Abstract
Several prebiotics, such as inulin, fructo-oligosaccharides and galacto-oligosaccharides, are widely used commercially in foods and there is convincing evidence, in particular for galacto-oligosaccharides, that prebiotics can modulate the microbiota and promote bifidobacterial growth in the intestinal tract of infants and adults. In this study we describe the identification and functional characterization of the genetic loci responsible for the transport and metabolism of purified galacto-oligosaccharides (PGOS) by Bifidobacterium breve UCC2003. We further demonstrate that an extracellular endogalactanase specified by several B. breve strains, including B. breve UCC2003, is essential for partial degradation of PGOS components with a high degree of polymerization. These partially hydrolysed PGOS components are presumed to be transported into the bifidobacterial cell via various ABC transport systems and sugar permeases where they are further degraded to galactose and glucose monomers that feed into the bifid shunt. This work significantly advances our molecular understanding of bifidobacterial PGOS metabolism and its associated genetic machinery to utilize this prebiotic.
Introduction
Neonates possess an immature intestinal immune system, a gastrointestinal tract devoid of a microbiota and a stomach with a relatively high pH, which is not effective in eliminating pathogens, thereby making newborns highly susceptible to infection. Colonization of the human gut commences at birth, when the mother's vaginal and faecal microbiota are considered to be the most important inocula (Vaishampayan et al., 2010). It has furthermore been suggested that mother's milk serves as an inoculum for breastfeeding infants as bacteria from the maternal gut are believed to reach the mammary gland through an endogenous route, the so-called ‘entero-mammary pathway’ that involves maternal dendritic cells and macrophages (Martín et al., 2003; Perez et al., 2007). Evidence is accumulating on the importance of the human gut microbiota in educating the immune system at this very early stage of extrauterine life (Martín et al., 2010). There is a broad consensus that breastfeeding is the best option for optimal development of the infant's intestinal microbiota, so as to promote a healthy, balanced immune system (reviewed by Marques et al., 2010). Human milk is a functional food; it contains semi-essential nutrients, free amino acids, enzymes, hormones, growth factors, polyamines, nucleotides and oligosaccharides (Walker, 2007b). Research has indicated that human milk has a protective effect against diarrhoeal diseases, respiratory tract infections, bacteremia, meningitis and necrotizing enterocolitis, all presumed to be mediated through its bioactive components (Martín et al., 2010). Human milk oligosaccharides (HMOs) are the main prebiotic factors in human milk and, while they do not directly nourish infants, HMOs are thought to enrich for specific gut commensals capable of utilizing these diverse substrates, in particular bifidobacteria that are abundant in and characteristic of the breastfed baby's intestinal microbiota (Sela and Mills, 2010; Sela, 2011).
Bifidobacteria are anaerobic, saccharolytic, high GC-content, Gram-positive bacteria that are non-motile and do not sporulate or produce gas through their fermentative metabolism (reviewed by Turroni et al., 2011). Bifidobacteria are frequently reported as being numerically dominant in the faecal microbiota of breastfed infants while their numbers decline on weaning and thereafter resemble those of the adult microbiota. The many beneficial effects associated with a breastfed infant microbiota, being high in bifidobacterial numbers, has resulted in major efforts being directed at manipulating infant food formulations so as to achieve in bottle-fed infants an intestinal microbiota composition resembling that found in breastfed babies. HMOs are very diverse in composition and structure (Niñonuevo et al., 2006; Lo Cascio et al., 2009; Niñonuevo and Lebrilla, 2009; Nwosu et al., 2012) and as yet these are not available for incorporation into infant food formulations. Consequently research has focused on the development of alternative prebiotic, non-human milk-derived oligosaccharides, such as galacto-oligosaccharides (GOS).
GOS are non-digestible oligosaccharides that are produced by enzymatic transglycosylation of lactose using β-galactosidases, thereby generating a mixture of oligomers of varying chain length ranging from DP2 to DP15 (Barboza et al., 2009). The raw material for the production of GOS is whey-derived lactose which is a by-product of the dairy industry. The composition of GOS is dependent on the source of the enzyme, substrate concentration and pH, furthermore, since GOS production involves a transglycosylation process which competes with the hydrolytic activity of the enzymes, GOS always contain a significant amount of lactose as well as its constituent monosaccharides galactose and glucose (Coulier et al., 2009).
Since 2002, several infant formulations containing specific mixtures of short-chain GOS (scGOS) and long-chain fructo-oligosaccharides (lcFOS) have been on the market. These formulations have been designed to elicit a prebiotic/bifidogenic effect comparable to that of human milk, as demonstrated in numerous clinical trials (reviewed by MacFarlane and Macfarlane, 2007; Sangwan et al., 2011). Furthermore, there is an increasing body of evidence that prebiotic supplementation of food formulations with GOS provides protection against pathogen infection (Eversbach et al., 2010; Searle et al., 2010; Quintero et al., 2011).
To date several β-galactosidases of bifidobacterial origin have been described in the literature and much research has focused on the ability of these enzymes to hydrolyse and synthesize GOS (Van Laere et al., 2000; Barboza et al., 2009; Goulas et al., 2009; Osman et al., 2010; 2012). In the present study we evaluated the transcriptional response of the human (infant) gut commensal Bifidobacterium breve UCC2003 during growth on a purified Vivinal® GOS (PGOS) formulation that contains very low levels of monosaccharides and lactose. Our results indicate that PGOS metabolism by B. breve UCC2003 is the result of the concerted action of several β-galactosidases and their associated transport systems that internalize GOS components. We furthermore demonstrate that an extracellular endogalactanase specified by several B. breve strains, including B. breve UCC2003, is essential for metabolism of GOS components with a long retention time and high degree of polymerization. The resulting endogalactanase-mediated breakdown products are transported into the bifidobacterial cell via various ABC transport systems and permeases where they are further metabolized to galactose and glucose, the latter feeding directly into the so-called bifid shunt, while the former likely being channelled through the Leloir pathway and converted to glucose 6-phosphate before entering the bifid shunt.
