Secondary metabolites (or natural products) are often synthesized by multi‐modular, multi‐domain proteins called non‐ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS). Various well‐known metabolites produced by microorganisms are listed in Table 1, and examples of structures are shown in Fig. 1. In particular, Streptomyces species are known for their ability to produce a wide variety of secondary metabolites such as antibiotics, herbicides, parasitocides, siderophores and pharmacologically active substances including antitumour agents and immunosuppressants. Genome sequencing of Streptomyces coelicolor (Bentley et al., 2002) and S. avermitilis (Omura et al., 2001) revealed over 20 gene clusters for biosynthesis of secondary metabolites, while only a few of their natural products were known prior to sequencing. High‐throughput genome sequencing of hundreds of other bacterial species and strains is now rapidly increasing the repertoire of identified gene clusters for biosynthesis of natural products (Donadio et al., 2007). Here we give a brief update of the current status of genome mining and bioinformatic tools to identify novel NRPS and PKS systems.
Table 1.
Examples of microbial natural products produced by NRPS/PKS systems.
| Natural product | Microorganism | NRP/PK |
|---|---|---|
| Antibiotics | ||
| Penicillin | Penicillium chrysogenum (fungi) | NRP |
| Bacitracin | Bacillus licheniformis | NRP |
| Tyrocidin | Bacillus brevis | NRP |
| Cephalosporin | Streptomyces clavuligerus | NRP |
| Erythromycin | Saccharopolyspora erythrea | PK |
| Tetracycline | Streptomyces aureofaciens | PK |
| Actinomycin | Streptomyces chrysomallus | NRP |
| Antitumour agents | ||
| Dolastatin 10 | Symploca species (cyanobacteria) | NRP |
| Bleomycin | Streptomyces verticillus | Hybrid NRP/PK |
| Chondramide | Chondromyces crocatus | Hybrid NRP/PK |
| Epothilone | Sorangium cellulosum | Hybrid NRP/PK |
| Immunosuppressants | ||
| Cyclosporin | Tolypocladium inflatum (fungi) | NRP |
| Rapamycin | Streptomyces hygroscopicus | PK |
| FK506 | Streptomyces sp. | PK |
| FK520 | Streptomyces hygroscopicus | PK |
| Protease inhibitors | ||
| Anabaenopeptin | Anabaena flos‐aquae (cyanobacteria) | NRP |
| Oscillamide | Oscillatoria agardhii (cyanobacteria) | NRP |
| Siderophores | ||
| Mycobactin | Mycobacterium tuberculosis | NRP |
| Bacillibactin | Bacillus subtilis | NRP |
| Enterobactin | Escherichia coli | NRP |
| Yersiniabactin | Yersinia pestis | Hybrid NRP/PK |
| Toxins | ||
| Mycolactone | Mycobacterium ulcerans | PK |
| Naphthazarins | Fusarium oxysporum (fungi) | PK |
| HC‐toxin | Cochliobolus carbonum (fungi) | NRP |
Bacteria unless otherwise indicated.
Figure 1.
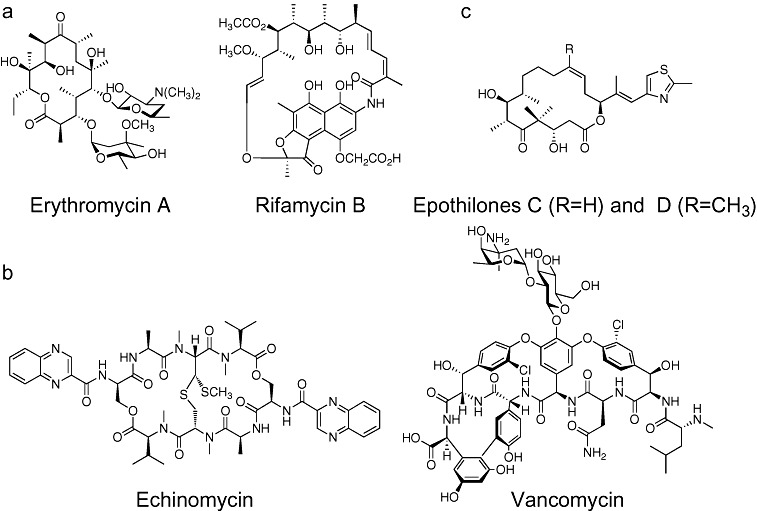
Examples of some chemical structures of (A) polyketides, (B) non‐ribosomal peptides and (C) mixed NRP‐PK compounds. Reprinted with permission from Watanabe and Oikawa (2007). Copyright Royal Society of Chemistry.
