Abstract
Insecticidal Cry proteins produced by Bacillus thuringiensis are use worldwide in transgenic crops for efficient pest control. Among the family of Cry toxins, the three domain Cry family is the better characterized regarding their natural evolution leading to a large number of Cry proteins with similar structure, mode of action but different insect specificity. Also, this group is the better characterized regarding the study of their mode of action and the molecular basis of insect specificity. In this review we discuss how Cry toxins have evolved insect specificity in nature and analyse several cases of improvement of Cry toxin action by genetic engineering, some of these examples are currently used in transgenic crops. We believe that the success in the improvement of insecticidal activity by genetic evolution of Cry toxins will depend on the knowledge of the rate-limiting steps of Cry toxicity in different insect pests, the mapping of the specificity binding regions in the Cry toxins, as well as the improvement of mutagenesis strategies and selection procedures.
Introduction
Bacillus thuringiensis (Bt) is a Gram-positive bacterium that produces insecticidal proteins as crystal inclusions during its sporulation phase of growth, known as Cry or Cyt toxins, which have been proven to be effective against important crop pests and also against mosquitoes that are vectors of human diseases such as dengue and malaria (Bravo et al., 2011). Bt was first discovered in 1901 in Japan by Shigetane Ishiwatari when the causal agent of wilt disease in silk worm (Bombyx mori) was isolated. Few years later Bt was rediscovered in Germany by Ernst Berliner from a Mediterranean fluor moth (Ephestia kuehniella) (reviewed by Sanahuja et al., 2011). The first Bt formulation was developed using the Bt strain isolated by Berliner in 1938. However, the success of Bt as bioinsecticide came with the development of Bt-crops that express the cry gene resulting in crops that resist insect attack including borers that were difficult to control with topical Bt-formulations leading to the commercial release of Bt-crops in 1995 (Sanahuja et al., 2011). Bt toxins are specific to a limited number of insect species with no toxicity against humans or other organisms (Bravo et al., 2011). In 2010, more than 58 million hectares were grown worldwide with Bt-Maize or Bt-cotton (James, 2010).
Many Bt strains that show activity towards Lepidoptera, Diptera, Coleoptera, Hymenoptera, Homoptera, Orthoptera and Mallophaga insect orders have been reported (Schnepf et al. 1998). In addition, Bt strains active against nematodes, mites and protozoa have also been isolated (Crickmore et al., 1998; Schnepf et al., 1998; de Maagd et al., 2001; Wei et al., 2003). However, still there are many insect pests that show no susceptibility to Cry toxins or that are poorly controlled by the Cry proteins identified so far. On the other hand, a major threat for the use of Cry toxins in transgenic plants is the appearance of insect resistance. Evolution of resistance to Bt-crops in the field has been documented for at least five different insect species (van Rensburg, 2007; Tabashnik et al., 2008; Bagla, 2010; Storer et al., 2010; Gassmann et al., 2011). Therefore, an alternative for the screening and isolation of novel Cry toxin protein in nature, is the in vitro genetic evolution of Cry toxins with the aim of enhancing toxicity against specific pests, to kill novel targets or to recover toxicity in the case of the appearance of resistance in the field (Pardo-López et al., 2009).
Natural evolution of Cry toxins
Extensive screening of Bt strains and cry gene sequencing has led to the identification of more than 700 cry gene sequences (Crickmore et al., 2011). These sequences have been classified according to their amino acid sequence identity in at least 70 different cry gene groups (Cry1, Cry2… Cry70) where toxins belonging to each Cry group share less than 40% amino acid identity with proteins from other groups (Crickmore et al., 1998). Within each group, a capital letter (Cry1A, Cry1B etc) is given when they share less that 70% identity. A small letter (Cry1Aa, Cry1Ab etc) is given when toxins share more than 70% but less than 95% identity. Phylogenetic analysis of Cry protein sequences showed that the whole family of Cry proteins belong to four non-phylogenetically related protein families, the family of three domain Cry toxins (3D), the family of mosquitocidal Cry toxins (Mtx), the family of the binary-like (Bin) and the Cyt family of toxins (reviewed in Bravo et al., 2005). Some Bt strains produce additional insecticidal toxins named VIP during vegetative growth, these proteins do not form parasporal crystals, thus were not named Cry toxins. Three VIP toxins have been characterized as VIP1/VIP2, which together compose a binary toxin, and VIP3 (Estruch et al., 1996; Warren, 1997).
The 3D family group is the largest group of Cry toxins with over 50 different Cry groups. At least seven 3D-Cry proteins have been crystallized and their three-dimensional structures were solved, Cry1Aa, Cry2Aa, Cry3Aa, Cry3Ba, Cry4Aa, Cry4Ba and Cry8Ea (reviewed in Bravo et al., 2011). All 3D-Cry proteins resolved structures show a similar fold, composed of three domains despite the fact that some of these proteins share very little amino acid sequence identity (less than 20%). Domain I is a seven α-helix bundle comprised by six amphipatic helices surrounding the hydrophobic helix α-5. This domain has been shown to be involved in toxin oligomerization, membrane insertion and pore formation. Domain II is composed of eleven beta sheets with exposed loop regions involved in binding to specific larval midgut proteins while domain III is a beta sandwich that is also involved in receptor recognition. Thus domains II and III are the specificity determinant domains of Cry toxins (Bravo et al., 2011) (Fig. 1). This group of 3D-Cry proteins is characterized by their production during sporulation of the bacteria as protoxins, with some members producing large protoxins of 130 kDa, such as the Cry1Aa protoxin, while other members are synthesized as short protoxins of 65–70 kDa, such as the Cry11Aa protoxin. In the case of large protoxins, they are processed by insect midgut proteases loosing half of the protein at the C-terminal end, approximately 600 amino acids. The large protoxins are also processed at the N-terminal end, where 20–50 amino acid residues were cleaved out, depending on the toxin. The short protoxins are only processed at the N-terminal end (de Maagd et al., 2001). The proteolytical activation of both, large or short Cry protoxins resulted in a protease resistant core of approximately 60 kDa that is biological active and is comprised by the three-dimensional structure (de Maagd et al., 2001).
Fig. 1.
