Summary
Biofilm‐grown bacteria are refractory to antimicrobial agents and show an increased capacity to evade the host immune system. In recent years, studies have begun on biofilm formation by Streptococcus pneumoniae, an important human pathogen, using a variety of in vitro model systems. The bacterial cells in these biofilms are held together by an extracellular matrix composed of DNA, proteins and, possibly, polysaccharide(s). Although neither the precise nature of these proteins nor the composition of the putative polysaccharide(s) is clear, it is known that choline‐binding proteins are required for successful biofilm formation. Further, many genes appear to be involved, although the role of each appears to vary when biofilms are produced in batch or continuous culture. Prophylactic and therapeutic measures need to be developed to fight S. pneumoniae biofilm formation. However, much care needs to be taken when choosing strains for such studies because different S. pneumoniae isolates can show remarkable genomic differences. Multispecies and in vivo biofilm models must also be developed to provide a more complete understanding of biofilm formation and maintenance.
Introduction
It is well recognized that ‘wild‐living’ bacteria organize themselves within biofilms and that their growth rate, metabolism, gene expression and protein production are different to those of planktonic cultures. Biofilms are sessile microbial communities in which cells are attached to a surface or an air–liquid interface, and enveloped within an extracellular polymeric matrix (Costerton et al., 1995). From a medical perspective the importance of biofilms lies in the reduced susceptibility of the participating bacteria to antimicrobial agents (Lewis, 2008) and their ability to evade host immune defence systems (Jensen et al., 2010). Biofilm‐associated growth has been associated with a high percentage of patients with chronic and persistent infections, the biofilms acting as pathogen reservoirs (Wolcott and Ehrlich, 2008).
Streptococcus pneumoniae is an important human respiratory pathogen that causes a variety of serious diseases such as community‐acquired pneumonia, meningitis and sepsis. It is also the main causal agent of otitis media in children. Several authors have recently detected pneumococcal biofilms on the surface of adenoid and mucosal epithelial tissues in children with recurrent middle‐ear infections and otitis media with effusion (Hall‐Stoodley et al., 2006; Coates et al., 2008; Hoa et al., 2009; Nistico et al., 2011), as well as on the sinus mucosa of patients with chronic rhinosinusitis (Sanderson et al., 2006). Biofilm‐like structures detected in the lungs of mice infected with S. pneumoniae are similar to those produced in a continuous flow‐through biofilm model (Sanchez et al., 2010).
Although the earliest reports on pneumococcal biofilms go back 10 years or more, the last 5 years have seen an increase in the number of studies examining pneumococcal biofilms at the structural and genetic level. Different laboratories have used different approaches for growing biofilms of human pathogens in vitro with the aim of producing an appropriate model that mimics in vivo environments. The first system designed for pneumococcal biofilms, described in 1997 and developed as a means of assessing susceptibility to antibiotics, was based on steady‐state growth on cellulose Sorbarod filters (Budhani and Struthers, 1997). It was later shown that pneumococcal growth on these filters in a continuous culture‐like system resembles the nasopharyngeal carriage of the pathogen (Waite et al., 2001). Other research groups developed biofilm reactor systems for analysing pneumococcal biofilms and studying biofilm processes in situ and in real time (Donlan et al., 2004; Goeres et al., 2005). Using a continuous‐culture one‐through flow cell, Allegrucci and colleagues (2006) showed that S. pneumoniae adopts multiple phenotypes over the course of biofilm development. Our group has been involved in developing an in vitro biofilm model for S. pneumoniae using polystyrene microtitre plates or glass‐bottom dishes as a support. This system allowed the effects of several factors (e.g. nutrients, pH changes, osmolarity, temperature) on biofilm development to be examined (Moscoso et al., 2006), as well as the rapid screening of mutants defective in biofilm formation (Muñoz‐Elías et al., 2008).
The present review summarizes the recent genetic, biochemical and structural data reported for S. pneumoniae biofilms.
Environmental factors affecting biofilm formation
The carbon source, the flow velocity and the physical properties of the surface to which bacteria adhere, such as its hydrophobicity and roughness, can lead to differences in the structure and composition of the biofilms produced (O'Toole and Kolter, 1998; Stoodley et al., 1999). Our group analysed the influence of several environmental factors in pneumococcal biofilm formation (Moscoso et al., 2006). The ability of S. pneumoniae to form biofilms on abiotic surfaces was tested on a range of materials including glass, polyvinylchloride and polystyrene, the last of which was associated with the strongest biofilm formation. The intense biofilm production observed on chemically defined (Cden or CDM) and semisynthetic (C) media indicates that biofilm formation represents a survival strategy in a nutritionally limited environment; pneumococcal cells growing in rich media showed poor biofilm formation in polystyrene (or glass) dishes. Enriching C medium with additives such as yeast extract or bovine serum albumin led to no significant change in biofilm formation on polystyrene microplates. Increasing the osmolarity above 0.2 M, however, drastically inhibited pneumococcal growth and biofilm formation. Variations in the starting pH also influenced biofilm formation; optimal development was seen when the initial pH of the culture medium was 7.0–8.0 (Moscoso et al., 2006).
Sialic acid (at a concentration equivalent to that of free sialic acid in human saliva) enhances pneumococcal biofilm formation in vitro, and a causal association has been established between free sialic acid and nasopharyngeal colonization and spread to the lungs in mice (Trappetti et al., 2009). Preliminary data from our group support the notion that the addition of some sugars, such as melicitose, tagatose, melibiose or pullulan can improve in vitro biofilm formation by S. pneumoniae (M. Domenech, unpublished). The oxidative stress caused by the production of hydrogen peroxide through the activity of pneumococcal SpxB pyruvate oxidase (Pericone et al., 2003) appears to be responsible for the development of non‐phase‐variable colony variants that appear consistently when encapsulated pneumococci are incubated under biofilm‐forming conditions (Allegrucci and Sauer, 2008). It is well known that non‐encapsulated S. pneumoniae mutants are better biofilm formers than their encapsulated parental strains (see below). Moreover, a TIGR4 ΔspxB mutant showed no variation in biofilm formation capacity compared with the wild‐type progenitor (Lizcano et al., 2010).
It has recently been reported that biofilm formation by pneumococci is favoured by a CO2‐enriched atmosphere (Camilli et al., 2011). It should be mentioned, however, that although the growth rate of S. pneumoniae was reported higher under anaerobic conditions than in ambient air enriched with 5% CO2, biofilm formation was less strong [although this could not be confirmed in our laboratory (M. Moscoso, unpublished)].