Results
PGOS utilization by B. breve strains
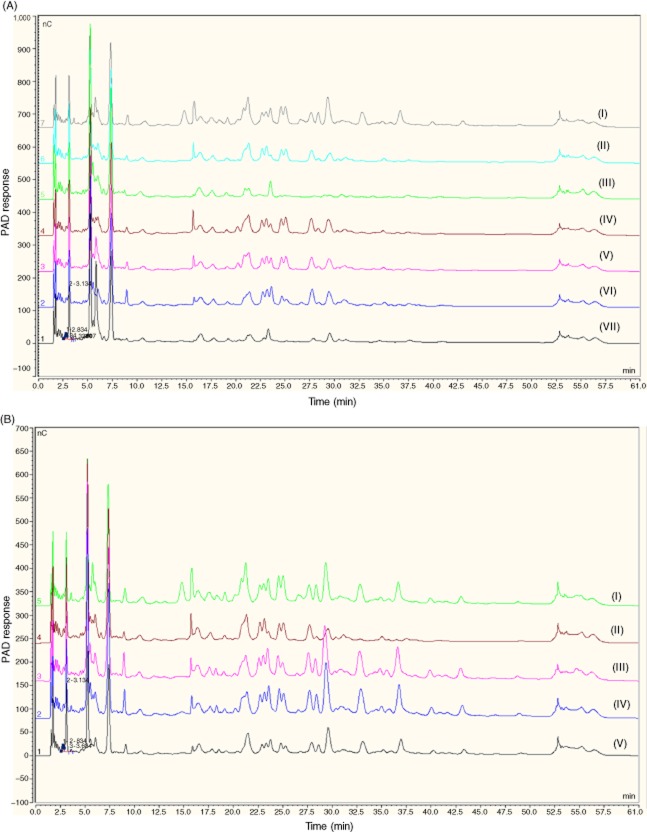
We previously established that not all B. breve strains possess endogalactanase activity and by comparative genome hybridization were able to establish that the presence of endogalactanase activity is strictly correlated with the presence of the galA gene in the B. breve strains examined (O'Connell Motherway et al., 2011b). In order to investigate the importance of endogalactanase activity for PGOS metabolism, growth of 13 B. breve strains, namely UCC2003, UCC2005, JCM7017, JCM7019, NCFB2257, NCFB2258, NCIMB8815, NCIMB11815, LMG13208, NIZO658, UCC2008, UCC2009 and UCC2010, was profiled in Modified Rogosa with 0.5% lactose or PGOS as the sole carbohydrate source. All strains grew well on lactose reaching an OD 600nm ≥ 1.0 within 8 h of growth, while growth on PGOS separated the B. breve strains into two groups: the first group all grew well on PGOS reaching an OD 600nm ≥ 1.0 within 9 h of growth while for the second group a final OD 600nm of ≤ 1.0 was achieved in the 24 h of the growth profile (Fig. 1). These two distinct bifidobacterial growth profiles correlated with the endogalactanase-positive (GalA+) and endogalactanase-negative (GalA−) strains respectively. Post-fermentation, cell-free supernatants were analysed by HPAEC-PAD and demonstrated that the GalA+ B. breve strains utilized almost all of the GOS components (Fig. 2A), while the GalA− strains exhibited more variability in the GOS fractions that were metabolized. The GalA− strains did not utilize PGOS components with a long retention time (between 30 and 48 min), which represent galacto-oligosaccharides with a high degree of polymerization (DP) (Coulier et al., 2009) (Fig. 2B), indicating that GalA is responsible for the extracellular degradation of these high DP GOS components.
Fig. 1.
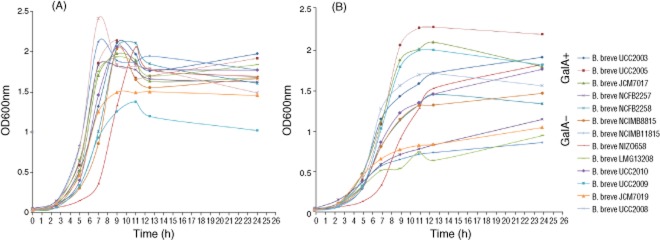
Growth profiles of B. breve strains on lactose (A) and PGOS (B). Data presented are averages of duplicate growth experiments.
Fig. 2.
HPAEC-PAD analysis of post-fermentation cell-free supernatants of B. breve strains. Modified Rogosa supplemented with 0.5% GOS (AI and BI); post-fermentation supernatants of B. breve UCC2003 (AII and BII), endogalactanase positive strains: B. breve UCC2005 (AIII); NCFB2258 (AIV); NCIMB8815 (AV); NIZO658 (AVI) and UCC2008 (AVII) and endogalactanase negative strains: B. breve NCFB2257 (BIII); NCIMB11815 (BIV) and LMG13208 (BV).
