Summary
The two‐component system TmoS/TmoT controls the expression of the toluene‐4‐monooxygenase pathway in Pseudomonas mendocina RK1 via modulation of PtmoX activity. The TmoS/TmoT system belongs to the family of TodS/TodT like proteins. The sensor kinase TmoS is a 108 kDa protein composed of seven different domains. Using isothermal titration calorimetry we show that purified TmoS binds a wide range of aromatic compounds with high affinities. Tightest ligand binding was observed for toluene (KD = 150 nM), which corresponds to the highest affinity measured between an effector and a sensor kinase. Other compounds with affinities in the nanomolar range include benzene, the 3 xylene isomers, styrene, nitrobenzene or p‐chlorotoluene. We demonstrate that only part of the ligands that bind to TmoS increase protein autophosphorylation in vitro and consequently pathway expression in vivo. These compounds are referred to as agonists. Other TmoS ligands, termed antagonists, failed to increase TmoS autophosphorylation, which resulted in their incapacity to stimulate gene expression in vivo. We also show that TmoS saturated with different agonists differs in their autokinase activities. The effector screening of gene expression showed that promoter activity of PtmoX and PtodX (controlled by the TodS/TodT system) is mediated by the same set of 22 compounds. The common structural feature of these compounds is the presence of a single aromatic ring. Among these ligands, toluene was the most potent inducer of both promoter activities. Information on the TmoS/TmoT and TodS/TodT system combined with a sequence analysis of family members permits to identify distinct features that define this protein family.
Introduction
An important mechanism by which environmental signals are sensed and their changes translated into modulatory responses is based on the action of two‐component systems (TCS) (Galperin, 2005; Krell et al., 2010). Bacteria contain a large number of TCSs and were found to harbour on average 52 TCS genes (Cock and Whitworth, 2007). TCSs are involved in the regulation of virtually all types of processes including virulence, sporulation, metabolism, quorum sensing, chemotaxis, transport or nitrogen fixation, which has been reviewed in Krell and colleagues (2010) and Mascher and colleagues (2006). A prototypal TCS contains a sensor kinase (SK) and a response regulator (RR) and its mechanism is based on a simple His to Asp transphosphorylation. Signal recognition by the SK leads to a modulation of its autokinase activity and/or its phosphatase activity towards its cognate RR (Krell et al., 2010). The resulting changes in the SK phosphorylation state reflect on the rate of phosphoryltransfer towards the receiver domain of the RR, which in turn alters the properties of its output domain. This domain has in most cases DNA‐binding properties and modulates gene expression by binding to promoter regions.
During evolution genes of prototypal TCSs have fused to more complex systems referred to as phosphorelay TCS (Whitworth and Cock, 2009), which employ a His1‐Asp1‐His2‐Asp2 phosphorylation cascade. Phosphorelay systems so far studied include for example the ArcBA, TorSR and EvgSA systems of Escherichia coli (Perraud et al., 1999; Malpica et al., 2006), the BvgST system of Bordetella sp. (Beier and Gross, 2008) or the sporulation phosphorelay of Bacillus subtilis (Burbulys et al., 1991). All of these systems were classified as TRPR systems (Fig. 1), implying that the relay involves consecutive phosphorylation of a transmitter module, followed by a first receiver domain, a histidine containing phosphotransfer domain and a second receiver domain (Williams and Whitworth, 2010). A transmitter module is composed of a dimerization and histidine phosphotransfer domain and a catalytic domain that catalyses transmitter module autophosphorylation using ATP as phosphoryldonor. However, genome analyses indicated that there is a different type of phosphorelay system, termed TRTR, which contain two transmitter modules and two receiver domains (Fig. 1). At the genetic level it was shown that TRTR systems are as abundant as the TRPR systems (Williams and Whitworth, 2010). However, at the functional level there is little data available on this type of system.
Figure 1.
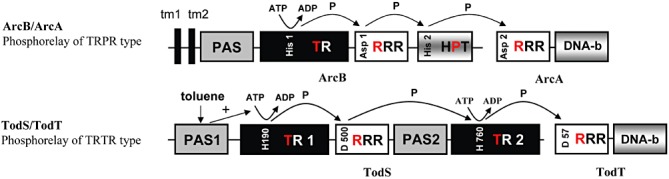
Schematic representation of domain organization and mode of action of phosphorelay two‐component systems. A. The ArcB/ArcA phosphorelay. This system belongs to the TRPR type of phosphorelay according to the classification proposed by Williams and Whitworth (2010). B. The TodS/TodT system that belongs to the TRTR type of phosphorelays. The sequence of phosphoryl group transfer is indicated. tm, transmembrane region; PAS, Per‐Arnt‐Sim type of sensor domain; TR, transmitter module comprised of a dimerization/histidine phosphotransfer domain and a catalytic domain; RRR, response regulator receiver domain; HPT, histidine containing phosphotransfer domain; DNA‐b, DNA‐binding domain.
The TodS/TodT TCS is the first TRTR system that has been functionally characterized (Fig. 1, Lacal et al., 2006; Busch et al., 2007; 2009; Lacal et al., 2008a,b). This system modulates the expression of the tod (toluene dioxygenase) operon encoding the enzymes of the TOD pathway, which permits the mineralization of benzene, toluene and ethylbenzene in strains of Pseudomonas putida (Zylstra and Gibson, 1989; Lau et al., 1997; Mosqueda et al., 1999). Although the TOD pathway uses toluene, benzene and ethylbenzene as substrates, our group was previously able to demonstrate that TodS/TodT mediates regulation of PtodX expression in response to a wide range of different aromatic compounds, including different methyl‐, amino‐, nitro‐ and halogen‐substituted benzene and toluene derivatives (Lacal et al., 2006). The TodS SK is unusually large (108 kDa), lacks transmembrane regions and is entirely located in the cytosol (Lacal et al., 2006). It is composed of two transmitter modules, two PAS domains and an internal RR receiver domain (Fig. 1). Both transmitter modules were found to possess autokinase activity (Busch et al., 2009). Effector molecules, such as toluene, bind to the PAS 1 domain with high affinity (Lacal et al., 2006). Effector binding was found to stimulate exclusively the activity of the N‐terminal transmitter domain (Busch et al., 2009). The phosphoryl group is then transferred to Asp500 of the internal receiver domain and subsequently to the C‐terminal transmitter module (Fig. 1, Busch et al., 2009) before the phosphorylation of Asp57 at the TodT RR (Busch et al., 2009). We have shown that TodT, monomeric in solution, binds to five different sites at the PtodX promoter in a sequential and cooperative manner (Lacal et al., 2008a,b). In addition, we have demonstrated that the activity of TodS/TodT is mediated by agonists and antagonists (Busch et al., 2007). Agonists (for example toluene or m‐xylene) are structural very similar to antagonists (o‐ xylene o o‐chlorotoluene). Both classes of compounds compete for binding at the PAS sensor 1 domain. However, only agonists cause an increase in TodS autophosphorylation activity and consequently induce gene expression. Although antagonists bind to TodS with comparable affinities, these compounds fail to increase TodS autophosphorylation and consequently gene expression (Busch et al., 2007). The physiological relevance of these observations is not understood.
There are a number of other TCSs that share significant similarities in sequence and domain arrangement with TodS/TodT. Members of this family are for example StyS/StyR, involved in styrene degradation (Velasco et al., 1998; O'Leary et al., 2001; Leoni et al., 2003; 2005; Rampioni et al., 2008) or the control of phenylacetyl‐coenzyme A ligase expression (del Peso‐Santos et al., 2008), TutC/TutB for anaerobic toluene degradation in Thauera aromatica (Coschigano and Young, 1997) or TmoS/TmoT involved in toluene degradation in Pseudomonas mendocina (Ramos‐González et al., 2002). Furthermore, genome sequencing projects have revealed that TodS/TodT like proteins are found in other bacteria such as Dechloromonas aromatica (Coates et al., 2001; Salinero et al., 2009) and Methylibium petroleiphilum (Kane et al., 2007) or in nonobligate predator of soil bacteria Cupriavidus necator N‐1 (Poehlein et al., 2011). These systems form thus the family of TodS/TodT like TCS (Fig. S1) and the particular domain arrangement of TodS appears to be a specific feature of this family.