Biofilm ultrastructure
Biofilms formed by a non‐encapsulated pneumococcal strain on abiotic surfaces were found to have a three‐dimensional organization with complex structures about 25–30 µm in thickness, as revealed by confocal laser scanning microscopy (CLSM) (Moscoso et al., 2006). The spatial distribution of the adherent bacteria was later examined by CLSM using a non‐encapsulated S. pneumoniae strain that synthesizes green fluorescent protein, or by staining the bacteria with fluorescent dyes after biofilm formation on glass dishes following 10–12 h of incubation at 34°C (Moscoso et al., 2006; Domenech et al., 2009). These studies detected the presence of small voids and channels separating the microcolonies within the pneumococcal biofilm. Using a continuous‐flow biofilm reactor system, Allegrucci and colleagues (2006) reported that the architecture of mature biofilms (those grown at 37°C in 5% CO2 for 6–9 days) differed significantly among the serotypes tested.
The honeycomb‐like structures observed by low‐temperature scanning electron microscopy (Moscoso et al., 2006) may provide mechanical stability to pneumococcal biofilms and might serve as an important virulence factor, helping to ward off host defences, as described for other microbial communities (Schaudinn et al., 2007; Moscoso et al., 2009). Figure 1 show pneumococcal cells associated with the walls of these honeycomb‐like structures, as well as their connection to the wall and one another by thin filaments (Fig. 1). Some areas free of bacterial cells may represent channels between cell clusters.
Figure 1.
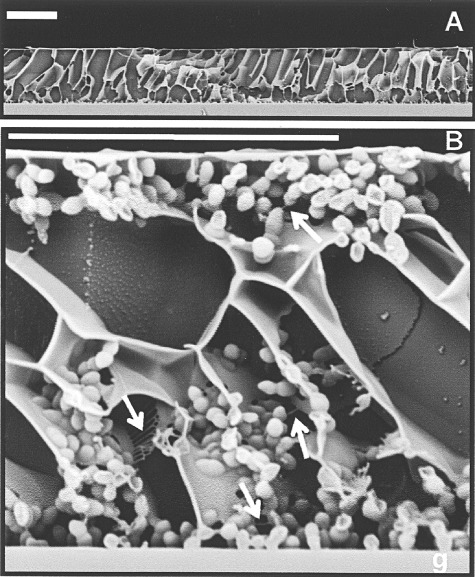
Low‐temperature scanning electron micrographs of a S. pneumoniae R6 biofilm. A. General view of the biofilm formed on the surface of a glass coverslip (g). B. In the magnification, arrows pointed to filamentous material linking pneumococcal cells to each other and to the intercellular matrix. Bar, 20 µm. Reprinted from Moscoso et al. (2006) with permission.
The extracellular matrix of pneumococcal biofilms
Within a biofilm, bacterial cells are embedded in an extracellular matrix composed of different extracellular polymeric substances (EPS), including exopolysaccharides, proteins, nucleic acids and lipids. EPS often determine the scaffold for the three‐dimensional architecture of the biofilm and provide it structural integrity and cohesion. They also contribute to antimicrobial resistance and host defences mediated by the biofilm, and allow the accumulation of nutrients from the environment and the release of post‐death cellular material. The presence of extracellular DNA in the matrix may facilitate horizontal gene transfer (HGT) between biofilm cells (for a review, see Flemming and Wingender, 2010).
Nucleic acids
Extracellular DNA (but apparently not RNA) and extracytoplasmic and surface‐exposed proteins appear to be critical elements of the matrix required for the initial attachment and maintenance of pneumococcal biofilms. Significant inhibitory and disintegrating effects on pneumococcal biofilms are seen when DNase I or proteases is added before or after biofilm development respectively (Fig. 2) (Moscoso et al., 2006). These findings have been independently confirmed (Hall‐Stoodley et al., 2008; Carrolo et al., 2010). It has recently been proposed that the spontaneous induction of temperate bacteriophages might constitute an important source of extracellular DNA for the pneumococcal biofilm matrix (Carrolo et al., 2010). This agrees with an early report showing that lysogenized S. pneumoniae strains are better biofilm formers than the corresponding cured strains (Loeffler and Fischetti, 2006). Other authors, however, report that DNase treatment does not significantly affect biofilm formation in vitro and suggest that DNA is likely not an essential constituent of biofilms formed under the experimental conditions they used (Muñoz‐Elías et al., 2008). It should be mentioned that competence induction and concomitant DNA release in pneumococcus strongly depends on the medium (Moscoso and Claverys, 2004).
Figure 2.

Inhibition of biofilm development in S. pneumoniae cultures in the presence of nucleases or proteases. A. S. pneumoniae R6 was distributed in the wells of a microtitre plate, which was then incubated for 6 h at 34°C (cross‐hatched bars). Other samples received either RNase (stippled bars), DNase I (hatched bars), trypsin (blackened bars) or proteinase K (open bars) at the indicated concentrations and were incubated as above. B. After biofilm development, nucleases or proteases were added at 100 µg ml–1 and incubation allowed for an additional 1 h at 34°C before staining with crystal violet to quantify biofilm formation. Slightly modified and reprinted from Moscoso et al. (2006) with permission.
Extracellular polysaccharides
It is generally thought that polysaccharides make up a major fraction of the extracellular matrix, providing mechanical stability to the biofilm. Extracellular polysaccharides have been classified as capsular polysaccharides (CPS) when closely associated with the cell surface, and exopolysaccharides when loosely associated (Branda et al., 2005). However, this distinction is inappropriate for biofilms because many of the extracellular polysaccharides they contain are insoluble and cannot be easily separated from cells. In fact, the presence of CPS reduces pneumococcal biofilm development; both clinical pneumococcal isolates and isogenic encapsulated transformants form significantly less biofilm than non‐encapsulated strains (Moscoso et al., 2006). With only one exception (Oggioni et al., 2006) the adherence of non‐encapsulated pneumococcal mutants to human bronchial epithelial cells or to abiotic surfaces is reported more efficient than that of encapsulated parent cells. Thus, non‐encapsulated pneumococci show a high capacity to form in vitro biofilms (Waite et al., 2001; 2003; Allegrucci and Sauer, 2007; Hiller et al., 2010; Camilli et al., 2011). Whether the discrepancy reported by Oggioni and colleagues (2006) is related to the particular conditions these authors used for growing their biofilms, i.e. tryptic soy broth, incubation of microplates in a CO2‐enriched atmosphere, and the addition of competence stimulating peptide, is not known. It has been shown that the emergence of non‐encapsulated genotypic and phenotypic variants enhance S. pneumoniae biofilm development. Different types of mutation (single nucleotide polymorphisms, deletions, tandem sequence duplications, etc.), mainly involving the cap3A/cps3A gene, have been seen among spontaneous capsular mutants of S. pneumoniae type 3 biofilms grown on microtitre plates, Sorbarod filters, flow cells and membrane filters (Waite et al., 2001; Allegrucci and Sauer, 2007; McEllistrem et al., 2007; Domenech et al., 2009). The existence of an inverse relationship between the ability of the non‐encapsulated variants to form biofilms and the amount of CPS has also been found (Domenech et al., 2009) (Fig. 3). Thus, the non‐encapsulated mutants of S. pneumoniae type 3, as good biofilm formers, might be essential in the attachment stage of biofilm formation, and that those variants producing reduced quantities of CPS might only appear in later stages (Allegrucci and Sauer, 2007; Domenech et al., 2009). These results are in keeping with the proposal that pneumococci regulate capsule expression in the transition from nasopharyngeal carriage associated with biofilm development to invasive disease (Waite et al., 2003), as recently shown in Neisseria meningitidis (O'Dwyer et al., 2009). In addition to one study showing that phenotypic variation of the polysaccharide capsule occurs in the initial phase of pneumococcal infections (Hammerschmidt et al., 2005), real‐time quantitative PCR results have indicated that cpsA, the first gene of the pneumococcal capsule operon, is downregulated (by up to 10‐fold) during biofilm growth compared with that seen in planktonic cultures (Hall‐Stoodley et al., 2008). Further, in situ capsule immunofluorescence staining is brighter in biofilm towers of encapsulated S. pneumoniae strains that in adherent cells, suggesting that surface‐attached pneumococci have a reduced amount of capsule (Hall‐Stoodley et al., 2008). It should be noted here that the factors involved in the regulation of S. pneumoniae CPS biosynthesis remain essentially unknown (Moscoso and García, 2009).
