Summary
For possible control of fire blight affecting apple and pear trees, we characterized Erwinia amylovora phages from North America and Germany. The genome size determined by electron microscopy (EM) was confirmed by sequence data and major coat proteins were identified from gel bands by mass spectroscopy. By their morphology from EM data, φEa1h and φEa100 were assigned to the Podoviridae and φEa104 and φEa116 to the Myoviridae. Host ranges were essentially confined to E. amylovora, strains of the species Erwinia pyrifoliae, E. billingiae and even Pantoea stewartii were partially sensitive. The phages φEa1h and φEa100 were dependent on the amylovoran capsule of E. amylovora, φEa104 and φEa116 were not. The Myoviridae efficiently lysed their hosts and protected apple flowers significantly better than the Podoviridae against E. amylovora and should be preferred in biocontrol experiments. We have also isolated and partially characterized E. amylovora phages from apple orchards in Germany. They belong to the Podoviridae or Myoviridae with a host range similar to the phages isolated in North America. In EM measurements, the genome sizes of the Podoviridae were smaller than the genomes of the Myoviridae from North America and from Germany, which differed from each other in corresponding nucleotide sequences.
Introduction
Bacteriophages occur in many environments and may even outnumber their host cells. They need an appropriate receptor for infection, which restricts the host range. After docking to the cell surface, bacteriophages inject their genome to multiply inside the cell. At the end of their life cycle viral proteins lyse the host cells. Efficient destruction of a pathogen can be beneficial to prevent infections of the host tissue (Jones et al., 2007). Bacteriophages have been applied in controlling bacterial populations on plants such as onion, tomato and potato (McKenna et al., 2001; Obradovic et al., 2004; Lang et al., 2007). The Gram‐negative bacterium Erwinia amylovora is the causal agent of fire blight, a necrotic disease that affects rosaceous plants and can lead to high commercial losses in production of the economically important fruit crops apple, pear and quince (Momol and Aldwinckle, 2000).
Bacteriophages have been classified by their electron microscopy (EM) morphotype, plaque morphology and restriction fragment pattern. MALDI‐TOF mass spectroscopy (MS) was used to identify structural proteins. The genomes of five E. amylovora phages have been fully sequenced. Phage φEra103 (Accession No. EF160123; Vandenbergh and Cole, 1986), φEa1h and φEa100 (Müller et al., 2011) belong to the short‐tailed Podoviridae, and phages φEa21‐4 (Accession No. EU710883, Lehman et al., 2009) and φEa104 (Müller et al., 2011) to the Myoviridae with a long contractile tail structure. The Podoviridae package an EPS depolymerase into their coat (Bernhard et al., 1996), which degrades the amylovoran capsules of the pathogen and exposes E. amylovora to plant defence mechanisms (Kim and Geider, 2000). The enzyme expressed in plant cells under control of the strong 35S promoter reduced fire blight symptoms on apple (Flachowsky et al., 2008) and pear (Malnoy et al., 2005). Attempts have been described to apply bacterio‐phages for control of fire blight (Erskine, 1973; Schnabel and Jones, 2001), although details about their interaction with E. amylovora and bacterial populations in flowers are missing. We have concentrated our efforts to growth requirements of E. amylovora phages and symptom reduction in fire blight tissue, such as apple flowers and pear slices.
Results
Morphology of four E. amylovora phages and their genome size
We have characterized four E. amylovora phages isolated in North America and three phages from Germany. The phages were allocated into morphotype groups according to Ackermann (2007). The American phages show an icosahedral head (size 60–73 nm, Table 1). The first group carries a short tail which was not seen in the contracted form (Fig. 1), and a second group of phages carries long tails of 114 nm (Table 1). This tail is contractile as shown in Fig. 1G for phage φEa104 and others. Erwinia amylovora phages φEa1h and φEa100 (Fig. 1A and D) are compact particles without an extended tail, whereas φEa104 and φEa116 have a well‐visible tail. In negative staining of the phage particles small tail fibres were detected, and genes encoding tail fibre proteins were identified on the genomes.
Table 1.
