Summary
Bbr_0838 from Bifidobacterium breve UCC2003 is predicted to encode a 683 residue membrane protein, containing both a permease domain that displays similarity to transporters belonging to the major facilitator superfamily, as well as a CBS (cystathionine beta synthase) domain. The high level of similarity to bile efflux pumps from other bifidobacteria suggests a significant and general role for Bbr_0838 in bile tolerance. Bbr_0838 transcription was shown to be monocistronic and strongly induced upon exposure to bile. Further analysis delineated the transcriptional start site and the minimal region required for promoter activity and bile regulation. Insertional inactivation of Bbr_0838 in B. breve UCC2003 resulted in a strain, UCC2003:838800, which exhibited reduced survival upon cholate exposure as compared with the parent strain, a phenotype that was reversed when a functional, plasmid‐encoded Bbr_0838 gene was introduced into UCC2003:838800. Transcriptome analysis of UCC2003:838800 grown in the presence or absence of bile demonstrated that transcription of Bbr_0832, which is predicted to encode a macrolide efflux transporter gene, was significantly increased in the presence of bile, representing a likely compensatory mechanism for bile removal in the absence of Bbr_0838. This study represents the first in‐depth analysis of a bile‐inducible locus in bifidobacteria, identifying a key gene relevant for bifidobacterial bile tolerance.
Introduction
Commensal bacteria inhabiting the human gastrointestinal tract have developed strategies to tolerate the presence of deleterious substances of gastric and intestinal origin that include bile. Bile is a mixture of organic and inorganic compounds containing bile salts as the major organic constituents, which are secreted into the duodenum at a concentration ranging from 0.2% to 2% for a given bile salt (Hofman, 1998). These compounds are detergent‐like substances that exert strong antimicrobial activity (Begley et al., 2005), representing a key challenge for enteric bacterial survival.
Members of the genus Bifidobacterium are inhabitants of the human colon (Ventura et al., 2004; 2009). In recent years, scientific evidence supporting the purported health‐promoting effects of bifidobacteria has been increasing. Indeed, positive effects associated with the consumption of certain bifidobacterial strains is not only observed at the intestinal level, but also at systemic level (Lomax and Calder, 2009). This correlation has led to certain Bifidobacterium strains being exploited as probiotics, and being widely included in functional foods (Stanton et al., 2005). For bifidobacteria to establish in the lower gastrointestinal tract they need to survive gastrointestinal transit, and at least transiently colonize the gut. This requirement makes it very important to obtain a fundamental understanding of the molecular mechanisms that confer bile tolerance to bifidobacteria.
Bifidobacterial mechanisms of bile tolerance have received considerable scientific attention and strains with enhanced bile resistance have been obtained by progressive adaptation to increasing concentrations of bile (Margolles et al., 2003; Noriega et al., 2004). Remarkably, some bile‐resistant derivatives displayed cross‐resistance to other environmental stresses (Sánchez et al., 2007a,b; 2008). Among specific bile resistance mechanisms described in enteric bacteria, bile transport and bile salt‐hydrolysing enzymes are the most frequently described (Piddock, 2006; Jones et al., 2008). In this respect multidrug resistance pumps have been demonstrated to play a significant role in bile tolerance (Piddock, 2006). These transporters are usually involved in detoxification of a wide variety of structurally unrelated compounds, and have been a main focus of biomedical research due to their clinical relevance in antibiotic resistance. However, they are ubiquitously present in all organisms and multidrug efflux pumps (COG 0534) are among the gene families that are over‐represented in the gut microbiome (Kurokawa et al., 2007; Qin et al., 2010).
Among Gram‐positive bacteria, bile efflux transporters have been described in species of Lactococcus and Lactobacillus (Yokota et al., 2000; Elkins and Savage, 2003; Zaidi et al., 2008; Pfeiler and Klaenhammer, 2009), including both bile‐inducible and constitutively expressed genes. Relatively few studies to date have focused on multidrug resistance transporters from Bifidobacterium. Transporters conferring antimicrobial resistance, BbmAB and BbmR, have been identified in Bifidobacterium breve (Margolles et al., 2005; 2006), and bile efflux systems, Ctr and BetA, have been described in Bifidobacterium longum (Price et al., 2006; Gueimonde et al., 2009). However, a significant knowledge gap remains with respect to the molecular mechanisms underlying bile‐induced regulation of specific bile protection systems in members of the genus Bifidobacterium, as well as their potential role in gut survival and colonization.
Here, we report on bile‐inducible genes from B. breve UCC2003, with particular focus on Bbr_0838, and its role in bile tolerance. Bile‐induced transcriptional regulation of Bbr_0838 was characterized, while insertional inactivation of Bbr_0838 coupled with microarray and phenotypic analysis demonstrated the importance of Bbr_0838 as a key bile tolerance protein. To the best of our knowledge this work represents the first in depth report on transcriptional regulation of a bile protection system in the genus Bifidobacterium.
Results and discussion
Insertional inactivation of Bbr_0838 causes reduced bile tolerance in B. breve UCC2003
In this study, a bile‐inducible membrane protein belonging to the MFS superfamily, Bbr_0838, which had previously been identified on the B. breve UCC2003 genome (Gueimonde et al., 2009), was further characterized. The Bbr_0838 gene is located upstream of a gene predicted to encode a serine/threonine protein kinase and downstream of a gene encoding a hypothetical protein (Fig. 1A). Bbr_0838 shows extensive homology to transporters from other bifidobacteria. Moreover, comparative analysis of the surrounding areas indicates that extensive reorganization has occurred in this region (Gueimonde et al., 2009). The gene product of Bbr_0838 is a 683 amino acid protein with an estimated molecular mass of approximately 71 kDa. Analysis of this protein sequence at the Expasy proteomic server (Gasteiger et al., 2003) predicts 12 to 14 transmembrane helices, constituting a permease domain (represented by the N‐terminal 417 amino acids), and a large C‐terminal hydrophilic loop that contains a cystathionine beta synthase (CBS) pair domain (represented by the C‐terminal 166 amino acids). A schematic representation of the putative membrane topology according to TMHMM prediction (Krogh et al., 2001) is shown in Fig. 1B.
Figure 1.
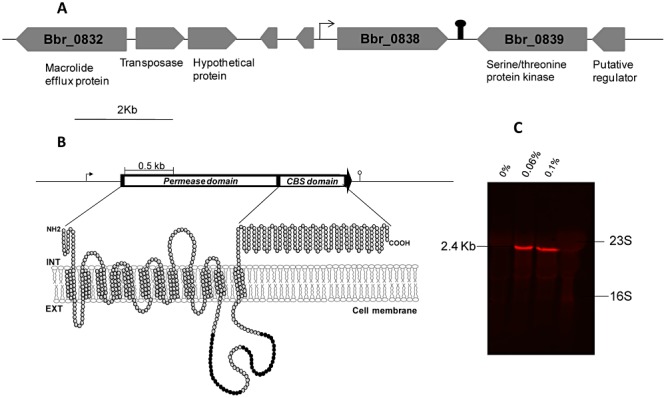
Representation of the Bbr_0838 gene and surrounding region of the B. breve UCC2003 chromosome (A). Schematic representation of the putative membrane topology of Bbr_0838, according to TMHMM prediction. Black circles represent amino acids that constitute additional transmembrane domains predicted by other algorithms (B). Northern hybridization analysis showing induction of monocistronic Bbr_0838 transcription by 0.06% and 0.1% of sodium cholate (C). Pin‐like symbols indicate putative terminators.