Figure 3.

Colony morphology and biofilm‐forming capacity of a type 3 encapsulated S. pneumoniae strain and of non‐encapsulated mutants appearing in biofilm‐grown cultures. A. Colonies of the encapsulated M23 strain and of several non‐mucous colonies of different morphology (indicated by arrows or triangles). Bar, 2.5 mm. B and C. CLSM of the biofilms formed by the M23 strain and one of the non‐encapsulated mutants respectively. Bar, 20 µm. Reprinted from Domenech et al. (2009) with permission.
Several methods have been used to try to identify the EPS putatively forming the structural scaffolding of the pneumococcal biofilm matrix. In one experiment, real‐time monitoring of S. pneumoniae in a biofilm reactor system led to the spectroscopic detection and quantification of proteins and polysaccharides during biofilm formation (Donlan et al., 2004). In addition, ‘EPS clouds’ were observed in some thick biofilm areas after staining with fluorescently labelled wheat germ agglutinin, suggesting that N‐acetylglucosamine residues are one of the biofilm matrix components. However, a clinical, encapsulated pneumococcal isolate was used and its serotype not specified (Donlan et al., 2004). Thus, whether the N‐acetylglucosamine residues belong to a previously unidentified polysaccharide or to the cell wall peptidoglycan and/or the CPS remains to be determined. More recently, strain‐related variability in the EPS distribution of pneumococcal biofilms was demonstrated using a cocktail of five fluorescently conjugated lectins (Hall‐Stoodley et al., 2008). However, further experiments are required to determine the composition and distribution of the carbohydrate(s) in the matrix.
The use of calcofluor white M2R to stain non‐encapsulated pneumococcal cells has revealed that only biofilm‐growing cells (Fig. 4), but not planktonic cells (not shown), were able to bind calcofluor. This indicates that S. pneumoniae biofilms are composed of aggregates of microbial cells encased in an extracellular polysaccharide matrix (different to the CPS) that contains – at least –β‐linked d‐glycopyranosyl units (M. Domenech, M. Moscoso, E. García, in preparation), because calcofluor white M2R is a compound that binds to β‐1,3 and β‐1,4 polysaccharides (Harrington and Hageage, 2003).
Figure 4.
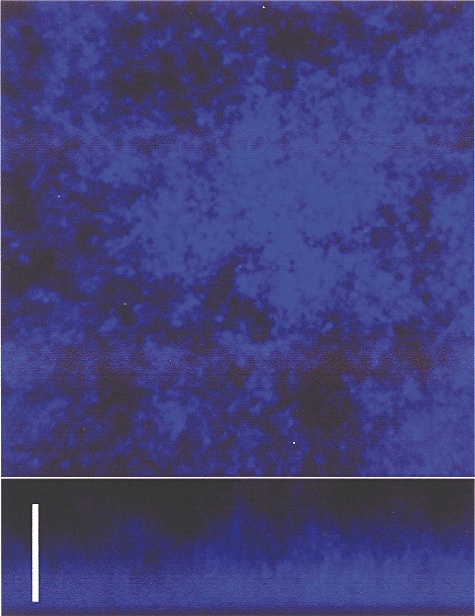
CLSM of the biofilm formed by the S. pneumoniae R6 strain stained with calcofluor white. Bar, 25 µm.
Gene expression patterns and protein production
It is well known that bacteria growing in biofilms show physiological and metabolic differences to their planktonic counterparts. The different stages of biofilm development, such as initial attachment and biofilm maturation, likely require the expression of genes different to those expressed by planktonic cells. Gene expression patterns of S. pneumoniae strain TIGR4 recovered from the tissues of mice with pneumonia or meningitis are similar to that of pneumococci growing in biofilms for nearly all the genes studied (Oggioni et al., 2006). An increase in the expression of neuraminidase‐coding genes (nanA/SP_1693 and nanB/SP_1687), competence genes (comA/SP_0042 and comX/SP_0014) and the virulence gene regulator mgrA/SP_1800 in lung and brain tissue isolates and biofilm bacteria was also reported. These authors also assert that sessile cells grown in a biofilm were more effective at inducing meningitis and pneumonia than planktonic cells (Oggioni et al., 2006). Certainly it has been reported that biofilm formation occurs at a slightly higher frequency (P = 0.04) among S. pneumoniae isolated from respiratory samples provided by patients with cystic fibrosis than among those from blood provided by subjects without cystic fibrosis (García‐Castillo et al., 2007). However, a correlation between the ability to form in vitro biofilms and the origin of pneumococcal isolates (either from the nasopharynx, middle‐ear effusion or blood) (Tapiainen et al., 2010), or the clinical presentation of pneumococcal disease (Lizcano et al., 2010), does not appear to exist.