The information available on the remaining TCS of the TodS/TodT family is primarily of genetic or microbiological nature. However, the functional characterization of the TodS/TodT system has revealed a number of intriguing features like: (i) the TodS effector range is much larger than the substrate range, (ii) contrary to most SKs, TodS lacks transmembrane regions and is located in the cytosol, (iii) ligand binding occurs with very high affinity, (iv) effectors can be classified into agonists or antagonists or (v) only agonists and not antagonists increase SK autokinase activity.
To elucidate whether these properties are specific to the TodS/TodT system or correspond to feature common to the protein family, we report here the analysis of a second member of this protein family, the TmoS/TmoT TCS (Ramos‐González et al., 2002). This system regulates the expression of the toluene‐4‐monooxygenase (T4MO) pathway in P. mendocina KR1 (Whited and Gibson, 1991a,b). The T4MO of this strain, the initial pathway enzyme, has been studied extensively over the last decades and is now considered a paradigm for monooxygenases (Mitchell et al., 2002; 2003; Bailey et al., 2008). We would like to note that P. putida DOT‐T1E, which harbours todST, was isolated in Spain (Ramos et al., 1995), whereas P. mendocina KR1 was isolated in Texas (Whited and Gibson, 1991a). The functional analysis of the TmoS/TmoT TCS combined with sequence analysis of family members permit the proposition of common functional features.
Results
Sequence analysis of family members
The sequence alignment of representative members of the TodS/TodT like family is shown in Fig. S1. In this alignment the four amino acids that participate in the TodS/TodT phosphorelay (H190, D500, H760 of TodS, and D57 of TodT, Busch et al., 2009) are strictly conserved. We have also identified phenylalanine 79 of TodS (located in the PAS 1 domain) as essential for effector binding. Mutation of this residue abolished recognition of agonists and antagonists (Busch et al., 2007). In the alignment this residue is also fully conserved. In addition, three other amino acids (F46, I74 and I114) were found to be involved in TodS effector recognition. As shown in Fig. S1 the hydrophobic nature of these amino acids is also conserved. These data are consistent with the notion that other family members also employ a phosphorelay mechanism and that similar effector molecules are recognized by the respective SKs.
Most SKs are either transmembrane (tm) proteins or are membrane anchored via tm regions (Krell et al., 2010). In this respect TodS was found to differ as it had no predicted tm regions, which was then confirmed by the soluble nature of purified TodS (Lacal et al., 2006). Sequences of the remaining family members were submitted to the tm prediction algorithm DAS (Cserzöet al., 1997). This analysis predicted all family members to be devoid of tm regions and thus to be soluble proteins, consistent with a cytosolic location.
The 3D structure of StyR RR from Pseudomonas fluorescens has been solved (Milani et al., 2005). StyR is composed of an N‐terminal RR receiver domain that is connected via a long helical linker to the DNA‐binding domain harbouring a winged helix–turn–helix (HTH) motif. Milani and colleagues (2005) propose a model for the StyR–DNA interaction in which the recognition helix α 8 of the HTH inserts into the major groove of the DNA. According to this model protein–DNA contact is primarily mediated by contacts with helix α 8. The sequence alignment of RRs of this family (Fig. S2) revealed a significant conservation of this helix for which the following consensus sequence can be derived: E‐x‐T‐[I/V]‐KVHRH‐[N/R]‐x‐[M/T]‐K.
The alignment of the promoters PtodX and PtmoX (Fig. S3) show 68% of sequence identity. The binding of TodT to PtodX has been studied extensively for P. putida strains F1 (Lau et al., 1997) and DOT‐T1E (Lacal et al., 2006; 2008a,b). In total five monomers were found to bind to boxes 1–3, of which boxes 1 and 2 are pseudopalindromes whereas box 3 corresponds to a half‐palindrome. In the alignment of promoters PtodX and PtmoX both these binding sites are well conserved and are thus proposed to form TmoT operator sites. In addition, the StyR operator has been experimentally identified for the protein from P. fluorescens[Leoni and colleagues (2005) and Rampioni and colleagues (2008)] and operator sites were predicted for the for Pseudomonas sp. strain Y2 (Velasco et al., 1998). The alignment of either experimentally determined or predicted operator sites of family members is shown in Fig. 2. From this alignment the following consensus can be derived: AAA‐x2‐[AT]‐x2‐GT‐[TA]‐[TC].
Figure 2.

Alignment of operator sites for members of the TodT family.1 Experimentally determined TodT binding sites for P. putida F1 (Lau et al., 1997) and P. putida DOT‐T1E (Lacal et al., 2006).2 Experimentally determined StyR sites for P. fluorescens (Leoni et al., 2005; Rampioni et al., 2008).3 Proposed StyR binding sites for Pseudomonas sp. strain Y2 (Velasco et al., 1998).4 Proposed TmoT binding site. The consensus sequence is indicated.
It has been shown that the systems TodS/TodT, TmoS/TmoT, TutC/TutB and StyS/StyR are involved in the regulation of aromatic hydrocarbon degradation routes. Close TodS and TodT homologues have also been detected in D. aromatica (Salinero et al., 2009) and M. petroleiphilum (Kane et al., 2007) or in the nonobligate predator of soil bacteria C. necator N‐1 (Poehlein et al., 2011). Figure S4 shows the genetic environment of the todS/todT genes in these microorganisms. Interestingly, in all three cases these genes are associated with gene clusters encoding different subunits of either T4MO or methane/phenol/toluene hydroxylase as well as TodX homologues. To obtain initial information as to whether the TodS/TodT like proteins are involved in the regulation of the expression of these catabolic genes, the DNA segment before the initial gene of the catabolic gene were aligned with the well‐studied promoter PtodX of P. putida DOT‐T1E. These segments were in detail (see also Fig. S4) the segments upstream of YP_287018.1 (methane/phenol/tolune hydroxylase of D. aromatica), YP_001020011.1 (toluene monooxygenase α‐subunit of M. petroleiphilum) and tbuX (encoding a TodX homologue) in C. necator. The alignment (Fig. S5) shows significant sequence conservation in D. aromatica and C. necator. In both cases a TodT operator and the IHF binding site appear to be conserved. In D. aromatica the presumed TodT binding site matched the consensus sequence described above. In M. petroleiphilum sequence similarities are less obvious. Taken together, these observations are consistent with the notion that the family of TodS/TodT like proteins controls the expression of aromatic hydrocarbon degradation pathways.
TmoS recognizes a wide range of effectors with very high affinity
To initiate the functional analysis of the TmoS/TmoT TCS, the corresponding genes were cloned into the expression plasmid pET28. Both proteins were expressed in E. coli and then purified by affinity chromatography from the soluble fraction of the cell lysate. Despite significant solvent engineering attempts, purified TmoT could not be stabilized in an active conformation. In contrast active TmoS could be obtained but was found to have a reduced stability. All biochemical studies of this protein had to be conducted on the two days following of its purification. Reduced protein stability was also a characteristic for TodS/TodT and might account for the scarceness of functional studies of TRTR type phosphorelay systems as noted by Williams and Whitworth (2010). All subsequent analyses were conducted using the experimental conditions used for the study of TodS.
Isothermal titration calorimetry (Krell, 2008) experiments were conducted to study the interaction of purified TmoS with effector molecules. For these experiments 14 compounds were chosen, which bind with different affinities to TodS and which had either agonistic or antagonistic effects on its activity (Busch et al., 2007, Table 1). For these experiments purified TmoS was titrated with aliquots of effector molecules and resulting heat changes are measured.
Table 1.