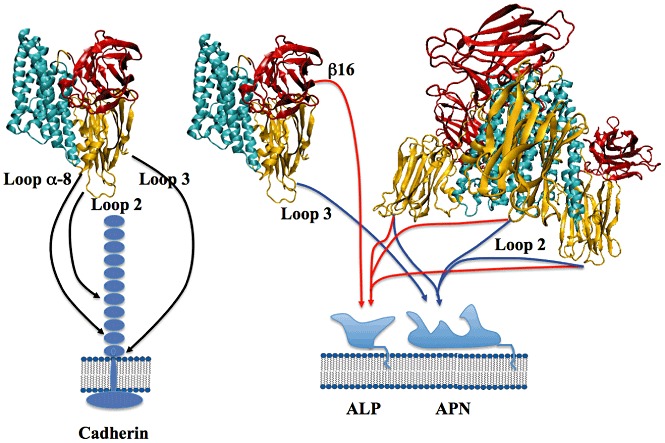
Binding regions of monomeric and oligomeric forms mapped in Cry1Ab toxin to Manduca sexta receptors, cadherin, alkaline phosphatase (ALP), and aminopeptidase-N (APN). The monomeric form depicted corresponds to the three-dimensional structure of Cry1Aa (pdb 1CIY) and the oligomeric structure corresponds to Cry4Ba trimeric structure (pdb 1W99).
The phylogenetic analysis of the 3D-Cry protein family revealed a different topology when the protoxin or the mature toxin protein fragments were analysed (Bravo, 1997; Crickmore, 2000; de Maagd et al., 2001). Cry toxins are classified by the amino acid sequence similarity of protoxin sequences. Phylogenetic analysis of toxin fragments revealed different evolutionary relationships of certain Cry toxins than the analysis of protoxin sequences (Bravo, 1997; Crickmore, 2000). For instance, Cry9Aa toxin fragment shows no evolutionary relationship with Cry9Ba or Cry9Ca toxin fragments indicating that the high sequence identity at the C-terminal protoxin fragment was responsible for clustering these toxins together (Bravo, 1997; Crickmore, 2000). The phylogenetic analysis of toxin fragments revealed interesting clustering of Cry proteins accordingly with their different insect specificity. However, some exceptions were found as exemplified by Cry1B, Cry1I that show toxicity against lepidopteran insects that clustered together with Cry3, Cry7 and Cry8 that are toxic to coleopteran insects (Bravo, 1997; Crickmore, 2000). This observation suggests that Cry1B and Cry1I proteins may have toxicity against some coleopteran as was latter shown for Cry1B toxin (López-Pazos et al., 2009). Thus, the phylogenetic relationships of whole protoxin did not revealed how Cry toxins evolved insect specificity. Interestingly, the analysis of the evolutionary relationships of single domains revealed that domains I and II co-evolved as the different toxins showed similar clustering when domain I or domain II sequences were analysed independently (Bravo, 1997; Crickmore, 2000). Interestingly, phylogenetic analysis of domain III sequences revealed several examples of domain III swapping among different toxins. For example, Cry1Ac and Cry1Bd share a similar domain III while Cry1Be, Cry1Cb and Cry1Eb share a related domain III amino acid sequence. Thus domain III swapping among different Cry toxins is likely to be an active evolutionary process for determining insect specificity (Bravo, 1997; de Maagd et al., 2001).
The analysis of adaptive evolution revealed several residues of 3D-Cry toxins that are under positive selection (Wu et al., 2007). Positive selection favours the retention of mutations that are beneficial to an individual or a population. Twenty-four residues were identified to be under positive selection and most of them located either in domain II loop regions or domain III, suggesting that these amino acid regions are likely to be involved in receptor recognition. Based on this result, it was proposed that the high divergence found in these regions could promote rapid evolution to their targets insect receptors (Wu et al., 2007). It was also proposed that the diversity of Cry toxins found in nature is the result of two fundamental evolutionary process, the independent evolution of the three structural domains with domain II and domain III regions under positive selection for insect receptor recognition and domain III swapping. These evolutionary processes had led to the selection of proteins with similar mode of action but with different insect specificity.
Mode of action of 3D-Cry toxins
As mention previously, different members of the 3D-Cry toxins share a similar three-dimensional fold suggesting that they share a similar mode of action. 3D-Cry toxins are recognized as pore forming toxins that kill larval epithelium midgut cells by causing an osmotic shock leading to cell lysis. To induce the pore formation of 3D-Cry toxins, the parasporal crystals have to be ingested by susceptible larvae, solubilized by the pH conditions of the insect gut, alkaline in the case of lepidopteran and dipteran insects and acidic in the case of coleopteran, and activated by midgut proteases to yield the three-dimensional resistant core of the activated toxin. In the case of Cry1A toxins that are active against lepidopteran insects, it has been shown that Cry1A toxins undergo a sequential binding mechanism with glycosyl-phosphatidyl-inositol anchored proteins such as alkaline phosphatase (ALP) or aminopeptidase-N (APN) and cadherin-like protein resulting in the formation of a pre-pore oligomeric structure that is proficient in membrane insertion and pore formation (Bravo et al., 2011). Receptor recognition by Cry toxins has been recognized as a key step of Cry toxicity that is fundamental for insect specificity (Schnepf et al., 1998; Bravo et al., 2011). However, protoxin activation by insect proteases has been shown to be also a limiting step either due to un-efficient toxin activation or rapid proteolytic degradation that in some cases has been shown to determine insect specificity (de Maagd et al., 2001). For instance, Cry7Aa is active against the coleopteran Colorado potato beetle only after in vitro solubilization and trypsin activation of the protoxin, suggesting that solubilization and activation of Cry7Aa is a limiting step of the insecticidal activity of this Cry protein (Lambert et al., 1992).
The mode of action of Cry1Ab toxin has been described to some detail in the tobacco hornworm (Manduca sexta) larvae. It is proposed that Cry1Ab binds with low affinity (Kd 100–200 nM) to glycosyl-phosphatidyl-inositol anchored ALP or APN receptors. This binding step is believed to concentrate the monomeric toxin in the surface of the brush border membrane. Following this binding step Cry1Ab binds with high affinity (Kd 1 nM) to cadherin; this binding step facilitates the proteolytic removal of helix α1 of domain I, inducing toxin oligomerization. Cry1Ab toxin oligomers gain binding affinity to both ALP and APN (Kd 0.6 nM) and this final binding step facilitates oligomer membrane insertion and pore formation (reviewed in Bravo et al., 2011). However, a different model of the mode of action of Cry toxins proposed that binding to cadherin is sufficient to trigger an intracellular signal transduction pathway that leads to cell death without the involvement of oligomer formation nor pore formation (Zhang et al., 2006). Nevertheless, as discussed below, the construction of modified Cry toxins that skip cadherin interaction has shown that binding to cadherin is not sufficient for toxicity (Soberón et al., 2007). The regions involved in the binding of Cry1Ab monomeric and oligomeric forms to these receptor molecules were mapped and shown to include domain II loop regions and exposed surface of domain III (Gómez et al., 2006; Pacheco et al., 2009; Arenas et al., 2010). Cry1Ab monomeric toxin binds ALP or APN through domain II loop 3 and domain III β16. In contrast, the oligomeric form of the Cry1Ab toxin binds ALP or APN through domain II loop 2 (Pacheco et al., 2009; Arenas et al., 2010). Binding to cadherin is mediated by three binding sites that include domain II loop α8, loop 2 and loop 3 (Gómez et al., 2003; 2006). Figure 1 shows the binding epitopes of Cry1Ab toxin that are involved in receptor recognition.