Proteomic studies have revealed an increase in the number of proteins synthesized de novo and differences in protein production patterns over the course of S. pneumoniae biofilm development (Allegrucci et al., 2006). A number of proteins differentially produced during biofilm development were identified by mass spectrometry as proteins involved in virulence, adhesion and resistance. Pneumolysin and pyruvate oxidase, two proteins associated with virulence, were the most abundant S. pneumoniae serotype 3 proteins obtained from 3‐ and 6‐day‐old biofilms. A discrepancy noted in the overexpression of pneumolysin in pneumococcal biofilms, possibly due to the production methods used, has been discussed elsewhere (Moscoso et al., 2009). In addition, high concentrations of the α‐subunit of ATP synthetase F1, of fructose‐stimulated pyruvate kinase and of several surface‐associated proteins (such as enolase, peptide methionine sulfoxide reductase MsrA and glyceraldehyde‐3‐phosphate dehydrogenase) were found after 3 days of biofilm growth (Allegrucci et al., 2006). Enzymes involved in glycolysis, gluconeogenesis and starch metabolism, such as NADP‐specific glutamate dehydrogenase, glucose‐6‐phosphate isomerase and phosphoglycerate kinase, were the most abundant under planktonic growth conditions (Allegrucci et al., 2006).
Muñoz‐Elías and colleagues (2008) used a collection of transposon insertion S. pneumoniae mutants to identify pneumococcal genes required for the initiation of biofilm development, and, in some cases, for the nasopharyngeal colonization of mice. The ability of these mutants to form biofilms was determined by their attachment to polystyrene plates. Mutations in the genes coding for the choline‐binding proteins (lytC/SP_1593, cbpA/SP_2190, cbpF/SP_0391), the neuraminidases (nanB/SP_1687), a putative cardiolipin synthase (SP_0199), the synthases of membrane and cell wall components (fibA/SP_0615, murE/SP_1531, murB/SP_1390), the ABC and PTS transporters (aliB/SP_1527, SP_1682, SP_0137), the proteolytic and ATP‐binding subunits of the Clp proteases family (clpP/SP_0746, clpX/SP_1569, clpC/SP_1294), the components of the shikimate pathway for synthesis of isochorismate (aroK/SP_1370, SP_1745), and other conserved proteins of unknown function, all contributed towards biofilm formation (Muñoz‐Elías et al., 2008). Moreover, the RrgA subunit of the pili (which are present in some but not all pneumococcal strains), but not the pilus structure per se, was reported to function as an adhesin in biofilm formation. Two genes involved in signal transduction (i.e. SP_2192 and ciaH/SP_0799) were also found to affect biofilm growth, as were two insertions in putative transcriptional regulators (SP_2131, LacR2/SP_1182), which led to biofilm hyperformation (Muñoz‐Elías et al., 2008; Trappetti et al., 2011b). In addition, pneumococcal rgg mutants, which are deficient in the putative transcriptional regulator Rgg, are reported more susceptible to oxidative stress and to show a reduced ability to form biofilms (Bortoni et al., 2009). The latter authors found this mutant to be sensitive to oxygen and paraquat, but not to H2O2. Interestingly, as mentioned above, they also described a role for pyruvate oxidase SpxB and its product, hydrogen peroxide, in the emergence of biofilm‐derived variants of S. pneumoniae type 19.
The noticeable differences in the subset of biofilm‐related genes identified by mutagenic approaches and proteomic analysis may be due to differences in the sensitivity of these systems, in the levels of transcription and translation, the genetic background of the strains used and/or the biofilm model used.
The roles of pneumococcal surface proteins in biofilm formation have been investigated, especially of those involved in nasopharyngeal colonization and/or adherence to the host cell (Hammerschmidt, 2006). Choline‐binding proteins (López and García, 2004; López et al., 2004), which bind the choline residues in cell wall teichoic acids, cell wall hydrolase LytA (the major autolysin), LytB (a glucosaminidase involved in daughter cell separation) and LytC (a lysozyme acting as an autolysin at 30°C) were all shown to contribute to S. pneumoniae biofilm formation by non‐encapsulated strains. Moreover, the inactivation of the genes coding for pneumococcal surface protein A or of the putative adhesins PcpA and CbpA leads to reduced biofilm formation on polystyrene plates (Moscoso et al., 2006). Although the implication of CbpA in biofilm formation has been confirmed in non‐encapsulated laboratory mutants, cpbA mutants in an encapsulated background showed levels of biofilm formation comparable with that of the parental wild‐type strain (Muñoz‐Elías et al., 2008; Lizcano et al., 2010). The reasons for the discrepancies between encapsulated and non‐encapsulated strains remain unclear.
The choline residues in cell wall teichoic acids were found to play an essential role in pneumococcal biofilm development when, after incubating pneumococci in the presence of high concentrations of choline or ethanolamine – at which some choline‐binding proteins are inhibited or released from the surface of pneumococcal cells (López et al., 2004) – a notable reduction in biofilm formation was observed (Moscoso et al., 2006). In addition, Trappetti and colleagues (2011b) recently proposed that the lic operon involved in choline metabolism (Hakenbeck et al., 2009) also contributes to the formation of the matrix.
Roles for neuraminidase NanA and the pneumococcal serine‐rich repeat protein PsrP in biofilm maturation have also been proposed. Using a modified in vitro biofilm model in which pneumococci had previously interacted with human airway epithelial cells, it was shown that NanA neuraminidase, which is conserved in all pneumococcal strains tested, albeit with a high level of diversity (King et al., 2005), releases terminal sialic acid residues from glycoconjugates, thus contributing to biofilm formation (Parker et al., 2009). Similarly, the Basic Region domain of PspR, located in a pathogenicity island present in a number of strains of S. pneumoniae, is reported involved in mature biofilm formation and promotes the formation of bacterial aggregates in the nasopharynx and lungs of infected mice (Sanchez et al., 2010).
Prevention of biofilm formation and therapy
One of the most important and persistent problems posed by biofilms is the inherent tolerance of their associated communities to antibiotic therapy and host defence mechanisms. Different strategies have been developed for the prevention and treatment of biofilm‐related infections, such as the use of enzymes that degrade the biofilm matrix, inhibitors of quorum‐sensing signals, antimicrobial and anticoagulant agents, surfactants and specific bacteriophages (Kaplan, 2010). Many bacteriophages produce depolymerases, i.e. enzymes that hydrolyse the polysaccharides of the biofilm matrix. The topical application of a mixture of phages on the surface of medical devices to prevent biofilm formation has also been proposed (Azeredo and Sutherland, 2008; Fu et al., 2010). In addition, phages (or their depolymerases) followed by disinfectants may be more effective in the control of biofilm formation than either alone (Flemming and Wingender, 2010).
