al. [DOI] [PubMed] [Google Scholar]
- Griffiths B.S., Caul S., Thompson J., Birch A.N.E., Cortet J., Andersen M.N., Krogh P.H. Microbial and microfaunal community structure in cropping systems with genetically modified plants. Pedobiologia. 2007;51:195–206. [Google Scholar]
- Gyamfi S., Pfeifer U., Stierschneider M., Sessitsch A. Effects of transgenic glufosinate‐tolerant oilseed rape (Brassica napus) and the associated herbicide application on eubacterial and Pseudomonas communities in the rhizosphere. FEMS Microbiol Ecol. 2002;41:181–190. doi: 10.1111/j.1574-6941.2002.tb00979.x. [DOI] [PubMed] [Google Scholar]
- Heuer H., Kroppenstedt R.M., Lottmann J., Berg G., Smalla K. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere relative to communities are negligible natural factors. Appl Environ Microbiol. 2002;68:1325–1335. doi: 10.1128/AEM.68.3.1325-1335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höflich G., Tauschke M., Kuhn G., Werner K., Frielinghaus M., Hohn W. Influence of long‐term conservation tillage on soil and rhizosphere microorganisms. Biol Fertil Soils. 1999;29:81–86. [Google Scholar]
- James C. ISAAA. ISAAA; 2007. Global status of commercialized biotech/gm crops: 2007; p. 143p. [Google Scholar]
- Kent A.D., Triplett E.W. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu Rev Microbiol. 2002;56:211–236. doi: 10.1146/annurev.micro.56.012302.161120. [DOI] [PubMed] [Google Scholar]
- Kieft T.L., Ringelberg D.B., White D.C. Changes in ester‐linked phospholipid fatty acid profiles of subsurface bacteria during starvation and desiccation in a porous medium. Appl Environ Microbiol. 1994;60:3292–3299. doi: 10.1128/aem.60.9.3292-3299.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc P.M., Hamelin R.C., Filion M. Alteration of soil rhizosphere communities following genetic transformation of white spruce. Appl Environ Microbiol. 2007;73:4128–4134. doi: 10.1128/AEM.02590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zeng Q., Yan F.M., Xu H.G., Xu C.R. Effects of transgenic plants on soil microorganisms. Plant Soil. 2005;271:1–13. [Google Scholar]
- Liu W., Lu H.H., Wu W.X., Wei Q.K., Chen Y.X., Thies J.E. Transgenic Bt rice does not affect enzyme activities and microbial composition in the rhizosphere during crop development. Soil Biol Biochem. 2008;40:475–486. [Google Scholar]
- Lottmann J., Berg G. Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and non‐transgenic potato plants. Microbiol Res. 2001;156:75–82. doi: 10.1078/0944-5013-00086. [DOI] [PubMed] [Google Scholar]
- Lottmann J., Heuer H., Smalla K., Berg G. Influence of transgenic T4‐lysozyme‐producing potato plants on potentially beneficial plant‐associated bacteria. FEMS Microbiol Ecol. 1999;29:365–377. [Google Scholar]
- Lukow T., Dunfield P.E., Liesack W. Use of the T‐RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non‐transgenic potato plants. FEMS Microb Ecol. 2000;32:241–247. doi: 10.1111/j.1574-6941.2000.tb00717.x. [DOI] [PubMed] [Google Scholar]
- Lynch J.M., Benedetti A., Insam H., Nuti M.P., Smalla K., Torsvik V., Nannipieri P. Microbial diversity in soil: ecological theories, the contribution of molecular techniques and the impact of transgenic plants and transgenic microorganisms. Biol Fertil Soils. 2004;40:363–385. [Google Scholar]
- Mansouri H., Petit A., Oger P., Dessaux Y. Engineered rhizosphere: the trophic bias generated by opine‐producing plants is independent of the opine type, the soil origin, and the plant species. Appl Environ Microbiol. 2002;68:2562–2566. doi: 10.1128/AEM.68.5.2562-2566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P., Crowley D., Yang C.H. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil. 2004;261:199–208. [Google Scholar]
- Matilla M.A., Espinosa‐Urgel M., Rodriguez‐Herva J.J., Ramos J.L., Ramos‐Gonzalez M.I. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Gen Biol. 2007;8:R179.1–R179.3. doi: 10.1186/gb-2007-8-9-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M., Funnell D.L., Raaijmakers J.M. Wheat cultivar‐specific selection of 2,4‐diacetylphloroglucinol‐producing fluorescent Pseudomonas species from resident soil populations. Microb Ecol. 2004;48:338–348. doi: 10.1007/s00248-003-1067-y. [DOI] [PubMed] [Google Scholar]
- Mendelsohn M., Kough J., Vaituzis Z., Matthews K. Are Bt crops safe? Nat Biotechnol. 2003;21:1003–1009. doi: 10.1038/nbt0903-1003. [DOI] [PubMed] [Google Scholar]
