Summary
Composting is the major technology in the treatment of animal manure and is a source of nitrous oxide, a greenhouse gas. Although the microbiological processes of both nitrification and denitrification are involved in composting, the key players in these pathways have not been well identified. Recent molecular microbiological methodologies have revealed the presence of dominant Bacillus species in the degradation of organic material or betaproteobacterial ammonia‐oxidizing bacteria on nitrification on the surface, and have also revealed the mechanism of nitrous oxide emission in this complicated process to some extent. Some bacteria, archaea or fungi still would be considered potential key players, and the contribution of some pathways, such as nitrifier denitrification or heterotrophic nitrification, might be involved in composting. This review article discusses these potential microbial players in nitrification–denitrification within the composting pile and highlights the relevant unknowns through recent activities that focus on the nitrogen cycle within the animal manure composting process.
Introduction
Composting is the simplest traditional animal manure management technology that depends on the degradation of organic matter by the microbial community within manure itself (Bernal et al., 2009). Easily degradable organic matter would be utilized as the energy source, and CO2, NH3 and moisture would be emitted and would generate large amounts of heat; the temperature inside compost piles is about 70°C. The mass of the pile decreases significantly, and the process also reduces odorous compounds and pathogens, while killing weed seeds. Because the mature product can be reused as organic fertilizer, composting is a very important technology from the viewpoint of the circulation of resources or environmental protection.
Because the organic or inorganic state of nitrogen contained within the compost is an important nutrient for crops, the available amount of nitrogen content in composted material is a precious component. Through the composting process, the organic nitrogen contained within initial fresh manure is degraded into ammonium by a wide variety of microorganisms including bacteria and fungi. Part of this nitrogen is lost as NH3 by volatilization, or through conversion into gases content such as N2O or N2 through the nitrification/denitrification process (Fig. 1). The range of nitrogen loss can vary between 19% and 77%, which mainly occur through NH3 volatilization and N2 emission (Martins and Dewes, 1992; Mahimairaja et al., 1995; Eghball et al., 1997; Tiquia and Tam, 2000; Tiquia et al., 2002). In addition, 0.2–9.9% of initial nitrogen content can be emitted as N2O, the intermediate of denitrification or by‐products of nitrification (Kuroda et al., 1996; Hao et al., 2001; 2004; Fukumoto et al., 2003; El Kader et al., 2007; Szanto et al., 2007) (Table 1). The loss of nitrogen during animal manure composting processes is affected by various parameters, such as the animal species, diet, bulking agents, moisture content, turning frequency, carbon/nitrogen ratio and initial nitrogen content.
Figure 1.
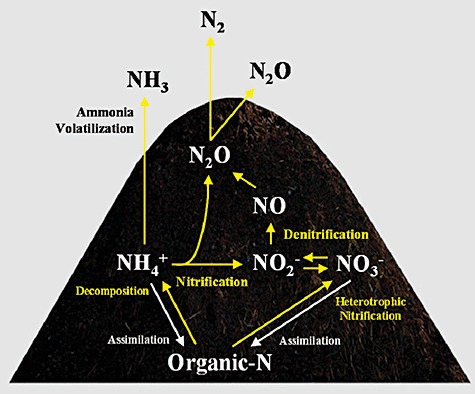
Nitrogen conversion and emission during the composting process.
Table 1.
Nitrous oxide emission from manure composting process.
| Animal | Process type | Unit | Reference | |
|---|---|---|---|---|
| Dairy | 0.582 | g‐N2O per kg DM | Pattey et al. (2005) | |
| Beef | 0.162 | g‐N2O per kg DM | ||
| Pig | Forced aeration | 1.9–71.9 | g‐N2O‐N per m3 | Osada and Fukumoto (2001) |
| Cattle | static | 1.1 | kg‐N per Mg manure | Hao et al. (2001) |
| Cattle | turned | 1.9 | kg‐N per Mg manure | |
| Cattle | Woodchip | 0.39 | %N | Hao et al. (2004) |
| Cattle | Straw | 0.68 | %N | |
| Dairy | static | 1.2 | %N | El Kader et al. (2007) |
| Dairy | turned | 1.9 | %N | |
| Turkey | 0.2–0.4 | %N | ||
| Pig | turned | 3.7–4.6 | %N | Fukumoto et al. (2003) |
| Pig | turned | 2.5 | %N | Szanto et al. (2007) |
| Pig | static | 9.9 | %N |
Nitrous oxide is an important greenhouse gas with strong global warming potential (300 times as that of CO2 (IPCC, 2001). Moreover, because N2O is greatly responsible for ozone depletion, reduction of its emission would be important for environmental protection (Ravishankara et al., 2009). Therefore, an important issue in the study of composting is the nitrifier/denitrifier microbial community, which plays a significant role in nitrogen conversion within the composting pile. In this review article, we deal with recent research activities that focus on the nitrifier/denitrifier microbial community in composting while referring to similar studies in other environments.
Overall microbial and fungal community in the composting process
There are many studies about microbial community structures in the composting process. Most of them focus on the bacteria mainly responsible for the degradation of organic matter. In order to identify microbes present in the compost process, besides the classical isolation technique, new approaches based on culture‐independent techniques, such as the extraction of DNA from the compost and amplification of 16S rRNA gene by PCR, followed by DNA sequencing are commonly used (Muyzer et al., 1993). The approach based on DNA sequencing provides relevant information on microbes that are difficult to culture.
It has been reported that some Bacillus species are important in the composting pile in the thermophilic stage, when active degradation of organic compounds occurs (Blanc et al., 1997; 1999; Ishii et al., 2000; Peters et al., 2000; Dees and Ghiorse, 2001; Zhang et al., 2002; Ishii and Takii, 2003; Schloss et al., 2003; Iida et al., 2005; Kim et al., 2006; Wang et al., 2007; Yamamoto et al., 2009). These Bacillus can grow and degrade organic compounds under thermophilic conditions up to 65°C, and Thermus species are dominant instead of Bacillus species above 70°C (Beffa et al., 1996). While this is relevant, it should be noted that the bacterial community structure changes dramatically even in the maturing phase, when active degradation of organic compounds has almost ended. In the maturing phase, mesophilic Proteobacteria or Actinobacteria are known to be dominant (Danon et al., 2008), and these bacterial groups are considered responsible for the maturation process.