The literature only contains a few reports on the activity of antibiotics against pneumococcal biofilms, and the data are inconclusive. No protection against the activity of benzylpenicillin, ampicillin, amoxicillin‐clavulanic acid or cefuroxime was found when a penicillin‐susceptible S. pneumoniae isolate was grown as a biofilm on Sorbarod filters (Budhani and Struthers, 1997). However, it has been reported that amoxicillin, erythromycin and levofloxacin at supra‐MIC concentrations are less active against biofilm‐associated pneumococci than against planktonic cells (del Prado et al., 2010), and it has been suggested that isolates forming biofilms from patients with cystic fibrosis may have become highly adapted to the presence of antibiotics such as penicillin and tetracycline (García‐Castillo et al., 2007). In addition, cefditoren, an oral third‐generation cephalosporin, better interferes with S. pneumoniae biofilm development than does amoxicillin‐clavulanic acid (Maestre et al., 2010). Interestingly, moxifloxacin, a fourth‐generation oral fluoroquinolone, can inhibit the formation of, and indeed disrupt, biofilms produced by respiratory pathogens, including S. pneumoniae, at concentrations easily achieved in the bronchial mucosa (Roveta et al., 2007).
The information on the effect of other agents such as N‐acetyl‐l‐cysteine (NAC) or xylitol on pneumococcal biofilm formation is also limited. NAC, a thiol‐containing antioxidant that disrupts disulfide bonds in mucus, is used in the treatment of chronic bronchitis, cancer and paracetamol poisoning, but it also has antibacterial properties. Contrary to previous findings with staphylococci (O'Dwyer et al., 2009), it has been reported that NAC alone has very little activity against either planktonic (MIC 4–10 mg ml–1) or biofilm‐grown clinical S. pneumoniae isolates (del Prado et al., 2010). Combining NAC with amoxicillin, erythromycin and/or levofloxacin barely enhanced the antibacterial activity of either compound. However, intramuscular‐to‐aerosol sequential therapy using NAC plus thiamphenicol had a noticeable clinical success rate (84–100%) in patients with recurrent rhinosinusitis and other upper respiratory tract infections in which biofilms have been proven present (Macchi et al., 2006). At our laboratory, NAC inhibited the formation a biofilm by a non‐encapsulated S. pneumoniae strain and partially disintegrated a previously formed biofilm at concentrations around the MIC (Fig. 5A and B). As discussed elsewhere (Riise et al., 2000), the concentrations of NAC that showed inhibition in these assays came close to that which theoretically can be obtained in oropharyngeal secretions when a normal dose of oral NAC medication is administered.
Figure 5.
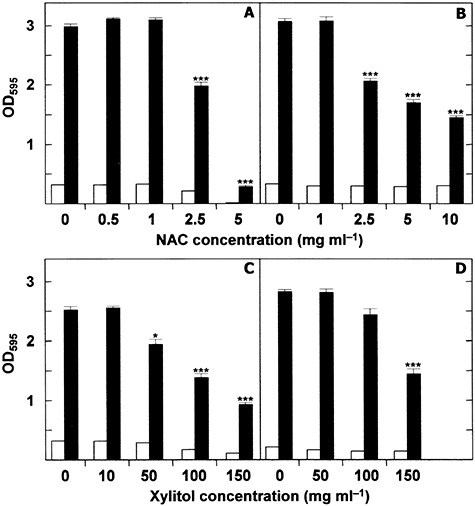
Inhibition of biofilm development and biofilm dispersal in S. pneumoniae cultures in the presence of N‐acetyl‐cysteine or xylitol. A. S. pneumoniae R6 was distributed in the wells of a microtitre plate, which was then incubated for 6 h at 34°C in the presence of different concentrations of NAC. B. S. pneumoniae R6 was distributed in the wells of a microtitre plate, which was then incubated for 6 h at 34°C. Biofilms were washed with fresh CpH 8 medium and incubated with NAC for 2 h at 37°C. C and D. As in (A) and (B) respectively, but with xylitol instead of NAC. Biofilm formation was quantified by staining with crystal violet. Open and blackened bars indicate growth and biofilm formation respectively. In all panels the results represent the mean ± standard error of at least four independent experiments, each performed in triplicate. Asterisk‐marked results are statistically significant (*P < 0.05; ***P < 0.001) compared with the control.
Xylitol, which is non‐fermentable by oral bacteria, is known to inhibit growth, metabolism and polysaccharide production in cariogenic Streptococcus mutans. In addition to growth inhibition, the reduction of insoluble extracellular polysaccharides is probably important in xylitol‐induced reductions of both bacterial numbers and transmission (Söderling, 2009). Xylitol may also be useful for the prophylaxis of acute otitis media in children, although in tests it did not reduce the nasopharyngeal carriage of pneumococci (reviewed by Danhauer et al., 2010). When clinical S. pneumoniae isolates were incubated with xylitol at concentrations of up to 50 mg ml–1 the inhibition of biofilm formation was reported as either insignificant (Ruiz et al., 2011) or only small (Kurola et al., 2011). In contrast, when the growth medium was supplemented with xylitol plus glucose or fructose (5 mg ml–1), biofilm formation was enhanced (Kurola et al., 2011). A significant reduction in biofilm formation by non‐encapsulated, laboratory strains of S. pneumoniae has been observed in our laboratory at concentrations of ≥ 50 mg ml–1) (Fig. 5C). However, biofilm disintegration was not observed when xylitol was added at concentrations of < 50 mg ml–1 (Fig. 5D).
Ceragenin CSA‐13, a cholic acid derivative that mimics the activity of antimicrobial peptides, is capable of actively destroying the biofilms formed by S. pneumoniae (M.M. Esteban, M. Moscoso, E. García, in preparation), in a manner similar to that previously reported for young and mature Pseudomonas aeruginosa biofilms (Nagant et al., 2010).
An alternative strategy for eradicating pneumococcal biofilms is the use of cell wall hydrolases encoded by S. pneumoniae and its phages. The antimicrobial activity of these enzymes on S. pneumoniae planktonic cultures, as well as its therapeutic effects in animal models, have been reported (reviewed by Hermoso et al., 2007). Recently, promising results regarding the destruction of pneumococcal biofilms in vitro have been obtained (M. Domenech, E. García, M. Moscoso submitted).
Future perspectives
Although great advances in understanding S. pneumoniae biofilm development and biofilm‐related infections have been made in the last 5 years, many basic aspects of biofilm formation remain to be investigated. The genetic grounds upon which pneumococcal biofilm formation and maturation is based are still largely unknown, probably because the genes essential for biofilm formation are yet to be identified. Further, the diversity of the polysaccharide and protein components of the pneumococcal biofilm matrix still needs to be characterized. Future studies should investigate the molecular mechanisms underlying the regulation of the synthesis and/or degradation of the matrix.