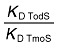
Thermodynamic parameters derived from the microcalorimetric titrations of TmoS with effectors
| Ligand | Effect on TodS | KA (M−1) | KD (µM) | ΔH kcal mol−1 | TΔS kcal mol−1 |
 a a
|
|---|---|---|---|---|---|---|
| Benzene | Agonist | (5.96 ± 0.4) 106 | 0.17 ± 0.01 | −5.3 ± 0.1 | 3.6 ± 0.1 | 4.5 |
| Toluene | Agonist | (6.41 ± 0.5) 106 | 0.15 ± 0.01 | −6.5 ± 0.1 | 2.4 ± 0.1 | 4.6 |
| Ethylbenzene | Agonist | (6.39 ± 0.4) 105 | 1.56 ± 0.1 | −3.5 ± 0.1 | 4.1 ± 0.1 | 2.0 |
| o‐xylene | Antagonist | (3.48 ± 0.1) 106 | 0.29 ± 0.01 | −8.9 ± 0.1 | −0.3 ± 0.1 | 2.0 |
| m‐xylene | Agonist | (1.41 ± 0.1) 106 | 0.71 ± 0.09 | −7.0 ± 0.8 | 1.1 ± 0.6 | 1.7 |
| p‐xylene | Agonist | (2.17 ± 0.3) 106 | 0.46 ± 0.05 | −3.4 ± 0.3 | 4.9 ± 0.29 | 1.7 |
| o‐Chlorotoluene | Antagonist | (4.68 ± 0.9) 105 | 2.14 ± 0.4 | −5.1 ± 0.9 | 2.4 ± 0.9 | 0.3 |
| m‐Chlorotoluene | Agonist | (4.82 ± 0.1) 105 | 2.07 ± 0.6 | −5.9 ± 0.9 | 1.5 ± 0.8 | 4.0 |
| p‐Chlorotoluene | Agonist | (1.24 ± 0.1) 106 | 0.80 ± 0.09 | −2.7 ± 0.1 | 5.3 ± 0.1 | 0.4 |
| Chlorobenzene | Agonist | (7.98 ± 0.4) 105 | 1.25 ± 0.07 | −6.9 ± 0.3 | 0.9 ± 0.3 | 1.0 |
| Nitrobenzene | Agonist | (4.32 ± 0.5) 106 | 0.23 ± 0.03 | −4.1 ± 0.1 | 4.5 ± 0.1 | 29.0 |
| Styrene | Agonist | (1.24 ± 0.2) 106 | 0.81 ± 0.1 | −4.8 ± 0.3 | 3.3 ± 0.3 | 0.7 |
| 1,2,4 Trimethylbenzene | Antagonist | (2.89 ± 0.4) 105 | 3.46 ± 0.5 | −1.5 ± 0.2 | 5.6 ± 0.2 | 0.5 |
| cyclohexane | No binding | No binding | ||||
The KD values for the binding of effectors to TodS are taken from Busch and colleagues (2007).
Figure 3 shows the titration of TmoS with benzene, toluene and ethylbenzene and the derived thermodynamic parameters are given in Table 1. In all cases downwards going peaks are observed indicative of exothermic heat changes implying favourable enthalpy changes. Binding curves for benzene and toluene were very similar and KD values of 170 and 150 nM were determined respectively. These affinities are around 4.5 times higher than the corresponding values obtained for TodS. To our knowledge, these values correspond to the highest affinities observed for an interaction of a SK with effector molecules. Ethylbenzene bound with a weaker affinity of 1.5 µM, which, however, was also tighter than its binding at TodS.
Figure 3.
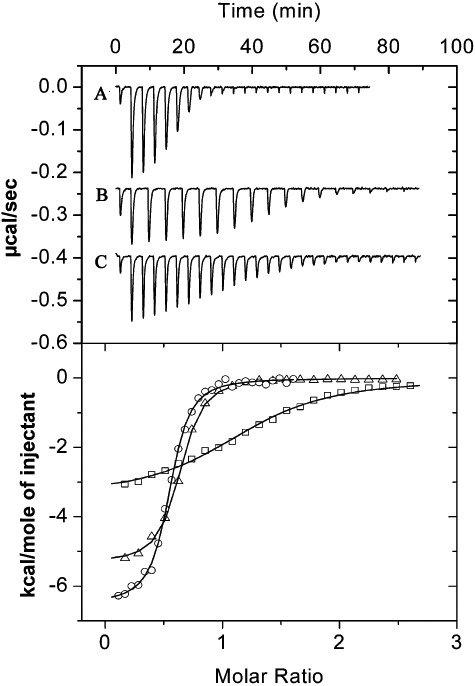
Binding of effector molecules to the purified TmoS. Shown are microcalorimetric titrations of 10 µM TmoS with 500 µM benzene (A), toluene (B) and ethylbenzene (C). Injection volumes were 1.6 µl for (B) and 3.2 µl for (A) and (C). Upper panel: titration raw data. Lower panel: Integrated and dilution‐corrected peak areas of raw data. Data were fitted with the ‘One binding site model’ of the MicroCal version of ORIGIN. ▵: benzene; ο: toluene; □, ethylbenzene. The derived thermodynamic parameters are given in Table 1.
Subsequently the binding of methyl‐ and chloro‐substituted toluene isomers were studied. The three xylene isomers bound with affinities in the range of 290–710 nM, which were again higher than the TodS affinities (Table 1). The three chlorotoluene isomers also bound with high affinity. As in the case of TodS, the para‐substituted isomer bound with highest affinity. The affinities of chlorobenzene and styrene were comparable with the values determined for TodS, whereas nitrobenzene binding to TmoS was 29 times tighter than to TodS. Finally, the triply substituted benzene derivative 1,2,4‐trimethylbenzene also showed tight binding. The predominant binding mode was characterized by favourable enthalpy changes supported by equally favourable entropy changes, which is a binding mode frequently observed for hydrophobic interactions. In analogy to TodS, cyclohexane, the only ligand that lacks an aromatic ring, failed to bind to TmoS. It can be concluded that TmoS recognizes a very wide range of different aromatic effector molecules.
TmoS ligands can be classified into agonists and antagonists
A feature of TodS that remains poorly understood is its differential response to agonists and antagonists (Busch et al., 2007). Agonists, which stimulate kinase activity, increase also gene expression, whereas antagonists, which bind but which do not stimulate kinase activity, fail to upregulate gene expression. The presence of antagonists was shown to reduce the magnitude of agonist‐mediated upregulation of gene expression (Busch et al., 2007).
Subsequent experiments were aimed at identifying whether such behaviour is also observed for TmoS. To this end purified TmoS was subjected to autophosphorylation assays with four TodS agonists (toluene, benzene, chlorobenzene and ethylbenzene) and three antagonists (o‐xylene, o‐chlorotoluene, 1,2,4‐trimethylbenzene). All of these compounds have been found to bind to with affinities between 0.15 and 3.5 µM to TmoS (Table 1). Agonists were chosen to cover the magnitudes of regulatory responses as observed in beta‐galactosidase measurements and include the most efficient agonist (toluene), the weakest agonist (ethylbenzene) and two effectors with an intermediate activity (benzene and chlorobenzene) (Busch et al., 2007). TmoS was incubated during 30 min with [γ32P]ATP in the absence and presence of saturating concentrations of effector molecules prior to SDS‐PAGE. The first lane in Fig. 4A shows the phosphorylation state of TmoS in the absence of effector. Interestingly, the four TodS agonists increased phosphorylation of TmoS. In contrast, the TmoS phosphorylation state in the presence of the three TodS antagonists was very similar to the control. These data indicate that the existence of agonists/antagonists is a feature common to TodS and TmoS.
Figure 4.