In the composting process, the temperature in the core zone of the pile reaches 60–70°C, and there are temperature gradient effects within the pile (Fernandes et al., 1994). In addition, there is an oxygen gradient and anoxic conditions deep inside the piles (Hao et al., 2001), especially in passively aerated composting systems. In these various complicated environments, bacterial communities differ significantly between the surface and the core zone (Maeda et al., 2010a). In the dairy cattle manure composting process, nitrite and nitrate accumulate on the surface layer of the pile even in the initial stage of the process, when there are still easily degradable organic compounds (Maeda et al., 2010b). In the surface layer, 16S rRNA‐dependent bacterial community analysis suggests that some Proteobacteria or Bacteroidetes are dominant, which is significantly different from the case in thermophilic core zones. Some part of these bacterial species are thought to contribute to the nitrification or denitrification that actively occurs in the surface layer.
Although Dees and Ghiorse (2001) reported that they failed to detect archaea in the compost piles, while they found many fungal species in the compost samples whose temperatures did not exceed 50°C. In this regard, Anastasi and colleagues (2005) reported the isolation of 194 fungal species, the Acremonium, Aspergillus, Cladosporium, Malbranchea, Penicillium, Pseudallescheria and Thermomyces species from compost. In another study, Hultman and colleagues (2009) reported that fungal biomass can represent between 6.3% and 38.5% of total biomass in municipal waste compost based on phospholipid fatty acid analysis. They also found that the fungal community suffers dramatic changes during the composting process, as does the bacterial community, and that a fungal community succession differed between a full‐scale composting facility and a laboratory‐scale small reactor. Studies are needed on the function of the fungal community in the degradation of organic matter in the huminification process, or potential interaction with the bacterial community and its contribution to the nitrification/denitrification pathway.
Microorganisms relevant to the nitrogen cycle in composting
Nitrifiers
Nitrification is known to be carried out by bacteria, archaea and fungi (De Boer and Kowalchuk, 2001; Leininger et al., 2006; Laughlin et al., 2008). In the bacterial process, nitrification consists of two steps, ammonia oxidation and nitrite oxidation, and each of these reactions is performed by an individual microbial group: ammonia‐oxidizing bacteria (AOB) (Kowalchuk and Stephen, 2001) and nitrite‐oxidizing bacteria (NOB) respectively. Nitrous oxide is known to be produced as a by‐product of hydroxylamine oxidation. betaproteobacterial AOB or Thaumarchaeota ammonia‐oxidizing archaea (AOA) are considered important in ammonia oxidation (Brochier‐Armanet et al., 2008), and the major NOB in the environment are alphaproteobacterial Nitrobacter or Nitrospira. Nitrifiers grow slowly under laboratory conditions, and their cultivation or isolation is very time‐consuming. In order to speed the tracking of these microbes, a molecular biology approach using primers specific to the 16S rRNA genes and the ammonia monooxygenase gene of betaproteobacterial AOB have been developed and used to track nitrifiers in the environment (Innerebner et al., 2006; Junier et al., 2010).
In the composting process, temporal nitrite accumulation in the middle stage and high accumulation of nitrate in the mature phase were observed (He et al., 2000; 2001; Fukumoto et al., 2006; Fukumoto and Inubushi, 2009; Maeda et al., 2010c), and it is evident that nitrification occurs in the compost pile. However, it remains unclear which microbes are responsible for this process. Some studies have detected sequences similar to known AOB species Nitrosomonas europaea‐eutropha or Nitrosospira in the composting process (Kowalchuk et al., 1999; Jarvis et al., 2009; Maeda et al., 2010b). Jarvis and colleagues (2009) also detected Nitrosomonas in the theromophilic stage and Nitrosospira in the maturation phase of household waste composting, while Maeda and colleagues (2010b) detected Nitrosomonas throughout the process, especially from the surface layer of a cattle manure composting pile. Some previous studies also detected Nitrosomonas from landfill cover, an organic‐rich environment similar to the composting process (Mertoglu et al., 2006; Zhu et al., 2007). Betaproteobacterial AOB are chemoautotrophic and generate energy from the hydroxylamine oxidation step, the ATP produced is used to fix CO2 as a carbon source. Therefore, the presence of these AOB indicates that these bacteria oxidize ammonia in the composting process. However, the extent of the contribution to net nitrification is yet unknown.
To clarify the contribution of AOB to ammonia oxidation in composting, the contribution of AOA must be studied. Although AOA is known to be responsible to some extent for nitrification in environments containing less organic matter, such as soil, ocean or river sediment (Leininger et al., 2006; Santoro et al., 2010), there are some reports that AOB's contribution is much more important than AOA's for actual nitrification in organic‐rich environments such as wastewater from treatment plants (Park et al., 2006; Wells et al., 2009). Another report shows that ammonia oxidation in zinc‐contaminated soil is restored not by AOA but by AOB (Mertens et al., 2009). Many heavy metals are included in livestock manure, especially in swine manure (Nicholson et al., 1999; Ko et al., 2008). AOB might be more important than AOA under these conditions. Although a previous report failed to detect the AOA in compost piles (Maeda et al., 2010b), Yamamoto et al. (2010) reported the existence of AOA from cattle manure compost in the later stage of the process. The relative contribution of AOB and AOA on actual nitrification needs to be clarified.
Heterotrophic nitrification is a reaction in which heterotrophic bacteria oxidize ammonia or degrade organic matter to nitrate directly (Papen and Von Berg, 1998). Many bacterial species are known to undergo this reaction, and species such as Paracoccus denitrificans or Pseudomonas putida are known to possess amoA sequences distinct from those of autotrophic nitrifiers (Moir et al., 1996; Daum et al., 1998). Although these bacteria have potential to contribute net nitrification in the compost, its actual contribution is not known at all. This reaction by heterotrophs has not been considered in depth because these species do not generate energy from this process, nor do they accumulate high concentrations of nitrite; however, this futile reaction may be of relevance in an environmental setting. These heterotrophic nitrifiers assimilate more ammonium than chemoautotrophic AOB, which leads to higher biomass, and they have been considered not useful for wastewater treatment systems (Podmirseg et al., 2010). Efforts to unveil nitrification process include the development of new culture media for thermophilic nitrifiers in compost under heterotrophic conditions is ongoing (Shimaya and Hashimoto, 2008). In summary, it can be said that to understand the nitrogen cycle in the composting process, we need to learn more about the role of the heterotrophic nitrifiers in the process.