Upper respiratory tract infections are caused by the synergistic and/or antagonistic interactions between the commensal microbiota, respiratory viruses and potential pathogens such as S. pneumoniae, Haemophilus influenzae or Moraxella catarrhalis (Murphy et al., 2009; Laufer et al., 2011). However, most of our current knowledge about biofilm‐related infections is derived from monospecific studies. It would be very interesting to use simple continuous culture biofilm systems to investigate the potential indirect pathogenicity of mixed S. pneumoniae and M. catarrhalis infections (Budhani and Struthers, 1998) and to study bacterial interactions under, for example, antibiotic stress. It has recently been reported that the presence of H. influenzae increases pneumococcal biofilm formation in vitro as well as the persistence of pneumococci on the mucosal surface of the middle ear (Weimer et al., 2010). Non‐typeable H. influenzae appear to provide passive protection against pneumococcus in the chinchilla model through two mechanisms: the production of β‐lactamase and the formation of biofilm communities (Weimer et al., 2011). Studying interspecies interactions in biofilms may be a new way to gain insight into the events underlying the formation and maintenance of mixed biofilms and pneumococcal disease such as otitis media, pneumonia and meningitis. Moreover, it would be interesting to test the capacity of S. pneumoniae to form biofilms, ideally using more than one model system. In this context, efforts to develop in vivo models of pneumococcal biofilms may represent an important technical step forward (Chaney et al., 2011).
Many bacteria use intercellular, cell density‐dependent communication systems (quorum sensing systems) to coordinate the expression of genes involved in the regulation of their interactions with one another and their environment. Information on the communication systems used by S. pneumoniae is limited. It has been shown that the induction of the pneumococcal competence system by competence‐stimulating (quorum sensing) peptide (CSP) promotes stable biofilm formation in vitro (Oggioni et al., 2006), although its impact varies depending on the experimental biofilm model used (Trappetti et al., 2011a). Therefore, in the search for new therapies, the inter‐ and intracellular signals that regulate the formation and/or dispersal of S. pneumoniae biofilms need to be identified.
Few studies have documented strategies to prevent biofilm development in human infections and any benefits it might bring. Given the important role of NanA neuraminidase in biofilm formation and nasopharyngeal colonization, efforts must be made to identify inhibitors targeting pneumococcal neuraminidase. Competition experiments using neuraminidase inhibitors have been performed (Trappetti et al., 2009), and the in silico docking studies reported by Parker and colleagues (2009) look promising. These authors identified a potent inhibitor of NanA neuraminidase activity (known as XX1) that acts at concentrations in the low‐micromolar range. It also inhibits biofilm formation. Studies to characterize the dispersal mechanisms of S. pneumoniae biofilms would also help in the development of agents that promote their eradication. Finally, biofilms show resistance to host phagocytic defences (Bryers, 2008), but at the moment this has received little research attention in S. pneumoniae. Most of our information on the immune response to bacteria has been obtained using planktonic cultures; studies focused on the interaction between biofilm‐associated pneumococci and the host immune system are therefore necessary.
The available data clearly show that many genes fulfilling quite diverse functions are involved in the formation and dispersal of S. pneumoniae biofilms. It is currently recognized that pneumococcus is a genetically diverse species capable of evolving over short‐time scales, mainly by intra‐ and inter‐species HGT (Donati et al., 2010). Biofilms provide the ideal environment for facilitating HGT in S. pneumoniae as this naturally transformable bacterium may easily encounter extracellular DNA that forms part of the biofilm matrix (Moscoso et al., 2006). Ehrlich and colleagues (2010) indicated that to be capable of promoting HGT, biofilms should be polyclonal in nature, being formed by different strains of the same species and/or different species. The real‐time in vivo generation of pneumococcal genetic diversity has recently been documented (Hiller et al., 2010). Over a period of 7 months, a high degree of HGT was found within the S. pneumoniae strains isolated from a child suffering from chronic upper respiratory and middle‐ear infections. Interestingly, sequencing showed four of the six isolates to have non‐typeable genomes, which were all capable of forming more biofilm than their encapsulated progenitor(s) (Hiller et al., 2010). This appears to be important for the establishment and maintenance of chronic otitis media. Given the enormous variability seen among S. pneumoniae isolates, studies on pneumococcal biofilms should be performed with strains differing in just one gene (or only a few) for reliable conclusions to be drawn.
Acknowledgments
We thank R. López and P. García for helpful comments and for critically reading the manuscript, and A. Burton for revising the English version. Partial support for this research was provided by Grant SAF2009‐10824 from the Dirección General de Investigación Científica y Técnica of Spain. Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CIBERES) is an initiative of the ISCIII. M.D. was supported by an FPI fellowship from the Spanish Ministerio de Ciencia e Innovación.
References
- Allegrucci M., Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M., Sauer K. Formation of Streptococcus pneumoniae non‐phase‐variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol. 2008;190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M., Hu F.Z., Shen K., Hayes J., Ehrlich G.D., Post J.C., Sauer K. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeredo J., Sutherland I.W. The use of phages for the removal of infectious biofilms. Curr Pharm Biotechnol. 2008;9:261–266. doi: 10.2174/138920108785161604. [DOI] [PubMed] [Google Scholar]
- Bortoni M.E., Terra V.S., Hinds J., Andrew P.W., Yesilkaya H. The pneumococcal response to oxidative stress includes a role for Rgg. Microbiology. 2009;155:4123–4134. doi: 10.1099/mic.0.028282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S.S., Vik Å., Friedman L., Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Bryers J.D. Medical biofilms. Biotechnol Bioeng. 2008;100:1–18. doi: 10.1002/bit.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhani R.K., Struthers J.K. The use of Sorbarod biofilms to study the antimicrobial susceptibility of a strain of Streptococcus pneumoniae. J Antimicrob Chemother. 1997;40:601–602. doi: 10.1093/jac/40.4.601. [DOI] [PubMed] [Google Scholar]