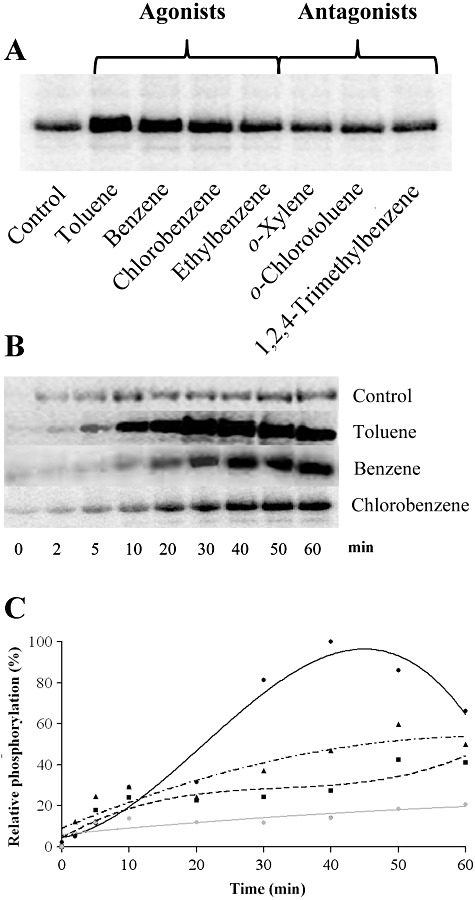
Autophosphorylation of TmoS in the absence and presence of agonists and antagonists. Assays were conducted as described in Experimental procedures. A. Autophosphorylation of TmoS in the absence and presence of 100 µM effector molecules. Autophophorylation reactions were stopped after 30 min. B. Kinetics of autophosphorylation of TmoS in the presence of 100 µM of the effector molecules indicated. C. Densitometric analysis of data presented in B: ●, toluene; ▴, benzene;  , chlorobenzene; grey line: buffer control.
, chlorobenzene; grey line: buffer control.
Differential impact of agonists on the stimulation of TmoS autophosphorylation
A feature that has not been assessed for TodS concerns the question whether sensor protein saturated with different agonists differs in its capacity to stimulate autokinase activity. Fig. 4A also reveals that the amount of autokinase stimulation in the presence of the four agonists differs significantly. Toluene caused the most pronounced increase followed by benzene, chlorobenzene and ethylbenzene. Under the experimental conditions used TmoS is entirely saturated with effector molecules that were added at a concentration of 100 µM, which is largely superior to the respective KD values. To study the effect of toluene, benzene and chlorobenzene on TmoS in more detail, the kinetics of its autophosphorylation was analysed (Fig. 4B and C). In the presence of toluene maximal phosphorylation was observed after 40 min and the reduction in autophosphorylation is due to the depletion of ATP in the reaction mixture and the intrinsic dephosphorylation activity. The stimulation of autokinase activity in the presence of benzene and chlorotoluene was significantly inferior to the experiments conducted with toluene. In both cases a steady increase in TmoS autophosphorylation was noted, which was slightly more pronounced for benzene.
The TmoS/TmoT system activates transcription from PtmoX in response to a wide range of aromatic compounds
To compare the capacities of the TodS/TodT and the TmoS/TmoT systems to induce gene expression in response to a wide range of different aromatic compounds, beta‐galactosidase measurements were conducted. To this end, fusions of promoters PtodX and PtmoX with the lacZ gene were introduced into P. putida DOT‐T1E and P. mendocina KR1 respectively. In total 54 mono‐ and biaromatic compounds were selected for screening. The initial enzymes of both pathways, toluene 2,3‐dioxygenase and T4MO, are characterized by a wide substrate range and most compounds selected are substrate of one or the other enzyme (Gibson and Parales, 2000; Tao et al., 2004a,b,Boyd et al., 2006; Feingersch et al., 2008)
The data initially reported on the PtodX expression were obtained using a protocol in which effector at a final concentration of 1.5 mM is added to the bacterial cultures (Lacal et al., 2006). Initial experiments with P. mendocina KR1 revealed that this effector concentration is toxic for this strain. Therefore, the effector concentration was reduced to 0.5 mM for experiments with P. mendocina KR1, whereas all experiments with P. putida DOT‐T1E were conducted with 1.5 mM effector. It was shown previously that the solvent resistance of strain DOT‐T1E is largely due to the action of the plasmid‐encoded efflux pump TtgGHI (Rojas et al., 2001).
The basal activities of promoters PtodX and PtmoX was of 2 ± 1 and 15 ± 4 Miller units respectively. Effectors that caused activities larger than two times the basal activities were considered as compounds that induce gene expression. From the 54 compounds analysed, 22 compounds induced gene expression in both systems. A plot of the activities of both promoters in response to this set of 22 compounds is shown in Fig. 5. The effector profile of PtodX was only marginally larger than that of PtmoX as only two compounds that were weak inducers of PtodX, namely catechol (13 ± 2 MU) and m‐bromotoluene (9 ± 2 MU), failed to induce PtmoX. The remaining 30 compounds, which are listed in the legend to Fig. 5, neither induced PtodX nor for PtmoX. Among these 30 compounds were o‐xylene, o‐chlorotoluene and 1,2,4‐trimethylbenzene, which were shown to bind tightly to purified TmoS (Table 1) but which failed to increase its phosphorylation state (Fig. 4). To elucidate whether compounds that bind but do not induce gene expression reduce the toluene‐mediated upregulation in gene expression, beta‐galactosidase measurements with mixtures of toluene with o‐xylene or o‐chlorotoluene were conducted. Cultures of P. mendocina KR1 harbouring pMIR38 were grown in Luria–Bertani (LB) to an OD600 of 0.2, at which point 0.25 mM o‐chlorotoluene, o‐xylene or the corresponding volume of buffer was added to three cultures. At an OD600 of 0.5 0.25 mM toluene was added to these cultures and β‐galactosidase activity was measured after another 2 h of growth. Similarly to the analogous experiments reported for the TodS/TodT system (Busch et al., 2007) the presence of o‐xylene and o‐chlorotoluene reduced the toluene mediated gene expression to 55 ± 7% and 32 ± 6%, respectively, of the value obtained for the culture containing toluene only.
Figure 5.
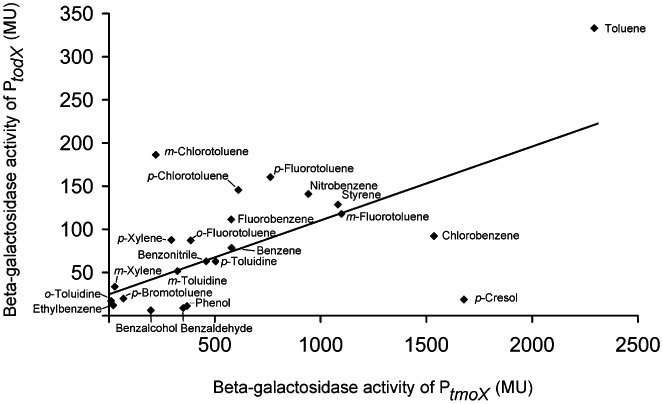
Expression from promoters PtodX and PtmoX. Shown is a plot of beta‐galactosidase activity of PtodX (y‐axis) against the corresponding value of PtmoX (x‐axis) for different compounds. Beta‐galactosidase measurements were carried our as described in Experimental procedures. Note that 1.5 mM of each compound was added to P. putida DOT‐T1E harbouring pMIR77 (PtodX::′lacZ), whereas 0.5 mM of each compound were added to P. mendocina harbouring pMIR38 (PtmoX::'lacZ). Expression in the absence of any added compound was found to be 2 ± 1 and 15 ± 4 MU, respectively, for PtodX and PtmoX. Shown in this graph are the compounds for which an activity of at least twice the basal rate is observed for both promoters. Compounds, which were analysed but which did not induce any of the promoters, are: 1‐hexanol, cyclohexane, propyl‐, butyl‐ and isopropylbenzene, the 3 trimethylbenzene isomers, 1,2,4 trihydroxybenzene, 1,2,4 trichlorobenzene, o‐xylene, o‐chlorotoluene, the 3 iodotoluene isomers, the 3 nitrotoluene isomers, m‐ethyltoluene, benzenesulfonic acid, benzamide, p‐hydroxybenzaldehyde, p‐toluylaldehyde, 2,3 dimethylphenol, resorcinol, hydroquinone, benzoate, p‐hydroxybenzoate, naphthalene, 1,2,3,4 tetrahydroxynaphthalene. Experiments were means of at least three independent experiments conducted in triplicates. The derived standard deviations are in all cases below 25 % of the mean. Part of the measurements of PtodX activity have been reported in Busch et al. (2007).