The permease domain shares high homology with permeases found in other intestinal microorganisms, suggesting specific relevance of this domain in this particular environment (Gueimonde et al., 2009). CBS domains are widely distributed in all divisions of life and are commonly present as tandem repeats (Ignoul and Eggermont, 2005). CBS domains are frequently found to be associated with transporters and, in some cases, a regulatory role has been described in protein trafficking and activity, through binding of adenosyl ligands or sensing ionic strength (Mahmood et al., 2009), although their precise function remains unknown.
The bile‐inducibility of Bbr_0838 (Fig. 1C) coupled with its homology to reported bile efflux systems in other bifidobacteria (> 98% identity to betA from B. longum NCC2705) suggests a role for Bbr_0838 in supporting B. breve UCC2003 bile tolerance. In support of this theory, our previous work demonstrated the ability of Bbr_0838 to enhance bile survival of recombinant Lactococcus lactis clones expressing the Bbr_0838 (Ruiz et al., 2011). In order to further investigate the physiological role of Bbr_0838 in B. breve, two Bbr_0838 insertion mutant strains designated UCC2003::838800 and UCC2003::8381000, were constructed (see Experimental procedures). Southern hybridization using EcoRI‐digested genomic DNA and pORI19‐8381000‐tetW as a probe unequivocally confirmed that the disruptions of Bbr_0838 were the result of site‐specific homologous recombination events (Fig. 2). Moreover, the stability of the integrated plasmid in UCC2003::838800 and UCC2003::8381000 strains was analysed following 50 generations in the absence of the selective antibiotic, showing that at least 96% of the cells retained the insertion.
Figure 2.
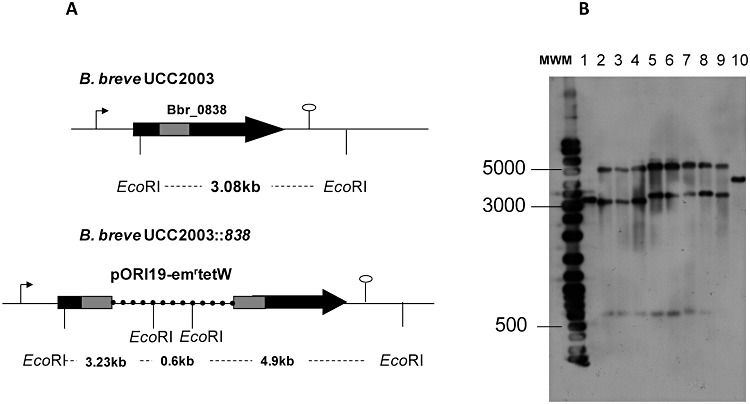
Schematic representation of relevant regions of Bbr_0838 gene in B. breve UCC2003 and B. breve UCC2003::838 chromosomes. A. Chromosomal DNA is represented by a thin line, Bbr_0838 by a black arrow, Bbr_0838 fragment employed for homologous recombination by a grey bar and pORI19 by a dashed line. Representative EcoRI sites for Southern hybridization are indicated. B. Southern hybridization of EcoRI digested chromosomal DNA from B. breve UCC2003 (lane 1) and 8 representative insertional mutants (lanes 2 to 4 correspond to UCC2003::838800 clones and lanes 5 to 9 correspond to UCC2003::8381000clones) are shown in B. Sizes of hybridized fragments are shown at the left of the panel. pORI19‐8381000‐tetW was used as a probe and positive control of hybridization was performed on lane 10.
Growth characteristics of UCC2003::838800 and UCC2003::8381000 under unstressed conditions (i.e. no added bile) showed no difference in growth rate or final optical density (OD) as compared with B. breve UCC2003. In addition, preliminary phenotypic analysis of UCC2003::838800 and UCC2003::8381000 showed that between them they did not exhibit obvious differences with regards to their bile resistance phenotype (data not shown). For this reason, further experiments were performed using B. breve UCC2003::838800.
To examine the effect of the Bbr_0838 mutation on bile tolerance, a series of survival and growth experiments were carried out in MRSc supplemented with ox‐gall, or particular components of bile. The minimal inhibitory concentrations of a complex mixture of bile salts, ox‐gall and individual bile salts, were determined during 24 h growth experiments. The main bile acids in humans are represented by glycocholate and taurocholate, while bile acids resulting from metabolism of bile in the intestine, are represented by cholate, and deoxycholate. Differences in MIC between both strains were only detected in MRSc supplemented with cholate, where a MIC value of 0.2% (w/v) was observed for the wild‐type strain, while the UCC2003::838800mutant was more sensitive with a corresponding MIC value of 0.1% (w/v). MICs were identical for ox‐gall (> 10%), glycocholate (0.25%), taurocholate (> 1%) and glycodeoxycholate (0.625%). This is in accordance with our recent results, in which maximum transcriptional induction levels of Bbr_0838 were observed in the presence of cholate (Ruiz et al., 2011) and were further substantiated by growth experiments. In the presence of 0.025%, 0.05% or 0.1% (w/v) of cholate, UCC2003::838800exhibited slower growth and a lower final OD at 600 nm after 24 h, as compared with UCC2003 (Fig. 3). Enumeration of viable cells in overnight cultures by plate counts and Dead/Live labelling also pointed to reduced growth and cell viability of UCC2003::838800 grown in the presence of cholate (data not shown).
Figure 3.
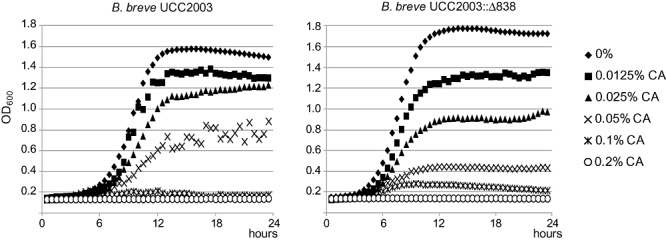
Growth of B. breve UCC2003 and B. breve UCC2003::838800 in medium containing different sodium cholate (CA) concentrations, ranging from 0% to 0.1% (w/v), measured through OD at 600 nm. Data are mean of three independent experiments.
Insertional inactivation of Bbr_0838 results in a strain that has both erythromycin and tetracycline resistance cassettes on the genome. Although a potential role of antibiotic resistance markers in bile tolerance would not be expected an additional alternative insertion mutant, UCC2003::bbr_1552 (M. O'Connell‐Motherway, unpubl. data) in a non‐bile‐inducible gene, encoding a predicted β‐galactosidase (Bbr_1552), was employed as a control for the phenotypic assays. This allowed us to confirm that the observed phenotype of low tolerance to cholate in UCC2003::838800 was not as a consequence of the antibiotic resistance genes carried by the insertion mutant strains (data not shown) but due to the insertional inactivation of Bbr_0838. These observations demonstrated a significant contribution of Bbr_0838 in bile detoxification in B. breve UCC2003, in accordance with previous results from the B. longum homologue, BetA (Gueimonde et al., 2009), and suggest that cholate is the main bile component detoxified by this system.
