Crude oil (petroleum) is a highly complex mixture of organic compounds of which some 1.3 million litres enters the environment each year. More then anything else, the numerous oil‐shipping disasters, such as of the Exxon Valdez (1989), the Erika (1999) and the Prestige (2003), have captured the public attention to this environmental problem (Fig. 1). However, these accidents account for only a small part of the annual global release of crude oil, as most enters the environment from deliberate discharge and processing sites. Around three million tons of oil enters the sea each year, of which about 20% originates from oil‐pumping operations, transport and refining activities and 25% from non‐tanker shipping and natural seepages. More than half (55%) originates from illegal activities that include the dumping of ballast water and oil residues as well as accidents (Golyshin et al., 2003). Hydrocarbons are also produced continuously by living cells as natural oils and fats (de Lorenzo, 2006). The observation that the oceans are not covered with an oily layer is a testimony to the activity of the hydrocarbon‐degrading microorganisms (Head et al., 2006). Several bacteria are even known to feed exclusively on hydrocarbons (Yakimov et al., 2007). For these (facultative) hydrocarbon degraders the occasional supertanker oil spill forms an occasional carbon banquet. They play an important role in the clean‐up after an oil spill and form the biological basis for the natural oil‐degrading capacity of the ecosystem. Studies have focused on identifying and characterizing these oil‐eating microbes, as well as how they cope with the oil/water interface, and how to improve this capacity. Here we highlight some of the recent genomics advances in this field.
Figure 1.

Left: Oil spill in the ocean (http://www.pacificariptide.com/pacifica_riptide/oil_spill/). Right: Shipping disaster of Exxon Valdez (http://symonsez.wordpress.com/2009/03/24/blame‐exxon‐valdez‐on‐captain‐bligh‐midwest‐storms/).
Degradation of hydrocarbons
Over 17 000 organic compounds have been identified in crude oil, and subdivided into four main classes: the saturates, aromatics, asphaltenes and resins (Marshall and Rodgers, 2004). The susceptibility of hydrocarbons to microbial degradation can be generally ranked as follows: linear alkanes > branched alkanes > small aromatics > cyclic alkanes (Leahy and Colwell, 1990). The bioremediation efforts of the Exxon Valdez oil spill have indeed shown that the (light) alkanes are depleted first and that some compounds, such as the high‐molecular weight polycyclic aromatic hydrocarbons (PAHs), may not be degraded at all (Atlas and Bragg, 2009).
Many environmental factors influence the breakdown of carbohydrates by microorganisms. Specifically in marine environments, low phosphorous and nitrogen levels may limit growth of oil‐degrading microorganisms and thus rapid oil consumption. There have been attempts to ‘fertilize’ oil‐spill areas to create a more optimal C : N : P balance of 100:10:1 (referred to as biostimulation) (Nikolopoulou and Kalogerakis, 2008). In open‐sea environments, dilution of soluble nutrients quickly occurs, and the administration of insoluble or hydrophobic (oil‐soluble) fertilizers is thought to enhance biostimulation effectiveness. Biosurfactants seem another promising form of biostimulation. Biosurfactants increase the oil‐surface area and with that the amount of oil actually available for attack by bacteria (Nikolopoulou and Kalogerakis, 2009). When oil washes up on beaches and is sequestered in the sediments, the bio‐availabilty can be severely reduced, significantly slowing down or even preventing biodegradation. Depending on the particular local circumstances, tilling of beaches may be applied to expose the sequestered oil. However, tilling itself is disruptive for many coastal plants and animals and, as the oil was beyond the reach of the biota in the first place, may do more harm than good. Local conditions, in general, have a big effect on the efficiency of oil breakdown, such as temperature, waves (mixing), availability of oxygen, and of course the composition of the spilled oil. These should be taken into account when devising an intervention strategy.