On the other hand, there have not been many molecular ecological studies on NOB. Because NOB has a diverse taxonomy, including Nitrobacter (α‐Proteobacteria), Nitrosococcus mobilis (γ‐Proteobacteria), Nitrospina gracilis (δ‐Proteobacteria) and Nitrospira (Nitrospira), it is difficult to detect all these strains by methods such as FISH (fluorescent in situ hybridization) or PCR, which depend on 16S rRNA sequences. Few studies have focused on functioning gene sequences of nitrite oxidoreductase of Nitrobacter (Poly et al., 2008; Wertz et al., 2008). Even though these methods may be effective for understanding nitrification in the composting process, they have not been used for studies of nitrite oxidation yet, and NOB in the composting process has not been characterized well.
Denitrifiers
Nitrite or nitrate generated by nitrifiers would usually be reduced by heterotrophic denitrifiers and emitted into the atmosphere as N2O or N2. Denitrification by bacteria has been well studied and the details of its molecular mechanisms have been characterized. The reaction consists of four reduction steps, namely, NO3→ NO2→ NO → N2O → N2. The genes nar, nir, nor and nos are coding the catalysing enzymes (Rudolf and Kroneck, 2005; Tavares et al., 2006). Denitrifying bacteria are known to be phylogenetically diverse, with at least 50 genera (Zumft, 1997). Therefore, the study of denitrifiers that depend on 16S rRNA gene sequences is very difficult, and functioning genes that code each enzyme catalysing each denitrification step are frequently used for studies of environmental denitrifiers (Sharma et al., 2005; Wertz et al., 2009). Because of the relative abundance of information in public databases, nitrite reductase (nirS and nirK) or nitrous oxide reductase (nosZ) have been used frequently.
There are two types of nitrite reductase: nirS, cytochrome c nitrite reductase, which has haem iron in its active centre (Einsle et al., 1999; 2000), and nirK, a copper‐containing nitrite reductase (Murphy et al., 1997; Antonyuk et al., 2005). It is possible to distinguish these nitrite reductases in denitrifiers by using diethyldithiocarbamate (DDTC), which chelates copper of the nirK denitrifier and prevents the process. DGGE primers targeting nitrite reductase gene sequences have been developed (Throback et al., 2004) and used to study denitrifiers in various environments. It is frequently discussed which types of nitrite reducers would be dominant in the environment. For example, one report shows that nirS denitrifiers are dominant in subtropical macrotidal estuaries (Abell et al., 2009). Because the horizonal transfer of denitrifying genes may occur within the environment, the incidence of nirK or nirS does not always agrees with the 16S rRNA gene phylogenetic sequences. Heylen and colleagues (2006a) concluded that nir genes may not be suitable to evaluate microbial diversity of denitrifiers in the environment. Thus, interpretation of biodiversity based on nir sequence analysis need to be interpreted with care.
Nitric oxide reductase (NOR) catalyses the reduction of NO to N2O. Nitric oxide, produced by the reduction of nitrite, is known to be toxic to microorganisms, and they need to metabolize it to protect themselves. Three kinds of NOR have been reported, namely cNOR, qNOR and qCuNOR (Tavares et al., 2006). The cytochrome c‐dependent nitric oxide reductase (cNOR) of P. denitrificans has been studied well. It is a component of cytochrome bc complex with two non‐haem irons in its active centre. Braker and Tiedje (2003) were the first to study denitrifying communities using norB as a functional marker, and others have used it for studies on environmental samples (Dandie et al., 2007) or isolates (Heylen et al., 2006b), but not yet for compost samples.
Nitrous oxide reductase is the terminal oxidoreductase of denitrification that transforms N2O to N2 (Brown et al., 2000). Because this multi‐copper‐containing enzyme prevents the accumulation of a potent greenhouse gas, it plays an important role in the nitrogen cycle (Zumft, 2005). Nitrous oxide reductase is most sensitive to molecular oxygen among the enzymes involved in denitrification, and its function is inhibited under aerobic conditions. This enzyme can also be inhibited by C2H2 easily (Yoshinari and Knowles, 1976) and is frequently used for the study of the denitrification potential of environmental samples (Teissier and Torre, 2002). Moreover, the nosZ gene that codes this enzyme is used as a biomarker in molecular ecological studies (Scala and Kerkhof, 1999; Stres et al., 2004).
Some bacterial denitrifiers and fungi are known not to possess nitrous oxide reductase (Takaya, 2009). Although N2O reduction is thermodynamically favourable and N2O is suitable for an electron acceptor, some denitrifiers produce N2O as the final product of the denitrification process. This might be explained by the fact that nitrous oxide is not toxic to microorganisms, whereas NO is toxic to bacterial cells. The lack of N2O reduction makes ∼20% difference to the bioenergetics of the bacterium (Richardson et al., 2009).
Diversity of denitrifiers in the environments
A study about denitrifier communities in the composting process revealed an initial variation of nirK diversity and stability after that (Maeda et al., 2010a). Hallin and colleagues (2006) also reported that the addition of methanol or ethanol to activated sludge significantly affected the diversity of nirS but not that of nirK. On the other hand, the addition of mature compost that contains NO2 or NO3‐N did not affect nirK diversity but significantly affected nosZ diversity, suggesting that denitrifiers possessing the nosZ gene in the compost would be more sensitive to environmental conditions. It is necessary to isolate the major denitrifier revealed by molecular methods in order to understand the actual denitrification occurring in the environment. In a denitrifier community study of rice paddy soil, Ashida and colleagues (2010) successfully isolated a major denitrifier through the enhancement of denitrification activity with succinate amendment and molecular methods such as the 16S rRNA gene clone library approach (Ishii et al., 2009) or the stable isotope probing approach (Saito et al., 2008). Moreover, Ishii and colleagues (2011) proved that denitrifiers with different 16S rRNA gene phylogeny possess same nirS or nirK gene in the same environment (Fig. 2). Their data show previously unknown complex relationship between 16S rRNA gene and functional gene possession. To understand the denitrifier community completely, it is necessary to combine independent approaches such as molecular and conventional cultivation approaches. The molecular methods used to characterize the unknown and uncultivated denitrifier communities, and the subsequent single‐cell isolation strategy would be effective for the denitrifiers that are truly functioning for actual denitrification in the environment (Ishii et al., 2010).
Figure 2.