Genome response of B. breve UCC2003 to growth on PGOS
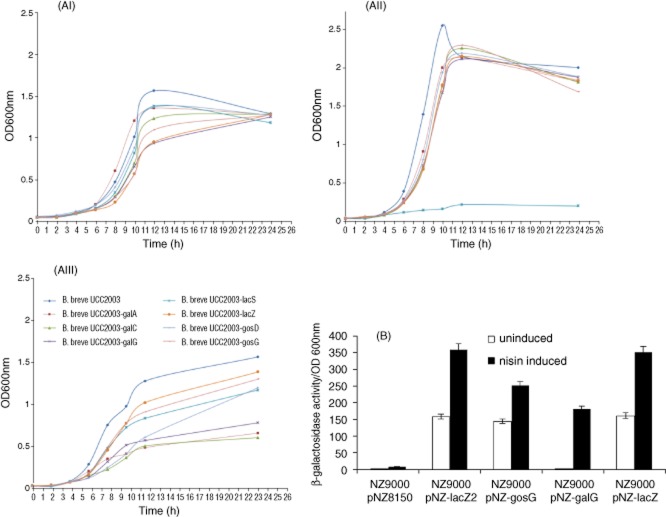
In order to investigate which genes may be involved in PGOS metabolism in B. breve UCC2003, global gene expression was determined by microarray analysis during growth of this bifidobacterial strain on PGOS and this was compared with its expression pattern when grown on ribose as sole carbohydrate source. Total RNA was isolated from mid-log phase cultures of B. breve UCC2003 grown on PGOS or ribose as the sole carbohydrate source. Analysis of the DNA microarray data revealed that the expression of four gene clusters was significantly upregulated when UCC2003 was grown on PGOS (fold change > 3.0, P < 0.001; Table 1 and Fig. 3). The first GOS-inducible cluster of six genes, galC, galD, galE galG, galR and galA, constitute the previously identified galactan metabolism cluster (O'Connell Motherway et al., 2011b), while the second cluster, Bbr_1551 to Bbr_1553, consists of three genes specifying a predicted galactoside symporter and a GH2 family β-galactosidase in divergent orientations, designated here as lacS and lacZ respectively, and an associated gene encoding a putative LacI-type transcriptional regulator. The third GOS-inducible cluster comprises five genes, Bbr_0526 to Bbr_530, that are predicted to specify a LacI-type transcriptional regulator, two ABC-type permeases, a GH42 family β-galactosidase and a solute-binding protein, which are designated gosR, gosD, gosE gosG and gosC respectively. The fourth set of upregulated genes are the presumed galT and galK genes that encode Galactose-1-phosphate uridyltransferase and galactokinase respectively (Kitaoka et al., 2005; Fushinobu, 2010), and represent key enzymes of the Leloir pathway that converts galactose to d-glucose 6-phosphate that can feed into the bifid shunt (Fushinobu, 2010; Pokusaeva et al., 2011a) (Fig. 3).
Table 1.
Effect of PGOS on the transcriptome of B. breve UCC2003
| Locus tag_gene | Putative function | PGOSa |
|---|---|---|
| Bbr_0417_galC | Solute-binding protein | 285.61 |
| Bbr_0418_galD | Sugar permease protein | 9.08 |
| Bbr_0419_galE | Sugar permease protein | 10.17 |
| Bbr_0420_galG | β-Galactosidase GH 42 family | 99.99 |
| Bbr_0421_galR | Transcriptional regulator, LacI family | 6.43 |
| Bbr_0422_galA | Endogalactanase | 45.25 |
| Bbr_1551_lacS | Galactoside symporter | 174.68 |
| Bbr_1552_lacZ | β-Galactosidase GH 2 family | 128.14 |
| Bbr_1553_lacI | Transcriptional regulator, LacI family | 2.9 |
| Bbr_0526_gosR | Transcriptional regulator, LacI family | 5.62 |
| Bbr_0527_gosD | Sugar permease protein | 8.75 |
| Bbr_0528_gosE | Sugar permease protein | 9.99 |
| Bbr_0529_gosG | β-Galactosidase GH 42 family | 6.33 |
| Bbr_0530_gosC | Solute-binding protein | 5.83 |
| Bbr_0491_galT | Galactose-1-phosphate uridyltransferase | 3.51 |
| Bbr_0492_galK | Galactokinase | 3.22 |
Expression ratios presented have a Bayesian P-value < 0.001 according to the Cyber-T-test (Long et al., 2001).
Fig. 3.
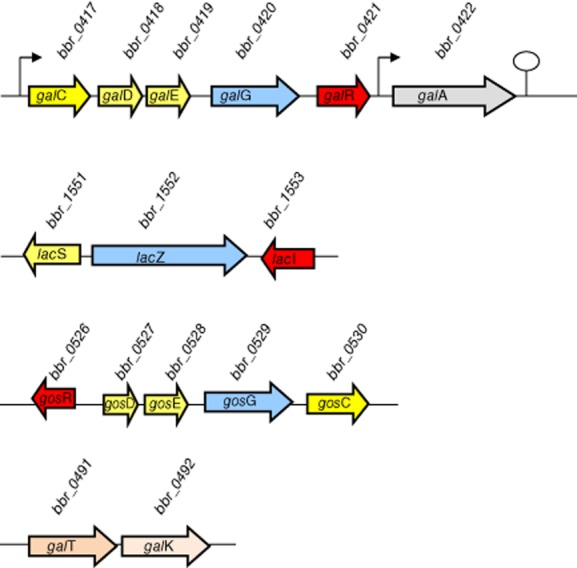
Schematic representation of B. breve UCC2003 gene clusters upregulated during growth of PGOS as sole carbohydrate source. The lengths of the arrows are proportional to the length of the predicted ORF and the gene locus name, which is indicative of its putative function, is indicated within the arrow. The bent arrows indicate the galC and galA promoters; the lollipop sign designates putative rho-independent terminator region. β-Galactosidase-encoding genes are indicated by blue shading, while genes encoding proteins with transport functions are shaded in yellow. Putative or proven genes encoding LacI-type transcriptional regulators are indicated by red shading.