Diversity and metabolism of oil‐degrading bacteria
Notwithstanding all the factors that influence the oil‐biodegrading capacity, it all comes down to the metabolic veracity of bacteria that do not mind to get their ‘hands’ dirty (or greasy to be more precise). A great number of bacteria have been identified that help clean up the hydrocarbon compounds in the aftermath of oil spills. But a significant part of the hydrocarbon content in seawater has a biological origin. Lipids and fatty acids from plants, animals and microbes and the products of their conversion in anoxic zones are ubiquitous. Evolution has created some bacteria that dine exclusively on hydrocarbons, including obligate hydrocarbon degraders of the genera Oleispira, Oleiphilus, Thalassolituus, Alcanivorax and Cycloclasticus (Fig. 2).
Figure 2.
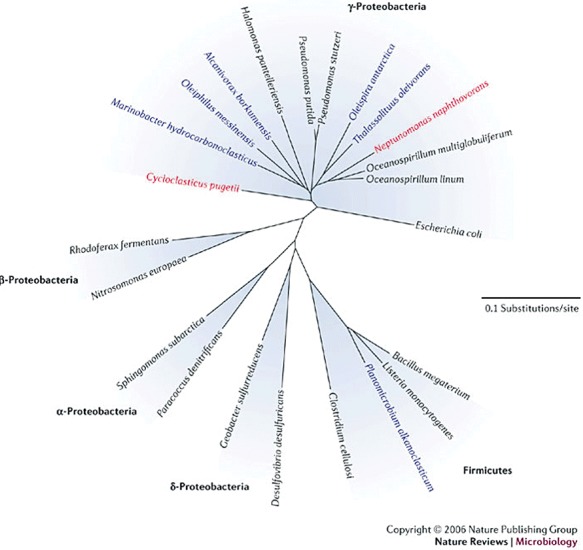
A phylogenetic tree illustrating the diversity of aerobic hydrocarbon‐degrading bacteria. Organisms shown in blue can degrade saturated hydrocarbons, whereas those in red can degrade polycyclic aromatic hydrocarbons. The organisms shown in black do not degrade hydrocarbons. Reprinted from Head and colleagues (2006) by permission from Macmillan Publishers Ltd, copyright 2006.
In typical carbon utilization tests, these bacteria only catabolize hydrocarbons such as Tween 40 and 80, leaving other carbon sources, including sugars and acids, untouched (Yakimov et al., 2007). Current interest in the metabolism of (obligate) hydrocarbon‐degrading bacteria has spurred on various genome sequencing projects (Table 1 and Fig. 3).
Table 1.
Genome sequencing projects of oil/hydrocarbon‐degrading bacteria (adapted from the GOLD database: http://www.genomesonline.org).
| Species | Genome size (kb) | GC% | Habitat | goldstampa/Reference |
|---|---|---|---|---|
| Completed | ||||
| Alcanivorax borkumensis SK2 | 3120 | 54.7 | Marine | Gc00411/b |
| Marinobacter hydrocarbonoclasticus VT8 | 4326 | 57.3 | Marine | Gc00504/c |
| Arthrobacter sp. FB24 | 4698 | 65.5 | Soil | Gc00445 |
| Geobacillus thermodenitrificans NG80‐2 | 3607 | 49 | Fresh water, oil fields | Gc00532/d |
| Ongoing | ||||
| Alcanivorax sp. DG881 | 3789 | 58 | Marine | Gi01400 |
| Cycloclasticus pugetii PS‐1 | Marine, sediment | Gi03249/e | ||
| Cycloclasticus sp.TUi26 | 2300 | Fresh water | Gi02200 | |
| Marinobacter hydrocarbonoclasticus ATCC 49840 | Fresh water | Gi01519 | ||
| Marinobacter algicola DG893 | 4413 | 57 | Marine | Gi01420 |
| Methylomicrobium album BG8 | Soil | Gi02102 | ||
| Oceanicaulis alexandrii HTCC2633 | 3166 | 64 | Marine | Gi00864 |
| Oleispira antarctica RB‐8 | 4400 | 43 | Marine | Gi02491/f |
| Rhodococcus opacus B4 PD630 | Soil | Gi03264 |
Links to sequence information and genome database can be retrieved with the goldstamp ID.