In vitro evolution of Cry toxin insecticidal activity
Proteolytic activation of Cry toxins
As mentioned previously activation by insect midgut proteases could be a limiting step of Cry toxicity in different insect species. In the case of Cry3Aa that has insecticidal activity against coleopteran larvae like Colorado potato beetle (Leptinotarsa decemlineata) it shows very low toxicity against Western corn rootworm (Diabrotica virgifera virgifera). The low toxicity of Cry3Aa against Western corn rootworm was proposed to be due to the low solubility of the protease activated Cry3Aa that yield a 67 kDa fragment. However, activation with chimiotrypsin was shown to increase the yield of a fully processed 55 kDa Cry3A fragment that showed increased solubility and toxicity (Carrrol et al., 1997). The 55 kDa form was shown to be nicked at the α3-α4 domain I loop region (Carrrol et al., 1997). The introduction of a chymiotrypsin/cathepsin G proteolytic site in the Cry3Aa α3-α4 loop (named mCry3Aa) resulted in increased yields of the 55 kDa form and increased toxicity towards Western corn rootworm (Walters et al., 2008). The increased toxicity of mCry3Aa also correlated with increased solubility of the 55 kDa processed form and also with the specific binding of the processed 55 kDa to Western corn root worm brush border membrane vesicles (BBMV) (Walters et al., 2008). Interestingly, mCry3Aa showed a similar insecticidal activity towards Colorado potato beetle larvae as Cry3Aa indicating the engineered protease site in mCry3Aa broadened the insecticidal activity rather changing its insect specificity. mCry3Aa has been expressed in transgenic maize and shown to be effective in controlling Western corn rootworm (Hibbard et al., 2011).
Appearance of insect resistance threatens the use of Cry toxins in transgenic plants (Tabashnik et al., 2008). In different lepidopteran insect colonies it has been shown that resistance to Cry1Ab or Cry1Ac toxins is linked to mutations in the cadherin gene (reviewed in Bravo and Soberón, 2008). Cadherin binding is a limiting step of Cry1A toxins action as it facilitates further proteolytic processing of the toxin removing helix α1 necessary for toxin oligomerization. Cry1Ab and Cry1Ac toxins that were genetically modified to delete helix α1 (Cry1AbMod or Cry1AcMod) were shown to form oligomers in vitro when activated with proteases in the absence of cadherin protein in contrast to native toxins that only formed the oligomeric structure when activated in the presence of a cadherin binding site (Soberón et al., 2007). Cry1AbMod and Cry1AcMod were shown to counter resistance of the Pink bollworm (Pectinophora gossypiella) Cry1Ac resistant colony linked to mutations in the cadherin gene (Soberón et al., 2007). This result suggested that Cry1AMod toxins have the potential to counter insect resistance when resistance is linked to mutations affecting cadherin expression. Interestingly, a follow-up study analysed the insecticidal activity of Cry1AbMod and Cry1AcMod against seven different lepidopteran insect resistant colonies that in some cases were not linked to mutations affecting cadherin expression (Tabashnik et al., 2011). These insect colonies included a laboratory selected tobacco budworm (Heliothis virescens) Cry1Ac resistant colony whose resistance was recently shown to be genetically linked to a mutant allele of an ABC transporter (ABCC2) (Gahan et al., 2010). The ABCC2 mutation affected the binding of Cry1Ab and Cry1Ac to BBMV. It was proposed that the ABCC2 protein could facilitate oligomer membrane insertion (Gahan et al., 2010). Also, the toxicity analysis of Cry1AMod toxins included two field evolved Cry1Ac resistant colonies of Diamondback moth (Plutella xylostella) and the cabbage looper (Trichoplusia ni) that were recently shown to be also linked to mutations in the ABCC2 transporter (Baxter et al., 2011). The data revealed that Cry1AMod also counters resistance in these two insect resistant lines and also in tobacco budworm resistant colony that contains both the ABCC2 and cadherin mutant alleles. Interestingly, Cry1AbMod and Cry1AcMod were not very effective against the tobacco budworm single mutant cadherin or ABCC2 insect colonies (Tabashnik et al., 2011). It is important to mention that Cry1AbMod and Cry1Ac toxins were not very effective against the susceptible line of tobacco budworm, showing an important reduction in toxicity, suggesting that the low efficacy of Cry1AMod toxins against insect lines with low resistance ratios, such as the tobacco budworm colonies affected only in cadherin gene or in ABCC2 gene, could be due to the reduce activity of Cry1AMod toxins when compared with the native Cry1A toxin. These results suggest that the mode of action of Cry1A toxins is more complex than explained above and will include additional proteins such as ABCC2 transporter and that although that Cry1AMod toxins may have lower efficacy against some insect pests, they have the potential to counter resistance based on different mechanisms (Tabashnik et al., 2011). The basis of the low efficacy of CryMod toxins against particular insect pests remains to be analysed. This could lead to the improvement of Cry1AMod in the future.