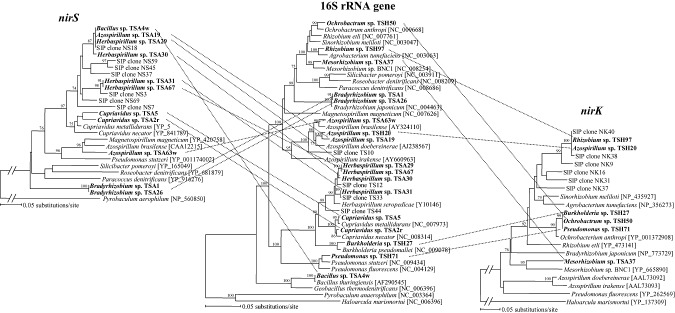
The relationship of 16S rRNA gene and functional denitrifier genes (nirS, nirK) possession of denitrifiers isolated from rice paddy soil (Redrawn from Ishii et al., 2011). Isolated strains were written in bold, and the possession relationship was connected by dotted lines.
The relationship between denitrifying gene diversity or abundance and potential denitrification activity in soil has been well studied. Potential denitrification activity, N2O/(N2O + N2) and the denitrifier community would be affected by pH (Palmer et al., 2010). Cuhel and colleagues (2010) reported that nirS diversity correlates with soil pH. On the other hand, Hallin and colleagues (2009) reported that denitrification activity did not correlate with denitrifier gene composition, but did correlate with the size of the total bacterial community or nosZ abundance. Another study reports that the nosZ ratio to total bacterial community is much more important than denitrifying gene abundance for potential N2O production (Philippot et al., 2009). Although much effort has been made in these environmental studies, it is still difficult to explain denitrification and nitrous oxide production by denitrifying gene abundance or diversity. Moreover, a previous paper reported that AOA possesses a novel nirK sequence (Bartossek et al., 2010), which had not been covered by previous denitrifier studies, and much effort should be made to learn about this unknown archaean denitrification.
It is also known that some autotrophic nitrifiers have the ability to denitrify (Wrage et al., 2001; Wrage et al., 2004; Shaw et al., 2006). These autotrophic nitrifiers possess nirK‐type nitrite reductase with distinct DNA sequences from those of heterotrophic denitrifiers. It is not well understood yet how these autotrophic nitrifiers acquired the nirK gene, which might have occurred by horizontal gene transfer; and how they became tolerant to nitrite produced by themselves (Casciotti and Ward, 2001). Because nitrifiers produce nitrite, they have many advantages for the utilization of nitrite as the substrate for denitrification. Therefore, nitrifier denitrification may contribute much more than heterotrophic denitrifiers, but it is difficult to distinguish these pathways with the current analytic techniques.
Who is responsible for nitrous oxide emission?
It is hard to understand how to share NO in nitrification and denitrification procedures. As stated above, nitrification would be performed by AOB, AOA, heterotrophic nitrifiers or fungi, whereas denitrification would be performed by heterotrophic denitrifiers, denitrifying fungi and autotrophic/heterotrophic nitrifiers. However, the relative contribution to net nitrification or denitrification of each group is not yet clear. Studies that focus on nitrifying genes are only about AOB or AOA, and those for denitrifying gene analysis are only about bacteria. The development of a tool for such study will be needed.
Stable isotope analysis of N2O is an alternative approach to studying its production processes because the relative abundance of stable isotopes is a function of their abundance in source materials and the isotope fractionation factor of each physical/chemical process. In particular, intramolecular 15N distribution within the N2O molecule (site preference, SP) has been found to depend only on enzymatic reaction processes and not on substrates (Toyoda and Yoshida, 1999; Yoshida and Toyoda, 2000; Toyoda et al., 2005; Sutka et al., 2006). Nitrous oxide, which originates from bacterial nitrification (hydroxylamine oxidation) and denitrification (nitrite reduction), can be distinguished by using SP. However, SP cannot distinguish between nitrifier denitrification and heterotrophic denitrification, and a recent study showed that fungal denitrification produces N2O with SP similar to that of bacterial nitrification (Sutka et al., 2008). In addition, isotope abundance is affected by nitrous oxide reduction (Ostrom et al., 2007; Jinuntuya‐Nortman et al., 2008). Although this analytical technique has some limitations as stated above, it would be a powerful tool by using all available isotopic data (N and O isotope ratios and SP) in a complementary style (e.g. Koba et al., 2009) or by combining other analytical approaches, such as a wide range of molecular methods. Isotopomer analysis of N2O directly collected from a composting pile by the dynamic chamber method (Osada and Fukumoto, 2001) revealed that bacterial denitrification is the most important and responsible nitrous oxide production pathway (Maeda et al., 2010b). This study relied on the isotopic characteristics of N2O produced by isolates not from compost but from other environments (such as soil). Therefore, in future studies, we need to isolate the major nitrifier, denitrifier, denitrifying fungi and isotope signature of their producing N2O.
Future perspective
Because of the ease with which it is managed, composting will continue to be a major technology for treating animal manure. However, analysing the techniques developed previously cannot explain the nitrogen cycle and nitrous oxide emission yet. In the future, combining distinct approaches such as molecular methods, stable isotope analysis and classical isolation techniques will help us to understand the nitrogen cycle during the composting process in detail. The results should lead to the development of relevant mitigation strategies, which will include: identification of the main players in the nitrification–denitrification process in the composting piles; isolation of these key players; and analysis of their physiological, biochemical and ecological properties. It would be also of interest to identify the nitrous oxide reducers and to study their function in the composting piles.
Acknowledgments
This work was supported by the National Agriculture and Food Research Organization (NARO), Japan. The authors would like to acknowledge Dr. Ishii for providing Figure 2.