Figure 3.
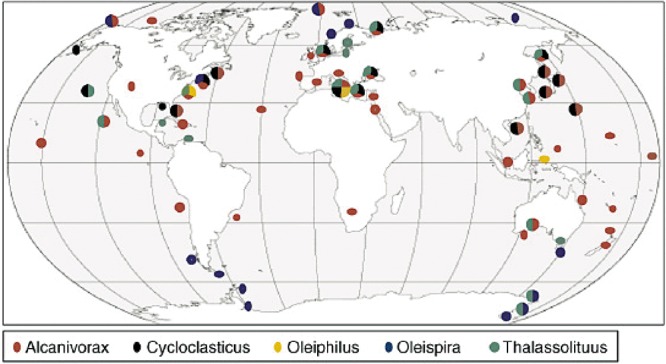
Geographic distribution of isolates of retrieved 16S rRNA sequences of marine obligate hydrocarbonoclastic γ‐proteobacteria. Reprinted from Yakimov and colleagues (2007) with permission from Elsevier.
The genome of the obligate hydrocarbon‐degrading bacterium (or hydro‐carbonoclastic bacterium) Alcanivorax borkumensis was recently discussed in some detail (Schneiker et al., 2006). This bacterium is of particular interest as it is a cosmopolitan marine bacterium that blooms in oil‐contaminated areas, where it can constitute up to 80% of the bacterial population. The features of its genome reveal how this bacterium grows efficiently on alkanes. As hydrocarbons are poorly soluble in water, this bacterium encodes extensive exopolyssacharide production and pili through which it can attach to the oil‐water interface. Biosurfactants can be produced that emulsify the alkanes and increase the oil‐water surface area. The genome contains several regions of atypical GC content, one of which carries a complete gene set for alkane degradation (alkSB1GJH), and more genes were found that encode proteins putatively involved in alkane degradation, such as alkK, alkL and alkN (Fig. 4). These encode proteins such as alkane hydroxylases, alcohol dehydrogenases, oxidoeductases, P450 cytochromes, rubredoxin and a rubredoxin reductase (rubA and rubB). As expected for an obligate hydrocarbon degrader, functional genes that encode typical sugar uptake mechanisms such as PEP‐dependent sugar/phosphotransferase systems are not found in the genome. The marine origin of A. borkumensis is evident, as many genes encode efficient scavenging systems, such as high‐affinity ABC‐transporters, to sequester essential nutrients (Fe, Zn, Co, Mg, Mn and Mo). Furthermore, the Na+‐dependent NADH‐quinone dehydrogenase (encoded by nqrABCDEF) and a variety of H+/Na+ antiporters (encoded by mnhABCDEFG, nhaD, nhaB, nhaP and nhaC) enables many transport processes to use a sodium gradient.
Figure 4.