Domain III swapping
As mentioned above domain III swapping has been recognized as a natural mechanism involved in the evolution of Cry toxins. In vitro domain III swapping among different Cry toxins have resulted in some cases in hybrid toxins with improved toxicities against certain insect species (Bosch et al., 1994). One of the first examples was the construction of a hybrid toxin containing domains I and II from Cry1Ab toxin and domain III of Cry1C that showed more than sixfold higher toxicity against beet armyworm (Spodoptera exigua) compared with Cry1C (de Maagd et al., 2000). Improvement of toxin activity by domain III swapping was also shown for the coleopteran Colorado potato beetle. In this case both Cry1Ba and Cry1Ia showed low toxicity against Colorado potato beetle. Construction of a hybrid toxin consisting of domains I and II of Cry1Ia and domain III of Cry1Ba showed three- and sevenfold higher insecticidal activity against Colorado potato beetle than the parental toxins Cy1Ia and Cry1Ba respectively (Naimov et al., 2001). An interesting example of toxin improvement by domain III swapping was the construction of a hybrid toxin containing domains I and II from Cry3Aa and domain III from Cry1Ab (eCry3.1Ab). eCry3.1Ab was shown to be toxic to Western corn rootworm in contrast to Cry3Aa and Cry1Ab that showed no toxicity to this insect (Walters et al., 2010). Interestingly, Cry1Ab is a lepidopteran specific toxin but its domain III could improve the toxicity of a coleopteran specific toxin such as the Cry3Aa. eCry3.1Ab expressed in transgenic maize was shown to be effective in controlling Western corn rootworm (Hibbard et al., 2011). Overall, these results show that domain III swapping could be an interesting strategy to improve toxicity of Cry toxins or to create novel hybrid toxins with toxicity against pests that show no susceptibility to the parental Cry toxins. Strategies for shuffling the three different domains among large numbers of cry genes (Knight et al., 2004) and high through output bioassay screening methods are likely to provide novel Cry toxins with improved or novel toxicities.
Domain II and Domain III mutations with enhanced insecticidal activity
Exposed loop regions in domain II have been shown to be important determinants of insect specificity (Bravo et al., 2005). Cry4Ba shows no toxicity against Culex sp while Cry4Aa is active against this mosquito species (Abdullah et al., 2003). Introducing domain II loop 3 Cry4Aa amino acid sequence into loop 3 of domain II of Cry4Ba resulted in a mutant toxin (4BL3GAV) with toxicity against Culex sp, retaining its insecticidal activity against Aedes aegypti (Abdullah et al., 2003). In the same line of ideas, the lepidopteran Cry1Aa toxin was engineered in domain II loop regions to mimic the Cry4Ba mutant 4BLGAV showing that the engineered Cry1Aa (1AaMosq) gained activity against Culex pipiens larvae, although the activity was three orders of magnitude lower than 4BLGAV (Liu and Dean, 2006). These results suggest that domain II loop swapping could provide a strategy for improving or changing the specificity of Cry toxins.
Site-directed mutagenesis of domain II loop sequences has in some cases resulted in mutant toxins with increased insecticidal activity. The first example of domain II loop mutants with increased insecticidal activity was Cry1Ab toxin where mutations in loop 2 resulted in higher insecticidal activity against Gypsy moth (Limantria dispar) (Rajamohan et al., 1996). A single Cry1Ab mutation in loop 2, N372A, or a triple loop 2 mutant in residues N372A, A282G and L283S showed 8- and 36-fold higher toxicity to Gypsy moth larvae respectively. Interestingly, the increased insecticidal activity correlated with increased binding affinities to BBMV isolated from Gypsy moth (Rajamohan et al., 1996). Similarly, it was shown that mutations of domain II loop regions in the coleopteran Cry3Aa resulted in enhanced toxicity to yellow mealworm (Tenebrio molitor) (Wu et al., 2000). A triple domain II loop 1 mutant R345, Y350F, Y351F showed tenfold higher toxicity to yellow mealworm than Cry3Aa and twofold higher toxicity against Colorado potato beetle that correlated with twofold higher binding affinity to Colorado potato beetle BBMV (Wu et al., 2000). These results show that domain II loop regions are key binding regions of Cry toxins that are suitable targets for mutagenesis and selection of Cry toxins with improved insecticidal properties.
In the case of domain III, there are just few examples of mutations in two different exposed loop regions with some mutants showing a moderate non-significant increase in toxicity against different insect species (Shu et al., 2007; Xiang et al., 2009; Lv et al., 2011; Shan et al., 2011). Nevertheless, there are few studies that have mapped the domain III binding epitopes with ALP or APN receptors (Atsumi et al., 2005; Gómez et al., 2006; Arenas et al., 2010). Mutagenesis of these domain III binding regions is likely to provide means for increasing Cry toxins insecticidal activity such as domain III swapping that has been shown to create novel toxins with improved toxicities (de Maagd et al., 2000).
Other mutations in Cry toxins with enhanced insecticidal activity
Besides the domain I mutations introducing protease cleavage sites described above, two other modifications in domain I of Cry1Ab or Cry2Aa have shown to increase insecticidal activity. Cry1Ac helix α5 mutant V171C was shown to have 25-fold higher insecticidal activity against Gypsy moth without affecting its toxicity to the tobacco hornworm (Alzate et al., 2010). The increased toxicity of Cry1Ac V171C was proposed to be due to a higher unfolding rate allowing the more rapidly partitioning of the toxin into the membrane (Alzate et al., 2010). In the case of Cry2Aa, two modifications in domain I resulted in a Cry2Aa mutant with four- to six-fold higher toxicity against cotton leaf worm (Spodoptera litura), cotton bollworm (Helicoverpa armigera) and black cutworm (Agrotis ipsilon) (Mandal et al., 2007). The first modification that enhanced threefold the toxicity of Cry2Aa, consisted in the deletion of the first 42 amino acid residues at the N-terminal end. Cry2Ab three-dimensional structure revealed the first 49 amino acids precedes domain I helix α1 and these residues are normally cleaved out during protease activation of Cry2Ab protoxin. This N-terminal fragment was shown to occlude a domain II hydrophobic patch proposed to be involved in receptor interaction (Morse et al., 2001). Thus, the proteolytic cleavage of the N-terminal protoxin fragment could be a rate-limiting step that is avoided with the 42 amino acid deletion (Mandal et al., 2007). Two additional point mutations in Cry2Aa helix α1 residues K63F and K64P were introduced based on enhancing the hydrophobic nature of a putative transmembrane region identified in silico (Mandal et al., 2007). Finally, a threefold increased in the insecticidal activity of Cry3Aa against Asian longhorn beetle (Anoplophora glabripennis) was achieved by fusion of an eight amino acid residue peptide that was shown to specifically bound longhorn beetle midgut Cx-cellulase (Guo et al., 2012). It was proposed that the fused peptide increased the toxin retention in the gut by binding to the gut cellulase (Guo et al., 2012). Figure 2 shows the three-dimensional structure of Cry1Aa toxin highlighting the regions where modifications had led to increased insecticidal activity in different Cry toxins.
Fig. 2.
Representation of Cry toxin regions where mutations enhanced insecticidal activity in different Cry toxins. The three-dimensional structure of Cry1Aa toxin (pdb 1CIY) is depicted.