References
- Abell G., Revill A., Smith C., Bissett A., Volkman J., Robert S. Archaeal ammonia oxidizers and nirS‐type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 2009;4:286–300. doi: 10.1038/ismej.2009.105. [DOI] [PubMed] [Google Scholar]
- Anastasi A., Varese G., Marchisio V.F. Isolation and identification of fungal communities in compost and vermicompost. Mycologia. 2005;97:33–44. doi: 10.3852/mycologia.97.1.33. [DOI] [PubMed] [Google Scholar]
- Antonyuk S., Strange R., Sawers G., Eady R., Hasnain S. Atomic resolution structures of resting‐state, substrate‐and product‐complexed Cu‐nitrite reductase provide insight into catalytic mechanism. Proc Natl Acad Sci USA. 2005;102:12041–12046. doi: 10.1073/pnas.0504207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida N., Ishii S., Hayano S., Tago K., Tsuji T., Yoshimura Y. Isolation of functional single cells from environments using a micromanipulator: application to study denitrifying bacteria. Appl Microbiol Biotechnol. 2010;85:1211–1217. doi: 10.1007/s00253-009-2330-z. et al. [DOI] [PubMed] [Google Scholar]
- Bartossek R., Nicol G., Lanzen A., Klenk H., Schleper C. Homologues of nitrite reductases in ammonia‐oxidizing archaea: diversity and genomic context. Environ Microbiol. 2010;12:1075–1088. doi: 10.1111/j.1462-2920.2010.02153.x. [DOI] [PubMed] [Google Scholar]
- Beffa T., Blanc M., Lyon P., Vogt G., Marchiani M., Fischer J., Aragno M. Isolation of Thermus strains from hot composts (60 to 80 degrees C) Appl Environ Microbiol. 1996;62:1723–1727. doi: 10.1128/aem.62.5.1723-1727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M., Alburquerque J., Moral R. Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour Technol. 2009;100:5444–5453. doi: 10.1016/j.biortech.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Blanc M., Marilley L., Beffa T., Aragno M. Notes: rapid identification of heterotrophic, thermophilic, spore‐forming bacteria isolated from hot composts. Int J Syst Evol Microbiol. 1997;47:1246–1248. doi: 10.1099/00207713-47-4-1246. [DOI] [PubMed] [Google Scholar]
- Blanc M., Marilley L., Beffa T., Aragno M. Thermophilic bacterial communities in hot composts as revealed by most probable number counts and molecular (16S rDNA) methods. FEMS Microbiol Ecol. 1999;28:141–149. [Google Scholar]
- Braker G., Tiedje J.M. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl Environ Microbiol. 2003;69:3476–3483. doi: 10.1128/AEM.69.6.3476-3483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier‐Armanet C., Boussau B., Gribaldo S., Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Brown K., Tegoni M., Prudêncio M., Pereira A., Besson S., Moura J. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat Struct Mol Biol. 2000;7:191–195. doi: 10.1038/73288. et al. [DOI] [PubMed] [Google Scholar]
- Casciotti K., Ward B. Dissimilatory nitrite reductase genes from autotrophic ammonia‐oxidizing bacteria. Appl Environ Microbiol. 2001;67:2213–2221. doi: 10.1128/AEM.67.5.2213-2221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuhel J., Simek M., Laughlin R., Bru D., Cheneby D., Watson C., Philippot L. Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl Environ Microbiol. 2010;76:1870–1878. doi: 10.1128/AEM.02484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandie C., Miller M., Burton D., Zebarth B., Trevors J., Goyer C. Nitric oxide reductase‐targeted real‐time PCR quantification of denitrifier populations in soil. Appl Environ Microbiol. 2007;73:4250–4258. doi: 10.1128/AEM.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon M., Franke‐Whittle I., Insam H., Chen Y., Hadar Y. Molecular analysis of bacterial community succession during prolonged compost curing. FEMS Microbiol Ecol. 2008;65:133–144. doi: 10.1111/j.1574-6941.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- Daum M., Zimmer W., Papen H., Kloos K., Nawrath K., Bothe H. Physiological and molecular biological characterization of ammonia oxidation of the heterotrophic nitrifier Pseudomonas putida. Curr Microbiol. 1998;37:281–288. doi: 10.1007/s002849900379. [DOI] [PubMed] [Google Scholar]
- De Boer W., Kowalchuk G. Nitrification in acid soils: micro‐organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- Dees P.M., Ghiorse W.C. Microbial diversity in hot synthetic compost as revealed by PCR‐amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol Ecol. 2001;35:207–216. doi: 10.1111/j.1574-6941.2001.tb00805.x. [DOI] [PubMed] [Google Scholar]
- Eghball B., Power J., Gilley J., Doran J.W. Nutrient, carbon, and mass loss during composting of beef cattle feedlot manure. J Environ Qual. 1997;26:189–193. [Google Scholar]
- Einsle O., Messerschmidt A., Stach P., Bourenkov G., Bartunik H., Huber R., Kroneck P. Structure of cytochrome c nitrite reductase. Nature. 1999;400:476–480. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- Einsle O., Stach P., Messerschmidt A., Simon J., Kröger A., Huber R., Kroneck P. Cytochrome c nitrite reductase from Wolinella succinogenes. J Biol Chem. 2000;275:39608–39616. doi: 10.1074/jbc.M006188200. [DOI] [PubMed] [Google Scholar]
- El Kader N., Robin P., Paillat J., Leterme P. Turning, compacting and the addition of water as factors affecting gaseous emissions in farm manure composting. Bioresour Technol. 2007;98:2619–2628. doi: 10.1016/j.biortech.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Fernandes L., Zhan W., Patni N., Jui P. Temperature distribution and variation in passively aerated static compost piles. Bioresour Technol. 1994;48:257–264. [Google Scholar]
- Fukumoto Y., Inubushi K. Effect of nitrite accumulation on nitrous oxide emission and total nitrogen loss during swine manure composting. Soil Sci Plant Nutr. 2009;55:428–434. [Google Scholar]
- Fukumoto Y., Osada T., Hanajima D., Haga K. Patterns and quantities of NH3, N2O and CH4 emissions during swine manure composting without forced aeration – effect of compost pile scale. Bioresour Technol. 2003;89:109–114. doi: 10.1016/s0960-8524(03)00060-9. [DOI] [PubMed] [Google Scholar]
- Fukumoto Y., Suzuki K., Osada T., Kuroda K., Hanajima D., Yasuda T., Haga K. Reduction of nitrous oxide emission from pig manure composting by addition of nitrite‐oxidizing bacteria. Environ Sci Technol. 2006;40:6787–6791. doi: 10.1021/es0611801. [DOI] [PubMed] [Google Scholar]