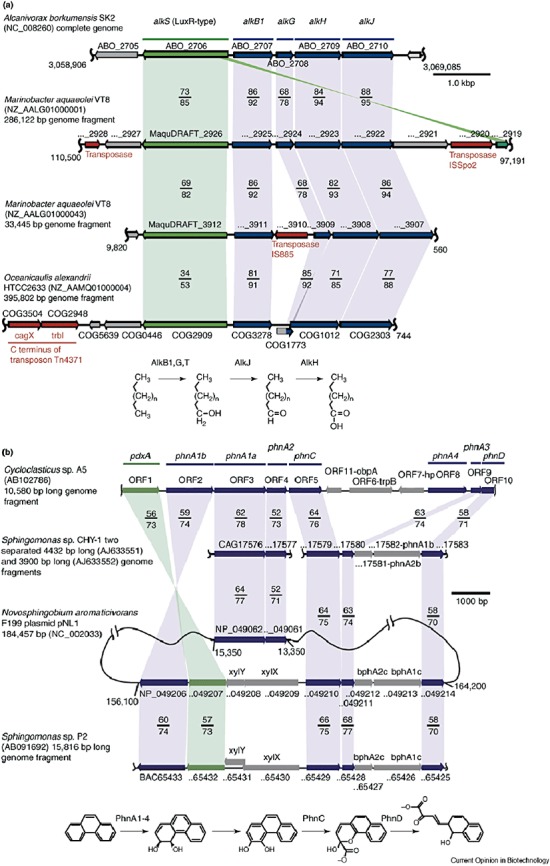
Ubiquity of gene clusters for the degradation of (A) aliphatic and (B) (poly) aromatic fractions of oil. A. Organization of genes homologous to the A. borkumensis alk gene cluster in the hydrocarbon‐degrading marine proteobacteria. Homologous genes are highlighted by shaded areas: sequences predicted to code for LuxR‐type transcriptional activators of the alkane genes, AlkS, are marked in green, genes for the alkane degradation pathway are indicated in blue. Percentages of protein identity/similarity of polypeptides from A. borkumensis with those of M. aqaeolei and O. alexandrii are shown. Gene designations: alkB1, alkane monooxygenase; alkG, rubredoxin; alkJ, alcohol dehydrogenase; alkH, aldehyde dehydrogenase. B. Organization of gene clusters of polycyclic aromatic hydrocarbons (PAH) degradation pathways of Cycloclasticus sp. A5 and PAH‐degrading α‐proteobacteria. pdxA genes predicted to code for the pyridoxal‐phosphate biosynthesis enzyme are colored in green, PAH degradation genes are in blue. Percentages of protein identity/similarity of the polypeptides from Cycloclasticus sp. A5 with those of the PAH‐degrading α‐proteobacteria are shown. Gene designations: phnA1, iron‐sulfur protein (ISP) α subunit of PAH dioxygenase; phnA2, ISP β‐subunit of PAH dioxygenase; phnA3, Rieske‐type [2Fe‐2S] ferredoxin, phnA4, NADH‐ferredoxin oxidoreductase; phnC, extradiol [3,4‐dihydrophenanthrene] dioxygenase; phnD, 2‐hydroxy‐2H‐benzo[h]chromene‐2‐carboxylate isomerase. Reprinted from Yakimov and colleagues (2007) with permission from Elsevier.
Biodegradation of crude oil
The PAHs found in oil are particularly resistant to microbial degradation, by the intrinsic stability of the aromatic ring. Polycyclic aromatic hydrocarbons form a great health hazard as many are toxic or carcinogenic and persist in the oil‐polluted environments long after the (linear) alkanes are degraded. Luckily, PAHs also have biological origins and are, for example, formed by the combustion of organic matter in forest fires. A natural occurrence of compounds usually means that some bacteria can be found in Nature that can feed on them. Polycyclic aromatic hydrocarbons comprise of two or more fused aromatic rings with a diverse range of branching types and aromatic groups. Not surprisingly therefore, effective PAH breakdown involves whole communities of both bacteria and eukaryotes. Within these communities the PAH‐degrading capabilities of Arthrobacter, Burkholderia, Mycobacterium, Pseudomonas, Sphingomonas and Rhodococcus are best studied, which is supplemented by sequencing efforts (Table 2) (Seo et al., 2007; Vandermeer and Daugulis, 2007; Haritash and Kaushik, 2009; Martinkova et al., 2009).
Table 2.