Polyketide and non‐ribosomal peptide biosynthesis
Both NRPS and PKS systems are molecular assembly lines for successive linking of multiple‐amino/hydroxy acids or acyl‐CoA precursors, respectively, into complex polymers which are often further modified into unique structures (Table 1, Fig. 1). The basic steps of both systems are initiation, elongation and termination performed by separate modules of the synthases (Fig. 2). These modules and others are usually encoded in large gene clusters (Khosla et al., 1999; Crosa and Walsh, 2002; Donadio et al., 2007; Rokem et al., 2007).
Figure 2.
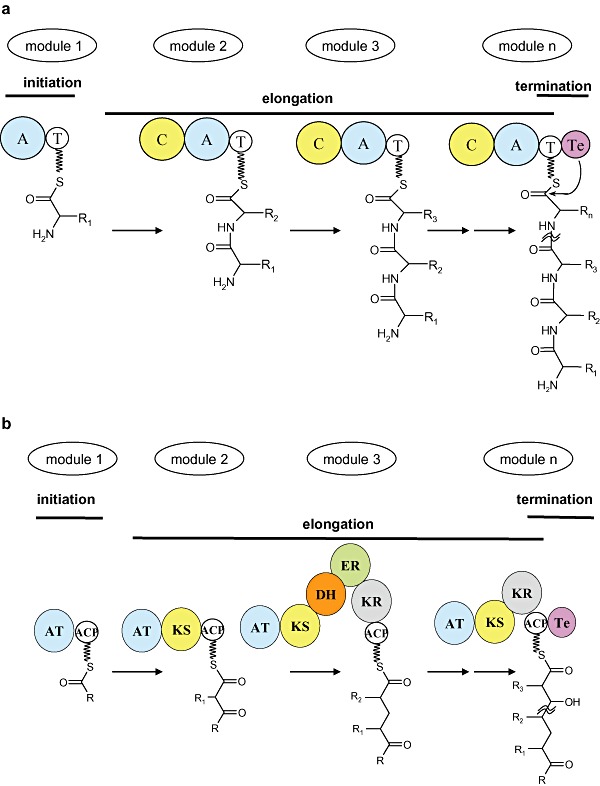
Basic steps during (A) non‐ribosomal peptide synthesis and (B) polyketide synthesis. Adapted with permission from Donadio and colleagues (2007). Copyright Royal Society of Chemistry.
Non‐ribosomal peptide synthetase modules can contain four principal domains (Fig. 2A): an adenylation domain (A) that selects, activates and loads the building blocks (proteinogenic and non‐proteinogenic amino acids or carboxylic acids), a thiolation domain (T), also known as peptidyl carrier protein (PCP) that covalently fixes the amino acid on the synthetase, a condensation domain (C) that catalyses the peptide bond formation, and a thio‐esterase domain (Te) that releases the assembled peptide from the synthetase (Sieber and Marahiel, 2005; Wenzel and Muller, 2005). The diversity in structure and composition of the products is achieved due to different specificities of the A domains and further modifications by gene cluster‐embedded or stand‐alone additional domains such as methyltransferase (MT), epimerization (E), cyclization (Cy) and others (Walsh et al., 2001). The assembled final peptide structures range from linear [such as the pentadecapeptide gramicidin (Kessler et al., 2004)], to branched [such as vibriobactin (Keating et al., 2000)], partially cyclic [such as daptomycin (McHenney et al., 1998)], cyclic [such as gramicidin S (Erlanger and Goode, 1960)] or bicyclic [such as actinomycin (Pfennig et al., 1999)].