Phenotypes of mutants carrying disruptions in the lacS, lacZ, gosD or gosG genes
In order to determine how disruption of particular genes from the galCDEGR, lacSZI and gosRDEGC gene clusters would impact on the ability of B. breve UCC2003 to metabolize PGOS, the previously constructed mutant strains B. breve UCC2003-galA, UCC2003-galC and UCC2003-galG were employed (O'Connell Motherway et al., 2011b). Additional insertion mutants in the B. breve lacS, lacZ, gosD and gosG genes were generated, resulting in strains B. breve UCC2003-lacS, UCC2003-lacZ, UCC2003-gosD and UCC2003-gosG respectively (Table 2). To determine the components of PGOS that were metabolized by each of these insertion mutants as compared with B. breve UCC2003, strains UCC2003 (wild type), and these seven insertion mutant strains were analysed for their ability to grow in mMRS supplemented with PGOS, lactose, lactulose or ribose (positive control) as the sole carbon source and the cell-free, post-fermentation supernatants of PGOS fermentation were analysed for remaining PGOS fractions by HPAEC-PAD. As expected, and in contrast to the wild-type, the B. breve UCC2003-lacS insertion mutant was shown to be incapable of growth on lactose (Fig. 4AII) or lactulose (results not shown) as the sole carbon source thereby identifying LacS as the main lactose/lactulose transporter in this strain. Intriguingly, B. breve UCC2003-lacZ is capable of growth on lactose (Fig. 4AII) and lactulose (results not shown) as the sole carbohydrate source provided. This growth could be attributed to the presence of one or more additional β-galactosidases that can hydrolyse lactose in the absence of functional LacZ. Candidates for such alternative β-galactosidase-encoding genes are lacZ2, which is predicted to encode a GH2 family β,1-4-galactosidase, gosG or galG (O'Connell Motherway et al., 2011b). To test this possibility these four prospective β-galactosidases, LacZ2, GosG, GalG and LacZ, were expressed in Lactococcus lactis NZ9000 using the nisin-inducible expression system (de Ruyter et al., 1996) after which β-galactosidase activity was measured in cell lysates of the various strains. Following nisin induction, cells of L. lactis NZ9000 expressing each of the four predicted bifidobacterial β-galactosidases exhibited significantly increased β-galactosidase activity thereby demonstrating that LacZ2, GosG, GalG and LacZ, can indeed hydrolyse lactose (Fig. 4B). The observed growth of the UCC2003-lacZ mutant strain in mMRS supplemented with lactose as the sole carbohydrate source is therefore likely due to the activity of one or more of the β-galactosidase enzymes specified by lacZ2, gosG or galG.
Table 2.
Bacterial Strains and Plasmids used in this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| E. coli EC101 | Cloning host, repA+ kmr | Law et al. (1995) |
| E. coli EC101-pNZ-M.BbrII + M.BbrIII | EC101 harbouring pNZ8048 derivative containing bbrIIM and bbrIIIM | O'Connell Motherway et al. (2009) |
| Bifidobacterium sp. strains | ||
| B. breve UCC2003 | Isolate from nursling stool | Maze et al. (2007) |
| B. breve UCC2003-pBC1.2 | UCC2003 harbouring pBC1.2 | This study |
| B. breve UCC2003-galA | pORI19-tet-galA-967 insertion mutant of UCC2003 | O'Connell Motherway et al. (2011b) |
| B. breve UCC2003-galA-pBC1.2-galA | pORI19-tet-galA-967 insertion mutant of UCC2003 harbouring complementationconstruct pBC1.2-galA | This study |
| B. breve UCC2003-galG | pORI19-tet-galG-410 insertion mutant of UCC2003 | O'Connell Motherway et al. (2011b) |
| B. breve UCC2003-galC | pORI19-tet-galC-701 insertion mutant of UCC2003 | O'Connell Motherway et al. (2011b) |
| B. breve UCC2003-gosD | pORI19-tet-bbr_0527 insertion mutant of UCC2003 | This study |
| B. breve UCC2003-gosG | pORI19-tet-bbr_0529 insertion mutant of UCC2003 | This study |
| B. breve UCC2003-lacS | pORI19-tet-bbr_1551 insertion mutant of UCC2003 | This study |
| B. breve UCC2003-lacZ | pORI19-tet-bbr_1552 insertion mutant of UCC2003 | This study |
| B. breve UCC2005 | Isolate from human faeces | UCC |
| B. breve JCM7017 | Isolate from human faeces | JCM |
| B. breve JCM7 019 | Isolate from infant faeces | JCM |
| B. breve NCFB2257 | Isolate from infant intestine | NCFB |
| B. breve NCFB2257-pBC1.2 | NCFB2257 harbouring plasmid pBC1.2 | This study |
| B. breve NCFB2257-pBC1.2-galA | NCFB2257 harbouring complementation construct pBC1.2-galA | This study |
| B. breve NCFB2258 | Isolate from infant intestine | NCFB |
| B. breve LMG13208 | Isolate from infant intestine | LMG |
| B. breve NCIMB11815 | Isolate from infant intestine | NCTC |
| B. breve NCIMB8815 | Isolate from human faeces | NCTC |
| B. breve NIZO658 | Isolate from human faeces | NIZO |
| B. breve UCC2008 | Isolate from infant intestine | UCC |
| B. breve UCC2009 | Isolate from infant intestine | UCC |
| B. breve UCC2010 | Isolate from human milk | UCC |
| Lactococcus lactis strains | ||
| L. lactis NZ9000 | MG1363, pepN::nisRK, nisin-inducible overexpression host | de Ruyter et al. (1996) |
| L. lactis NZ9000-pNZ-Bbr_0010 | NZ9000 containing pNZ-lacZ2 | This study |
| L. lactis NZ9000-pNZ-gosG | NZ9000 containing pNZ-gosG | This study |
| L. lactis NZ9000-pNZ-galG | NZ9000 containing pNZ-galG | O'Connell Motherway et al. (2011b) |
| L. lactis NZ9000-pNZ-lacZ | NZ9000 containing pNZ-lacZ | This study |
| Plasmids | ||
| pAM5 | pBC1-puC19-Tcr | Alvarez-Martín et al. (2007) |
| pBC1.2 | pBC1-pSC101-Cmr | Alvarez-Martín et al. (2007) |
| pBC1.2-galA | pBC1-pSC101-Cmr harbouring galA including its own promoter | This study |
| pORI19 | Emr, repA−, ori+, cloning vector | Law et al. (1995) |
| pORI19-tet-gosD | Internal 325 bp fragment of bbr_0527 and tetW cloned in pORI19 | This study |
| pORI19-tet-gosG | Internal 395 bp fragment of bbr_0529 and tetW cloned in pORI19 | This study |
| pORI19-tet-lacS | Internal 479 bp fragment of bbr_1551 and tetW cloned in pORI19 | This study |
| pORI19-tet-lacZ | Internal 609 bp fragment of bbr_1552 and tetW cloned in pORI19 | This study |
| pNZ8150 | Cmr, nisin inducible translational fusion vector | Mierau and Kleerebezem (2005) |
| pNZ-lacZ2 | Cmr, pNZ8150 derivative containing translational fusion of Bbr_0010 encoding DNA fragment to nisin inducible promoter | This study |
| pNZ-gosG | Cmr, pNZ8150 derivative containing translational fusion GosG encoding DNA fragment to nisin inducible promoter | This study |
| pNZ-galG | Cmr, pNZ8150 derivative containing translational fusion of GalG encoding DNA fragment to nisin inducible promoter | O'Connell Motherway et al. (2011b) |
| pNZ-lacZ | Cmr, pNZ8150 derivative containing translational fusion of LacZ encoding DNA fragment to nisin inducible promoter | This study |
JCM, Japan Collection of Microorganisms; LMG, Belgian Co-ordinated Collection of Microorganisms; NCFB, National Collection of Food Bacteria; NCTC, National Collection of Type Cultures; UCC, University College Cork Culture Collection.