Genome sequencing projects of PAH‐degrading bacteria (adapted from the GOLD database: http://www.genomesonline.org).
| PAH degraders | Genome size (kb) | GC% | Habitat | goldstampa/Reference |
|---|---|---|---|---|
| Completed | ||||
| Arthrobacter chlorophenolicus A6 | 4395 | 65.9 | Soil | Gc00930 |
| Mycobacterium flavenscens PYR‐GCK | 5619 | 67 | Soil | Gc00534 |
| Mycobacterium sp. JLS | 6048 | 68 | Soil | Gc00516 |
| Mycobacterium sp. MCS | 5705 | 68 | Soil | Gc00392 |
| Mycobacterium vanbaalenii PYR‐1 | 6491 | 67.8 | Sediment | Gc00479 |
| Polaromonas naphthalenivorans CJ2 | 4410 | 62.5 | Fresh water | Gc00486/c |
| Rhodococcus erythropolis PR4 | 6516 | 62 | Marine | Gc00980 |
| Rhodococcus opacus B4 | 7913 | 67 | Gc00982 | |
| Sphingomonas wittichii RW1 | 5382 | 67 | Fresh water | Gc00571 |
| Pseudomonas sp.b | ||||
| Ongoing | ||||
| Arthrobacter phenanthrenovorans Sphe3 | Soil | Gi01675 | ||
| Burkholderia sp. Ch1‐1 | 3789 | 58 | Soil | Gi03275 |
| Burkholderia sp. Cs1‐4 | Rhizosphere | Gi03276 | ||
| Cycloclasticus sp. TU126 | 2300 | Fresh water | Gi02200 | |
| Cycloclasticus pugetii PS‐1 | Marine, sediment | Gi03249 | ||
| Mycobacterium sp. Spyr1 | 66 | Soil | Gi02013 |
Links to sequence information and genome database can be retrieved with the goldstamp ID.
There are genome sequencing projects of over 50 Pseudomonas species/strains, of which 17 are complete, many of which are capable of PAH degradation.
These microbes involved in the second‐phase clean‐up require different approaches to biostimulation. For example, although aeration and availability of water remain crucial, the addition of fertilizers actually had a negative impact on the PAH degradation (Vinas et al., 2005). The study of PAH‐degrading bacteria may provide insights how to get a more complete oil clean‐up (Lafortune et al., 2009). The metabolism required for efficient breakdown of PAHs has been best studied in species of Pseudomonas with relatively simple model PAHs, such as napthalenes, phenanthrene and anthracene (Habe and Omori, 2003). In Pseudomonas species, the genes that are required for the breakdown of PAHs (nah, pah, dox phn, phd and nag operons) (Fig. 4) can often be found on plasmids, facilitating genetic transfer and acquisition in oil‐polluted environments.
In light of the ever‐increasing world demand for efficient oil‐cleanup measures, we should mention the ‘flip‐side of the coin’ of oil‐degrading bacteria. The growth of hydrocarbon‐degrading bacteria in oil fields and oil tanks in general affects not so much the quantity, but certainly the quality of oil. Oil fields occur typically at several thousand metres below ground and, at temperatures of 60–90°C, are considered sterile. However, as soon as oil is extracted, hydrocarbon‐degrading bacteria capable of oxidizing hydrocarbons start to grow (Yemashova et al., 2007). Moreover, when the self‐flow of oil stops, a common practice to extract the oil is to flood the stratum with (non‐sterile) water. Bacteria (and fungi) that grow in the oil fields produce peroxides, pitch acids and in some cases H2S that decrease the thermostability and volatility of the fuel. Although many different species are involved, a notorius group of bacteria in this regard are the sulfate‐reducing bacteria. These bacteria are able to use the sulfate in the oil as alternative to oxygen and produce H2S. Moreover, they stimulate growth of other bacteria that can subsequently use the H2S (Muyzer and Stams, 2008). Their ability to use sulfate combined with their significant ecological (and economic) impact has made them the focus of current research and several sulfate‐reducing bacteria have been completely sequenced (and published) in the last few years. These include the Desulfovibrio vulgaris Hildenborough, the arctic Desulfotalea psychrophila and the archeal Archaeoglobus fulgidus (Klenk et al., 1997; Heidelberg et al., 2004; Rabus et al., 2004). Their ability to degrade oil in anaerobic conditions may prove useful for the natural oil‐degrading capacity in certain types of ecosystems. Moreover, the lessons learnt on biostimulation can be applied (in reverse) to control microbial deterioration of oil, such as removal of water, oxygen and addition of biocides.