Apart from the observation that the set of effectors, which induce the expression from both promoters in vivo, is almost identical, several other conclusions can be drawn. First, toluene was the compound that had in both cases the most pronounced effect of gene expression. Toluene is one of the three pathway substrates for the TOD pathway and also the substrate of the T4MO pathway, but it is not known whether the T4MO pathway mineralizes further compounds. However, the case of toluene demonstrates a clear link between the potency in pathway expression and degradability by the corresponding pathway. Second, despite the fact that the effector concentration added to P. mendocina was only a third of the concentration used for P. putida, the induction in gene expression of PtmoX was largely superior to that of PtodX. The data shown in Fig. 5 were fitted by a linear regression model, resulting in the dependency y = 0.085x + 28.0 (r2 = 0.42). This implies that the regulatory response of the PtmoX system is on average around 12 times stronger than that of PtodX. Third, the linear regression shown in Fig. 5 is characterized by an r2 = 0.42, indicative of a modest correlation between both sets of measurements. This is consistent with the idea that the capacity of different compounds to induce gene expression has been conserved during evolution to a certain degree. Fourth, the minimal structural requirement of an effector capable of inducing both promoters appears to be the presence of a single aromatic ring as experiments in the presence of 1‐hexanol, cyclohexane and the biaromatic naphthalene and 1,2,3,4 tetrahydroxynaphthalene showed no promoter activity. Finally, benzene was found to be more efficient than chlorobenzene to increase TmoS autophosphorylation (Fig. 4). This, however, was not reflected in the gene expression studies (Fig. 5) where a superior activity was observed for chlorobenzene. We are currently unable to provide an explanation for this observation. To characterize the dependency of binding affinity of agonists to purified TodS and the induced increase in gene expression, the KA values (Table 1) were plotted against the beta‐galactosidase measurements (Fig. S6). The resulting r2 values of the corresponding linear fits were – 0.26 (for the TodS/T system) and 0.11 (for the TmoS/T system). This indicates that there is only a weak correlation between the binding affinity and the response observed in vivo.
Discussion
The combined interpretation of data available on the TmoS/TmoT and the TodS/TodT system can be used to propose features, which might be common to the protein family. The majority of SKs possess either one or several tm regions (Mascher et al., 2006; Krell et al., 2010). Purified TodS and TmoS are soluble proteins in the absence of detergents. In addition, the prediction of tm regions for the TodS like SKs (Fig. S1) resulted in all cases in an absence of such regions. Taken together, it appears that the family of TodS like SKs forms part of the small group of soluble SKs with cytosolic location (Mascher et al., 2006).
TodS like proteins are involved in the regulation of degradation pathways of toxic compounds. It has been well documented that the major bacterial resistance mechanism towards organic solvents consists in its expulsion into the medium (Ramos et al., 2009). This implies that there are significant differences between the cytosolic and extracytoplasmic concentration of these compounds. In this context the detection of signal molecules in the periplasm might not reflect well the availability of cytosolic pathway substrates and the regulatory events triggered may be of little precision. In contrast, cytosolic substrate sensing reflects well the concentration of available substrates and the resulting responses are more precise. Many soluble cytosolic SKs possess a complex domain arrangement with frequently multiple sensor domains (Krell et al., 2009). All TodS like SK sequences harbour two PAS domains of which the PAS 1 domain was found to sense aromatic effectors in the case of TodS (Lacal et al., 2006). Based on sequence similarities with FixL Lau and colleagues (1997) proposed that the PAS 2 is involved in oxygen sensing. The sensing of a secondary signal molecule present in the cytosol might thus result in a fine‐tuning of the regulatory response.
Another striking feature is the high affinity by which signal molecules are recognized by TmoS and TodS. Because toluene is toxic to the cell at higher concentrations, the degradation pathways have evolved to respond to low substrate concentrations. The kinetic characterization of the initial enzyme of the T4MO pathway, the T4MO, resulted in a KM of 4 µM for toluene (Mitchell et al., 2002). TmoS and TodS bind many agonists with nanomolar affinity. Busch and colleagues (2007) have shown that tight toluene binding translates into an increase in protein autophosphorylation at low toluene concentrations, as evidenced by a half‐maximal autophosphorylation at 10 µM. The discrepancy between the affinity of TodS for toluene (0.7 µM) and the concentration at which half‐maximal autokinase stimulation is achieved (10 µM) shows that initial ligand recognition must not trigger subsequent molecular events in the signalling cascade in an immediate manner. However, data show that transcriptional activation sets in at low toluene concentrations, which in turn permits toluene utilization at sublethal concentrations. The toluene degradation mechanism has co‐evolved with toluene resistance mechanisms. The action of the TtgGHI efflux pump was found to be the primary determinant for toluene resistance (Rojas et al., 2001). Pump expression is under the control of the TtgV repressor, which recognizes toluene and derepresses pump expression (Guazzaroni et al., 2005). However, TtgV was found to bind toluene with a relatively weak affinity of 118 µM (Guazzaroni et al., 2005), which is far above the concentration at which transcriptional activation occurs (Busch et al., 2007).
For most TCSs the cognate signals are unknown (Krell et al., 2010). For some TCS with known signal molecule the corresponding binding constants have been determined. For example, citrate binds to the CitA SK with a KD of 5.5 µM (Kaspar et al., 1999, also determined by ITC), nitrate to NarX with a KD of 35 µM (Lee et al., 1999, as determined by autophosphorylation stimulation), oxygen to FixL with a KD of 50 µM (Gilles‐Gonzalez et al., 1994, determined by spectroscopic techniques) or Mg2+ to PhoQ with a KD of 300 µM (Lesley and Waldburger, 2001, determined by circular dichroism and fluorescence spectroscopy). In this context the affinities of toluene for TmoS (0.15 µM) and TodS (0.69 µM) are the highest affinities reported. We suggest that the toxic nature of these effectors is one reason for the high affinity.
Gene expression studies of promoters PtodX and PtmoX in the presence of 54 different effectors showed that the same set of 22 compounds (agonists) activated transcription in both cases (Fig. 5), whereas 30 other compounds did not induce any of the two promoters. The magnitude of transcriptional activation of these 22 compounds appeared to correlate for both promoters because the linear regression showed a r2 of 0.42. In both cases toluene was the most potent inducer. The common structural feature of these 22 activating compounds was the presence of a single aromatic ring. We have shown previously that phenylalanine 79 is essential for effector binding as its mutation abolished effector recognition (Busch et al., 2007). Effector molecules are likely to interact via pi stacking with this residue. Interestingly, this phenylalanine is conserved among all family members (Fig. S1). However, many of the 22 agonists share striking structural similarities with the 30 compounds that did not induce transcription. In both cases the m‐ and p‐isomers of xylene and chlorotoluene induced transcription whereas both o‐isomers were inactive. We show that o‐xylene and o‐chlorotoluene bind to TmoS with affinities comparable with the other isomers (Table 1). However, the binding of these compounds did not trigger an increase in TmoS autophosphorylation. The data presented thus suggest that the existence of agonists and antagonists is not a feature specific to TodS but might correspond to a characteristic of the entire family. Environmental pollutants can be understood as a mixture of agonists and antagonists. Bacteria exposed to complex mixtures were found to express degradation pathways inefficiently (Cases and de Lorenzo, 2005), which might be partly due to the combined action of agonists and antagonists.
We have shown that effector binding to TodS and TmoS can either cause an agonistic or antagonistic effect. In this work we have also compared the magnitudes of the effects of the agonists toluene, benzene and chlorobenzene on the TmoS phosphorylation state (Fig. 4). The measurement of protein autophosphorylation under conditions guaranteeing complete saturation with effectors showed significant differences (Fig. 4). Exposure to toluene caused the most pronounced increase, followed by benzene and chlorobenzene. Therefore, TmoS effector molecules can be classified into agonists and antagonists and, in addition, agonists differ in their capacity to increase TmoS gene expression. This might be due to different binding modes of agonists to the effector binding pocket, which has multidrug binding properties. Similar observations have been made for the binding of aromatic compounds to TtgV (Guazzaroni et al., 2007). In summary, data presented here permit to initiate the definition of a family of TCS and advance the understanding of the so far poorly characterized phosphorelay systems of the type TRTR. The work provides also important insight into entirely cytosolic TCS, for which only very scarce information is available.