Fig. 4.
Growth profiles of B. breve UCC2003 and insertion mutant strains on ribose (AI), lactose (AII), and PGOS (AIII). Data presented are averages of duplicate growth experiments.
B. β-Galactosidase activity assays of uninduced and nisin induced L. lactis cultures, NZ9000-pNZ8150, NZ9000-pNZ-lacZ2, NZ9000-pNZ-gosG, NZ9000-pNZ-galG and NZ9000-pNZ-lacZ.
When growth was examined using PGOS as the sole carbohydrate source, B. breve UCC2003-galA, B. breve UCC2003-galC and B. breve UCC2003-galG reached lower final optical densities as compared with UCC2003 or indeed the other four insertion mutant strains (Fig. 4AIII). These data suggest key roles for the endogalactanase encoded by galA, the ABC transport system specified by galCDE and the β-galactosidase encoded by galG in the metabolism of PGOS by B. breve UCC2003.
HPAEC-PAD analysis of the cell-free supernatants identified that the B. breve UCC2003-galA strain did not metabolize PGOS components with a predicted high degree of polymerization (retention time 30–48 min) (Fig. 5C). This finding was verified by Mass Spectroscopy analysis that demonstrated that B. breve UCC2003 consumed all detectable PGOS fractions, while B. breve UCC2003-galA could only metabolize PGOS fractions with a DP of up to and equal to three (Fig. 6). Interestingly, a GOS component present in fractions eluted from 22.5 to 25 min, of the HPAEC analysis, was shown to increase in the CFS of B. breve UCC2003-galC (Fig. 5D). This GOS component is likely β,1-4 linked galactotriose, which is the main product of the endogalactanase activity (Hinz et al., 2005; O'Connell Motherway et al., 2011b), and which is apparently not transported by this strain due to the galC mutation. When B. breve UCC2003-lacS was grown on PGOS, it was incapable of internalizing lactose, a GalA-mediated PGOS degradation product, which therefore accumulates in the growth medium, and which elutes at approximately 14 min under the conditions tested (Fig. 5F). The CFS post-fermentation profiles obtained following growth of UCC2003-galG (Fig. 5E), UCC2003-lacZ (Fig. 5G), or UCC2003-gosG (Fig. 5I) in PGOS-containing medium, closely resembled that of B. breve UCC2003, a result that was not unexpected given that GalG, LacZ and GosG are all intracellular β-galactosidases, while the extracellular endogalactanase activity and transport functions are fully functional in these mutant strains.
Fig. 5.
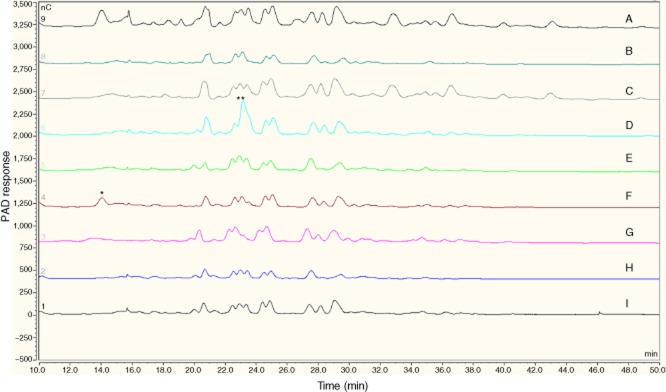
HPAEC-PAD analysis of post-fermentation cell-free supernatants of B. breve UCC2003 and insertion mutant strains. Modified Rogosa supplemented with 0.5% GOS (A); Post-fermentation supernatants of B. breve UCC2003 (B), UCC2003-galA (C), UCC2003-galC (D), UCC2003-galG (E), UCC2003-lacS (F), UCC2003-lacZ (G), UCC200-gosD (H), UCC2003-gosG (I). The position of lactose and galactotriose are indicated by asterisk (*) and double-asterisk (**) respectively.
Fig. 6.
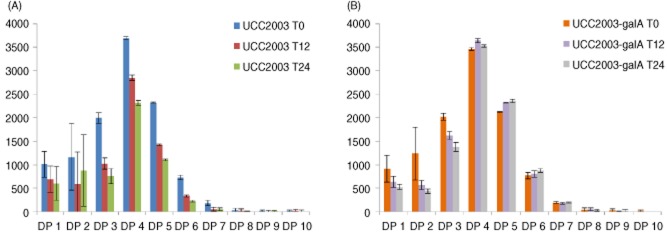
Mass Spectroscopy analysis of cell-free supernatants of B. breve UCC2003 (A) and B. breve UCC2003-galA (B). Samples were retained for analysis at 0, 12 and 24 h. Samples from duplicate experiments were run in triplicate and the data presented are averages of these six data sets with standard deviations.