Genome features and morphology of the E. amylovora phages investigated.
| Podoviridae | Myoviridae | ||||||
|---|---|---|---|---|---|---|---|
| φEa1ha | φEa100b | φEaJ08T | φEaK08T | φEa104c | φEa116 | φEaJ08C | |
| Size from EM measurement (bp) | 45 741 ± 880 | 45 642 ± 1 340 | 39 600 ± 629 | 39 865 ± 518 | 84 522 ± 1 014 | 85 578 ± 755 | 84 189 ± 1 117 |
| Genome size by sequencing (bp) | 45 522 | 45 554 | 84 564 | 70% sequenced | |||
| GC content (%) | 49.7 | 49.7 | 43.9 | ||||
| Predicted ORFs | 50 | 50 | 118 | ||||
| Predicted tRNAs | 0 | 0 | 24 | ||||
| Head diameter (nm) | 59.89 ± 1.49 | 61.42 ± 2.14 | 59.35 ± 1.39 | 59.83 ± 3.03 | 71.56 ± 2.20 | 73.36 ± 1.89 | 72.98 ± 3.14 |
| Tail length (nm) | 114.42 ± 2.51 | 114.62 ± 2.28 | 115.54 ± 5.17 | ||||
| Tail diameter (nm) | 17.75 ± 1.58 | 20.41 ± 1.06 | 22.42 ± 2.08 | ||||
Figure 1.
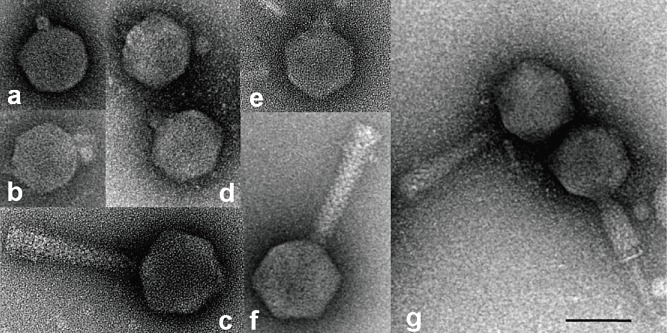
Electron microscopic visualization of Erwinia amylovora phages. (A) φEa1h; (B) φEaJ08T; (C) φEaJ08C; (D) φEa100; (E) φEa08KT; (F) φEa116; (G) φEa104. The bar represents a size of 60 nm.
The four American bacteriophages showed different lysis properties (Fig. 2). Phages φEa1h and φEa100 formed turbid plaques. φEa1h produced large plaques with a distinct turbid halo as described before by Ritchie and Klos (1977). φEa100 formed smaller plaques than φEa1h and no halos were observed. The phages φEa104 and φEa116 produced clear plaques of about 1 mm in diameter on E. amylovora strain Ea1/79Sm.
Figure 2.
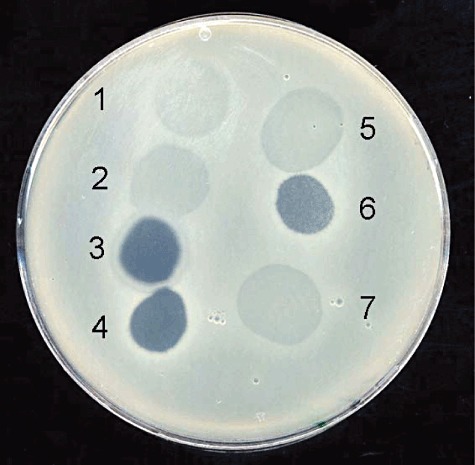
Bacteriophage drop test with American and German phages on E. amylovora strain Ea1/79Sm: 1: φEa1h, 2: φEa100, 3: φEa104, 4: φEa116, 5: φEaK08T, 6: φEaJ08C, 7: φEaJ08T.
For measuring the size of their genome the phages were lysed on a grid. Phages φEa1h and φEa100 were close to 46 kb, whereas φEa104 and φEa116 were larger with a size of 85 kb (Table 1). EM measurements for the lengths of the genomes of φEa1h, φEa100 and φEa104 were in good agreement with sequence data (Table 1).
Genomic properties of bacteriophages φEa1h, φEa100, φEa104, φEa116
A striking difference of the phages φEa1h and φEa100 to the related phage φEra103 was an insertion within the DNA polymerase gene. The insertion was confirmed by resequencing this region twice in both directions. In an amino acid alignment both domains reconstitute a DNA polymerase similar to the enzyme of bacteriophage φEra103.
Restriction digests of the Podoviridae genomes with BamHI, BglII, ClaI, HindIII, SpeI and XbaI agreed with the expected positions. For EcoRI a 1.7 kb band instead of a 1.3 kb fragment was detected on a 1% agarose gel.