Experimental procedures
Strains and plasmids used in this study
The strains and plasmids used are summarized in Table 2.
Table 2.
Strains and plasmids used
| Strain/plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| E. coli BL21 (DE3) | F‐, ompI, hsdSB (r‐B m‐B) | Studier and Moffat (1986) |
| P. putida DOT‐T1E | Prototroph, Tol+ (tod pathway) | Ramos and colleagues (1995) |
| P. mendocina KR1 | Prototroph, Tol+ (tmo pathway) | Whited and Gibson (1991a) |
| Plasmids | ||
| pMIR77 | TcR, PtodX ::′lacZ | Ramos‐González and colleagues (2002) |
| pMIR38 | TcR, PtmoX::′lacZ | Ramos‐González and colleagues (2002) |
| pMAX‐47‐2 | GmR, derivative of pBBR1MCS‐5 containing tmoST genes | Ramos‐González and colleagues (2002) |
| pET28b | Protein expression plasmid | Novagen |
| pET28b‐TmoS | pET28b containing tmoS | This work |
| pET28b‐TmoT | pET28b containing tmoT | This work |
Construction of TmoS and TmoT expression plasmid
The tmoS and tmoT genes were cloned into the expression vector pET28b+ (Novagen). The DNA fragment of tmoS was amplified by PCR using the primers 5′‐CTTGTGAGTCATTCCATATGAGCTCCTTGG‐3′ and 5′‐ATTCATGTGCTGGGATCCTAACTGACCGG‐3′ and plasmid pMAX‐47‐2 as template (Ramos‐González et al., 2002). The tmoT fragment was amplified using the primers 5′‐TTAGGACGCCAGCATATGAATGATCAAGAG‐3′ and 5′‐GCCAAAAGCAGGATCCTATTTCAGACTATCCT‐3′ and genomic DNA of P. mendocina KR1 as template. The resulting PCR products were digested with NdeI and BamHI and cloned into pET28b(+) (Novagen) linearized with the same enzymes. The plasmids pET28b‐TmoS and pET28b‐TmoT were verified by sequencing the inserts and flanking regions.
Overexpression and purification of TmoS
Escherichia coli BL21 (DE3) was transformed with plasmid pET28b‐TmoS. Cultures were grown in 2l Erlenmeyer flasks containing 500 ml of LB medium supplemented with 50 µg ml−1 kanamycin at 30°C. At an OD660 of 0.6, protein expression was induced by the addition of 0.1 mM IPTG. Growth was continued at 16°C overnight before cell harvest by centrifugation at 10 000 g for 30 min. Cell pellets were resuspended in buffer A [20 mM Tris, 0.1 mM EDTA, 500 mM NaCl, 10 mM imidazole, 5 mM β‐mercaptoethanol, and 5% (vol/vol) glycerol, pH 8.0] and broken using a French press at 1000 psi. After centrifugation at 20 000 g for 1 h, the supernatant was loaded onto a 5 ml HisTrap column (Amersham Bioscience), washed with 10 column volumes of buffer A and eluted with an imidazole gradient of 45–500 mM in buffer A. Fractions containing TmoS were dialysed against analysis buffer 50 mM Tris, 200 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, and 10% (vol/vol) glycerol, pH 7.5 for immediate analysis. Purified protein had to be analysed within 2 days of this dialysis. Protein freezing led to complete loss of activity.
Isothermal titration calorimetry
ITC experiments were conducted using freshly purified protein and a VP‐microcalorimeter (Microcal, Amherst, MA). Protein was dialysed into analysis buffer (50 mM Tris‐HCl, 200 mM KCl, 2 mM MgCl2, 2 mM DTT, 0.1 mM EDTA, 10% glycerol, pH 7.5) and placed into the sample cell. Typically, 10 µM TmoS was titrated with 500 µM effector solutions. Because these compounds are volatile and hydrophobic, these solutions were made up in glass vessels immediately before use. The mean enthalpies measured from the injection of effectors into the buffer were subtracted from raw titration data before data analysis with the MicroCal version of ORIGIN. Data were fitted with the ‘One binding site model’.
Phosphorylation assay of TmoS
Assays were done in 50 mM Tris‐HCl, 200 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 10% (vol/vol) glycerol, 2 mM DTT, pH 7.5. The autophosphorylation assay was performed at 4°C with 10 µM purified TmoS in a final reaction volume of 100 µl in the presence or absence of different effectors at a concentration of 100 µM. Reactions were initiated by adding radiolabelled ATP (200 µM ATP containing 4 µCi [γ32P]ATP), and samples were removed at different times. The reaction was stopped by adding 4 × SDS sample buffer. Samples were then stored on ice before analysis by SDS‐PAGE using 7.5% (wt/vol) gels.
Beta‐galactosidase measurements
Pseudomonas putida DOT‐T1E bearing pMIR77 (Ramos‐González et al., 2002, containing a PtodX::lacZ fusion) and P. mendocina KR1 containing pMIR38 (Ramos‐González et al., 2002) were used. Cells were grown overnight on LB medium supplemented with 10 µg ml−1 tetracycline. Cultures were diluted 100 times with the same medium and the different compounds were added at a concentration of 1.5 mM to P. putida cultures and a concentration of 0.5 mM to P. mendocina cultures. When the cultures reached an OD600 of 0.8 ± 0.05, β‐galactosidase activity was determined in permeabilized cells as described in Ramos‐González and colleagues (2002).
Acknowledgments
The work was financed by grants from the BBVA Foundation, the FEDER grants of the Andalusian regional government Junta de Andalucía (Grant P09‐RNM‐4509) and the Spanish Ministry for Science and Innovation (Grant Bio2010‐16937, BIO2006‐05668, BIO2010‐17227). We acknowledge funding for B.H.C. from TUBITAK (The Scientific and Technological Research Council of Turkey, Fellowship 2214‐2007‐1) and EMBO (Fellowship 184.00–2010).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Sequence alignment of TodS like sensor kinases. The following sequences were used for this alignment: TodS of Pseudomonas putida DOT‐T1E; TmoS of P. mendocina KR1, NodV of Cupriavidus necator N‐1; A2SDY5 of Methylibium petroleiphilum PM1; TutC of Thauera aromatica; Q479E0 of Dechloromonas aromatica RCB; StyS of P. fluorescens (StyS Pf) and StyS of Pseudomonas sp. Y2 (StyS Ps). The amino acids that form part of the intra‐TodS phosphorelay (Busch et al., 2009) are highlighted by green arrows. The amino acids that are involved in effector recognition (Busch et al., 2007) are marked with red arrows. Sequences were aligned using the Clustal W multiple sequence alignment algorithm (Thompson et al. (1994) Nucleic Acids Res 22: 4673–4680) of the NPS@ server (http://npsa‐pbil.ibcp.fr/cgi‐bin/npsa_automat.pl?page=/NPSA/npsa_server.html). The GONNET protein weight matrix, a gap opening penalty of 10 and a gap extension penalty of 0.2 were used. Fully conserved amino acids are shown in red, highly conserved residues in green and weakly conserved amino acids in blue.
Fig. S2. Sequence alignment of TodT like response regulators. The following sequences were used for this alignment: TodT of Pseudomonas putida DOT‐T1E; TmoT of P. mendocina KR1, StyR of Pseudomonas sp. Y2 (StyR Ps); StyR of P. fluorescens (StyR Pf); TutB of Thauera aromatica; NodW of Cupriavidus necator N‐1; A2SDY4 of Methylibium petroleiphilum PM1 and Q479E1 of Dechloromonas aromatica RCB. The same alignment procedure as detailed in Fig. S1 was used. The phosphoryl group accepting aspartate in TodS is highlighted by a green arrow. The region shaded in yellow corresponds to the recognition helix of the helix–turn–helix DNA binding motif of the StyR structure (Milani et al., 2005).