Future perspectives
In the last few years, considerable progress has been made in getting to know the bacteria that eat oil and how they do it. The next step is to actually use all this information to enhance and control the capability of oil degradation by bio‐augmentation. Bio‐augmentation, the addition of specific microorganisms, could considerably speed‐up the natural biodegradation progress. Ideally, as alkanes are relatively quickly degraded, bio‐augmentation should focus on microbes that are specifically engineered to tackle compounds that are still resistant to microbial attack. The search for such a super‐bug has culminated in the first ever patent of a living organism, an engineered Pseudomonas sp., capable of degrading camphor, octane, salicylate and naphthalene (US Patent 4259444). When such bacteria are introduced into the environment, they typically quickly disappear. Their introduction into already colonized niches is very difficult, as most fall prey to protozoa (Cases and de Lorenzo, 2005). In summary, the oil spills demonstrate the immense adaptive capabilities of the bacterial community to their environment. In time, like all natural hydrocarbons, crude oil is completely degraded by the microorganisms or made inaccessible. Ironically, when speeding up the biodegradation of oil‐spill sites via bacteria, we should probably take most care not to do disturb the environment, again.
Acknowledgments
This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
References
- Atlas R., Bragg J. Bioremediation of marine oil spills: when and when not – the Exxon Valdez experience. Microb Biotechnol. 2009;2:213–221. doi: 10.1111/j.1751-7915.2008.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I., De Lorenzo V. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int Microbiol. 2005;8:213–222. [PubMed] [Google Scholar]
- Dyksterhouse S.E., Gray J.P., Herwig R.P., Lara J.C., Staley J.T. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon‐degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- Feng L., Wang W., Cheng J., Ren Y., Zhao G., Gao C. Genome and proteome of long‐chain alkane degrading Geobacillus thermodenitrificans NG80‐2 isolated from a deep‐subsurface oil reservoir. Proc Natl Acad Sci USA. 2007;104:5602–5607. doi: 10.1073/pnas.0609650104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golyshin P.N., Martins Dos Santos V.A., Kaiser O., Ferrer M., Sabirova Y.S., Lunsdorf H. Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon‐degrading bacterium that plays a global role in oil removal from marine systems. J Biotechnol. 2003;106:215–220. doi: 10.1016/j.jbiotec.2003.07.013. et al. [DOI] [PubMed] [Google Scholar]
- Habe H., Omori T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci Biotechnol Biochem. 2003;67:225–243. doi: 10.1271/bbb.67.225. [DOI] [PubMed] [Google Scholar]
- Haritash A.K., Kaushik C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- Head I.M., Jones D.M., Roling W.F. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- Heidelberg J.F., Seshadri R., Haveman S.A., Hemme C.L., Paulsen I.T., Kolonay J.F. The genome sequence of the anaerobic, sulfate‐reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. et al. [DOI] [PubMed] [Google Scholar]
- Huu N.B., Denner E.B., Ha D.T., Wanner G., Stan‐Lotter H. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil‐producing well. Int J Syst Bacteriol. 1999;49:367–375. doi: 10.1099/00207713-49-2-367. [DOI] [PubMed] [Google Scholar]
- Klenk H.P., Clayton R.A., Tomb J.F., White O., Nelson K.E., Ketchum K.A. The complete genome sequence of the hyperthermophilic, sulphate‐reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. et al. [DOI] [PubMed] [Google Scholar]