High through output systems for evolution of Cry toxins
As indicated above, the improvement of the insecticidal activity by site-directed mutagenesis of the binding epitopes found in domains II and III has a lot of potential for selection of Cry toxins with improved activity against different insect pests as shown by the different examples of Cry toxins with modifications in these amino acid regions that have resulted in toxins with improved insecticidal properties. However, those examples have been the result of analysing few mutants in different insect species but not from a high through output system that could detect improved mutants from a large pool of variants. Gene shuffling was reported to be a useful method for the generation of cry gene mutant libraries to select improved variants with increased insecticidal activity. The first example was the evolution of Cry1Ca protein with increased insecticidal activity against fall armyworm. Improved Cry1Ca variants were detected by bioassay screening where mutants showing more than sixfold higher toxicity against tobacco budworm than the parental Cry1Ca toxin were selected (Lassner and Bedbrook, 2001). The regions that resulted in the enhanced insecticidal activity of these Cry1Ca proteins were not reported (Lassner and Bedbrook, 2001). This last approach is likely to provide better Cry mutant toxins as they are selected from many variants. Nevertheless, the identification of improved variants by bioassays is a challenging task when many Cry toxin variants are screened. Thus, a desirable method should be a method that screens Cry mutant libraries for binding to BBMV from the selected insect or to receptor molecules purified from the target insect. In the latter case, it will be desirable to restrict the mutagenesis to the Cry toxin binding regions that are important for binding with specific receptor molecules. Below we will discuss recent work showing that gene shuffling combined with a binding screening selection using phage display could lead to selection of improved Cry toxins.
Phage display
Phage display allows a rapid selection of variants with improved binding characteristics (Azzazy and Highsmith, 2002; Mullen et al., 2006). For this purpose, the foreign protein DNA sequence is fused to a coat protein gene enabling the fusion protein to be displayed on the surface of the phage that can be then screened by enabling the phage to interact with ligands, a process known as biopanning. Filamentous M13 phage libraries are constructed by fusing the protein of interest with a capsid protein such as capsid proteins p3 or p8. The fusion protein can be incorporated to the phage genome or into molecular vectors called phagemids. Phagemid vectors contain a coat protein gene usually with a polilinker site for protein fusion, an antibiotic resistance gene as ampicillin and are capable of replicating in Escherich coli cells due to a plasmid replication site and be packed into the phage particle due to a M13 DNA sequence that is recognized by the packaging machinery when a helper M13 phage is used to infect E. coli cells harbouring the phagemid constructs (Azzazy and Highsmith, 2002; Mullen et al., 2006). To display the fusion protein, it has to be translocated to the host periplasm by a leader-peptide sequence while a helper phage provides all the necessary components for phage assembly. This is a powerful technique as the selected phages maintain a physical link between the displayed protein (phenotype) and the encoding gene (genotype) and further mutagenesis and selection by biopanning allows an in vitro high through output molecular evolution of proteins (Droge et al., 2003).
Cry1A toxins have been displayed in three different phages M13, T7 and λ but, as discussed below, these systems have shown not to be optimal systems for displaying Cry1A toxins (Marzari et al., 1997; Kasman et al., 1998; Vílchez et al., 2004; Pacheco et al., 2006. M13 phage-display systems have an intrinsic problem in displaying big proteins as fusion proteins have to be transported into the E. coli periplasm where phage assembly occurs. The first display of Cry1Aa toxin in M13 (Marzari et al., 1997; Kasman et al., 1998), showed important deletions in the displayed fused toxin protein (Marzari et al., 1997). Nevertheless, in another report it was shown that Cry1Ac was displayed in M13 showing toxicity to tobacco budworm larvae, but the displayed Cry1Ac protein did not bind to functional APN receptor in vitro suggesting structural constraints of the displayed toxin (Kasman et al., 1998). Later it was shown that phages λ and T7 could be much better systems to display Cry1A toxins as the assembling of phage particles in both systems occurs in the cytoplasm of bacterial cells, allowing the display of big proteins (Vílchez et al., 2004; Pacheco et al., 2006). In the case of λ phage, the Cry1Ac protein was fused with the capsid protein D and displayed on the surface of phage particles. The displayed Cry1Ac toxin retained similar toxicity as the wild-type toxin to tobacco hornworm and the capacity to interact with the APN receptor (Vílchez et al., 2004). In another report the cry1Ac gene was fused to the 3′ end of the T7 10B capsid protein gene and the chimeric protein was displayed on the surface of T7 phage. The T7-Cry1Ac binds Cry1Ac-receptors and BBMV isolated from tobacco hornworm and retained toxicity against tobacco hornworm larvae, suggesting the successful display of Cry1Ac toxin in T7 phages (Pacheco et al., 2006). Nevertheless, a problem with both λ and T7 displaying systems is that for displaying the fusion protein both systems rely on in vitro packaging systems that under the best scenario allow for the production of up to 107 recombinant phage particles mg−1 of DNA, making the construction of libraries with large number of variants inefficient.
Despite the problems mentioned above on the efficiency to display certain Cry toxins in phage particles, at least three successful examples of selection of improved variants of Cry toxins selected by biopanning of phage display libraries have been described (Ishikawa et al., 2007; Craveiro et al., 2010; Oliveira et al., 2011). The first example was the display of Cry1Aa toxin using T7 phage system. A library of mutants in domain II loop 2 region was constructed and used to recover toxin variants with increased binding affinity to a silkworm (Bombyx mori) cadherin fragment. After five rounds of selection using cadherin coated magnetic beads, a domain II loop 2 mutant with apparent similar binding affinity to cadherin, and a fourfold higher insecticidal activity against silkworm larvae than Cry1Aa was selected (Ishikawa et al., 2007). The two other examples relied on the construction of Cry toxin library variants by gene shuffling, on the display on M13 of these libraries and selection of improved variants by biopanning against BBMV from the selected target insect. The sugarcane giant borer (Telchin licus licus) is not susceptible to the known Cry toxins including Cry1Ia that was shown to be toxic to fall armyworm (Spodoptera frugiperda) (Craveiro et al., 2010). A library of Cry1Ia variants created by gene shuffling was cloned into a phagemid vector and display in the M13 phage. This library was used to select better Cry1Ia binders to sugarcane giant borer BBMV. Four variants with significant insecticidal activity against sugarcane borer were recover and shown to contain single point mutations in domains I and III (Craveiro et al., 2010). Finally, a similar strategy was used for selection of Cry8Ka mutants with increased insecticidal activity against cotton boll weevil (Anthonomus grandis). Cry8Ka gene was identified as a toxin showing moderate toxicity against cotton boll weevil. Gene shuffling and the selection of improved Cry8Ka binders against cotton boll weevil BBMV resulted in a mutant (Cry8Ka5) with threefold higher toxicity (Oliveira et al., 2011). Cry8Ka5 showed a 16 amino acid deletion in the N-terminal end and six additional amino acid changes, one located in what was predicted as domain II loop 3 amino acid region (Oliveira et al., 2011).