In order to show that the reduced bile survival phenotype in the insertion mutant was due to the absence of functional Bbr_0838, we complemented UCC2003::838800 strain with a plasmid containing the structural gene. Complementation of UCC2003::838800 with Bbr_0838 expressed from its own promoter on pBC1.2 restored the growth in the presence of cholate and the bile resistant phenotype to levels comparable with wild‐type B. breve UCC2003 (Fig. 4B).
Figure 4.
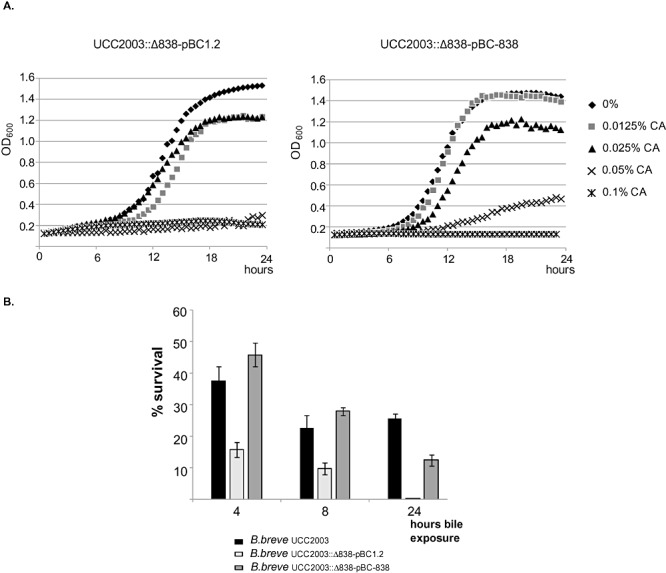
Restoration of wild‐type phenotype in the complemented mutant strain. A. Growth curves in the presence of different cholate concentrations are presented for the complemented strain (UCC2003::838800‐pBC838) and for the mutant strain containing pBC1.2 as a control (UCC2003::838800‐pBC1.2). Presented data are means of three independent experiments. B. Bile survival determined by colony counts of cells exposed to 0.1% sodium cholate.
To date several bile‐transport‐mediated bile‐detoxification systems, displaying different specificities for different bile salts, have been characterized in Gram‐positive microorganisms (Lin et al., 2003; Lubelski et al., 2006; Cerda‐Maira et al., 2008; Watson et al., 2008; 2009; Pfeiler and Klaenhammer, 2009; Quillin et al., 2011). However, this is the first report describing the inactivation of a gene encoding a bile detoxification system in bifidobacteria, and demonstrates the usefulness of this KO generation system as a means to functionally characterize transporters in B. breve, as has been shown previously for other proteins (O'Connell‐Motherway et al., 2008; Pokusaeva et al., 2010, 2011).
In order to determine if other genes were able to at least partially counteract the absence of a functional Bbr_0838 in the presence of bile salts, a comparative transcriptomic analysis of bile stress response in both wild‐type and UCC2003::838800strains was performed. No significant differences were revealed by whole‐genome microarray analysis, although the absence of transcriptional induction of Bbr_0838 was confirmed in the mutant strain. Analysis by q‐RT‐PCR showed that significant differences in bile‐induction levels of selected transporters (previously shown to be induced by bile, Ruiz et al., 2011), were not detected. Nevertheless, significant differences in basal expression levels between the wild‐type and the mutant strains were detected in the particular case of Bbr_0832, which exhibited basal expression levels that were 5.8 times higher in the mutant strain compared with the wild‐type UCC2003 (Fig. 5).
Figure 5.
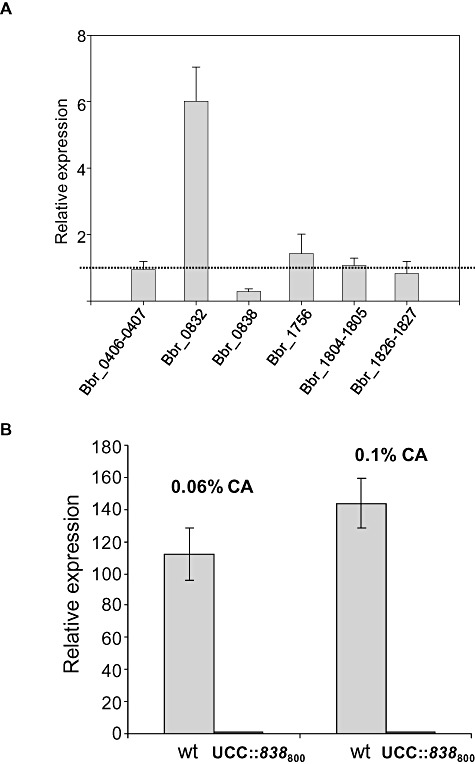
Transcriptional analysis of a selection of bile‐inducible transporters from B. breve UCC2003 genome. Relative expression of UCC2003::838800 mutant in relation to the wild type was analysed under non‐stressed conditions (absence of cholate), measured through q‐RT‐PCR (A) Dashed line indicates similar expression levels in the two compared conditions. B shows the absence of Bbr_0838 induction in UCC2003::838800 under cholate (CA) exposure.
The ORF Bbr_0832 is homologous to genes encoding macrolide efflux transporters, and has been demonstrated to be bile‐inducible (Ruiz et al., 2011). In view of this, we speculate that Bbr_0832 overexpression in the B. breve UCC2003::838800 mutant strain is contributing to some extent to the bile survival phenotype by counteracting the impact of Bbr_0838 inactivation on bile response.
Characterization of the Bbr_0838 promoter and analysis of the region involved in its bile‐inducibility
In order to characterize the Bbr_0838 promoter region and identify regulatory regions responsible for its bile‐inducible behaviour, various transcriptional fusions with gusA were constructed using the promoter probe vector pNZ272 (see Experimental procedures). pNZ272 was originally designed to analyse promoters in Lactobacillus and related microorganisms (Platteeuw et al., 1994) and has previously been successfully used to analyse promoters in B. breve UCC2003 (Ventura et al., 2005). The various fragments used by us to create gusA fusions represented DNA segments encompassing sequences of different length between 579 bp upstream of the start codon and 66 bp of the coding region of Bbr_0838. Bile‐induction of gene expression in the different gusA fusions was determined through measurement of GusA activity following overnight growth in the presence of sub‐inhibitory concentrations of sodium cholate. The region comprising 579 bp upstream from the start codon of Bbr_0838 was determined to be necessary for both efficient mRNA synthesis and bile‐inducibility. Within this region the ribosomal binding site, the transcription start site, the promoter region and also possible binding sites for putative regulatory proteins may be located.