Polyketide synthase modules can contain four core domains (Fig. 2B): an acyltransferase (AT) domain that selects and activates the acyl‐CoA building blocks (such as acetyl‐CoA, malonyl‐CoA, methylmalonyl‐CoA and ethylmalonyl‐CoA), an acyl carrier (ACP) domain, a keto‐acylsynthase (KS) condensation domain and a releasing thio‐esterase (Te) domain. The modules may contain other modification domains such as ketoreductase (KR), dehydratase (DH) and enoylreductase (ER). Polyketide synthases generate enzyme‐bound ketoacyl intermediates in stepwise decarboxylative condensations between the extender building blocks and the growing polyketide chain in a process similar to fatty acid synthesis. An example of such an assembly process is shown in Fig. 3.
Figure 3.

Biosynthetic pathway, module and domain organization of two polyketide synthases (Type I) (MlsA1 and MlsA2) responsible for mycolactone core biosynthesis in Mycobacterium ulcerans. Reprinted with permission from http://www.med.monash.edu.au/microbiology/research/stinear.html.
Prediction of structure of non‐ribosomally synthesized peptides
In most of the NRPS systems known so far, the order and structure of building blocks present in the secondary polypeptide product are reflected by the modular architecture of the NRPS. This relation between the template and the product is referred to as co‐linearity rule. The specificity of A domains as well as the role of the other modifying domains will specify the composition of the produced polypeptide. General rules for predicting substrate specificity of A domains were initially developed based on the crystal structure of an adenylation domain of gramicid in synthetase (Stachelhaus et al., 1999; Challis et al., 2000). The NRPSpredictor (http://www.ab.informatik.uni‐tuebingen.de/toolbox) uses transductive support vector machines (TSVMs) as a predictive tool for detecting substrate specificities of A domains (Rausch et al., 2005) based on the physicochemical properties of substrate‐binding pocket residues.
In silico genome screening for NRPS/PKS gene clusters
There are several bioinformatic tools available for searching NRPS/PKS systems in genome sequences. The NRPS‐PKS tool is web‐based software (http://www.nii.res.in/nrps‐pks.html) for analysing the large multi‐enzymatic, multi‐domain megasynthases (Ansari et al., 2004). The results of these analyses have been organized as four searchable databases for elucidating domain organization and substrate specificity of NRPS and PKS. These databases provide an interface to correlate chemical structures of these natural products with the domains and modules in the corresponding PKS or NRPS. ASMPKS is a web‐based tool (http://gate.smallsoft.co.kr:8008/~hstae/asmpks/index.html) for computational analysis of PKS systems against genome sequences (Tae et al., 2007). The ASMPKS can predict functional modules for each protein sequence, estimate the chemical composition of a polyketide synthesized from the modules, and display the carbon chain structure on the web interface. Another recent method to accurately predict PK/NRP structures from genome sequences is described by Minowa and colleagues (2007). Norine (http://bioinfo.lifl.fr/norine) is a platform that includes a database of non‐ribosomal peptides (currently more than 700) together with tools for their analysis. The Norine database stores peptide structures as well as various annotations such as the biological activity, producing organisms, bibliographical references and others (Caboche et al., 2008).
Analysis of over 220 completed bacterial genomes up to 2005 revealed that PKS and NRPS systems are mainly found in actinobacteria, β‐proteobacteria, γ‐proteobacteria, firmicutes and cyanobacteria (Donadio et al., 2007). We have now analysed the 140 most recently sequenced microbial genomes (July 2007–April 2008; GOLD database http://www.genomesonline.org/) using Hidden Markov Model profiles of all core domains of both NRPS and PKS. Many of these genomes are publicly accessible in the NCBI database but have not been described in the scientific literature yet (Siezen and Wilson, 2008). Numerous NRPS/PKS systems were found, and Table 2 lists the genomes with three or more systems; several are described in more detail below. They are mainly found in microorganisms with genomes larger than 4 Mb isolated from soil or aquatic environments. In addition, at least two NRPS or PKS systems are predicted in Yersinia pseudotuberculosis IP 31758, Azorhizobium caulinodans ORS 571, Marinomonas sp. MWYL1 and Bacillus cereus cytotoxis NVH 391‐98, while at least one system is predicted in Escherichia coli HS, Coxiella burnetii Dugway 7E9‐12, Enterobacter sakazakii ATCC BAA‐894, Staphylococcus aureus ssp. aureus Mu3, Vibrio harveyi BB120, Serratia proteamaculans 568, Delftia acidovorans SPH‐1, Salmonella enterica arizonae sv. 62:z4,z23 RSK2980, Klebsiella pneumonia MGH78578 and Kineococcus radiotolerans SRS30216. Quite a number of the latter bacteria are human pathogens.