A general strategy for evolution of Cry toxin insecticidal activity
We have revised several examples of evolved Cry toxins with improved performance in controlling different insect pests. Some of these Cry mutants show novel insecticidal activity, others improved toxicity against a specific target and others were shown to be active against resistant insects to Cry toxins. Nevertheless, as pointed before, most of these Cry mutants were the result of analysing few mutants in different insect species but not from a high through output system that could detect improved mutants from a large number of variants. We believe that the evolution of Cry toxicity using high through output systems is likely to provide toxins that will perform better in controlling insect pests. In Fig. 3 we propose a general strategy for in vitro evolution of toxicity of Cry toxins. The rational behind this strategy is that Cry toxin mutants with improved binding affinities to either BBMV from the target insect or isolated insect Cry-binding proteins, will provide mutants that are likely to show enhanced insecticidal activity. The first step is the construction of cry gene library of variants that could then be screened for binding to BBMV or toxin-receptors from the target insect. Several methods for creating variability could be exploited depending on the binding selection procedure. Using general mutagenesis strategies as gene shuffling or prone PCR can explore the whole gene cry sequence including domain I. As discussed earlier, there are examples of domain I mutations that enhance Cry toxicity presumably by enhancing membrane partioning into the membrane (Mandal et al., 2007; Alzate et al., 2010). Gene libraries could also be created by shuffling domain III among different Cry toxins or by shuffling domain II loop regions that are likely to provide Cry toxins with improved toxicity or altered specificity. Finally, gene libraries could be created by mutation of receptor binding epitopes like domain II loop regions or residues of domain III β16. In the second step the cry gene libraries are cloned into phagemid vectors for the display of Cry mutants in the phage particles. The third step is the screening of libraries by biopanning against BBMV or pure receptor molecules. The fourth step is the selection of Cry mutants with enhanced binding to BBMV or receptors. Finally, the fifth step is the determination of toxicity of Cry toxins with improved binding against the target insect. Improved variants could be the substrate for additional mutagenesis, binding selection and bioassays.
Fig. 3.
General strategy for in vitro evolution of toxicity of Cry toxins. Five steps are proposed for in vitro evolution of Cry toxins, 1. Construction of gene libraries with Cry variants obtained by different mutagenesis strategies (prone PCR, gene shuffling, domain III swapping, domain II loop 2 swapping and mutagenesis of receptor binding regions); 2. Display of gene libraries on phage; 3. Biopanning of phage display libraries using brush border membrane vesicles of insect of interest or purified receptors (cadherin is shown as example); 4. Selection of variants with improved binding characteristics; 5. Toxicity assays against the target insect to select Cry toxins with improved insecticidal activity.
Conflict of interest
None declared.
References
- Abdullah MA, Alzate O, Mohammad M, McNall RJ, Adang MJ, Dean DH. Introduction of Culex toxicity into Bacillus thuringiensis Cry4Ba by protein engineering. Appl Environ Microbiol. 2003;69:5343–5353. doi: 10.1128/AEM.69.9.5343-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzate O, Osorio C, Florez AM, Dean DH. Participation of valine 171 in alpha-Helix 5 of Bacillus thuringiensis Cry1Ab delta-endotoxin in translocation of toxin into Lymantria dispar midgut membranes. Appl Environ Microbiol. 2010;76:7878–7880. doi: 10.1128/AEM.01428-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas I, Bravo A, Soberon M, Gomez I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem. 2010;285:12497–12503. doi: 10.1074/jbc.M109.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Mizuno E, Hara H, Nakanishi K, Kitami M, Miura N, et al. Location of the Bombyx mori aminopeptidase N type I binding site on Bacillus thuringiensis Cry1Aa toxin. Appl Environ Microbiol. 2005;71:3966–3977. doi: 10.1128/AEM.71.7.3966-3977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzazy HME, Highsmith JWE. Phage display technology: clinical applications and recent innovations. Clin Biochem. 2002;35:425–445. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- Bagla P. Hardy cotton-munching pests are latest blow to GM crops. Science. 2010;327:1439. doi: 10.1126/science.327.5972.1439. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Badenes-Pérez FR, Morrison A, Vogel H, Crickmore N, Kain W, et al. Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genet. 2011;189:675–679. doi: 10.1534/genetics.111.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch D, Schipper B, van der Kleij H, de Maagd RA, Stiekema J. Recombinant Bacillus thuringiensis insecticidal proteins with new properties for resistance management. Biotechnology. 1994;12:915–918. doi: 10.1038/nbt0994-915. [DOI] [PubMed] [Google Scholar]
- Bravo A. Phylogenetic relationships of Bacillus thuringiensis delta-endotoxin family proteins and their functional domains. J Bacteriol. 1997;179:2793–27801. doi: 10.1128/jb.179.9.2793-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Soberón M. How to cope with resistance to Bt toxins? Trends Biotechnol. 2008;26:573–579. doi: 10.1016/j.tibtech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Bravo A, Gill SS, Soberón M. Bacillus thuringiensis mechanisms and use. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Oxford: ELSEVIER; 2005. pp. 175–206. [Google Scholar]
- Bravo A, Likitvivatanavong S, Gill SS, Soberón M. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem Mol Biol. 2011;41:423–431. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrrol J, Convents D, Van Damme J, Boets A, Van Rie J, Ellar DJ. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A δ-endotoxin may facilitate its coleopteran toxicity. J Invertebr Pathol. 1997;70:41–49. doi: 10.1006/jipa.1997.4656. [DOI] [PubMed] [Google Scholar]