Primer extension analysis on total RNA isolated from cells grown under sub‐inhibitory concentrations of cholate showed that the transcriptional start site (tss) is located at a Guanine residue 375 bp upstream of the start codon of the coding sequence. Sequences representing the putative −10 and −35 regions were located upstream from the tss (Fig. 6). Determination of this transcriptional start site is in accordance with the absence of gene expression in the gusA fusion containing only 162 bp upstream from the start codon (pNZ272‐short), as the transcriptional start site and −10 and −35 hexamers are not encompassed in this construct (Fig. 7).
Figure 6.

Schematic representation of the Bbr_0838 promoter region (B). Relevant regions and localization of primers used in Primer Extension experiment are highlighted. A shows the primer extension results from cultures grown in the absence and presence of sodium cholate.
Figure 7.
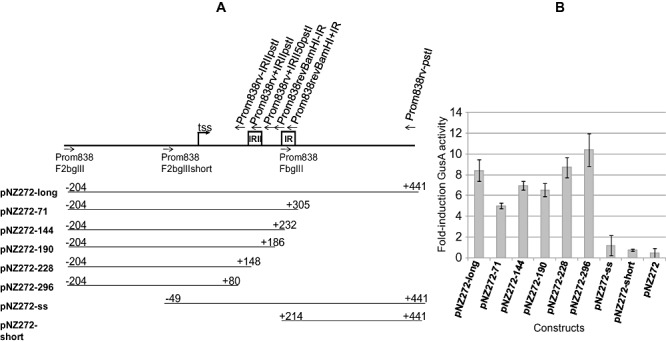
Schematic representation of Bbr_0838 promoter –gusA transcriptional fusions performed in pNZ272 (A) and determination of their inducibility by 0.025% of sodium cholate measured through β‐D‐glucuronidase activity (B).
Bioinformatic analysis of the promoter region showing transcriptional activity and bile‐induced gene expression, revealed two large inverted repeats that may have regulatory roles affecting either transcription or translation of the ORF they precede. Interestingly, the high evolutionary conservation of these repeats in different bifidobacterial species containing gene homologues of Bbr_0838 (i.e. Bifidobacterium bifidum, Bifidobacterium adolescentis and B. longum) might imply an important biological function for this long untranslated RNA segment. In this regard, there is increasing evidence about the role of certain RNA structures, including long 5′ untranslated regions (5′UTR), in transcriptional or translational regulation of gene expression in bacteria (Waters and Storz, 2009). In addition to putative binding sites for regulatory proteins, in some cases 5′UTR regions of mRNA have been reported to autoregulate transcription or translation of the coding region they precede. This is the case of some riboswitches, in which secondary structures of mRNA may act both as target site for certain ligands and transcriptional attenuators. In this way, binding of the specific ligand to the RNA riboswitch may result in structural changes that could turn a gene on or off (Coppins et al., 2007). Furthermore, some UP elements, DNA fragments located upstream from the tss and usually with a high A/T content, have also been involved in transcriptional regulation in Gram‐positive bacteria (Estrem et al., 1998; Ke et al., 2009). To determine the putative role of the large inverted repeats on Bbr_0838 expression, gusA fusions lacking DNA regions of variable length immediately upstream from Bbr_0838, but retaining the region encompassing the presumed −35 and −10 sequences and the transcriptional start site, were constructed and their bile‐inducibility measured through GusA activity (Fig. 7).
The obtained results showed that the DNA region encompassing base pairs 579 and 296 bp upstream from the start codon is sufficient for efficient mRNA synthesis and bile‐induction (construct pNZ272‐296 in Fig. 7). DNA regions located until 296 bp upstream from the start codon of Bbr_0838 do not seem to have any effect on cholate induction, as induction levels in the smaller constructs analysed were similar to the ones measured in the construct containing the whole promoter region (Fig. 7). According to these results, although a role of the DNA sequence comprising the 296 bp immediately upstream from the start codon of Bbr_0838 (and thus the two large inverted repeats detected within) in gene expression regulation may not be ruled out, results from gusA fusions do not show any significance of these sequences in Bbr_0838 bile‐induced expression. In addition, absolute values of GusA activity for constructs lacking the region encompassing both inverted repeats were found to be 9.93 ± 1.65 times lower compared with the construct encompassing the complete UTR, thus suggesting a potential involvement of these regions in mRNA stability. The full extent of the UTR is predicted to adopt secondary structures in RNA, where both inverted repeats significantly contribute to the presumed structure (Fig. S1). The potential involvement of certain secondary RNA structures in contributing to mRNA stability and, consequently affecting efficiency of translation, either protecting RNA against ribonucleases, affecting protein–DNA interactions or avoiding translation initiation has been extensively reported (Diwa et al., 2000; Hambraeus et al., 2002).
An additional gusA fusion in which a 155 bp DNA fragment (comprising nucleotides −50 to −204 upstream from the transcriptional start site) was removed from the promoter sequence, while the 375 bp encompassed among the tss and the start codon in the native sequence and the −10 and −35 hexamers were conserved (pNZ272‐ss), was found to have lost its bile‐inducibility (Fig. 7). From this result it was concluded that bile‐dependent regulatory sequences, possibly targets for a DNA binding regulatory protein, are present within the 155 bp region upstream from the tss. Further research is required to investigate the exact role of this region in controlling Bbr_0838 transcription. Within this DNA segment, an A/T rich region including two short direct repeats were detected, and their role in the regulation of Bbr_0838 expression regulation will be subject to future investigations.
Conclusions
Bile tolerance is an important feature for probiotic bacteria, as it influences their chances of reaching and colonizing the intestine. To date some mechanisms of bile tolerance in the genus Bifidobacterium have been reported, although no detailed information has been described on their genetic regulation. In this work, through construction of an insertional mutant, a bile‐inducible transporter was demonstrated to play a role in bile tolerance of B. breve UCC2003. In addition, preliminary characterization of the Bbr_0838 promoter sequence, revealed a long 5′UTR highly conserved among intestinal bifidobacteria, exerting an efficient, tight and complex regulatory mechanism in which both transcriptional regulators and RNA secondary structures may be involved. The apparent universal distribution of homologues to Bbr_0838 in intestinal bifidobacteria and the extensive homology they share, both in the coding sequence and corresponding promoter regions, seems to reflect the importance of this gene in the intestinal ecosystem. Further detailed investigations to reveal its precise regulatory mechanism(s) and inducing signals, as well as to research additional and apparently compensatory bile resistance genes and mechanisms, are ongoing and no doubt will expand our knowledge on bifidobacterial adaptation to the intestinal environment.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacterium breve UCC2003 was grown at 37°C in MRS broth (Difco, BD Diagnostic Systems, Sparks, MD, USA) supplemented with 0.05% l‐cysteine (w/v) (Sigma, St Louis, MO, USA) (MRSc) in an anaerobic chamber (Mac 500, Don Whitley Scientific, West Yorkshire, UK) with an atmosphere of 5% CO2– 5% H2– 90% N2. Analysis of Bifidobacterium tolerance to toxic compounds was performed by supplementing MRSc before inoculation with different concentrations of ox‐gall (Oxoid Limited, Hampshire, UK), or a particular bile salt (cholate, glycocholate, taurocholate, deoxycholate) supplied by Sigma. The Dead/Live labelling kit was supplied by Invitrogen (Invitrogen, Barcelona, Spain). Growth of Bifidobacterium strains was monitored using a PowerWave Microplate Spectrophotometer (Biotek, UK).