Complementation of B. breve UCC2003-galA and B. breve NCFB2257
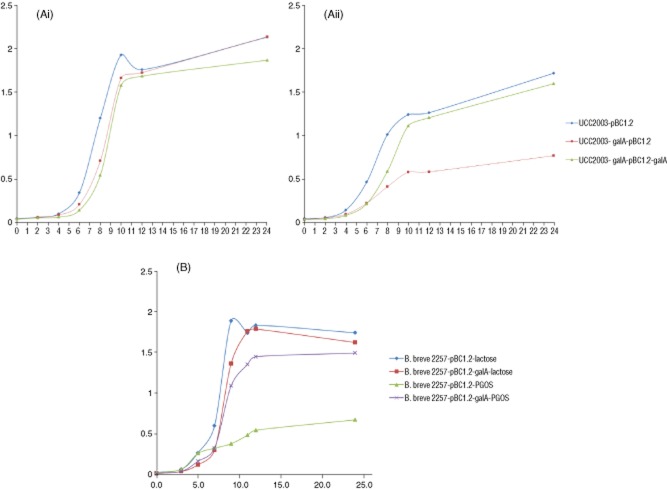
To demonstrate that the protein product of galA is uniquely necessary for the metabolism of galacto-oligosaccharides with a DP > 3, two complementation experiments were performed. In the first, the endogalactanase-encoding gene, galA, was cloned under the control of its own promoter on plasmid pBC1.2 and introduced into B. breve UCC2003-galA (to generate strain UCC2003-galA-pBC1.2-galA; see Experimental procedures). Expression of GalA in strain UCC2003-galA-pBC1.2-galA allowed this strain to grow on PGOS to an optical density comparable to B. breve UCC2003 (Fig. 7Aii), while HPAEC analysis of the post-fermentation CFS of UCC2003-galA-pBC1.2-galA clearly shows the consumption of the PGOS fractions with a high degree of polymerization (Fig. S1). For the second complementation experiment plasmid pBC1.2-galA was introduced into the GalA− strain B. breve NCFB2257 (to generate strain B. breve NCFB2257-pBC1.2-galA). Expression of GalA in B. breve NCFB2257-pBC1.2-galA enabled this strain to grow to a higher optical density compared with the wild type, B. breve NCFB2257, when grown on PGOS as the sole carbohydrate source. This PGOS-dependent growth profile of B. breve NCFB2257-pBC1.2-galA was comparable to those observed for GalA+ B. breve strains (Fig. 7B), while HPAEC analysis demonstrated that B. breve NCFB2257-pBC1.2-galA could metabolize PGOS components with a high degree of polymerization (Fig. S2). These complementation experiments unequivocally demonstrate the role of GalA in the metabolism of PGOS with a high degree of polymerization and long retention time.
Fig. 7.
Growth profiles of (A) B. breve UCC2003 pBC1.2, UCC2003-galA-pBC1.2 and UCC2003-galA-pBC1.2-galA on lactose (Ai) and PGOS (Aii) and (B) B. breve NCFB2257-pBC1.2 and NCFB2257-pBC1.2-galA on lactose and PGOS. Data presented are averages of duplicate experiments.
Discussion
Several studies have demonstrated that bifidobacteria dedicate a significant portion of their coding capacity to the metabolism of a wide variety of carbohydrates (Schell et al., 2002; Ventura et al., 2007a,b). Over 50 different bifidobacterial carbohydrases have been described in the literature to date (reviewed by Van den Broek et al., 2008; Pokusaeva et al., 2011a). Using B. breve UCC2003 as a model to study bifidobacterial carbohydrate metabolism, we previously characterized an operon encoding a β-fructofuranosidase (Ryan et al., 2005), an extracellular amylopullulanase which hydrolyses α-1,4 and α-1,6 glucosidic linkages in starch and related polysaccharides (Ryan et al., 2006; O'Connell Motherway et al., 2012), two α-glucosidases exhibiting hydrolytic activities towards panose, isomaltose, isomaltotriose and four sucrose isomers-palatinose, trehalulose, turanose and maltulose (Pokusaeva et al., 2009), as well as gene clusters dedicated to ribose (Pokusaeva et al., 2010), galactan (O'Connell Motherway et al., 2011b) and cellodextrin (Pokusaeva et al., 2011b) metabolism. In addition, a PEP-PTS system involved in fructose metabolism was identified and studied in this bacterium (Maze et al., 2007).
In this study, we describe the identification and functional characterization of genetic loci involved in the utilization of PGOS by B. breve UCC2003. To date several studies have demonstrated the presence of β-galactosidase isoenzymes in bifidobacteria (Van Laere et al., 2000; Barboza et al., 2009; Goulas et al., 2009; Osman et al., 2010); however, to our knowledge this is the first study adopting functional genomics approaches to understand the apparent synergistic relationship between β-galactosidases and their associated transport systems required for PGOS metabolism by bifidobacteria.
A range of B. breve strains was tested for their ability to grow on PGOS as the sole carbohydrate source, and while all strains were shown to grow well on PGOS, reaching final optical densities of ≥ 0.8, two distinct groups could be identified based on the growth profiles and analysis of the post-fermentation cell-free supernatants by HPAEC-PAD. The first group included B. breve UCC2003 and for this group HPAEC-PAD analysis showed that all strains metabolized most GOS components including those with a high degree of polymerization. Interestingly and consistent with our expectations, CGH analysis (O'Connell Motherway et al., 2011a) revealed that these strains each encoded an extracellular endogalactanase. The strains in the second group also grew well on PGOS but reached a lower final optical density; however, HPAEC-PAD analysis of the post-fermentation CFS clearly showed that oligosaccharides with a high degree of polymerization remained in the CFS of these strains and were not metabolized. This correlated with the CGH analysis where each of these strains was found to lack the galA gene. These data suggest a key role for GalA in the metabolism of galacto-oligosaccharides with a high degree of polymerization.