- Hallin S., Throback I.N., Dicksved J., Pell M. Metabolic profiles and genetic diversity of denitrifying communities in activated sludge after addition of methanol or ethanol. Appl Environ Microbiol. 2006;72:5445–5452. doi: 10.1128/AEM.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S., Jones C., Schloter M., Philippot L. Relationship between N‐cycling communities and ecosystem functioning in a 50‐year‐old fertilization experiment. ISME J. 2009;3:597–605. doi: 10.1038/ismej.2008.128. [DOI] [PubMed] [Google Scholar]
- Hao X., Chang C., Larney F.J., Travis G.R. Greenhouse gas emissions during cattle feedlot manure composting. J Environ Qual. 2001;30:376–386. doi: 10.2134/jeq2001.302376x. [DOI] [PubMed] [Google Scholar]
- Hao X., Chang C., Larney F.J. Carbon, nitrogen balances and greenhouse gas emission during cattle feedlot manure composting. J Environ Qual. 2004;33:37–44. doi: 10.2134/jeq2004.3700. [DOI] [PubMed] [Google Scholar]
- He Y., Inamori Y., Mizuochi M., Kong H., Iwami N., Sun T. Nitrous oxide emissions from aerated composting of organic waste. Environ Sci Technol. 2001;35:2347–2351. doi: 10.1021/es0011616. [DOI] [PubMed] [Google Scholar]
- He Y.W., Inamori Y., Mizuochi M., Kong H.N., Iwami N., Sun T.H. Measurements of N2O and CH4 from the aerated composting of food waste. Sci Total Environ. 2000;254:65–74. doi: 10.1016/s0048-9697(00)00439-3. [DOI] [PubMed] [Google Scholar]
- Heylen K., Gevers D., Vanparys B., Wittebolle L., Geets J., Boon N., De Vos P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ Microbiol. 2006a;8:2012–2021. doi: 10.1111/j.1462-2920.2006.01081.x. [DOI] [PubMed] [Google Scholar]
- Heylen K., Vanparys B., Gevers D., Wittebolle L., Boon N., De Vos P. Nitric oxide reductase (norB) gene sequence analysis reveals discrepancies with nitrite reductase (nir) gene phylogeny in cultivated denitrifiers. Environ Microbiol. 2006b;9:1072–1077. doi: 10.1111/j.1462-2920.2006.01194.x. [DOI] [PubMed] [Google Scholar]
- Hultman J., Vasara T., Partanen P., Kurola J., Kontro M., Paulin L. Determination of fungal succession during municipal solid waste composting using a cloning‐based analysis. J Appl Microbiol. 2009;108:472–487. doi: 10.1111/j.1365-2672.2009.04439.x. et al. [DOI] [PubMed] [Google Scholar]
- Iida K., Ueda Y., Kawamura Y., Ezaki T., Takade A., Yoshida S., Amako K. Paenibacillus motobuensis sp. nov., isolated from a composting machine utilizing soil from Motobu‐town, Okinawa, Japan. Int J Syst Evol Microbiol. 2005;55:1811–1816. doi: 10.1099/ijs.0.63636-0. [DOI] [PubMed] [Google Scholar]
- Innerebner G., Knapp B., Vasara T., Romantschuk M., Insam H. Traceability of ammonia‐oxidizing bacteria in compost‐treated soils. Soil Biol Biochem. 2006;38:1092–1100. [Google Scholar]
- Intergovernmental Panel on Climate Change. Cambridge University Press; 2001. [Google Scholar]
- Ishii K., Takii S. Comparison of microbial communities in four different composting processes as evaluated by denaturing gradient gel electrophoresis analysis. J Appl Microbiol. 2003;95:109–119. doi: 10.1046/j.1365-2672.2003.01949.x. [DOI] [PubMed] [Google Scholar]
- Ishii K., Fukui M., Takii S. Microbial succession during a composting process as evaluated by denaturing gradient gel electrophoresis analysis. J Appl Microbiol. 2000;89:768–777. doi: 10.1046/j.1365-2672.2000.01177.x. [DOI] [PubMed] [Google Scholar]
- Ishii S., Yamamoto M., Kikuchi M., Oshima K., Hattori M., Otsuka S., Senoo K. Microbial populations responsive to denitrification‐inducing conditions in rice paddy soil, as revealed by comparative 16S rRNA gene analysis. Appl Environ Microbiol. 2009;75:7070–7078. doi: 10.1128/AEM.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Tago K., Senoo K. Single‐cell analysis and isolation for microbiology and biotechnology: methods and applications. Appl Microbiol Biotechnol. 2010;86:1281–1292. doi: 10.1007/s00253-010-2524-4. [DOI] [PubMed] [Google Scholar]
- Ishii S., Ashida N., Otsuka S., Senoo K. 2011. , and ) Isolation of oligotrophic denitrifiers carrying previously uncharacterized functional gene sequences. Appl Environ Microbiol (in press): doi: 10.1128/AEM.0218910. [DOI] [PMC free article] [PubMed]
- Jarvis A., Sundberg C., Milenkovski S., Pell M., Smars S., Lindgren P., Hallin S. Activity and composition of ammonia oxidizing bacterial communities and emission dynamics of NH3 and N2O in a compost reactor treating organic household waste. J Appl Microbiol. 2009;106:1502–1511. doi: 10.1111/j.1365-2672.2008.04111.x. [DOI] [PubMed] [Google Scholar]
- Jinuntuya‐Nortman M., Sutka R., Ostrom P., Gandhi H., Ostrom N. Isotopologue fractionation during microbial reduction of N2O within soil mesocosms as a function of water‐filled pore space. Soil Biol Biochem. 2008;40:2273–2280. [Google Scholar]
- Junier P., Molina V., Dorador C., Hadas O., Kim O., Junier T. Phylogenetic and functional marker genes to study ammonia‐oxidizing microorganisms (AOM) in the environment. Appl Microbiol Biotechnol. 2010;85:425–440. doi: 10.1007/s00253-009-2228-9. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Lee S., Weon H., Kwon S., Go S., Park Y. Ureibacillus suwonensis sp. nov., isolated from cotton waste composts. Int J Syst Evol Microbiol. 2006;56:663–666. doi: 10.1099/ijs.0.63703-0. et al. [DOI] [PubMed] [Google Scholar]
- Ko H., Kim K., Kim H., Kim C., Umeda M. Evaluation of maturity parameters and heavy metal contents in composts made from animal manure. Waste Manag. 2008;28:813–820. doi: 10.1016/j.wasman.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Koba K., Osaka K., Tobari Y., Toyoda S., Ohte N., Katsuyama M., Suzuki N. Biogeochemistry of nitrous oxide in groundwater in a forested ecosystem elucidated by nitrous oxide isotopomer measurements. Geoch Cosmoch Acta. 2009;73:3115–3133. [Google Scholar]
- Kowalchuk G., Naoumenko Z., Derikx P., Felske A., Stephen J., Arkhipchenko I. Molecular analysis of ammonia‐oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalchuk G., Stephen J. Ammonia‐oxidizing bacteria: a model for molecular microbial ecology. Ann Rev Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Osada T., Yonaga M., Kanematu A., Nitta T., Mouri S., Kojima T. Emissions of malodorous compounds and greenhouse gases from composting swine feces. Bioresour Technol. 1996;56:265–271. [Google Scholar]