Table 1.
Strains and plasmids used
| Relevant phenotype or genotype | Reference or source | |
|---|---|---|
| Strains | ||
| E. coli EC101 | Cloning host, repA+, Kanr | Law and colleagues (1995) |
| B. breve UCC2003 | Infant isolate | MacConaill and colleagues (2003) |
| B. breve UCC2003::838800 | pORI19‐Bbr_0838 insertional mutant of B. breve UCC2003 | This work |
| B. breve UCC2003::8381000 | pORI19‐Bbr_0838 insertional mutant of B. breve UCC2003 | This work |
| L. lactis NZ9000 | L. lactis MG1363 pepN::nisRK | de Ruyter and colleagues (1996) |
| Plasmids | ||
| pORI19 | Emr RepA‐ Ori+; cloning vector | Law and colleagues (1995) |
| pORI19‐838800‐tetW | Internal 800 bp fragment of Bbr_0838 cloned in pORI19 | This work |
| pORI19‐8381000‐tetW | Internal 1000 bp fragment of Bbr_0838 cloned in pORI19 | This work |
| pNZ272 | Cmr; gusA promoter‐probe vector | Platteeuw and colleagues (1994) |
| pNZ272‐long | Cmr; pNZ272 derivative; containing 0.58 kb promoter region of Bbr_0838 translationally fused to gusA | This work |
| pNZ272‐short | Cmr; pNZ272 derivative | This work |
| pNZ272‐ss | Cmr; pNZ272 derivative | This work |
| pNZ272‐71 | Cmr; pNZ272 derivative | This work |
| pNZ272‐144 | Cmr; pNZ272 derivative | This work |
| pNZ272‐190 | Cmr; pNZ272 derivative | This work |
| pNZ272‐228 | Cmr; pNZ272 derivative | This work |
| pNZ272‐296 | Cmr; pNZ272 derivative | This work |
| pBC1.2 | Ampr; Cmr; E. coli‐Bifidobacterium shuttle cloning vector | Álvarez‐Martín and colleagues (2008) |
| pBC‐838 | pBC1.2 derivative; Bbr_0838 cloned under its native promoter | This work |
Escherichia coli was cultured in Luria–Bertani (LB) broth at 37°C with agitation, and L. lactis was cultivated in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) at 30°C. Where appropriate, the growth medium contained chloramphenicol (Cm; 5 µg ml−1 for L. lactis and 2 µg ml−1 for B. breve), erythromycin (Em; 100 µg ml−1 for E. coli), kanamycin (50 µg ml−1 for E. coli), ampicillin (100 µg ml−1 for E. coli) or tetracycline (10 µg ml−1 for E. coli or B. breve) when required for plasmid maintenance. Recombinant E. coli cells containing pORI19‐838 were selected on LB agar containing Em and supplemented with 40 µg ml−1 of Xgal and 1 mM of IPTG.
Restoration of bile survival phenotype in the complemented mutant strain was analysed by exposing early exponential growth phase cells to different concentrations of cholate in fresh MRSc, incubating samples at 37°C in anaerobic conditions and determining colony counts after several hours of bile exposure.
Nucleotide and protein sequence analysis
Sequence data were obtained from genome annotations of B. breve UCC2003 (O'Connell‐Motherway et al., 2011). Database searches were performed using BLAST at the National Center for Biotechnology Information internet site (‘http://www.ncbi.nlm.nih.gov’) and predictions of protein topology and domains were performed at ExPasy server (http://expasy.org) (Gasteiger et al., 2003).
DNA manipulations
Chromosomal DNA from B. breve UCC2003 was obtained according to previously described procedures (Margolles and de los Reyes‐Gavilán, 2003) and sequences for cloning were amplified from B. breve chromosomal DNA using high‐fidelity Platinum PfX polymerase (Invitrogen). Plasmid minipreparations from E. coli and L. lactis were performed using spin‐in‐column system (Sigma), according to the manufacturer's instructions, with the incorporation of an initial lysis step for L. lactis, involving suspension of the cells in lysis buffer (20% sucrose, 10 mM Tris‐HCl pH 8.1, 10 mM EDTA and 50 mM NaCl) supplemented with lysozyme (10 mg ml−1), followed by incubation at 37°C for 30 min. Plasmid preparations from B. breve were performed according to previously described procedures (O'Connell‐Motherway et al., 2009). Electroporation of plasmid DNA into E. coli was carried out as described by Sambrook and colleagues (1989), into L. lactis as described by Wells and colleagues (1993) and into B. breve UCC2003 was performed as reported by Mazé and colleagues (2007). The absence of PCR‐introduced mutations was verified in all plasmid constructs created in this study by sequencing both DNA strands on an ABI Prism sequencer (Applied Biosystems, Foster City, CA, USA). Southern blot transfer of EcoRI‐digested DNA from B. breve UCC2003 and insertion mutants was performed according to standard procedures (Sambrook et al., 1989). Hybridization and detection were carried out using the digoxigenin DNA‐labelling and detection kit (Roche Molecular Biochemicals, Lewes, UK) according to manufacturer's instructions.
Construction of B. breve UCC2003::838 mutant and functional complementation
Internal 800 and 1000 bp fragments of Bbr_0838 (which corresponds to amino acid residues 100 to 357, and 100 to 438 respectively) were PCR‐amplified using B. breve UCC2003 chromosomal DNA as a template and primer combinations 838F and 838800R, or 838F and 8381000R. Amplified products were digested with HindIII and XbaI, and cloned into similarly digested pORI19, to generate pORI19‐838800 and pORI19‐8381000 respectively. The tetracycline (Tet) resistance antibiotic cassette, tet(W), from pAM4 (Álvarez‐Martín et al., 2008) was then cloned as a SacI fragment into the unique SacI site on each of these pORI19 derivatives, yielding pORI19‐838800‐tetW and pORI19‐8381000‐tetW respectively. These latter two plasmids were then introduced into E. coli EC101 harbouring pNZ‐M.BbrII‐M.BbrIII in order to obtain methylated plasmid DNA as previously described (O'Connell‐Motherway et al., 2009) and such methylated plasmid preparations were used to transform B. breve UCC2003 by electroporation. Selection for transformants was made on Reinforced Clostridial Agar (RCA, Oxoid) plates containing tetracycline. Transformants were checked by PCR and Southern hybridization to confirm whether they contained either pORI19‐838800‐tetW or pORI19‐8381000‐tetW inserted into their chromosome, and one verified insertion mutant for each of these events was retained for further use and designated UCC2003::838800 and UCC2003::8381000 respectively. The stability of these insertions was analysed following 50 generations of growth in MRSc in the absence of selective agents, by spreading cells with and without antibiotic. Stability was expressed as the proportion of viable cells retaining the insertion.
For mutant complementation, primers pBC‐838‐f and pBC‐838‐r were used to PCR‐amplify the region comprising the coding sequence of Bbr_0838 under the control of its own promoter from B. breve UCC2003 chromosomal DNA. Purified PCR product was HindIII and BamHI digested, and ligated into similarly digested pBC1.2, yielding pBC‐838, which was transformed into B. breve UCC2003::838800 by electroporation.