Fig. S3. Alignment of the promoters PtodX and PtmoX . The three TodT binding sites and the ‐10 extended region are boxed (Lacal et al., 2008a,b). The transcription start is marked.
Fig. S4. Genetic environment of TodS/TodT homologues in Dechloromonas aromatica, Methylibium petroleiphilum and Cupriavidus necator. Figures were prepared using the genome data bank in ncbi (http://www.ncbi.nlm.nih.gov/genome). In D. aromatica and M. petroleiphilum genes were present on the chromosome. In the case of Cupriavidus necator genes were present on the plasmid pBB1p.
Fig. S5. Alignments of the PtodX promoter of P. putida DOT‐T1E with DNA regions preceding the toluene monooxygenase clusters in Dechloromonas aromatica, Methylibium petroleiphilum and Cupriavidus necator.
Fig. S6. Plot of beta‐galactosidase measurements for the promoter PtodX (A) and PtmoX (B) (taken from Fig. 5) against the association constants determined for the binding of different agonists to purified TodS (A) or TmoS (B). Values for TodS were taken from Busch et al. (2007) and values for TmoS were taken from Table 1 of this work.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bailey L.J., McCoy J.G., Phillips G.N., Jr, Fox B.G. Structural consequences of effector protein complex formation in a diiron hydroxylase. Proc Natl Acad Sci USA. 2008;105:19194–19198. doi: 10.1073/pnas.0807948105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D., Gross R. The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv Exp Med Biol. 2008;631:149–160. doi: 10.1007/978-0-387-78885-2_10. [DOI] [PubMed] [Google Scholar]
- Boyd D.R., Sharma N.D., Bowers N.I., Dalton H., Garrett M.D., Harrison J.S., Sheldrake G.N. Dioxygenase‐catalysed oxidation of disubstituted benzene substrates: benzylic monohydroxylation versus aryl cis‐dihydroxylation and the meta effect. Org Biomol Chem. 2006;4:3343–3349. doi: 10.1039/b608417f. [DOI] [PubMed] [Google Scholar]
- Burbulys D., Trach K.A., Hoch J.A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Busch A., Lacal J., Martos A., Ramos J.L., Krell T. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc Natl Acad Sci USA. 2007;104:13774–13779. doi: 10.1073/pnas.0701547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Guazzaroni M.E., Lacal J., Ramos J.L., Krell T. The sensor kinase TodS operates by a phosphorelay mechanism involving two autokinase domains. J Biol Chem. 2009;284:10353–10360. doi: 10.1074/jbc.M900521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I., de Lorenzo V. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int Microbiol. 2005;8:213–222. [PubMed] [Google Scholar]
- Coates J.D., Chakraborty R., Lack J.G., O'Connor S.M., Cole K.A., Bender K.S., Achenbach L.A. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature. 2001;411:1039–1043. doi: 10.1038/35082545. [DOI] [PubMed] [Google Scholar]
- Cock P.J., Whitworth D.E. Evolution of prokaryotic two‐component system signaling pathways: gene fusions and fissions. Mol Biol Evol. 2007;24:2355–2357. doi: 10.1093/molbev/msm170. [DOI] [PubMed] [Google Scholar]
- Coschigano P.W., Young L.Y. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. Prediction of transmembrane alpha‐helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Feingersch R., Shainsky J., Wood T.K., Fishman A. Protein engineering of toluene monooxygenases for synthesis of chiral sulfoxides. Appl Environ Microbiol. 2008;74:1555–1566. doi: 10.1128/AEM.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y. A census of membrane‐bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.T., Parales R.E. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol. 2000;11:236–243. doi: 10.1016/s0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- Gilles‐Gonzalez M.A., Gonzalez G., Perutz M.F., Kiger L., Marden M.C., Poyart C. Heme‐based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- Guazzaroni M.E., Krell T., Felipe A., Ruiz R., Meng C., Zhang X. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA: evidence from isothermal titration calorimetry. J Biol Chem. 2005;280:20887–20893. doi: 10.1074/jbc.M500783200. et al. [DOI] [PubMed] [Google Scholar]
- Guazzaroni M.E., Gallegos M.T., Ramos J.L., Krell T. Different modes of binding of mono‐ and biaromatic effectors to the transcriptional regulator TtgV: role in differential derepression from its cognate operator. J Biol Chem. 2007;282:16308–16316. doi: 10.1074/jbc.M610032200. [DOI] [PubMed] [Google Scholar]
- Kane S.R., Chakicherla A.Y., Chain P.S., Schmidt R., Shin M.W., Legler T.C. Whole‐genome analysis of the methyl tert‐butyl ether‐degrading beta‐proteobacterium Methylibium petroleiphilum PM1. J Bacteriol. 2007;189:1931–1945. doi: 10.1128/JB.01259-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar S., Perozzo R., Reinelt S., Meyer M., Pfister K., Scapozza L., Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- Krell T. Microcalorimetry: a response to challenges in modern biotechnology. Microb Biotechnol. 2008;1:126–136. doi: 10.1111/j.1751-7915.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell T., Busch A., Lacal J., Silva‐Jiménez H., Ramos J.L. The enigma of cytosolic two‐component systems: a hypothesis. Environ Microbiol Rep. 2009;1:171–176. doi: 10.1111/j.1758-2229.2009.00020.x. [DOI] [PubMed] [Google Scholar]
- Krell T., Lacal J., Busch A., Guazzaroni M.E., Silva H., Ramos J.L. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- Lacal J., Busch A., Guazzaroni M.E., Krell T., Ramos J.L. The TodS/TodT two‐component regulatory system recognizes a wide range of effectors and works with DNA‐bending proteins. Proc Natl Acad Sci USA. 2006;103:8191–8196. doi: 10.1073/pnas.0602902103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal J., Guazzaroni M.E., Busch A., Krell T., Ramos J.L. Hierarchical binding of the TodT response regulator to its multiple recognition sites at the tod pathway operon promoter. J Mol Biol. 2008a;376:325–337. doi: 10.1016/j.jmb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Lacal J., Guazzaroni M.E., Gutiérrez del Arroyo P., Busch A., Vélez M., Krell T., Ramos J.L. Two levels of cooperativeness in the binding of TodT to the tod operon promoter. J Mol Biol. 2008b;384:1037–1047. doi: 10.1016/j.jmb.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Lau P.C., Wang Y., Patel A., Labbé D., Bergeron H., Brousseau R. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.I., Delgado A., Gunsalus R.P. Signal‐dependent phosphorylation of the membrane‐bound NarX two‐component sensor‐transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J Bacteriol. 1999;181:5309–5316. doi: 10.1128/jb.181.17.5309-5316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni L., Ascenzi P., Bocedi A., Rampioni G., Castellini L., Zennaro E. Styrene‐catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA‐binding. Biochem Biophys Res Commun. 2003;303:926–931. doi: 10.1016/s0006-291x(03)00450-9. [DOI] [PubMed] [Google Scholar]
- Leoni L., Rampioni G., Di Stefano V., Zennaro E. Dual role of response regulator StyR in styrene catabolism regulation. Appl Environ Microbiol. 2005;71:5411–5419. doi: 10.1128/AEM.71.9.5411-5419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley J.A., Waldburger C.D. Comparison of the Pseudomonas aeruginosa and Escherichia coli PhoQ sensor domains: evidence for distinct mechanisms of signal detection. J Biol Chem. 2001;276:30827–30833. doi: 10.1074/jbc.M104262200. [DOI] [PubMed] [Google Scholar]
- Malpica R., Sandoval G.R., Rodríguez C., Franco B., Georgellis D. Signaling by the arc two‐component system provides a link between the redox state of the quinine pool and gene expression. Antioxid Redox Signal. 2006;8:781–795. doi: 10.1089/ars.2006.8.781. [DOI] [PubMed] [Google Scholar]
- Mascher T., Helmann J.D., Unden G. Stimulus perception in bacterial signal‐transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani M., Leoni L., Rampioni G., Zennaro E., Ascenzi P., Bolognesi M. An active‐like structure in the unphosphorylated StyR response regulator suggests a phosphorylation‐ dependent allosteric activation mechanism. Structure. 2005;13:1289–1297. doi: 10.1016/j.str.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Mitchell K.H., Studts J.M., Fox B.G. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4‐monooxygenase regiospecificity. Biochemistry. 2002;41:3176–3188. doi: 10.1021/bi012036p. [DOI] [PubMed] [Google Scholar]