Table 2.
Recently sequenced bacterial genomes (1 July 2007 to April 2008) with at least three predicted NRPS/PKS gene clusters.
| Species | Habitat | Genome size (Mb) | Gene clusters (predicted) | Reference and/or NCBI code |
|---|---|---|---|---|
| Sorangium cellulosum So ce56 | Soil | 13.0 | 3 NRPS | Schneiker et al. (2007) |
| 6 PKS | NC_010162 | |||
| 4 NRPS/PKS | ||||
| Salinispora tropica CNB‐440 | Marine, sediment | 5.2 | 3 NRPS | Udwary et al. (2007) |
| 6 PKS | NC_009380 | |||
| 4 NRPS/PKS | ||||
| Streptomyces griseus IFO13350 | Soil | 8.5 | 9 NRPS | Ohnishi et al. (2008) |
| 5 PKS | NC_010572 | |||
| 4 NRPS/PKS | ||||
| Salinispora arenicola CNS205 | Marine, sediment | 5.8 | 4 NRPS | NC_009953 |
| 2 NRPS/PKS | ||||
| 2 PKS | ||||
| 2 ambiguous PKS | ||||
| Frankia sp. EAN1pec | Plant symbiont, soil | 9.0 | 2 NRPS | NC_009921 |
| 3 PKS | ||||
| 5 ambiguous PKS | ||||
| Bacillus amyloliquefaciens FZB42 | Rhizosphere‐colonizing, soil | 3.9 | 4 NRPS | Chen et al. (2007) |
| 2 PKS | NC_009725 | |||
| 2 NRPS/PKS | ||||
| 1 ambiguous NRPS | ||||
| Herpetosiphon aurantiacus ATCC 23779 | Aquatic | 6.4 | 5 NRPS | NC_009972 |
| 4 NRPS/PKS | ||||
| Pseudomonas aeruginosa PA7 | Soil, aquatic, host (human) | 6.6 | 5 NRPS | NC_009656 |
| Xanthobacter autotrophicus Py2 | Soil, aquatic, sediment | 4.8 | 2 NRPS | NC_009720 |
| 2 ambiguous PKS | ||||
| 1 NRPS/PKS | ||||
| Clostridium kluyveri DSM 555 | Aquatic, mud | 4.0 | 1 NRPS | Seedorf et al. (2008) |
| 3 NRPS/PKS | NC_009706 | |||
| Bacillus pumilus SAFR‐032 | Soil | 3.7 | 2 NRPS | Gioia et al. (2007) |
| 1 NRPS/PKS | NC_009848 | |||
| Citrobacter koseri ATCC BAA‐895 | Soil, aquatic, food, human intestine | 4.7 | 3 NRPS/PKS | NC_009792 |
Recently sequenced microbial genomes with large potential for production of NRPS/PKS natural products
Sorangium cellulosum is a soil‐dwelling δ‐proteobacterium of the group myxobacteria. The genus Sorangium synthesizes approximately half of the secondary metabolites isolated from myxobacteria, including the anticancer metabolite epothilone. Seventeen secondary metabolite loci are encoded in the genome of strain So ce56 (Schneiker et al., 2007), mostly PKS and NRPS systems (Table 2). Known products are chivosazol, etnangien and myxochelin, while others are still unknown. Metabolites secreted by S. cellulosum known as epothilones have been noted to have antineoplastic activity. This has led to the development of analogues that mimic its activity. One such analogue, known as Ixabepilone, is a US Food and Drug Administration (FDA)‐approved chemotherapy agent for the treatment of metastatic breast cancer.
The soil actinomycete Streptomyces griseus produces the well‐known antituberculosis agent streptomycin. Recent sequencing of the genome of S. griseus IFO 13350 shows that it has 34 gene clusters or genes for biosynthesis of secondary metabolites, of which 14 PKS or NRPS gene clusters seem to be specific for this species (Ohnishi et al., 2008). These clusters presumably direct the synthesis of various as yet unknown secondary metabolites.