Transcriptional analysis
Differences in transcriptional response to bile stress between B. breve UCC2003 and its derivative B. breve UCC2003::838800 were determined by microarray experiments. Verification of microarray results was carried out by quantitative reverse transcription PCR (q‐RT‐PCR) (Table 2). Volumes of 50 ml of B. breve UCC2003 and B. breve UCC2003::838800 cultures, grown until the OD600 reached 0.5, were exposed to sub‐inhibitory concentrations of sodium cholate for 1 h, and samples for RNA isolation were collected. Methods for cell disruption, RNA isolation, cDNA synthesis and indirect labelling for microarray hybridizations were performed according to previously described procedures (Zomer et al., 2009). DNA microarray data were processed as previously described (Zomer et al., 2009). A gene was considered differentially expressed between a test condition and a control when an expression ratio of > 3 or < 0.33 relative to the result for the control was obtained with a corresponding P‐value that was < 0.001. Methods for gene expression analysis through q‐RT‐PCR were performed according to Gueimonde and colleagues (2009). For Northern hybridization analysis of Bbr_0838 expression in B. breve UCC2003, total RNA (20 µg) obtained from sodium cholate‐exposed and control cultures, was separated on a 1% (w/v) agarose/0.66 M formaldehyde gel. Blotting, hybridization and washing conditions were performed using a NorthernMax kit (Ambion – Applied Biosystems), according to manufacturer's instructions. A biotinylated antisense‐RNA probe encompassing 951 bp of the Bbr_0838 coding sequence was generated through in vitro transcription. To achieve this, an internal fragment of Bbr_0838 was amplified using primers N838f and N838r (Table 2). The amplified region was HindIII and XbaI digested, and ligated into pBluescript‐SK (Invitrogen). The resulting construct was isolated from E. coli, HindIII linearized, and used as a template for in vitro transcription with T7 RNA polymerase incorporating biotin‐labelled dUTPs (Roche). Post hybridization signal detection was accomplished using Streptavidin IRDye 680 conjugate and visualized using the Odyssey imaging system (Li‐Cor Biosciences, Lincoln, NE, USA).
Table 2.
Primers used in this study
| Sequence | Source | |
|---|---|---|
| Promoter fragments | ||
| Prom838F2bglII | 5′‐GCTCGTAGATCTGCCTCGCATGTTCAGCTTCAC‐3′ | This work |
| Prom838FbglII | 5′‐GCTCGTAGATCTCATAGCCATCAACTTACG‐3′ | This work |
| Prom838F2BglIIshort | 5′‐CGAGTCAGATCTGGCATGCGTGGGTTTCAC‐3′ | This work |
| Prom838revBamHI + IR | 5′‐CGCATAGGATCCGATGAGTCCGGACGTAAAGG‐3′ | This work |
| Prom838revBamHI‐IR | 5′‐CTGACTGGATCCGCGTAAGTTGATGGCTATG‐3′ | This work |
| Prom838rv + IRII50pstI | 5′‐CGTACAGCTGCAGGATGGGCCATCGGTACATC‐3′ | This work |
| Prom838rv + IRIIpstI | 5′‐CTATAGCCTGCAGCCGCTACACCAGCATCGAG‐3′ | This work |
| Prom838rv‐IRIIpstI | 5′‐GTATACTCTGCAGCACGGCAGGCGCGTGCGATG‐3′ | This work |
| Prom838rv‐pstI | 5′‐CTCACGCTGCAGGTGCGGATCAATAGGCTTTAC‐3′ | This work |
| Insertional mutation | ||
| 838F | 5′‐TGCTCTAAGCTTCTTTCATGGTCGGCACACTG‐3′ | This work |
| 838800R | 5′‐CGCGTCTAGAGAGATGGTCATGAGCGCAAGG‐3′ | This work |
| 8381000R | 5′‐TCAACTTCTAGACATGTGCGAGGCGGTCACG‐3′ | This work |
| Mutant complementation | ||
| pBC‐838‐f | 5′‐CGATCGAAGCCTCGCATGTTCAGCTTCAC‐3′ | This work |
| pBC‐838‐r | 5′‐CGATGCGGATCCTTATCACCGATCATCCTTTGGGAAC‐3′ | This work |
| Primer extension | ||
| 838pseqfor | 5′‐CATCTTGAACGCTTGAATGCCG‐3′ | This work |
| 838pseqrev | 5′‐GCGCCATCCGAAACTGTCAATC‐3′ | This work |
| PE3 | 5′‐GCGTAAGTTGATGGCTATGGTGAG‐3′ | This work |
| PE4 | 5′‐CGATTCATGCGACGTCGACTGC‐3′ | This work |
| Northern probe f | ||
| N838f | 5′‐CCTAGCAAGCTTCGCCGTAGTGCCGATGCT‐3′ | This work |
| N838r | 5′‐GCTCAGTCTAGAGAACGCGATGCCTCTGG‐3′ | This work |
| q‐RT‐PCR | ||
| 406‐fq | 5′‐CACGCTGCTGCATGTAATCG‐3′ | Ruiz and colleagues (2011) |
| 406‐rq | 5′‐GCCAGCCTCGGTCATTTGTA‐3′ | Ruiz and colleagues (2011) |
| 1805‐fq | 5′‐CCTCCAACCGTAACTTCCTGAA‐3′ | Ruiz and colleagues (2011) |
| 1805‐rq | 5′‐CGACCTTGGCCAGCAGTTC‐3′ | Ruiz and colleagues (2011) |
| 1827‐fq | 5′‐CGCGGCTGGCACCAT‐3′ | Ruiz and colleagues (2011) |
| 1827‐rq | 5′‐TCATTGGTCATGTCCTTGAGCTT‐3′ | Ruiz and colleagues (2011) |
| 832‐fq | 5′‐TGCTGCATCTGGTGTTGATCTG‐3′ | Ruiz and colleagues (2011) |
| 832‐rq | 5′‐AAGCCGACCTCCGCATACC‐3′ | Ruiz and colleagues (2011) |
| 1756‐fq | 5′‐CTTCAATGCCACCGTGTATGG‐3′ | Ruiz and colleagues (2011) |
| 1756‐rq | 5′‐CGCCTGCTACTCCGGAAAC‐3′ | Ruiz and colleagues (2011) |
| Bbr838F | 5′‐CGCCCTGATCATCTGCATCT‐3′ | Ruiz and colleagues (2011) |
| Bbr838R | 5′‐TTGCGTGCGGCCTTCTC‐3′ | Ruiz and colleagues (2011) |
Restriction sites are underlined.
Plasmid constructions for promoter characterization
The key plasmids constructed in this work for Bbr_0838 promoter characterization are listed in Table 2. DNA fragments of different lengths encompassing various segments of the Bbr_0838 promoter region were used to construct transcriptional fusions to the reporter gene gusA in the promoter‐less probe vector pNZ272 (Platteeuw et al., 1994). The DNA region comprising the complete Bbr_0838 promoter was amplified from B. breve UCC2003 chromosomal DNA using primers Prom838rv‐pstI and Prom838F2bglII. These primers are complementary to the DNA sequence located in the 5′ end of Bbr_0838 coding sequence and 579 bp further upstream from this point respectively. Amplified product was digested using unique BglII and PstI restriction sites (primer‐introduced) and ligated into the similarly digested promoter cloning vector pNZ272, resulting in the plasmid pNZ272‐long.