- Craveiro KI, Gomes Júnior JE, Silva MC, Macedo LL, Lucena WA, Silva MS, et al. Variant Cry1Ia toxins generated by DNA shuffling are active against sugarcane giant borer. J Biotechnol. 2010;145:215–221. doi: 10.1016/j.jbiotec.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Crickmore N. The diversity of Bacillus thuringiensis δ-endotoxins. In: Charles J-F, Delécluse A, Nielsen-LeRoux C, editors. Enthomopathogenic Bacteria: From Laboratory to Field Application. Dordrecht: Kluwer Academic Publishers; 2000. pp. 65–79. [Google Scholar]
- Crickmore N, Zeigler D, R Feitelson J, Schnepf E, Van Rie J, Lereclus D, et al. Revision of the nomenclture for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore N, Zeigler DR, Schnepf E, Van Rie J, Lereclus D, Baum J, et al. 2011. ‘Bacillus thuringiensis toxin nomenclature’ [WWW document]. URL http://www.lifesci.sussex.ac.uk/Home/Neil_Crickmore/Bt/
- Droge MJ, Ruggeberg CJ, van der Sloot AM, Schimmel J, Dijkstra DW, Verhaert RMD, et al. Binding of phage display Bacillus subtilis lipase A to a phosphonate suicide inhibitor. J Biotechnol. 2003;101:19–28. doi: 10.1016/s0168-1656(02)00289-4. [DOI] [PubMed] [Google Scholar]
- Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc Natl Acad Sci USA. 1996;93:5389–5394. doi: 10.1073/pnas.93.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6:e1001248. doi: 10.1371/journal.pgen.1001248. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann AJ, Petzold-Maxwell JL, Keweshan RS, Dunbar MW. Field-evolved resistance to Bt maize by western corn rootworm. PLoS ONE. 2011;6:e22629. doi: 10.1371/journal.pone.0022629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez I, Dean DH, Bravo A, Soberón M. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops α-8 and 2 in domain II of Cy1Ab toxin. Biochem. 2003;42:10482–10489. doi: 10.1021/bi034440p. [DOI] [PubMed] [Google Scholar]
- Gómez I, Arenas I, Benitez I, Miranda-Ríos J, Becerril B, Grande G, et al. Specific epitopes of Domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J Biol Chem. 2006;281:34032–34039. doi: 10.1074/jbc.M604721200. [DOI] [PubMed] [Google Scholar]
- Guo CH, Zhao ST, Ma Y, Hu JJ, Han XJ, Chen J, Lu MZ. Bacillus thuringiensis Cry3Aa fused to a cellulase-binding peptide shows increased toxicity against the longhorned beetle. Appl Microbiol Biotechnol. 2012;93:1249–1256. doi: 10.1007/s00253-011-3523-9. [DOI] [PubMed] [Google Scholar]
- Hibbard BE, Frank DL, Kurtz R, Boudreau E, Ellersieck MR, Odhiambo JF. Mortality impact of Bt transgenic maize roots expressing eCry3.1Ab, mCry3A, and eCry3.1Ab plus mCry3A on western corn rootworm larvae in the field. J Econ Entomol. 2011;104:1584–1591. doi: 10.1603/ec11186. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Hoshino Y, Motoki Y, Kawahara T, Kitajima M, Kitami M, et al. A system for the directed evolution of the insecticidal protein from Bacillus thuringiensis. Mol Biotechnol. 2007;36:90–101. doi: 10.1007/s12033-007-0001-9. [DOI] [PubMed] [Google Scholar]
- James C. 2010. Global Status of Commercialized Biotech/GM Crops: 2010 ISAAA Brief No. 42. ISAAA: Ithaca, NY.
- Kasman LM, Lukowiak AA, Garcynski SF, McNall RJ, Youngman P, Adang MJ. Phage display of a biologically active Bacillus thuringiensis toxin. Appl Environ Microbiol. 1998;64:2995–3003. doi: 10.1128/aem.64.8.2995-3003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Broadwell AH, Grant WN, Shoemaker CB. A strategy for shuffling numerous Bacillus thuringiensis crystal protein domains. J Econ Entomol. 2004;97:1805–1813. doi: 10.1093/jee/97.6.1805. [DOI] [PubMed] [Google Scholar]
- Lambert B, Höfte H, Annys K, Jansens S, Soetaert P, Peferoen M. Novel Bacillus thuringiensis insecticidal crystal protein with a silent activity against coleopteran larvae. Appl Environ Microbiol. 1992;58:2536–2542. doi: 10.1128/aem.58.8.2536-2542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassner M, Bedbrook J. Directed molecular evolution in plant improvement. Curr Opin Plant Biol. 2001;4:152–156. doi: 10.1016/s1369-5266(00)00152-7. [DOI] [PubMed] [Google Scholar]
- Liu XS, Dean DH. Redesigning Bacillus thuringiensis Cry1Aa toxin into a mosquito toxin. Protein Eng Des Sel. 2006;19:107–111. doi: 10.1093/protein/gzj009. [DOI] [PubMed] [Google Scholar]
- López-Pazos SA, Martínez JW, Castillo AX, Cerón-Salamanca JA. Cry1B and Cry3A are active against Hypothenemus hampei Ferrari (Coleoptera: Scolytidae) J Invertebr Pathol. 2009;101:242–245. doi: 10.1016/j.jip.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Lv Y, Tang Y, Zhang Y, Xia L, Wang F, Ding X, et al. The role of β20-β21 loop structure in insecticidal activity of Cry1Ac toxin from Bacillus thuringiensis. Curr Microbiol. 2011;62:665–670. doi: 10.1007/s00284-010-9760-9. [DOI] [PubMed] [Google Scholar]
- de Maagd RA, Weemen-Hendriks M, Stiekema W, Bosch D. Domain III substitution in Bacillus thuringiensis delta-endotoxin Cry1C domain III can function as a specific determinant for Spodoptera exigua in different, but not all, Cry1-Cry1C hybrids. Appl Environ Microbiol. 2000;66:1559–1563. doi: 10.1128/aem.66.4.1559-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maagd RA, Bravo A, Crickmore N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001;17:193–199. doi: 10.1016/s0168-9525(01)02237-5. [DOI] [PubMed] [Google Scholar]
- Mandal CC, Gayen S, Basu A, Ghosh KS, Dasgupta S, Maiti MK, Sen SK. Prediction-based protein engineering of domain I of Cry2A entomocidal toxin of Bacillus thuringiensis for the enhancement of toxicity against lepidopteran insects. Protein Eng Des Sel. 2007;20:599–606. doi: 10.1093/protein/gzm058. [DOI] [PubMed] [Google Scholar]
- Marzari R, Edomi P, Bhatnagar RK, Ahmad S, Selvapandiyan A, Bradbury A. Phage display of Bacillus thuringiensis CryIA(a) insecticidal toxin. FEBS Lett. 1997;411:27–31. doi: 10.1016/s0014-5793(97)00647-9. [DOI] [PubMed] [Google Scholar]
- Morse RJ, Yamamoto T, Stroud RM. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure. 2001;9:409–417. doi: 10.1016/s0969-2126(01)00601-3. [DOI] [PubMed] [Google Scholar]