Actinomycetes of the marine‐dwelling genus Salinispora are a rich source of drug‐like molecules. Salinispora strains are commonly isolated from tropical marine sediment, and many isolates produce compounds that inhibit cancer cells, such as salinosporamide A (Feling et al., 2003). The Salinispora tropica CNB‐440 genome dedicates nearly 10% of its genome to natural product assembly (Udwary et al., 2007), which is greater than S. coelicolor and S. avermitilis as well as other secondary metabolite‐producing actinomycetes. The S. tropica genome features PKS systems of every known formally classified family, NRPS systems and several hybrid clusters. The majority of the 17 biosynthetic loci are novel. Genome sequencing is ongoing of Salinispora arenicola CNS‐205, a producer of the bioactive compounds staurosporine and rifamycin which may be useful in the treatment of cancer. Other marine actinobacteria are also potential sources of bioactive natural products (Bull and Stach, 2007)
Frankia species form a separate lineage among the high % G+C Gram‐positive Actinobacteria. They are filamentous ‘euactinomycetes’ that grow by hyphal branching and tip extension and thus resemble the antibiotic‐producing Streptomyces species. Frankia species form a symbiotic nitrogen‐fixing association with a number of plants. These symbioses add a large proportion of new nitrogen to several ecosystems. The genome of Frankia sp. strain EAN1pec has all housekeeping genes necessary for saprophytic existence plus genes for sporulation, vesicle development, symbiosis, N2 fixation and secondary metabolite production. Ten putative NRPS/PKS clusters were identified in the genome sequence of strain EAN1pec.
Bacillus amyloliquefaciens is a Gram‐positive bacterium belonging to the firmicutes. It is member of a group of free‐living soil bacteria known to promote plant growth and suppress plant pathogenic bacteria and fungi. The B. amyloliquefaciens FZB42 genome reveals an unexpected potential to produce secondary metabolites, with more than 8.5% of the genome devoted to synthesizing antibiotics and siderophores by NRPS and PKS pathways (Chen et al., 2007). Besides five gene clusters known from Bacillus subtilis to mediate biosynthesis of secondary metabolites (surfactin, fengycin, bacillibactin, bacilysin, bacillaene), an additional four giant gene clusters were identified for biosynthesis of bacillomycin D, macrolactin, difficidin and a putative siderophore. Bacillus spores are notoriously resistant to unfavourable conditions such as UV radiation, γ‐radiation, H2O2, desiccation, chemical disinfection or starvation. Bacillus pumilus SAFR‐032 spores and vegetative cells exhibit elevated resistance to UV radiation and H2O2 compared with other Bacillus species, and its genome sequence provides insight into numerous DNA repair and oxidative stress pathways (Gioia et al., 2007). It also encodes three NRPS/PKS systems of unknown function.
Clostridium kluyveri DSM555, a strictly anaerobe Firmicute, was isolated from canal mud in the Netherlands. It is unique among clostridia in that it can grow on ethanol and acetate as sole energy sources, producing butyrate, caproate and H2 (Seedorf et al., 2008). Furthermore, it is biotechnologically interesting as the genome sequence predicts that it could ferment ethanol and glycerol to 1,3‐propanediol. Quite unexpected in an anaerobe Firmicute is the presence of three hybrid PKS‐NRPS clusters of unknown function, and one NRPS gene cluster which is predicted to synthesize a yersiniabactin/pyochelin‐like siderophore.
Chloroflexi are a class of eubacteria that produce energy through photosynthesis. They make up the bulk of the filamentous anoxygenic phototrophs (formerly known as green non‐sulfur bacteria). The phylum Chloroflexi accommodates additional genera, including filamentous but non‐phototrophic species. Herpetosiphon aurantiacus is a non‐phototrophic, strictly aerobic, gliding bacterium. Herpetosiphon spp. have been found in soil, freshwater and sewage treatment plants and grow in microbial mats. The genome of H. aurantiacus strain ATCC 23779 is predicted to encode nine NRPS/PKS systems of unknown function.