Both Podoviridae may use direct repeats for replication as concatemers, also proposed for the related Escherichia coli phage T7. The 54‐mer ‘GTCTATAGGTAGGCCCAGGTTATCCAGGTCTATAGGTAGGCCCAGGTTATCCAG’ is repeated twice and could serve as required direct repeat. However, an in silico tested alternative start of the Podoviridae genomes with the 26‐mer ‘GCAAGGTAATGGCTAGGCTATGTCCC’ was in full agreement with the EcoRI digest.
About 70% of the genome size of φEa116 as determined by EM were obtained by shotgun sequencing and primer walking using the complete genome sequence of the related phages φEa104 and φEa21‐4 for primer design. We assume strong viral promoters to cause lethal effects by cloning some insertions in E. coli. The relationship of φEa116 to φEa104 indicated a genome length of 85 kb, which is in agreement with EM data for the contour length of DNA from φEa116. The positions of the six contigs obtained are outlined in Fig. 3 and Table 2, with respect to the genome of the related phage φEa104. The contigs show 80–90% identity to the corresponding sequences of φEa104. Their annotation corresponds to parts of the annotated φEa104 genome. No EPS depolymerase gene was identified in the genome of both phages, which could indicate their EPS‐independent action for growth on E. amylovora strains without an amylovoran capsule.
Figure 3.
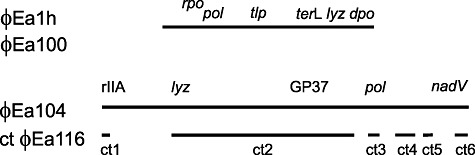
A map for the genomes of φEa1h and φEa100 and position of contigs of phage φEa116 aligned to the genome of φEa104. Abbreviations: rpo, RNA polymerase; pol, DNA polymerase; tlp, tail tubular protein; terL, large terminase subunit; lyz, lysozyme; dpo, EPS depolymerase; rIIA, membrane integrity protector; GP37, gene product 37; nadV, nicotinamide phosphoribosyl transferase.
Table 2.
Size and positions of φEa116 contigs (ct) within the genome of φEa104.
| ct | Size (in bp) | Position of φEa116 cts in the genome of φEa104 | Nucleotide identity (%) |
|---|---|---|---|
| 1 | 1 978 | 1 to 1 979 | 85 |
| 2 | 41 982 | 16 565 to 58 154 | 87 |
| 3 | 2 566 | 61 463 to 64 028 | 88 |
| 4 | 4 302 | 67 670 to 71 976 | 80 |
| 5 | 2 208 | 74 049 to 76 256 | 89 |
| 6 | 3 033 | 81 533 to 84 565 | 89 |
Identity on nucleotide level is indicated.
Identification of phage proteins by MALDI‐TOF MS analysis
Protein preparations of the bacteriophages φEa1h, φEa100, φEa104 and φEa116 were analysed on an SDS gel (Fig. 4). A size of 39–40 kDa was estimated for a dominant protein band, possibly the major capsid protein. MALDI‐TOF MS analysis indicated a nominal molecular mass of 40.0 kDa for the φEa1h and φEa100 protein and 40.8 kDa for the protein of φEa104 and φEa116, with intermediate to high sequence coverage of the cleavage peptides. A high amino acid similarity was found to the capsid proteins of E. amylovora phages φEra103 (gi 125999997) for φEa1h and φEa100 and of φEa21‐4 (gi 219681311) for φEa104 and φEa116. The molecular weight of the major capsid proteins calculated for φEa1h/φEa100 from the genome sequences is 39.95 kDa and for φEa104 and φEa116 40.79 kDa. In addition, a structural protein was identified for φEa104 and φEa116. This protein has similarity to a conserved structural protein from φEa21‐4 (gi 219681316). A band below 40 kDa may represent a bacterial protein.
Figure 4.
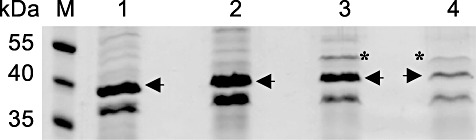
SDS‐PAGE of E. amylovora phage protein preparations. The proteins from bands marked with arrows and asterisks were identified by MALDI‐TOF MS analysis. The major capsid protein is marked for all four phages by an arrow, a structural protein for φEa104 and φEa116 by an asterisk. Lanes 1–4: φEa1h, φEa100, φEa104, φEa116 respectively; M: pre‐stained protein ladder (Fermentas, St. Leon‐Rot, Germany).