- Laughlin R., Stevens R., Müller C., Watson C. Evidence that fungi can oxidize NH4+ to NO3‐ in a grassland soil. Eur J Soil Sci. 2008;59:285–291. [Google Scholar]
- Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G. Archaea predominate among ammonia‐oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. et al. [DOI] [PubMed] [Google Scholar]
- Maeda K., Hanajima D., Morioka R., Osada T. Characterization and spatial distribution of bacterial communities within passively aerated cattle manure composting piles. Bioresour Technol. 2010a;101:9631–9637. doi: 10.1016/j.biortech.2010.07.057. [DOI] [PubMed] [Google Scholar]
- Maeda K., Toyoda S., Shimojima R., Osada T., Hanajima D., Morioka R., Yoshida N. The source of nitrous oxide emission from cattle manure composting process revealed by isotopomer analysis and amoA abundance of beta‐proteobacterial ammonia oxidizing bacteria. Appl Environ Microbiol. 2010b;76:1555–1562. doi: 10.1128/AEM.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Morioka R., Hanajima D., Osada T. The impact of using mature compost on nitrous oxide emission and the denitrifier community in the cattle manure composting process. Microb Ecol. 2010c;59:25–36. doi: 10.1007/s00248-009-9547-3. [DOI] [PubMed] [Google Scholar]
- Mahimairaja S., Bolan N.S., Hedley M.J. Denitrification losses of N from fresh and composted manures. Soil Biol Biochem. 1995;27:1223–1225. [Google Scholar]
- Martins O., Dewes T. Loss of nitrogenous compounds during composting of animal wastes. Bioresour Technol. 1992;42:103–111. [Google Scholar]
- Mertens J., Broos K., Wakelin S., Kowalchuk G., Springael D., Smolders E. Bacteria, not archaea, restore nitrification in a zinc‐contaminated soil. ISME J. 2009;3:916–923. doi: 10.1038/ismej.2009.39. [DOI] [PubMed] [Google Scholar]
- Mertoglu B., Calli B., Inanc B., Ozturk I. Evaluation of in situ ammonia removal in an aerated landfill bioreactor. Proc Biochem. 2006;41:2359–2366. [Google Scholar]
- Moir J., Crossman L., Spiro S., Richardson D. The purification of ammonia monooxygenase from Paracoccus denitrificans. FEBS Lett. 1996;387:71–74. doi: 10.1016/0014-5793(96)00463-2. [DOI] [PubMed] [Google Scholar]
- Murphy M., Turley S., Adman E. Structure of nitrite bound to copper‐containing nitrite reductase from Alcaligenes faecalis. J Biol Chem. 1997;272:28455–28460. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- Muyzer G., De Waal E., Uitterlinden A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction‐amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson F., Chambers B., Williams J., Unwin R. Heavy metal contents of livestock feeds and animal manures in England and Wales. Bioresour Technol. 1999;70:23–31. [Google Scholar]
- Osada T., Fukumoto Y. Development of a new dynamic chamber system for measuring harmful gas emissions from composting livestock waste. Wat Sci Technol. 2001;44:79–86. [PubMed] [Google Scholar]
- Ostrom N., Pitt A., Sutka R., Ostrom P., Grandy A., Huizinga K., Robertson G. Isotopologue effects during N2O reduction in soils and in pure cultures of denitrifiers. J Geophys Res. 2007;112:G02005. [Google Scholar]
- Palmer K., Drake H., Horn M. Association of novel and highly diverse acid‐tolerant denitrifiers with N2O fluxes of an acidic fen. Appl Environ Microbiol. 2010;76:1125–1134. doi: 10.1128/AEM.02256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papen H., Von Berg R. A most probable number method (MPN) for the estimation of cell numbers of heterotrophic nitrifying bacteria in soil. Plant Soil. 1998;199:123–130. [Google Scholar]
- Park H., Wells G., Bae H., Criddle C., Francis C. Occurrence of ammonia‐oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattey E., Trzcinski M.K., Desjardins R.L. Quantifying the reduction of greenhouse gas emissions as a result of composting dairy and beef cattle manure. Nutr Cycl Agroecosys. 2005;72:173–187. [Google Scholar]
- Peters S., Koschinsky S., Schwieger F., Tebbe C. Succession of microbial communities during hot composting as detected by PCR‐single‐strand‐conformation polymorphism‐based genetic profiles of small‐subunit rRNA genes. Appl Environ Microbiol. 2000;66:930–936. doi: 10.1128/aem.66.3.930-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L., Čuhel J., Saby N., Chèneby D., Chronáková A., Bru D. Mapping field‐scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol. 2009;11:1518–1526. doi: 10.1111/j.1462-2920.2009.01879.x. et al. [DOI] [PubMed] [Google Scholar]
- Podmirseg S., Schoen M., Murthy S., Insam H., Wett B. Quantative and qualitative effects of bioaugmentation on ammonia oxidizers at a two‐step WWTP. Wat Sci Technol. 2010;61:1003–1009. doi: 10.2166/wst.2010.016. [DOI] [PubMed] [Google Scholar]
- Poly F., Wertz S., Brothier E., Degrange V. First exploration of Nitrobacter diversity in soils by a PCR cloning‐sequencing approach targeting functional gene nxrA. FEMS Microbiol Ecol. 2008;63:132–140. doi: 10.1111/j.1574-6941.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- Ravishankara A., Daniel J., Portmann R. Nitrous oxide (N2O): the dominant ozone‐depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- Richardson D., Felgate H., Watmough N., Thomson A., Baggs E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle – could enzymic regulation hold the key? Trends Biotechnol. 2009;27:388–397. doi: 10.1016/j.tibtech.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Rudolf M., Kroneck P. The nitrogen cycle: its biology. Met Ions Biol Syst. 2005;43:75–104. [PubMed] [Google Scholar]
- Saito T., Ishii S., Otsuka S., Nishiyama M., Senoo K. Identification of novel betaproteobacteria in a succinate‐assimilating population in denitrifying rice paddy soil by using stable isotope probing. Microb Environ. 2008;23:192–200. doi: 10.1264/jsme2.23.192. [DOI] [PubMed] [Google Scholar]