The 5′ boundary of the promoter region was deduced using PCR‐derived sub‐fragments. Primers annealing 424 bp (Prom838F2BglIIshort) and 162 bp (Prom838FbglII) upstream from the 5′ end of Bbr_0838 respectively, were used in combination with Prom838rv‐pstI for PCR amplification, using B. breve UCC2003 chromosomal DNA as a template. Amplified fragments were BglII and PstI digested and ligated into similarly digested pNZ272, yielding pNZ272ss and pNZ272‐short respectively.
Employing a similar approach, the promoter region was minimized from 3′ end. Primers annealing 71 bp (Prom838revBamHI + IR), 144 bp (Prom838revBamHI‐IR), 190 bp (Prom838rv + IRII50pstI), 228 bp (Prom838rv + IRIIpstI), and 296 bp (Prom838rv‐IRIIpstI) upstream of Bbr_0838 start codon respectively, were used in combination with Prom838F2bglII for PCR amplification, using B. breve UCC2003 chromosomal DNA as a template. Amplified fragments were BglII and PstI or BamHI digested, and ligated into similarly digested pNZ272, yielding to pNZ272‐71, pNZ272‐144, pNZ272‐190, pNZ272‐228 and pNZ272‐296 respectively.
All these constructs were initially electroporated into L. lactis and DNA integrity of inserts was verified through sequencing of plasmid minipreparations from positive clones. Sequenced‐verified constructs isolated from the corresponding L. lactis clones were then introduced into electrocompetent cells of B. breve UCC2003.
Measurement of β‐D‐glucuronidase activity
β‐D‐glucuronidase (GusA) activity was routinely determined in cultures grown until mid‐exponential growth phase, in the presence of sub‐inhibitory concentrations of sodium cholate (0.025% w/v). Briefly, cells from 10 ml cultures were collected, resuspended in 0.5 ml of PBS and disrupted with glass‐beads (Sigma) using a Fast‐Prep, by five 15 s treatments, with cooling intervals of 45 s when samples were put on ice. After centrifugation, GusA activity was immediately determined: Appropriate dilutions of the extracts (4 µl) were mixed with 88 µl of sodium acetate 0.1 M and 8 µl of a 20 mM solution of para‐nitro‐β‐D‐glucuronid acid (Sigma), and the mixtures were incubated at 37°C. After 10 min, 100 µl of 1 M ice‐cold sodium carbonate was added to stop the reaction. Optical density at 420 nm was measured and GusA activity was determined measuring the para‐nitrophenol (pNP) released, quantified by a pNP standard curve. Protein concentration in extracts was measured by using the BCA protein assay kit (Pierce, Rockford, IL, USA) and used to determine GusA specific activity. One unit of enzymatic activity was defined as the amount of protein that releases 1 nmol of pNP per minute. Specific activities were expressed as units per mg of protein and were measured in triplicate for each extract. All data reported here represent the results of at least three independent biological replicates.
Primer extension analysis
Primer extension was performed by annealing 1 pmol of IRD800 synthetic oligonucleotides PE3 or PE4 to 20 µg of RNA as described by Ventura and colleagues (2005). Sequence ladders of the presumed Bbr_0838 promoter regions, which were run alongside the primer extension products, were produced using the same primer as the primer extension reaction and a PCR product template generated using 838pseqfor and 838pseqrev. Thermo Sequenase Primer Cycle Sequencing Kit (Healthcare Biosciences, Buckinghamshire, UK) was employed. Separation was achieved on a 6.5% Li‐Cor Matrix KB Plus acrylamide gel. Signal detection and image capture was performed by means of a Li‐Cor sequencing instrument (Li‐Cor Biosciences).
Acknowledgments
This work was financed by European Union FEDER funds and the Spanish Plan Nacional de I+D (project AGL2007‐61805). Lorena Ruiz was supported by an I3P predoctoral contract granted by CSIC and FEDER funds. Work at the University of Cork was supported through the Alimentary Pharmabiotic Centre by the Irish Government's National Development Plan Grant 07/CE/B1368.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Whole UTR secondary structure.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Álvarez‐Martín P., Flórez A.B., Margolles A., Solar G., Mayo B. Improved cloning vectors for bifidobacteria, based on the Bifidobacterium catenulatum pBC1 replicon. Appl Environ Microbiol. 2008;74:4656–4665. doi: 10.1128/AEM.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Gaham C.G., Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Cerda‐Maira F.A., Ringelberg C.S., Taylor R.K. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J Bacteriol. 2008;190:6809–6814. doi: 10.1128/JB.00584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppins R.L., Hall K.D., Groisman E.A. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwa A., Bricker A.L., Jain C., Belasco J.G. An evolutionarily conserved RNA stem‐loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- Elkins C.A., Savage D.C. CbsT2 from Lactobacillus johnsonii 100‐100 is a transport protein of the major facilitator superfamily that facilitates bile acid antiport. J Mol Microbiol Biotechnol. 2003;6:76–87. doi: 10.1159/000076738. [DOI] [PubMed] [Google Scholar]
- Estrem S.T., Gaal T., Ross W., Gourse R.L. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Baroch A. ExPASy: the proteomics server for in‐depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueimonde M., Garrigues C., van Sinderen D., de los Reyes‐Gavilán C.G., Margolles A. Bile‐inducible efflux transporter from Bifidobacterium longum NCC2705, conferring bile resistance. Appl Environ Microbiol. 2009;75:3153–3160. doi: 10.1128/AEM.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambraeus G., Karhumaa K., Rutberg B. A 5' stem‐loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology. 2002;148:1795–1803. doi: 10.1099/00221287-148-6-1795. [DOI] [PubMed] [Google Scholar]
- Hofman A.F. Bile secretion and the enterohepatic circulation of bile acids. In: Feldman M., Scharschmidt B.F., Sleisenger M.H., editors. W.B. Saunders; 1998. pp. 937–948. [Google Scholar]
- Ignoul S., Eggermont J. CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol. 2005;289:1369–1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- Jones B.V., Begley M., Hill C., Gahan C.G., Marchesi J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke W.J., Chang B.Y., Lin T.P., Liu S.T. Activation of the promoter of the fengycin synthetase operon by the UP element. J Bacteriol. 2009;191:4615–4623. doi: 10.1128/JB.00255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Hejine G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Itoh H., Kuwhara T., Oshima K., Toh H., Toyoda A. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiome. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Buist G., Haandrikman A., Kok J., Venema G., Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Sahin O., Michel L.O., Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 2003;71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax A.R., Calder P.C. Probiotics, immune function, infection and inflammation: a review of the evidence from studies conducted in humans. Curr Pharm Des. 2009;15:1428–1518. doi: 10.2174/138161209788168155. [DOI] [PubMed] [Google Scholar]
- Lubelski J., de Jong A., van Merkerk R., Agustiandari H., Kuipers O.P., Kok J., Driessen A.J. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol. 2006;61:771–781. doi: 10.1111/j.1365-2958.2006.05267.x. [DOI] [PubMed] [Google Scholar]