- Milling A., Smalla K., Maidl F.X., Schloter M., Munch J.C. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil. 2005;266:23–39. [Google Scholar]
- Nielsen K.M., Gebhard F., Smalla K., Bones A.M., Van Elsas J.D. Evaluation of possible horizontal gene transfer from transgenic plants to the soil bacterium Acinetobacter calcoaceticus BD413. Theor Appl Genet. 1997;95:815–821. [Google Scholar]
- Nielsen K.M., Van Elsas J.D., Smalla K. Dynamics, horizontal transfer and selection of novel DNA in bacterial populations in the phytosphere of transgenic plants. Ann Microbiol. 2001;51:79–94. [Google Scholar]
- O'Callaghan M., Gerard E.M., Waipara N.W., Young S.D., Glare T.R., Barrell P.J., Conner A.J. Microbial communities of Solanum tuberosum and magainin‐producing transgenic lines. Plant Soil. 2004;266:47–56. [Google Scholar]
- Oger P., Petit A., Dessaux Y. Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol. 1997;15:369–372. doi: 10.1038/nbt0497-369. [DOI] [PubMed] [Google Scholar]
- Oger P., Mansouri H., Dessaux Y. Effect of crop rotation and soil cover on alteration of the soil microflora generated by the culture of transgenic plants producing opines. Mol Ecol. 2000;9:881–890. doi: 10.1046/j.1365-294x.2000.00940.x. [DOI] [PubMed] [Google Scholar]
- Oger P.M., Mansouri H., Nesme X., Dessaux Y. Engineering root exudation of lotus toward the production of two novel carbon compounds leads to the selection of distinct microbial populations in the rhizosphere. Microb Ecol. 2004;47:96–103. doi: 10.1007/s00248-003-2012-9. [DOI] [PubMed] [Google Scholar]
- Olfs H.‐W., Scherer H.W. Estimating microbial biomass N in soils with and without living roots: limitations of a pre‐extraction step. Biol Fertil Soils. 1996;21:314–318. [Google Scholar]
- Palm C.J., Schaller D.L., Donegan K.K., Seidler R.J. Persistence in soil of transgenic plant producted Bacillus thuringiensis var kurstaki delta‐endotoxin. Can J Microbiol. 1996;42:1258–1262. [Google Scholar]
- Pinton R., Varanini Z., Nannipieri P. The rhizosphere as a site of biochemical interactions among soil components, plants, and microorganisms. In: Pinton R., Varanini Z., Nannipieri P., editors. Marcel Dekker, Inc; 2001. pp. 1–17. [Google Scholar]
- Radajewski S., Ineson P., Parekh N.R., Murrell J.C. Stable‐isotope probing as a tool in microbial ecology. Nat Biotechnol. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Rasche F., Hodl V., Poll C., Kandeler E., Gerzabek M.H., Van Elsas J.D., Sessitsch A. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild‐type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol Ecol. 2006;56:219–235. doi: 10.1111/j.1574-6941.2005.00027.x. [DOI] [PubMed] [Google Scholar]
- Raubuch M., Roose K., Warnstorff K., Wichern F., Joergensen R.G. Respiration pattern and microbial use of field‐grown transgenic Bt‐maize residues. Soil Biol Biochem. 2007;39:2380–2389. [Google Scholar]
- Robin A., Mougel C., Siblot S., Vansuyt G., Mazurier S., Lemanceau P. Effect of ferritin overexpression in tobacco on the structure of bacterial and pseudomonad communities associated with the roots. FEMS Microbiol Ecol. 2006a;58:492–502. doi: 10.1111/j.1574-6941.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Robin A., Vansuyt G., Corberand T., Briat J.F., Lemanceau P. The soil type affects both the differential accumulation of iron between wild‐type and ferritin overexpressor tobacco plants and the sensitivity of their rhizosphere bacterioflora to iron stress. Plant Soil. 2006b;283:73–81. [Google Scholar]
- Robin A., Mazurier S., Mougel C., Vansuyt G., Corberand T., Meyer J.M., Lemanceau P. Diversity of root‐associated fluorescent pseudomonads as affected by ferritin overexpression in tobacco. Environ Microbiol. 2007;9:1724–1737. doi: 10.1111/j.1462-2920.2007.01290.x. [DOI] [PubMed] [Google Scholar]
- Rondon M.R., August P.R., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R. Cloning the soil metanogenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y.K., Yi G.X., Zhao J., Wang B.M., Li Z.H., Zhai Z.X. Changes of Bt toxin in the rhizosphere of transgenic Bt cotton and its influence on soil functional bacteria. World J Microbiol Biotechnol. 2005;21:1279–1284. et al. [Google Scholar]