- Lafortune I., Juteau P., Deziel E., Lepine F., Beaudet R., Villemur R. Bacterial diversity of a consortium degrading high‐molecular‐weight polycyclic aromatic hydrocarbons in a two‐liquid phase biosystem. Microb Ecol. 2009;57:455–468. doi: 10.1007/s00248-008-9417-4. [DOI] [PubMed] [Google Scholar]
- Leahy J.G., Colwell R.R. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo V. Blueprint of an oil‐eating bacterium. Nat Biotechnol. 2006;24:952–953. doi: 10.1038/nbt0806-952. [DOI] [PubMed] [Google Scholar]
- Marshall A.G., Rodgers R.P. Petroleomics: the next grand challenge for chemical analysis. Acc Chem Res. 2004;37:53–59. doi: 10.1021/ar020177t. [DOI] [PubMed] [Google Scholar]
- Martinkova L., Uhnakova B., Patek M., Nesvera J., Kren V. Biodegradation potential of the genus Rhodococcus. Environ Int. 2009;35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Muyzer G., Stams A.J. The ecology and biotechnology of sulphate‐reducing bacteria. Nat Rev Microbiol. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Nikolopoulou M., Kalogerakis N. Enhanced bioremediation of crude oil utilizing lipophilic fertilizers combined with biosurfactants and molasses. Mar Pollut Bull. 2008;56:1855–1861. doi: 10.1016/j.marpolbul.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Nikolopoulou M., Kalogerakis N. Biostimulation strategies for fresh and chronically polluted marine environments with petroleum hydrocarbons. J Chem Technol Biotechnol. 2009;84:802–807. [Google Scholar]
- Rabus R., Ruepp A., Frickey T., Rattei T., Fartmann B., Stark M. The genome of Desulfotalea psychrophila, a sulfate‐reducing bacterium from permanently cold Arctic sediments. Environ Microbiol. 2004;6:887–902. doi: 10.1111/j.1462-2920.2004.00665.x. et al. [DOI] [PubMed] [Google Scholar]
- Schneiker S., Martins dos Santos V.A., Bartels D., Bekel T., Brecht M., Buhrmester J. Genome sequence of the ubiquitous hydrocarbon‐degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol. 2006;24:997–1004. doi: 10.1038/nbt1232. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.S., Keum Y.S., Harada R.M., Li Q.X. Isolation and characterization of bacteria capable of degrading polycyclic aromatic hydrocarbons (PAHs) and organophosphorus pesticides from PAH‐contaminated soil in Hilo, Hawaii. J Agric Food Chem. 2007;55:5383–5389. doi: 10.1021/jf0637630. [DOI] [PubMed] [Google Scholar]
- Vandermeer K.D., Daugulis A.J. Enhanced degradation of a mixture of polycyclic aromatic hydrocarbons by a defined microbial consortium in a two‐phase partitioning bioreactor. Biodegradation. 2007;18:211–221. doi: 10.1007/s10532-006-9056-8. [DOI] [PubMed] [Google Scholar]
- Vinas M., Sabate J., Espuny M.J., Solanas A.M. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote‐contaminated soil. Appl Environ Microbiol. 2005;71:7008–7018. doi: 10.1128/AEM.71.11.7008-7018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi J.M., Sims D., Brettin T., Bruce D., Madsen E.L. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar‐contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ Microbiol. 2009;11:2253–2270. doi: 10.1111/j.1462-2920.2009.01947.x. [DOI] [PubMed] [Google Scholar]
- Yakimov M.M., Giuliano L., Gentile G., Crisafi E., Chernikova T.N., Abraham W.R. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int J Syst Evol Microbiol. 2003;53:779–785. doi: 10.1099/ijs.0.02366-0. et al. [DOI] [PubMed] [Google Scholar]
- Yakimov M.M., Timmis K.N., Golyshin P.N. Obligate oil‐degrading marine bacteria. Curr Opin Biotechnol. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Yemashova N.A., Murygina V.P., Zhukov D.V., Zakharyantz A.A., Gladchenko M.A., Appanna V., Kalyuzhnyi S.V. Biodeterioration of crude oil and oil derived products: a review. Rev Environ Sci Biotechnol. 2007;6:315–337. [Google Scholar]