- MacConaill L.E., Butler D., O'Connell‐Motherway M., Fitzgerald G.F., van Sinderen D. Identification of two‐component regulatory systems in Bifidobacterium infantis by functional complementation and degenerate PCR approaches. Appl Environ Microbiol. 2003;69:4219–4226. doi: 10.1128/AEM.69.7.4219-4226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood N.A.B., Biemans‐Oldehinkel E., Poolman B. Engineering of ion sensing by the cystathionine β‐synthase module of the ABC transporter OpuA. J Biol Chem. 2009;284:14368–14376. doi: 10.1074/jbc.M901238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolles A., de los Reyes‐Gavilán C.G. Purification and functional characterization of a novel alpha‐L‐arabinofuranosidase from Bifidobacterium longum B667. Appl Environ Microbiol. 2003;69:5096–5103. doi: 10.1128/AEM.69.9.5096-5103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolles A., García L., Sánchez B., Gueimonde M., de los Reyes‐Gavilán C.G. Characterisation of a Bifidobacterium strain with acquired resistance to cholate – a preliminary study. Int J Food Microbiol. 2003;82:191–198. doi: 10.1016/s0168-1605(02)00261-1. [DOI] [PubMed] [Google Scholar]
- Margolles A., Moreno J.A., van Sinderen D., de los Reyes‐Gavilán C.G. Macrolide resistance mediated by a Bifidobacterium breve membrane protein. Antimicrob Agents Chemother. 2005;49:4379–4381. doi: 10.1128/AAC.49.10.4379-4381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolles A., Flórez A.B., Moreno J.A., van Sinderen D., de los Reyes‐Gavilán C.G. Two membrane proteins from Bifidobacterium breve UCC2003 constitute and ABC‐type multidrug transporter. Microbiology. 2006;152:3497–3505. doi: 10.1099/mic.0.29097-0. [DOI] [PubMed] [Google Scholar]
- Mazé A., O'Connell‐Motherway M., Fitgerald G.F., Deutscher J., van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;75:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega L., Gueimonde M., Sánchez B., Margolles A., de los Reyes‐Gavilán C.G. Effect of the adaptation to high bile salts concentrations on glycosidic activity, suvival at low pH and cross‐resistance to bile salts in Bifidobacterium. Int J Food Microbiol. 2004;94:79–86. doi: 10.1016/j.ijfoodmicro.2004.01.003. [DOI] [PubMed] [Google Scholar]
- O'Connell‐Motherway M., Fitzgerald G.F., Neirynck S., Ryan S., Steidler L., van Sinderen D. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell‐Motherway M., O'Driscoll J., Fitzgerald G.F., van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell‐Motherway M., Zomer A., Leahy S.C., Reunanen J., Bottacini F., Claesson M.J. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host‐colonization factor. Proc Natl Acad Sci USA. 2011;108:11217–11222. doi: 10.1073/pnas.1105380108. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiler E.A., Klaenhammer T.R. The role of transporter proteins in bile tolerance in Lactobacillus acidophilus. Appl Environ Microbiol. 2009;75:6013–6016. doi: 10.1128/AEM.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L.J.V. Multidrug‐resistance efflux‐pumps‐not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Platteeuw C., Simons G., de Vos W.M. Use of the Escherichia coli beta‐glucuronidase (gusA) as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K., Neves A.R., Zomer A., O'Connell‐Motherway M., MacSharry J., Curley P. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K., O'Connell‐Motherway M., Zomer A., MacSharry J., Fitzgerald G.F., van Sinderen D. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2011;77:1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.E., Reid S.J., Driessen A.J., Abratt V.R. The Bifidobacterium longum NCIMB 702259 ctr gene codes for a novel cholate transporter. Appl Environ Microbiol. 2006;72:78–87. doi: 10.1128/AEM.72.1.923-926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin S.J., Schwartz K.T., Leber J.H. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol Microbiol. 2011;81:129–142. doi: 10.1111/j.1365-2958.2011.07683.x. [DOI] [PubMed] [Google Scholar]
- Ruiz L., Zomer A., O'Connell‐Motherway M., van Sinderen D., Margolles A. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruyter P.G., Kuipers O.P., de Vos W.M. Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. 2nd. Cold String Harbor Laboratory; 1989. [Google Scholar]
- Sánchez B., Champomier‐Vergès M.C., Stuer‐Lauridsen B., Ruas‐Madiedo P., Anglade P., Baragie F. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl Environ Microbiol. 2007a;73:6757–6767. doi: 10.1128/AEM.00637-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez B., Champomier‐Vergès M.C., Collado M.C., Anglade P., Baraige F., Sanz Y. Low‐pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl Environ Microbiol. 2007b;73:6450–6459. doi: 10.1128/AEM.00886-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez B., Ruiz L., de los Reyes‐Gavilán C.G., Margolles A. Proteomics of stress response in Bifidobacterium. Front Biosci. 2008;13:6905–6919. doi: 10.2741/3198. [DOI] [PubMed] [Google Scholar]
- Stanton C., Ross R.P., Fitzgerald G.F., van Sinderen D. Fermented functional foods based on probiotics and their biogenic metabolites. Curr Opin Biotechnol. 2005;16:198–203. doi: 10.1016/j.copbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Ventura M., van Sinderen D., Fitzgerald G.F., Zink R. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek. 2004;86:205–223. doi: 10.1023/B:ANTO.0000047930.11029.ec. [DOI] [PubMed] [Google Scholar]
- Ventura M., Fitzgerald G.F., van Sinderen D. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2005;71:6282–6291. doi: 10.1128/AEM.71.10.6282-6291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., O'Flaherty S., Claesson M.J., Turroni F., Klaenhammer T.R., van Sinderen D., O'Toole P.W. Genome‐scale analyses of health‐promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Waters L.S., Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D., Sleator R.D., Hill C., Gahan C.G. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract. BMC Microbiol. 2008;8:176. doi: 10.1186/1471-2180-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D., Sleator R.D., Casey P.G., Hill C., Gahan C.G. Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes. Infect Immun. 2009;77:4895–4904. doi: 10.1128/IAI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Wilson P.W., Le Page R.W. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- Yokota A., Veenstra M., Kurdi P., van Veen H.W., Konings W.N. Cholate resistance in Lactococcus lactis is mediated by an ATP‐dependent multispecific organic anion transporter. J Bacteriol. 2000;182:5196–5201. doi: 10.1128/jb.182.18.5196-5201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi A.H., Bakkes P.J., Lubelski J., Agustiandari H., Kuipers O.P., Driessen A.J. The ABC‐type multidrug resistance transporter LmrCD is responsible for an extrusion‐based mechanism of bile acid resistance in Lactococcus lactis. J Bacteriol. 2008;190:7357–7366. doi: 10.1128/JB.00485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A., Fernández M., Kearney B., Fitzgerald G.F., Ventura M., van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191:7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole UTR secondary structure.