- Sabharwal N., Icoz I., Saxena D., Stotzky G. Release of the recombinant proteins, human serum albumin, beta‐glucuronidase, glycoprotein B from human cytornegalovirus, and green fluorescent protein, in root exudates from transgenic tobacco and their effects on microbes and enzymatic activities in soil. Plant Physiol Biochem. 2007;45:464–469. doi: 10.1016/j.plaphy.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Sanguin H., Remenant B., Dechesne A., Thioulouse J., Vogel T.M., Nesme X. Potential of a 16S rRNA‐based taxonomic microarray for analysing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl Environ Microbiol. 2006;72:4302–4312. doi: 10.1128/AEM.02686-05. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena D., Stotzky G. Bacillus thuringiensis (Bt) toxin released from root exudates and biomass of Bt corn has no apparent effect on earthworms, nematodes, protozoa, bacteria, and fungi in soil. Soil Biol Biochem. 2001;33:1225–1230. [Google Scholar]
- Schmalenberger A., Tebbe C.C. Bacterial community composition in the rhizosphere of a transgenic, herbicide‐resistant maize (Zea mays) and comparison to its non‐transgenic cultivar. FEMS Microbiol Ecol. 2002;40:29–37. doi: 10.1111/j.1574-6941.2002.tb00933.x. , and Bosphore. [DOI] [PubMed] [Google Scholar]
- Schmalenberger A., Tebbe C.C. Genetic profiling of noncultivated bacteria from the rhizospheres of sugar beet (Beta vulgaris) reveal field and annual variability but no effect of a transgenic herbicide resistance. Can J Microbiol. 2003;49:1–8. doi: 10.1139/w02-111. [DOI] [PubMed] [Google Scholar]
- Sessitsch A., Kan F.Y., Pfeifer U. Diversity and community structure of culturable Bacillus spp. populations in the rhizospheres of transgenic potatoes expressing the lytic peptide cecropin B. Appl Soil Ecol. 2003;22:149–158. [Google Scholar]
- Sessitsch A., Gyamfi S., Tscherko D., Gerzabek M.H., Kandeler E. Activity of microorganisms in the rhizosphere of herbicide treated and untreated transgenic glufosinate‐tolerant and wildtype oilseed rape grown in containment. Plant Soil. 2004;266:105–116. [Google Scholar]
- Shen R.F., Cai H., Gong W.H. Transgenic Bt cotton has no apparent effect on enzymatic activities or functional diversity of microbial communities in rhizosphere soil. Plant Soil. 2006;285:149–159. [Google Scholar]
- Siciliano S.D., Germida J.J. Taxonomic diversity of bacteria associated with the roots of field‐grown transgenic Brassica napus cv. Quest, compared to the non‐transgenic B. napus cv. Excel and B. rapa cv. FEMS Microbiol Ecol. 1999;29:263–272. , and Parkland. [Google Scholar]
- Siciliano S.D., Theoret C.M., De Freitas J.R., Hucl P.J., Germida J.J. Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can J Microbiol. 1998;44:844–851. [Google Scholar]
- Siddiqui Z.A. Springer; 2006. [Google Scholar]
- Tapp H., Stotzky G. Persistence of the insecticidal toxin from Bacillus thuringiensis subsp. kurstaki in soil. Soil Biol Biochem. 1998;30:471–476. [Google Scholar]
- Tesfaye M., Dufault N.S., Dornbusch M.R., Allan D.L., Vance C.P., Samac D.A. Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol Biochem. 2003;35:1103–1113. [Google Scholar]
- Torsvik V.L. Isolation of bacterial DNA from soil. Soil Biol Biochem. 1980;12:15–21. [Google Scholar]
- Travis E.R., Hannink N.K., Van der Gast C.J., Thompson I.P., Rosser S.J., Bruce N.C. Impact of Transgenic tobacco on trinitrotoluene (TNT) contaminated soil community. Environ Sci Technol. 2007;41:5854–5861. doi: 10.1021/es070507a. [DOI] [PubMed] [Google Scholar]
- De Vries J., Heine M., Harms K., Wackernagel W. Spread of recombinant DNA by roots and pollen of transgenic potato plants, identified by highly specific biomonitoring using natural transformation of an Acinetobacter sp. Appl Environ Microbiol. 2003;69:4455–4462. doi: 10.1128/AEM.69.8.4455-4462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M., Weller R., Bateson M.M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Wei X.D., Zou H.L., Chu L.M., Liao B., Ye C.M., Lan C.Y. Field released transgenic papaya affects microbial communities and enzyme activities in soil. Plant Soil. 2006;285:347–358. [PubMed] [Google Scholar]
- Zwahlen C., Hillbeck A., Gugerli P., Nentwig W. Degradation of the Cry1Ab protein within transgenic Bacillus thuringiensis corn tissue in the field. Mol Ecol. 2003;12:765–775. doi: 10.1046/j.1365-294x.2003.01767.x. [DOI] [PubMed] [Google Scholar]