- Mitchell K.H., Rogge C.E., Gierahn T., Fox B.G. Insight into the mechanism of aromatic hydroxylation by toluene 4‐monooxygenase by use of specifically deuterated toluene and p‐xylene. Proc Natl Acad Sci USA. 2003;100:3784–3789. doi: 10.1073/pnas.0636619100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueda G., Ramos‐González M.I., Ramos J.L. Toluene metabolism by the solvent‐tolerant Pseudomonas putida DOT‐T1 strain, and its role in solvent impermeabilization. Gene. 1999;232:69–76. doi: 10.1016/s0378-1119(99)00113-4. [DOI] [PubMed] [Google Scholar]
- O'Leary N.D., O'Connor K.E., Duetz W., Dobson A.D. Transcriptional regulation of styrene degradation in Pseudomonas putida CA‐3. Microbiology. 2001;147:973–979. doi: 10.1099/00221287-147-4-973. [DOI] [PubMed] [Google Scholar]
- Perraud A.L., Weiss V., Gross R. Signalling pathways in two‐component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- del Peso‐Santos T., Shingler V., Perera J. The styrene‐responsive StyS/StyR regulation system controls expression of an auxiliary phenylacetyl‐coenzyme A ligase: implications for rapid metabolic coupling of the styrene upper‐ and lower‐degradative pathways. Mol Microbiol. 2008;69:317–330. doi: 10.1111/j.1365-2958.2008.06259.x. [DOI] [PubMed] [Google Scholar]
- Poehlein A., Kusian B., Friedrich B., Daniel R., Bowien B. Complete genome sequence of the type strain Cupriavidus necator N‐1. J Bacteriol. 2011;193:5017. doi: 10.1128/JB.05660-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Huertas M.J., Haïdour A. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J Bacteriol. 1995;177:3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Krell T., Daniels C., Segura A., Duque E. Responses of Pseudomonas to small toxic molecules by a mosaic of domains. Curr Opin Microbiol. 2009;12:215–220. doi: 10.1016/j.mib.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Ramos‐González M.I., Olson M., Gatenby A.A., Mosqueda G., Manzanera M., Campos M.J. Cross‐regulation between a novel two‐component signal transduction system for catabolism of toluene in Pseudomonas mendocina and the TodST system from Pseudomonas putida. J Bacteriol. 2002;184:7062–7067. doi: 10.1128/JB.184.24.7062-7067.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G., Leoni L., Pietrangeli B., Zennaro E. The interplay of StyR and IHF regulates substrate‐dependent induction and carbon catabolite repression of styrene catabolism genes in Pseudomonas fluorescens ST. BMC Microbiol. 2008;8:92. doi: 10.1186/1471-2180-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., Duque E., Mosqueda G., Golden G., Hurtado A., Ramos J.L., Segura A. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT‐T1E. J Bacteriol. 2001;183:3967–3973. doi: 10.1128/JB.183.13.3967-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinero K.K., Keller K., Feil W.S., Feil H., Trong S., Di Bartolo G., Lapidus A. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life‐style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics. 2009;10:351. doi: 10.1186/1471-2164-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Moffat B.A. Use of bacteriophage T7 RNA polymerase to direct selective high‐level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tao Y., Fishman A., Bentley W.E., Wood T.K. Oxidation of benzene to phenol, catechol, and 1,2,3‐trihydroxybenzene by toluene 4‐monooxygenase of Pseudomonas mendocina KR1 and toluene 3‐monooxygenase of Ralstonia pickettii PKO1. Appl Environ Microbiol. 2004a;70:3814–3820. doi: 10.1128/AEM.70.7.3814-3820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Fishman A., Bentley W.E., Wood T.K. Altering toluene 4‐monooxygenase by active‐site engineering for the synthesis of 3‐methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J Bacteriol. 2004b;186:4705–4713. doi: 10.1128/JB.186.14.4705-4713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco A., Alonso S., García J.L., Perera J., Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited G.M., Gibson D.T. Separation and partial characterization of the enzymes of the toluene‐4‐monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol. 1991a;173:3017–3020. doi: 10.1128/jb.173.9.3017-3020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited G.M., Gibson D.T. Toluene‐4‐monooxygenase, a three component enzyme system that catalyzes the oxidation of toluene to p‐cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991b;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth D.E., Cock P.J. Evolution of prokaryotic two‐component systems: insights from comparative genomics. Amino Acids. 2009;37:459–466. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- Williams R.H., Whitworth D.E. The genetic organisation of prokaryotic two‐component system signalling pathways. BMC Genomics. 2010;11:720. doi: 10.1186/1471-2164-11-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G.J., Gibson D.T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence alignment of TodS like sensor kinases. The following sequences were used for this alignment: TodS of Pseudomonas putida DOT‐T1E; TmoS of P. mendocina KR1, NodV of Cupriavidus necator N‐1; A2SDY5 of Methylibium petroleiphilum PM1; TutC of Thauera aromatica; Q479E0 of Dechloromonas aromatica RCB; StyS of P. fluorescens (StyS Pf) and StyS of Pseudomonas sp. Y2 (StyS Ps). The amino acids that form part of the intra‐TodS phosphorelay (Busch et al., 2009) are highlighted by green arrows. The amino acids that are involved in effector recognition (Busch et al., 2007) are marked with red arrows. Sequences were aligned using the Clustal W multiple sequence alignment algorithm (Thompson et al. (1994) Nucleic Acids Res 22: 4673–4680) of the NPS@ server (http://npsa‐pbil.ibcp.fr/cgi‐bin/npsa_automat.pl?page=/NPSA/npsa_server.html). The GONNET protein weight matrix, a gap opening penalty of 10 and a gap extension penalty of 0.2 were used. Fully conserved amino acids are shown in red, highly conserved residues in green and weakly conserved amino acids in blue.
Fig. S2. Sequence alignment of TodT like response regulators. The following sequences were used for this alignment: TodT of Pseudomonas putida DOT‐T1E; TmoT of P. mendocina KR1, StyR of Pseudomonas sp. Y2 (StyR Ps); StyR of P. fluorescens (StyR Pf); TutB of Thauera aromatica; NodW of Cupriavidus necator N‐1; A2SDY4 of Methylibium petroleiphilum PM1 and Q479E1 of Dechloromonas aromatica RCB. The same alignment procedure as detailed in Fig. S1 was used. The phosphoryl group accepting aspartate in TodS is highlighted by a green arrow. The region shaded in yellow corresponds to the recognition helix of the helix–turn–helix DNA binding motif of the StyR structure (Milani et al., 2005).
Fig. S3. Alignment of the promoters PtodX and PtmoX . The three TodT binding sites and the ‐10 extended region are boxed (Lacal et al., 2008a,b). The transcription start is marked.
Fig. S4. Genetic environment of TodS/TodT homologues in Dechloromonas aromatica, Methylibium petroleiphilum and Cupriavidus necator. Figures were prepared using the genome data bank in ncbi (http://www.ncbi.nlm.nih.gov/genome). In D. aromatica and M. petroleiphilum genes were present on the chromosome. In the case of Cupriavidus necator genes were present on the plasmid pBB1p.
Fig. S5. Alignments of the PtodX promoter of P. putida DOT‐T1E with DNA regions preceding the toluene monooxygenase clusters in Dechloromonas aromatica, Methylibium petroleiphilum and Cupriavidus necator.
Fig. S6. Plot of beta‐galactosidase measurements for the promoter PtodX (A) and PtmoX (B) (taken from Fig. 5) against the association constants determined for the binding of different agonists to purified TodS (A) or TmoS (B). Values for TodS were taken from Busch et al. (2007) and values for TmoS were taken from Table 1 of this work.