- Budhani R.K., Struthers J.K. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of β‐lactamase‐producing moraxellae by use of a continuous‐culture biofilm system. Antimicrob Agents Chemother. 1998;42:2521–2526. doi: 10.1128/aac.42.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli R., Pantosti A., Baldassarri L. Contribution of serotype and genetic background to biofilm formation by Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis. 2011;30:97–102. doi: 10.1007/s10096-010-1060-6. [DOI] [PubMed] [Google Scholar]
- Carrolo M., Frias M.J., Pinto F.R., Melo‐Cristino J., Ramirez M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE. 2010;5:e15678. doi: 10.1371/journal.pone.0015678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney E.J., Nguyen C.T., Boppart S.A. Novel method for non‐invasive induction of a middle‐ear biofilm in the rat. Vaccine. 2011;29:1628–1633. doi: 10.1016/j.vaccine.2010.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates H., Thornton R., Langlands J., Filion P., Keil A.D., Vijayasekaran S., Richmond P. The role of chronic infection in children with otitis media with effusion: evidence for intracellular persistence of bacteria. Otolaryngol Head Neck Surg. 2008;138:778–781. doi: 10.1016/j.otohns.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin‐Scott H.M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Danhauer J.L., Johnson C.E., Corbin N.E., Bruccheri K.G. Xylitol as a prophylaxis for acute otitis media: systematic review. Int J Audiol. 2010;49:754–761. doi: 10.3109/14992027.2010.493897. [DOI] [PubMed] [Google Scholar]
- Domenech M., García E., Moscoso M. Versatility of the capsular genes during biofilm formation by Streptococcus pneumoniae. Environ Microbiol. 2009;11:2542–2555. doi: 10.1111/j.1462-2920.2009.01979.x. [DOI] [PubMed] [Google Scholar]
- Donati C., Hiller N.L., Tettelin H., Muzzi A., Croucher N., Angiuoli S. Structure and dynamics of the pan‐genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R.M., Piede J.A., Heyes C.D., Sanii L., Murga R., Edmonds P. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol. 2004;70:4980–4988. doi: 10.1128/AEM.70.8.4980-4988.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich G.D., Ahmed A., Earl J., Hiller N.L., Costerton J.W., Stoodley P. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol Med Microbiol. 2010;59:269–279. doi: 10.1111/j.1574-695X.2010.00704.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.‐C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fu W., Forster T., Mayer O., Curtin J.J., Lehman S.M., Donlan R.M. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010;54:397–404. doi: 10.1128/AAC.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Castillo M., Morosini M.I., Valverde A., Almaraz F., Baquero F., Cantón R., del Campo R. Differences in biofilm development and antibiotic susceptibility among Streptococcus pneumoniae isolates from cystic fibrosis samples and blood cultures. J Antimicrob Chemother. 2007;59:301–304. doi: 10.1093/jac/dkl482. [DOI] [PubMed] [Google Scholar]
- Goeres D.M., Loetterle L.R., Hamilton M.A., Murga R., Kirby D.W., Donlan R.M. Statistical assessment of a laboratory method for growing biofilms. Microbiology. 2005;151:757–762. doi: 10.1099/mic.0.27709-0. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R., Madhour A., Denapaite D., Brückner R. Versatility of choline metabolism and choline‐binding proteins in Streptococcus pneumoniae and commensal streptococci. FEMS Microbiol Rev. 2009;33:572–586. doi: 10.1111/j.1574-6976.2009.00172.x. [DOI] [PubMed] [Google Scholar]
- Hall‐Stoodley L., Hu F.Z., Gieseke A., Nistico L., Nguyen D., Hayes J. Direct detection of bacterial biofilms on the middle‐ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall‐Stoodley L., Nistico L., Sambanthamoorthy K., Dice B., Nguyen D., Mershon W.J. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt S. Adherence molecules of pathogenic pneumococci. Curr Opin Microbiol. 2006;9:12–20. doi: 10.1016/j.mib.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt S., Wolff S., Hocke A., Rosseau S., Müller E., Rohde M. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun. 2005;73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington B.J., Hageage G.J. Calcofluor white: a review of its uses and applications in clinical mycology and parasitology. Lab Med. 2003;34:361–367. [Google Scholar]
- Hermoso J.A., García J.L., García P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hiller N.L., Ahmed A., Powell E., Martin D.P., Eutsey R., Earl J. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 2010;6:e1001108. doi: 10.1371/journal.ppat.1001108. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa M., Tomovic S., Nistico L., Hall‐Stoodley L., Stoodley P., Sachdeva L. Identification of adenoid biofilms with middle ear pathogens in otitis‐prone children utilizing SEM and FISH. Int J Pediatr Otorhinolaryngol. 2009;73:1242–1248. doi: 10.1016/j.ijporl.2009.05.016. et al. [DOI] [PubMed] [Google Scholar]
- Jensen P.Ø, Givskov M., Bjarnsholt T., Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- Kaplan J.B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.J., Whatmore A.M., Dowson C.G. NanA, a neuraminidase from Streptococcus pneumoniae, shows high levels of sequence diversity, at least in part through recombination with Streptococcus oralis. J Bacteriol. 2005;187:5376–5386. doi: 10.1128/JB.187.15.5376-5386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurola P., Tapiainen T., Sevander J., Kaijalainen T., Leinonen M., Uhari M., Saukkoriipi A. Effect of xylitol and other carbon sources on Streptococcus pneumoniae biofilm formation and gene expression in vitro. APMIS. 2011;119:135–142. doi: 10.1111/j.1600-0463.2010.02703.x. [DOI] [PubMed] [Google Scholar]
- Laufer A.S., Metlay J.P., Gent J.F., Fennie K.P., Kong Y., Pettigrew M.M. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011;2:e00245–10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- Lizcano A., Chin T., Sauer K., Tuomanen E.I., Orihuela C.J. Early biofilm formation on microtiter plates is not correlated with the invasive disease potential of Streptococcus pneumoniae. Microb Pathog. 2010;48:124–130. doi: 10.1016/j.micpath.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler J.M., Fischetti V.A. Lysogeny of Streptococcus pneumoniae with MM1 phage: improved adherence and other phenotypic changes. Infect Immun. 2006;74:4486–4495. doi: 10.1128/IAI.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López R., García E. Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol Rev. 2004;28:553–580. doi: 10.1016/j.femsre.2004.05.002. [DOI] [PubMed] [Google Scholar]
- López R., García E., García P., García J.L. Cell wall hydrolases. In: Tuomanen E.I., Mitchell T.J., Morrison D.A., Spratt B.G., editors. ASM Press; 2004. pp. 75–88. [Google Scholar]
- Macchi A., Ardito F., Marchese A., Schito G.C., Fadda G. Efficacy of N‐acetyl‐cysteine in combination with thiamphenicol in sequential (intramuscular/aerosol) therapy of upper respiratory tract infections even when sustained by bacterial biofilms. J Chemother. 2006;18:507–513. doi: 10.1179/joc.2006.18.5.507. [DOI] [PubMed] [Google Scholar]