DNA microarrays that allowed comparison of gene expression of exponentially growing B. breve UCC2003 when grown on either PGOS or ribose as the sole carbohydrate source identified four gene clusters that are highly upregulated. These include the previously described galactan gene cluster, galBCDEGRA (O'Connell Motherway et al., 2011b), as well as three additional clusters gosRDEGC, lacSZI and galTK. Here we demonstrate that compared with the parent strain UCC2003, the galA, galC and galG mutant strains exhibited impaired growth on PGOS, thereby demonstrating that the endogalactanase, ABC transport system and β-galactosidase encoded by the gal locus significantly contribute to the extracellular metabolism, transport and subsequent intracellular metabolism of PGOS oligosaccharides respectively. Furthermore, analysis of the CFS of UCC2003-galA by HPAEC-PAD following growth on PGOS identified the presence of oligosaccharide fractions with a high degree of polymerization as observed for the galA− B. breve strains and further supporting the importance of GalA in the metabolism of GOS oligosaccharide fractions with a high degree of polymerization. Our recent studies on starch (O'Connell Motherway et al., 2012) and cellodextrin (Pokusaeva et al., 2011b) metabolism by B. breve UCC2003 have demonstrated that the associated ABC transport systems can internalize malto-oligosaccharides or cellodextrins with a DP of 2–5. We would therefore expect that the B. breve gal and gos locus encoded ABC transport systems would transport PGOS fractions with similar relatively low degrees of polymerization and that utilization of GOS oligosaccharide fractions with a high degree of polymerization requires initial extracellular degradation by endogalactanase. This expectation was confirmed by Mass Spectroscopy analysis which demonstrated that GalA enables bifidobacterial cells to metabolize PGOS components with a DP ≥ 4. It is plausible that other Bifidobacterium sp. may possess an extracellular β-galactosidase that may fulfil a similar role; however, to date just two extracellular β-galactosidases from bifidobacteria have been described in the literature: while Moller and colleagues (2001) did not evaluate the ability of the extracellular β-galactosidase, BIF3, from B. bifidum DSM20215 to hydrolyse GOS, BbgIII from B. bifidum NCIMB41171 was found to have a high affinity for lactose hydrolysis but exhibited relatively low GOS hydrolysis activity (Goulas et al., 2009).
Our results also demonstrate a significant role for the LacS symporter in transporting lactose, lactulose and possibly other GOS di-/oligo-saccharides into B. breve UCC2003. Collectively these data demonstrate that PGOS metabolism by B. breve UCC2003 can be attributed to the cooperative activity of multiple β-galactosidases and their associated transport systems. Our observations are consistent with those of Barboza and colleagues (2009) who examined PGOS consumption of four bifidobacterial strains, namely B. breve ATCC15700, B. longum subsp. infantis ATCC15697, B. adolescentis DSM20083 and B. longum subsp. longum DJ010A, and observed strain specific preferences for PGOS utilization. Interestingly, and in contrast to our findings for B. breve UUCC2003, Andersen and colleagues (2011) have recently reported that LacS is the sole transporter of lactose, lactulose, lactitol and GOS into Lactobacillus acidophilus NCFM although it has yet to be demonstrated if L. acidophilus NCFM can utilize GOS components with a high degree of polymerization.
The ability of probiotic strains to ferment particular oligo- and poly-saccharides has been the basis for their selection as prebiotics and incorporation into functional foods. The health benefits associated with GOS are well documented and many placebo controlled clinical trials have explored the health benefits of incorporating GOS in functional food formulations. It is now well documented that GOS promotes the proliferation of beneficial bacteria, in particular bifidobacteria, in the gut, which can subsequently provide protection against colonization with pathogens thereby reducing the incidence of infections. In addition, metabolism of GOS by the gut microbiota leads to the production of short-chain fatty acids (SCFAs). These SCFAs are reported to have numerous beneficial health effects that include reducing cancer risk, increased mineral absorption, improving bowel habit, stool consistency and reduction of IBD inflammation (Davis et al., 2011 reviewed by MacFarlane et al., 2008; Sangwan et al., 2011).
Future research will focus on characterizing the specific PGOS components that are metabolized by B. breve UCC2003, in particular identifying the PGOS structures that are metabolized by GalA, identifying the GOS components that are transported by the ABC-transport systems and LacS. It will also be necessary to determine if differences exist in substrate specificities of the identified β-galactosidases. The results presented here significantly advance our knowledge of GOS metabolism by bifidobacteria and, in due course, may contribute towards the development of targeted bifidogenic galacto-oligosaccharides, for specific probiotic strains. The incorporation of such galacto-oligosaccharides in foods has potential for the development of enhanced functional foods or infant food formulas.
Experimental procedures
The description of the Experimental procedures resides in Appendix S1 in Supporting information.
Acknowledgments
The authors wish to acknowledge FrieslandCampina for the gift of Purified GOS. The Alimentary Pharmabiotic Centre is a research centre funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan. The authors and their work were supported by SFI (Grant Nos 02/CE/B124 and 07/CE/B1368) and a HRB postdoctoral fellowship (Grant No. PDTM/20011/9) awarded to MOCM. The Mass Spectroscopy Instrument and analysis was funded under the Higher Education Authority Programme for Research in Third-Level Institutions (PRTLI) and co-funded under the European Regional Development Fund.
Conflict of interest
None declared.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. HPAEC-PAD analysis of post fermentation cell free supernatants of B. breve UCC2003, UCC2003-gal A and complemented strain. Modified Rogosa supplemented with 0.5 % GOS (A); post-fermentation supernatants of B. breve UCC2003 (B), UCC2003-galA-pBC1.2 (C) and UCC2003-galA-pBC1.2-galA (D).
Fig. S2. HPAEC-PAD analysis of post fermentation cell free supernatants of B. breve NCFB2257-pBC1.2, and GalA complemented derivative. Modified Rogosa supplemented with 0.5 % GOS (A); post-fermentation supernatants of B. breve NCFB2257-pBC1.2 (B) and B. breve NCFB2257-pBC1.2-galA (C).
Appendix S1. Experimental procedures.