Xanthobacter autotrophicus, an α‐proteobacterium, is a nitrogen‐fixing methylotroph, commonly isolated from organic‐rich soil, sediment and water. Xanthobacter autotrophicus strain Py2 is unique in that it can use propene as a sole carbon and energy source, converting it to epoxypropane using an alkene‐specific monooxygenase. The monooxygenase gene and other genes involved in alkene degradation are located on a 320 kb megaplasmid. The genome sequence provides further information on the production and regulation of the genes involved in alkene degradation. The genome also has five putative NRPS/PKS gene clusters of as yet unknown function.
High‐throughput experimental screening for NRPS/PKS gene clusters
The newly discovered gene clusters for NRP and PK synthesis represent a tremendous source of novel bioactive compounds, but in most cases the natural product is unknown. Classical methods to characterize the products include heterologous expression of gene clusters (Wenzel and Muller, 2005), metabolic profiling and assay‐guided fractionation (Zazopoulos et al., 2003; McAlpine et al., 2005). A novel ‘genomisotopic’ approach uses a combination of genomic sequence analysis and isotope‐guided fractionation to identify unknown compounds synthesized by NRPS gene clusters (Gross et al., 2007). A phage‐display method was developed for high‐throughput mining of gene clusters encoding PKS and NRPS systems, which can be applied to genomes of unknown sequence and metagenomes (Yin et al., 2007), providing opportunities for exploiting the potentially rich source of natural products from unculturable microbes.
Novel natural products and applications
The past decade has already seen numerous examples of genetic engineering, metabolic engineering, rational design, and directed evolution of NRPS and PKS systems to provide novel compounds based on known NRPS/PKS gene clusters for biosynthesis of natural products (Stachelhaus et al., 1996; Cane et al., 1998; Chartrain et al., 2000; Du and Shen, 2001; Du et al., 2001). The impact of systems biology to control and regulate secondary metabolite production has only recently been addressed (Rokem et al., 2007). The ever‐increasing pace of microbial genome sequencing is revealing a plethora of new NRPS/PKS gene clusters, mostly of unknown function. A major challenge for the next decade is to back this up with characterization of the chemical structures and biological activities of these secondary metabolites, so that we can chart Nature's unique repertoire of natural products and exploit them for the directed synthesis of novel molecules of biotechnological, agricultural and pharmaceutical utility.
Acknowledgments
We thank Kenji Watanabe, Stefano Donadio and Tim Stinear for permission to use Figs 1–3, respectively, and Greer Wilson for reading and correcting the manuscript. This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
References
- Ansari M.Z., Yadav G., Gokhale R.S., Mohanty D. NRPS‐PKS: a knowledge‐based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32:W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley S.D., Chater K.F., Cerdeno‐Tarraga A.M., Challis G.L., Thomson N.R., James K.D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. et al. [DOI] [PubMed] [Google Scholar]
- Bull A.T., Stach J.E. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Caboche S., Pupin M., Leclere V., Fontaine A., Jacques P., Kucherov G. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 2008;36:D326–D331. doi: 10.1093/nar/gkm792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane D.E., Walsh C.T., Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- Challis G.L., Ravel J., Townsend C.A. Predictive, structure‐based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Chartrain M., Salmon P.M., Robinson D.K., Buckland B.C. Metabolic engineering and directed evolution for the production of pharmaceuticals. Curr Opin Biotechnol. 2000;11:209–214. doi: 10.1016/s0958-1669(00)00081-1. [DOI] [PubMed] [Google Scholar]
- Chen X.H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I. Comparative analysis of the complete genome sequence of the plant growth‐promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. et al. [DOI] [PubMed] [Google Scholar]
- Crosa J.H., Walsh C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadio S., Monciardini P., Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- Du L., Shen B. Biosynthesis of hybrid peptide‐polyketide natural products. Curr Opin Drug Discov Devel. 2001;4:215–228. [PubMed] [Google Scholar]
- Du L., Sanchez C., Shen B. Hybrid peptide‐polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab Eng. 2001;3:78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- Erlanger B.F., Goode L. Antibacterial activity of acyclic decapeptide analogs of gramicidin S. Science. 1960;131:669–670. doi: 10.1126/science.131.3401.669. [DOI] [PubMed] [Google Scholar]