- McEllistrem M.C., Ransford J.V., Khan S.A. Characterization of in vitro biofilm‐associated pneumococcal phase variants of a clinically relevant serotype 3 clone. J Clin Microbiol. 2007;45:97–101. doi: 10.1128/JCM.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre J.R., Mateo M., Méndez M.L., Aguilar L., Gimenez M.J., Alou L. In vitro interference of β‐lactams with biofilm development by prevalent community respiratory tract isolates. Int J Antimicrob Agents. 2010;35:274–277. doi: 10.1016/j.ijantimicag.2009.10.020. et al. [DOI] [PubMed] [Google Scholar]
- Moscoso M., Claverys J.P. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol Microbiol. 2004;54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- Moscoso M., García E. Transcriptional regulation of the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae: a bioinformatic analysis. DNA Res. 2009;16:177–186. doi: 10.1093/dnares/dsp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso M., García E., López R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso M., García E., López R. Pneumococcal biofilms. Int Microbiol. 2009;12:77–85. [PubMed] [Google Scholar]
- Muñoz‐Elías E.J., Marcano J., Camilli A. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun. 2008;76:5049–5061. doi: 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.F., Bakaletz L.O., Smeesters P.R. Microbial interactions in the respiratory tract. Pediatr Infect Dis J. 2009;28:S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- Nagant C., Tré‐Hardy M., El‐Ouaaliti M., Savage P., Devleeschouwer M., Dehaye J.P. Interaction between tobramycin and CSA‐13 on clinical isolates of Pseudomonas aeruginosa in a model of young and mature biofilms. Appl Microbiol Biotechnol. 2010;88:251–263. doi: 10.1007/s00253-010-2748-3. [DOI] [PubMed] [Google Scholar]
- Nistico L., Kreft R., Gieseke A., Coticchia J.M., Burrows A., Khampang P. Adenoid reservoir for pathogenic biofilm bacteria. J Clin Microbiol. 2011;49:1411–1420. doi: 10.1128/JCM.00756-10. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dwyer C.A., Li M.‐S., Langford P.R., Kroll J.S. Meningococcal biofilm growth on an abiotic surface – a model for epithelial colonization? Microbiology. 2009;155:1940–1952. doi: 10.1099/mic.0.026559-0. [DOI] [PubMed] [Google Scholar]
- O'Toole G.A., Kolter R. Initiation of biofilms formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Oggioni M.R., Trappetti C., Kadioglu A., Cassone M., Iannelli F., Ricci S. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol. 2006;61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Soong G., Planet P., Brower J., Ratner A.J., Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun. 2009;77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone C.D., Park S., Imlay J.A., Weiser J.N. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol. 2003;185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Prado G., Ruiz V., Naves P., Rodríguez‐Cerrato V., Soriano F., Ponte M.C. Biofilm formation by Streptococcus pneumoniae strains and effects of human serum albumin, ibuprofen, N‐acetyl‐L‐cysteine, amoxicillin, erythromycin, and levofloxacin. Diagn Microbiol Infect Dis. 2010;67:311–318. doi: 10.1016/j.diagmicrobio.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Riise G.C., Qvarfordt I., Larsson S., Eliasson V., Andersson B.A. Inhibitory effect of N‐acetylcysteine on adherence of Streptococcus pneumoniae and Haemophilus influenzae to human oropharyngeal epithelial cells in vitro. Respiration. 2000;67:552–558. doi: 10.1159/000067473. [DOI] [PubMed] [Google Scholar]
- Roveta S., Schito A.M., Marchese A., Schito G.C. Activity of moxifloxacin on biofilms produced in vitro by bacterial pathogens involved in acute exacerbations of chronic bronchitis. Int J Antimicrob Agents. 2007;30:415–421. doi: 10.1016/j.ijantimicag.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Ruiz V., Rodríguez‐Cerrato V., Huelves L., del Prado G., Naves P., Ponte C., Soriano F. Adherence of Streptococcus pneumoniae to polystyrene plates and epithelial cells and the antiadhesive potential of albumin and xylitol. Pediatr Res. 2011;69:23–27. doi: 10.1203/PDR.0b013e3181fed2b0. [DOI] [PubMed] [Google Scholar]
- Sanchez C.J., Shivshankar P., Stol K., Trakhtenbroit S., Sullam P.M., Sauer K. The pneumococcal serine‐rich repeat protein is an intra‐species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson A.R., Leid J.G., Hunsaker D. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope. 2006;116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- Schaudinn C., Stoodley P., Kainović A., O'Keeffe T., Costerton B., Robinson D. Bacterial biofilms, other structures seen as mainstream concepts. Microbe. 2007;2:231–237. et al. [Google Scholar]
- Söderling E.M. Xylitol, mutans streptococci, and dental plaque. Adv Dent Res. 2009;21:74–78. doi: 10.1177/0895937409335642. [DOI] [PubMed] [Google Scholar]
- Stoodley P., Dodds I., Boyle J.D., Lappin‐Scott H.M. Influence of hydrodynamics and nutrients on biofilm structure. J Appl Microbiol. 1999;85:19S–28S. doi: 10.1111/j.1365-2672.1998.tb05279.x. [DOI] [PubMed] [Google Scholar]
- Tapiainen T., Kujala T., Kaijalainen T., Ikäheimo I., Saukkoriipi A., Renko M. Biofilm formation by Streptococcus pneumoniae isolates from paediatric patients. APMIS. 2010;118:255–260. doi: 10.1111/j.1600-0463.2010.02587.x. et al. [DOI] [PubMed] [Google Scholar]
- Trappetti C., Kadioglu A., Carter M., Hayre J., Iannelli F., Pozzi G. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis. 2009;199:1497–1505. doi: 10.1086/598483. et al. [DOI] [PubMed] [Google Scholar]
- Trappetti C., Gualdi L., Di Meola L., Jain P., Korir C., Edmonds P. The impact of the competence quorum sensing system on Streptococcus pneumoniae biofilms varies depending on the experimental model. BMC Microbiol. 2011a;11:75. doi: 10.1186/1471-2180-11-75. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappetti C., Ogunniyi A.D., Oggioni M.R., Paton J.C. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS ONE. 2011b;6:e19844. doi: 10.1371/journal.pone.0019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite R.D., Struthers J.K., Dowson C.G. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high‐frequency phase variation. Mol Microbiol. 2001;42:1223–1232. doi: 10.1046/j.1365-2958.2001.02674.x. [DOI] [PubMed] [Google Scholar]
- Waite R.D., Penfold D.W., Struthers J.K., Dowson C.G. Spontaneous sequence duplications within capsule genes cap8E and tts control phase variation in Streptococcus pneumoniae serotypes 8 and 37. Microbiology. 2003;149:497–504. doi: 10.1099/mic.0.26011-0. [DOI] [PubMed] [Google Scholar]
- Weimer K.E., Armbruster C.E., Juneau R.A., Hong W., Pang B., Swords W.E. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis. 2010;202:1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer K.E., Juneau R.A., Murrah K.A., Pang B., Armbruster C.E., Richardson S.H., Swords W.E. Divergent mechanisms for passive pneumococcal resistance to β‐lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis. 2011;203:549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolcott R.D., Ehrlich G.D. Biofilms and chronic infections. JAMA. 2008;299:2682–2684. doi: 10.1001/jama.299.22.2682. [DOI] [PubMed] [Google Scholar]