References
- Alvarez-Martín P, O'Connell-Motherway M, van Sinderen D, Mayo B. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol. 2007;76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
- Andersen JM, Barrangou R, Abou Hachem M, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. Transcriptional and functional analysis of galactooligosaccharide uptake by LacS in Lactobacillus acidophilus. Proc Natl Acad Sci USA. 2011;108:17785–17790. doi: 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza M, Sela DA, Pirim C, LoCascio RG, Freeman SL, German JB, et al. Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl Environ Microbiol. 2009;75:7319–7325. doi: 10.1128/AEM.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulier L, Timmermans J, Bas R, Van Den Dool R, Haaksman I, Klarenbeek B, et al. In-depth characterization of prebiotic galacto-oligosaccharides by a combination of analytical techniques. J Agric Food Chem. 2009;23:8488–8495. doi: 10.1021/jf902549e. [DOI] [PubMed] [Google Scholar]
- Davis LM, Martínez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS ONE. 2011;6:e25200. doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eversbach T, Jørgensen JB, Heegaard PM, Lahtinen SJ, Ouwehand AC, Poulsen M, et al. Certain dietary carbohydrates promote Listeria infection in a guinea pig model, while others prevent it. Int J Food Microbiol. 2010;140:218–224. doi: 10.1016/j.ijfoodmicro.2010.03.030. [DOI] [PubMed] [Google Scholar]
- Fushinobu S. Unique sugar metabolic pathways of bifidobacteria. Biosci Biotechnol Biochem. 2010;74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- Goulas T, Goulas A, Tzortzis G, Gibson GR. Expression of four beta-galactosidases from Bifidobacterium bifidum NCIMB41171 and their contribution on the hydrolysis and synthesis of galactooligosaccharides. Appl Microbiol Biotechnol. 2009;84:899–907. doi: 10.1007/s00253-009-2009-5. [DOI] [PubMed] [Google Scholar]
- Hinz SW, Pastink MI, van den Broek LA, Vincken JP, Voragen AG. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl Environ Microbiol. 2005;71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Niñonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microbiol Biotechnol. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr Opin Biotechnol. 2007;18:156–162. doi: 10.1016/j.copbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21:149–156. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Martín R, Langa S, Reviriego C, Jimínez E, Marín ML, Xaus J, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Martín R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1:367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- Maze A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, Van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- Moller P, Flemming J, Hansen OC, Madsen SM, Stougaard P. Intra- and extracellular B-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterisation. Appl Environ Microbiol. 2001;67:2276–2283. doi: 10.1128/AEM.67.5.2276-2283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niñonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Niñonuevo MR, Lebrilla CB. Mass spectrometric methods for analysis of oligosaccharides in human milk. Nutr Rev. 2009;67(Suppl. 2):S216–S226. doi: 10.1111/j.1753-4887.2009.00243.x. [DOI] [PubMed] [Google Scholar]
- Nwosu CC, Aldredge D, Lee H, Lerno LA, Jr, Zivkovic AM, German JB, Lebrilla CB. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11:2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, Van Sinderen D. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, O' Driscoll J, Fitzgerald GF, Van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011a;108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Fitzgerald GF, Van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol. 2011b;4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman A, Tzortzis G, Rastall RA, Charalampopoulos D. A comprehensive investigation of the synthesis of prebiotic galactooligosaccharides by whole cells of Bifidobacterium bifidum NCIMB 41171. J Biotechnol. 2010;150:140–148. doi: 10.1016/j.jbiotec.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Osman A, Tzortzis G, Rastall RA, Charalampopoulos D. BbgIV Is an important bifidobacterium β-galactosidase for the synthesis of prebiotic galactooligosaccharides at high temperatures. J Agric Food Chem. 2012 doi: 10.1021/jf204719w. (in press) [DOI] [PubMed] [Google Scholar]
- Perez PF, Doré J, LeClerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K, O'Connell-Motherway M, Zomer A, Fitzgerald GF, Van Sinderen D. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2009;75:1135–1143. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, Neves AR, Zomer A, O'Connell Motherway M, MacSharry J, Curley P, et al. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, Fitzgerald GF, Van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011a;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, O'Connell-Motherway M, Zomer A, MacSharry J, Fitzgerald GF, Van Sinderen D. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2011b;77:1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero M, Maldonado M, Perez-Munoz M, Jimenez R, Fangman T, Rupnow J, et al. Adherence inhibition of Cronobacter sakazakii to intestinal epithelial cells by prebiotic oligosaccharides. Curr Microbiol. 2011;62:1448–1454. doi: 10.1007/s00284-011-9882-8. [DOI] [PubMed] [Google Scholar]
- de Ruyter PG, Kuipers O, de Vos WM. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SM, Fitzgerald GF, Van Sinderen D. Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2005;71:3475–3482. doi: 10.1128/AEM.71.7.3475-3482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SM, Fitzgerald GF, Van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan V, Tomar SK, Singh RR, Singh AK, Ali B. Galactooligosaccharides: novel components of designer foods. J Food Sci. 2011;76:R103–R111. doi: 10.1111/j.1750-3841.2011.02131.x. [DOI] [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle LE, Cooley WA, Jones G, Nunez A, Crudgington B, Weyer U, et al. Purified galactooligosaccharide, derived from a mixture produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium adhesion and invasion in vitro and in vivo. J Med Microbiol. 2010;59:1428–1439. doi: 10.1099/jmm.0.022780-0. [DOI] [PubMed] [Google Scholar]
- Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek LA, Hinz SW, Beldman G, Vincken JP, Voragen AG. Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res. 2008;52:146–163. doi: 10.1002/mnfr.200700121. [DOI] [PubMed] [Google Scholar]
- Van Laere KM, Abee T, Schols HA, Beldman G, Voragen AG. Characterization of a novel beta-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl Environ Microbiol. 2000;66:1379–1384. doi: 10.1128/aem.66.4.1379-1384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007a;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, O'Connell-Motherway M, Leahy S, Moreno-Munoz JA, Fitzgerald GF, van Sinderen D. From bacterial genome to functionality; case bifidobacteria. Int J Food Microbiol. 2007b;120:2–12. doi: 10.1016/j.ijfoodmicro.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Walker A. Breast milk as the gold standard for protective nutrients. J Pediatr. 2010;156(2 Suppl):S3–S7. doi: 10.1016/j.jpeds.2009.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.