Phage growth and propagation on various E. amylovora strains and other bacteria
The four American bacteriophages were tested for their lysis efficiency on various E. amylovora strains, as well as on other Erwinia and Pantoea strains (Table 3). Apart from E. amylovora fruit tree isolates, two raspberry isolates were tested. The Podoviridae were not able to lyse all E. amylovora strains. The Myoviridae exhibited a broader host range than the Podoviridae. Strong lytic activity was visible for all E. amylovora and some Erwinia pyrifoliae strains. Erwinia pyrifoliae isolates from Japan were less sensitive than the Korean strains. Erwinia billingiae was slightly sensitive, but Erwinia tasmaniensis was resistant to all four E. amylovora phages. A low sensitivity was seen for Pantoea agglomerans MB96 and C9‐1, whereas Pantoea stewartii strain DC283 was highly sensitive to φEa104 and φEa116, but less to φEa1h and φEa100.
Table 3.
Host range of bacteriophages from North America and Germany on Erwinia and Pantoea host strains.
| Strain | Podoviridae | Myoviridae | |||||
|---|---|---|---|---|---|---|---|
| φEa1h | φEa100 | φEaJ08T | φEaK08T | φEa104 | φEa116 | φEaJ08C | |
| E. amylovora | |||||||
| CFBP1430 | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| CFBP1430Sm‐amsD | − | − | − | − | +++ | +++ | +++ |
| PMV6076 | ++ | +++ | +++ | +++ | +++ | +++ | ++(+) |
| Ea1/79 | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Ea1/79Sm | ++ | +++ | ++ | ++ | +++ | +++ | +++ |
| Ea1/79del100 | − | − | − | − | +++ | +++ | ++(+) |
| Ea322A | −a | −a | −a | −a | +++ | +++ | +++ |
| EaDS05 | +++ | +++ | +++ | +++ | ++(+) | +++ | ++(+) |
| EaDS08 | ++ | ++ | ++ | ++ | ++(+) | ++(+) | ++ |
| EaRW06 | ++ | ++(+) | +(+) | +(+) | +++ | +++ | ++(+) |
| EaOR1/07 | ++a | ++a | ++a | ++a | ++(+) | ++(+) | ++(+) |
| EaOR2/07 | (+)a | (+)a | (+)a | (+)a | +++ | +++ | +++ |
| IL6 | +++ | +++ | +++ | +++ | ++ | ++ | + |
| MR1 | +(+) | +(+) | ++ | ++ | ++ | ++ | +(+) |
| E. pyrifoliae | |||||||
| Ep1/96 | (+)a | +a | +a | +a | ++ | ++ | + |
| Ep2/97 | (+)a | +a | +a | +a | ++ | ++(+) | +(+) |
| Ejp557 | − | − | − | − | + | + | (+) |
| Ejp617 | (+) | (+) | (+) | (+) | (+) | + | − |
| E. billingiae | |||||||
| Eb661 | − | + | + | + | + | + | + |
| E. tasmaniensis | |||||||
| Et1/99 | − | − | − | − | − | − | − |
| P. stewartii | |||||||
| DC283 | +(+) | + | +++ | +++ | +++ | +++ | ++(+) |
| P. agglomerans | |||||||
| C9‐1 | − | − | (+) | (+) | + | +(+) | (+) |
| MB96 | − | + | + | + | + | + | (+) |
Strains grown on MM2C minimal agar; Pantoea strains were grown on LBglc agar.
The phages were assayed in soft agar drop tests. Lysis efficiency is rated from very good (+++) to no lysis (−). A tendency to a lower category is indicated by parentheses.
Propagation of the four American E. amylovora phages was also assayed on nitrocellulose filters with wild type E. amylovora strains isolated from 2005 to 2007. They multiplied up to 1010 pfu per disk with a slight decrease for EaOR1/07 (Table 4). The non‐pathogenic hrp mutant PMV6076 was also a good host strain, whereas Ea322A, a hrp mutant of a French strain, was less efficient for φEa116 in three independent assays. As seen in drop tests on lawns of E. amylovora, the EPS mutants Ea1/79‐del100 and CFBP1430Sm‐amsD did not propagate the phages φEa1h and φEa100, in contrast to φEa104 and φEa116. No increase of phage titre was found with E. billingiae strain Eb661. Only phage φEa1h grew to a high titre on P. stewartii strain DC283, in contrast to the low phage sensitivity of this strain in drop tests (Table 3).
Table 4.