- Feling R.H., Buchanan G.O., Mincer T.J., Kauffman C.A., Jensen P.R., Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus salinospora. Angew Chem Int Ed Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Gioia J., Yerrapragada S., Qin X., Jiang H., Igboeli O.C., Muzny D. Paradoxical DNA repair and peroxide resistance gene conservation in Bacillus pumilus SAFR‐032. PLoS ONE. 2007;2:e928. doi: 10.1371/journal.pone.0000928. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H., Stockwell V.O., Henkels M.D., Nowak‐Thompson B., Loper J.E., Gerwick W.H. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Keating T.A., Marshall C.G., Walsh C.T. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry. 2000;39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- Kessler N., Schuhmann H., Morneweg S., Linne U., Marahiel M.A. The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J Biol Chem. 2004;279:7413–7419. doi: 10.1074/jbc.M309658200. [DOI] [PubMed] [Google Scholar]
- Khosla C., Gokhale R.S., Jacobsen J.R., Cane D.E. Tolerance and specificity of polyketide synthases. Annu Rev Biochem. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- McAlpine J.B., Bachmann B.O., Piraee M., Tremblay S., Alarco A.M., Zazopoulos E., Farnet C.M. Microbial genomics as a guide to drug discovery and structural elucidation: ECO‐02301, a novel antifungal agent, as an example. J Nat Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- McHenney M.A., Hosted T.J., Dehoff B.S., Rosteck P.R., Jr, Baltz R.H. Molecular cloning and physical mapping of the daptomycin gene cluster from Streptomyces roseosporus. J Bacteriol. 1998;180:143–151. doi: 10.1128/jb.180.1.143-151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minowa Y., Araki M., Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J Mol Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Ishikawa J., Hara H., Suzuki H., Ikenoya M., Ikeda H. The genome sequence of the streptomycin‐producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008 doi: 10.1128/JB.00204-08. et al. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig F., Schauwecker F., Keller U. Molecular characterization of the genes of actinomycin synthetase I and of a 4‐methyl‐3‐hydroxyanthranilic acid carrier protein involved in the assembly of the acylpeptide chain of actinomycin in Streptomyces. J Biol Chem. 1999;274:12508–12516. doi: 10.1074/jbc.274.18.12508. [DOI] [PubMed] [Google Scholar]
- Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D.H. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokem J.S., Lantz A.E., Nielsen J. Systems biology of antibiotic production by microorganisms. Nat Prod Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- Schneiker S., Perlova O., Kaiser O., Gerth K., Alici A., Altmeyer M.O. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. et al. [DOI] [PubMed] [Google Scholar]
- Seedorf H., Fricke W.F., Veith B., Bruggemann H., Liesegang H., Strittmatter A. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci USA. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S.A., Marahiel M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- Siezen R.J., Wilson G. Unpublished but public microbial genomes with biotechnological relevance. Microb Biotechnol. 2008;1:202–207. doi: 10.1111/j.1751-7915.2008.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelhaus T., Schneider A., Marahiel M.A. Engineered biosynthesis of peptide antibiotics. Biochem Pharmacol. 1996;52:177–186. doi: 10.1016/0006-2952(96)00111-6. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T., Mootz H.D., Marahiel M.A. The specificity‐conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Tae H., Kong E.B., Park K. ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformatics. 2007;8:327. doi: 10.1186/1471-2105-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W. Genome sequencing reveals complex secondary metabolome in the marine actino mycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.T., Chen H., Keating T.A., Hubbard B.K., Losey H.C., Luo L. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. et al. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Oikawa H. Robust platform for de novo production of heterologous polyketides and nonribosomal peptides in Escherichia coli. Org Biomol Chem. 2007;5:593–602. doi: 10.1039/b615589h. [DOI] [PubMed] [Google Scholar]
- Wenzel S.C., Muller R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotechnol. 2005;16:594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Yin J., Straight P.D., Hrvatin S., Dorrestein P.C., Bumpus S.B., Jao C. Genome‐wide high‐throughput mining of natural‐product biosynthetic gene clusters by phage display. Chem Biol. 2007;14:303–312. doi: 10.1016/j.chembiol.2007.01.006. et al. [DOI] [PubMed] [Google Scholar]
- Zazopoulos E., Huang K., Staffa A., Liu W., Bachmann B.O., Nonaka K. A genomics‐guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. et al. [DOI] [PubMed] [Google Scholar]