- Santoro A., Casciotti K., Francis C. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Scala D., Kerkhof L. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol. 1999;65:1681–1687. doi: 10.1128/aem.65.4.1681-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P., Hay A., Wilson D., Walker L. Tracking temporal changes of bacterial community fingerprints during the initial stages of composting. FEMS Microbiol Ecol. 2003;46:1–9. doi: 10.1016/S0168-6496(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Sharma S., Aneja M.K., Mayer J., Munch J.C., Schloter M. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes. Appl Environ Microbiol. 2005;71:2001–2007. doi: 10.1128/AEM.71.4.2001-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L., Nicol G., Smith Z., Fear J., Prosser J., Baggs E. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ Microbiol. 2006;8:214–222. doi: 10.1111/j.1462-2920.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- Shimaya C., Hashimoto T. Improvement of media for thermophilic ammonia‐oxidizing bacteria in compost. Soil Sci Plant Nutr. 2008;54:529–533. [Google Scholar]
- Stres B., Mahne I., Avgustin G., Tiedje J.M. Nitrous oxide reductase (nosZ) gene fragments differ between native and cultivated Michigan soils. Appl Environ Microbiol. 2004;70:301–309. doi: 10.1128/AEM.70.1.301-309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutka R.L., Ostrom N.E., Ostrom P.H., Breznak J.A., Gandhi H., Pitt A.J., Li F. Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl Environ Microbiol. 2006;72:638–644. doi: 10.1128/AEM.72.1.638-644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutka R., Adams G., Ostrom N., Ostrom P. Isotopologue fractionation during N2O production by fungal denitrification. Rapid Commun Mass Spectrom. 2008;22:3989–3996. doi: 10.1002/rcm.3820. [DOI] [PubMed] [Google Scholar]
- Szanto G.L., Hamelers H.V.M., Rulkens W.H., Veeken A.H.M. NH3, N2O and CH4 emissions during passively aerated composting of straw‐rich pig manure. Bioresour Technol. 2007;98:2659–2670. doi: 10.1016/j.biortech.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Takaya N. Response to hypoxia, reduction of electron acceptors, and subsequent survival by filamentous fungi. Biosci Biotechnol Biochem. 2009;73:1–8. doi: 10.1271/bbb.80487. [DOI] [PubMed] [Google Scholar]
- Tavares P., Pereira A., Moura J., Moura I. Metalloenzymes of the denitrification pathway. J Inorg Biochem. 2006;100:2087–2100. doi: 10.1016/j.jinorgbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Teissier S., Torre M. Simultaneous assessment of nitrification and denitrification on freshwater epilithic biofilms by acetylene block method. Wat Res. 2002;36:3803–3811. doi: 10.1016/s0043-1354(02)00098-2. [DOI] [PubMed] [Google Scholar]
- Throback I.N., Enwall K., Jarvis A., Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Tiquia S.M., Tam N.F.Y. Fate of nitrogen during composting of chicken litter. Environ Pollut. 2000;110:535–541. doi: 10.1016/s0269-7491(99)00319-x. [DOI] [PubMed] [Google Scholar]
- Tiquia S., Richard T., Honeyman M. Carbon, nutrient, and mass loss during composting. Nutr Cycl Agroecosyst. 2002;62:15–24. [Google Scholar]
- Toyoda S., Yoshida N. Determination of nitrogen isotopomers of nitrous oxide on a modified isotope ratio mass spectrometer. Anal Chem. 1999;71:4711–4718. [Google Scholar]
- Toyoda S., Mutobe H., Yamagishi H., Yoshida N., Tanji Y. Fractionation of N2O isotopomers during production by denitrifier. Soil Biol Biochem. 2005;37:1535–1545. [Google Scholar]
- Wang C., Shyu C., Ho S., Chiou S. Species diversity and substrate utilization patterns of thermophilic bacterial communities in hot aerobic poultry and cattle manure composts. Microb Ecol. 2007;54:1–9. doi: 10.1007/s00248-006-9139-4. [DOI] [PubMed] [Google Scholar]
- Wells G., Park H., Yeung C., Eggleston B., Francis C., Criddle C. Ammonia‐oxidizing communities in a highly aerated full‐scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol. 2009;11:2310–2328. doi: 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- Wertz S., Poly F., Le Roux X., Degrange V. Development and application of a PCR‐denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter‐like nxrA sequences in soil. FEMS Microbiol Ecol. 2008;63:261–271. doi: 10.1111/j.1574-6941.2007.00416.x. [DOI] [PubMed] [Google Scholar]
- Wertz S., Dandie C.E., Goyer C., Trevors J.T., Patten C.L. Diversity of nirK denitrifying genes and transcripts in an agricultural soil. Appl Environ Microbiol. 2009;75:7365–7377. doi: 10.1128/AEM.01588-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrage N., Velthof G., Van Beusichem M., Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem. 2001;33:1723–1732. [Google Scholar]
- Wrage N., Velthof G., Laanbroek H., Oenema O. Nitrous oxide production in grassland soils: assessing the contribution of nitrifier denitrification. Soil Biol Biochem. 2004;36:229–236. [Google Scholar]
- Yamamoto N., Otawa K., Nakai Y. Bacterial communities developing during composting processes in animal manure treatment facilities. Asian Austral J Anim Sci. 2009;22:900–905. [Google Scholar]
- Yamamoto N., Otawa K., Nakai Y. Diversity and abundance of ammonia‐oxidizing bacteria and ammonia‐oxidizing archaea during cattle manure composting. Microb Ecol. 2010;60:807–815. doi: 10.1007/s00248-010-9714-6. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Toyoda S. Constraining the atmospheric N2O budget from intramolecular site preference in N2O isotopomers. Nature. 2000;405:330–334. doi: 10.1038/35012558. [DOI] [PubMed] [Google Scholar]
- Yoshinari T., Knowles R. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem Biophys Res Commun. 1976;69:705–710. doi: 10.1016/0006-291x(76)90932-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ronimus R., Turner N., Zhang Y., Morgan H. Enumeration of thermophilic Bacillus species in composts and identification with a random amplification polymorphic DNA (RAPD) protocol. Syst Appl Microbiol. 2002;25:618–626. doi: 10.1078/07232020260517760. [DOI] [PubMed] [Google Scholar]
- Zhu S., Chan G., Cai K., Qu L., Huang L. Leachates from municipal solid waste disposal sites harbor similar, novel nitrogen‐cycling bacterial communities. FEMS Microbiol Lett. 2007;267:236–242. doi: 10.1111/j.1574-6968.2006.00560.x. [DOI] [PubMed] [Google Scholar]
- Zumft W.G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W.G. Biogenesis of the bacterial respiratory Cu. J Mol Microbiol Biotechnol. 2005;10:154–166. doi: 10.1159/000091562. [DOI] [PubMed] [Google Scholar]