- Mullen LM, Nair SP, Ward JM, Rycroft A, Henderson N. Phage display in the study of infectious diseases. Trends Microbiol. 2006;14:141–147. doi: 10.1016/j.tim.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimov S, Weemen-Hendriks M, Dukiandjiev S, de Maagd RA. Bacillus thuringiensis delta-endotoxin Cry1 hybrid proteins with increased activity against the Colorado potato beetle. Appl Environ Microbiol. 2001;67:5328–5330. doi: 10.1128/AEM.67.11.5328-5330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira GR, Silva MC, Lucena WA, Nakasu EY, Firmino AA, Beneventi MA, et al. Improving Cry8Ka toxin activity towards the cotton boll weevil (Anthonomus grandis. BMC Biotechnol. 2011;9:11–85. doi: 10.1186/1472-6750-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco S, Gómez I, Sato R, Bravo A, Soberón M. Functional display of Bacillus thuringiensis Cry1Ac toxin on T7 phage. J Invertebr Pathol. 2006;92:45–49. doi: 10.1016/j.jip.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Pacheco S, Gomez I, Arenas I, Saab-Rincon G, Rodriguez-Almazan C, Gill SS, et al. Domain II loop 3 of Bacillus thuringiensis Cry1Ab toxin is involved in a ‘ping-pong’ binding mechanism with Manduca sexta aminopetidase-N and cadherin receptors. J Biol Chem. 2009;284:32750–32757. doi: 10.1074/jbc.M109.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-López L, Muñoz-Garay C, Porta H, Rodríguez-Almazán C, Soberón M, Bravo A. Strategies to improve the insecticidal activity of Cry toxins from Bacillus thuringiensis. Peptides. 2009;30:589–595. doi: 10.1016/j.peptides.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan F, Alzate O, Cotrill JA, Curtiss A, Dean DH. Protein engineering of Bacillus thuringiensis delta-endotoxin: mutations at domain II of CryIAb enhance receptor affinity and toxicity toward gypsy moth larvae. Proc Natl Acad Sci USA. 1996;93:14338–14343. doi: 10.1073/pnas.93.25.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rensburg JBJ. First report of field resistance by stem borer Busseola fusca (Fuller) to Bt-transgenic maize. S Afr J Plant Soil. 2007;24:147–151. [Google Scholar]
- Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P. Bacillus thuringiensis: a century of research development and commercial applications. Plant Biotechnol J. 2011;9:283–300. doi: 10.1111/j.1467-7652.2011.00595.x. [DOI] [PubMed] [Google Scholar]
- Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum JR, Feitelson J, et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:705–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S, Zhang Y, Ding X, Hu S, Sun Y, Yu Z, et al. A Cry1Ac toxin variant generated by directed evolution has enhanced toxicity against Lepidopteran insects. Curr Microbiol. 2011;62:358–365. doi: 10.1007/s00284-010-9714-2. [DOI] [PubMed] [Google Scholar]
- Shu C, Liu R, Wang R, Zhang J, Feng S, Huang D, Song F. Improving toxicity of Bacillus thuringiensis strain contains the cry8Ca gene specific to Anomala corpulenta larvae. Curr Microbiol. 2007;55:492–496. doi: 10.1007/s00284-007-9018-3. [DOI] [PubMed] [Google Scholar]
- Soberón M, Pardo-López L, López I, Gómez I, Tabashnik B, Bravo A. Engineering modified Bt toxins to counter insect resistance. Science. 2007;318:1640–1642. doi: 10.1126/science.1146453. [DOI] [PubMed] [Google Scholar]
- Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW, Huckaba RM. Discovery and characterization of field resistance to Bt Maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J Econ Entomol. 2010;103:1031–1038. doi: 10.1603/ec10040. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Gassman AJ, Crowdwer DW, Carriere Y. Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol. 2008;26:199–202. doi: 10.1038/nbt1382. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Huang F, Ghimire MN, Leonard BR, Siegfried BD, Randasamy M, et al. Efficacy of genetically modified Bt toxins against insects with different mechanism of resistance. Nat Biotechnol. 2011;29:1128–1131. doi: 10.1038/nbt.1988. [DOI] [PubMed] [Google Scholar]
- Vílchez S, Jacoby J, Ellar DJ. Display of biologically functional insecticidal toxin on the surface of λ phage. Appl Environ Microbiol. 2004;70:6587–6594. doi: 10.1128/AEM.70.11.6587-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters FS, Stacy CM, Lee MK, Palekar N, Chen JS. An engineered chymotrypsin/cathepsin G site in domain I renders Bacillus thuringiensis Cry3A active against Western corn rootworm larvae. Appl Environ Microbiol. 2008;74:367–374. doi: 10.1128/AEM.02165-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters FS, deFontes CM, Hart H, Warren GW, Chen JS. Lepidopteran-active variable-region sequence imparts coleopteran activity in eCry3.1Ab, an engineered Bacillus thuringiensis hybrid insecticidal protein. Appl Environ Microbiol. 2010;76:3082–3088. doi: 10.1128/AEM.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. Vegetative insecticidal proteins: novel proteins for control of corn pests. In: Carozzi N, Koziel M, editors. Advances in Insect Control: The Role of Transgenic Plants. London: Taylor & Francis; 1997. pp. 109–120. [Google Scholar]
- Wei J-Z, Hale K, Carta L, Platzer E, Wong C, Fang S-C, Arioan RV. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci USA. 2003;100:2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Y, Zhao F-Q, Bai J, Deng G, Qin S, Bao Q-Y. Adapatative evolution of cry gene in Bacillus thuringiensis: Implications for their specificity determination. FEBS Lett. 2007;5:102–110. doi: 10.1016/S1672-0229(07)60020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Koller CN, Miller DL, Bauer LS, Dean DH. Enhanced toxicity of Bacillus thuringiensis Cry3A delta-endotoxin in coleopterans by mutagenesis in a receptor binding loop. FEBS Lett. 2000;473:227–232. doi: 10.1016/s0014-5793(00)01505-2. [DOI] [PubMed] [Google Scholar]
- Xiang WF, Qiu XL, Zhi DX, Min ZX, Yuan L, Quan YZ. N546 in beta18-beta19 loop is important for binding and toxicity of the Bacillus thuringiensis Cry1Ac toxin. J Invertebr Pathol. 2009;101:119–123. doi: 10.1016/j.jip.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Taussig R, Bulla LA., Jr A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci USA. 2006;103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]