Multiplication of bacteriophages with E. amylovora and other bacteria on filter disks.
| Strain | Bacteriophages | |||
|---|---|---|---|---|
| φEa1h | φEa100 | φEa104 | φEa116 | |
| E. amylovora | ||||
| EaDS05 | 1.0 × 109 | 2.0 × 109 | 5.0 × 109 | 2.0 × 109 |
| EaRW06 | 2.0 × 109 | 5.0 × 108 | 2.0 × 109 | 2.3 × 108 |
| EaOR1/07 | 2.0 × 108 | 2.0 × 108 | 5.0 × 108 | 1.0 × 109 |
| EaOR2/07 | 5.0 × 109 | 1.0 × 1010 | 1.8 × 1010 | 1.5 × 1010 |
| PMV6076 | 5.5 × 109 | 2.2 × 1010 | 5.0 × 1010 | 2.0 × 1010 |
| Ea1/79 del100 | 2.2 × 105 | 3.3 × 102 | 2.0 × 108 | 2.5 × 108 |
| Ea322A | 1.1 × 109 | 2.4 × 108 | 2.9 × 108 | 4.3 × 105 |
| E. billingiae | ||||
| Eb661 | 1.0 × 104 | 0 | 7.0 × 103 | 3.5 × 103 |
| P. stewartii | ||||
| DC283a | 3.5 × 107 | 0 | 5.0 × 103 | 5.0 × 105 |
Grown on LBglc agar
EPS synthesis and phage sensitivity
The amount of EPS produced by E. amylovora strains from France, Germany and North America significantly differed (Table 5). The French strain CFBP1430 and its corresponding hrp mutant PMV6076 produced high amounts of EPS, similar to the Sm‐resistant strain Ea1/79Sm. The three German isolates EaRW1/06, EaDS05 and EaDS08 produced less EPS than the former strains. Two strains from Oregon synthesized low amounts of EPS, close to the EPS‐negative ams deletion mutant Ea1/79‐del100.
Table 5.
EPS synthesis of suspension cultures of various Erwinia strains and the P. stewartii strain DC283.
| Strain | EPS produced (µg ml−1) |
|---|---|
| CFBP1430 | 38.3 |
| PMV6076 | 71.6 |
| Ea1/79 | 0.8 |
| Ea1/79Sm | 46.0 |
| Ea1/79del100 | 0.4 |
| Ea322A | 0.7 |
| EaRW1/06 | 1.2 |
| EaOR1/07 | 0.6 |
| EaOR2/07 | 0.2 |
| EaDS05 | 6.7 |
| EaDS08 | 2.9 |
| Ep1/96 | 551.1 |
| Ep2/97 | 479.6 |
| Ejp557 | 0.4 |
| Ejp617 | 17.1 |
| Eb661 | 7.4 |
| Et1/99a | 1.3 |
| P. stewartii DC283a | 97.7 |
Grown in LBglc medium.
The height of EPS production was determined with the CPC assay and is expressed in µg ml−1
In general, the Myoviridae were not influenced by presence or absence of amylovoran capsules of the host cells (Table 3). The rubus strains IL6 and MR1 and also the German strain Ea1/79 showed an intermediate sensitivity. The French strains CFBP1430 and PMV6076 were highly susceptible to φEa1h and φEa100, as E. amylovora strain EaDS05 isolated in Germany. The German strain Ea1/79 with low EPS synthesis showed an intermediate sensitivity to the phages φEa1h and φEa100. The increased EPS synthesis of the spontaneous mutant Ea1/79Sm enhanced phage sensitivity for the Podoviridae. Generally, a correlation between high EPS production and enhanced phage sensitivity was noted for the Podoviridae.
Erwinia pyrifoliae strains, except Ejp557, showed high EPS synthesis, as well as the E. billingiae strain Eb661 and the P. stewartii strain DC283. Nevertheless, they were slightly sensitive to the Podoviridae, which might be due to unsuited phage receptors on the cell surface. Erwinia tasmaniensis Et1/99 without detectable EPS synthesis was insensitive to the phages. These strains were not or only partially sensitive to the Myoviridae.
Reduction of disease symptoms on apple flowers and immature pears
The Podoviridae had a weak effect on growth reduction of E. amylovora in flowers (Fig. 5A). φEa1h and φEa100 reduced the recovered pathogen by 40% compared with the control flowers. The MyoviridaeφEa104 and φEa116 reduced the recovered cells by 90%.
Figure 5.
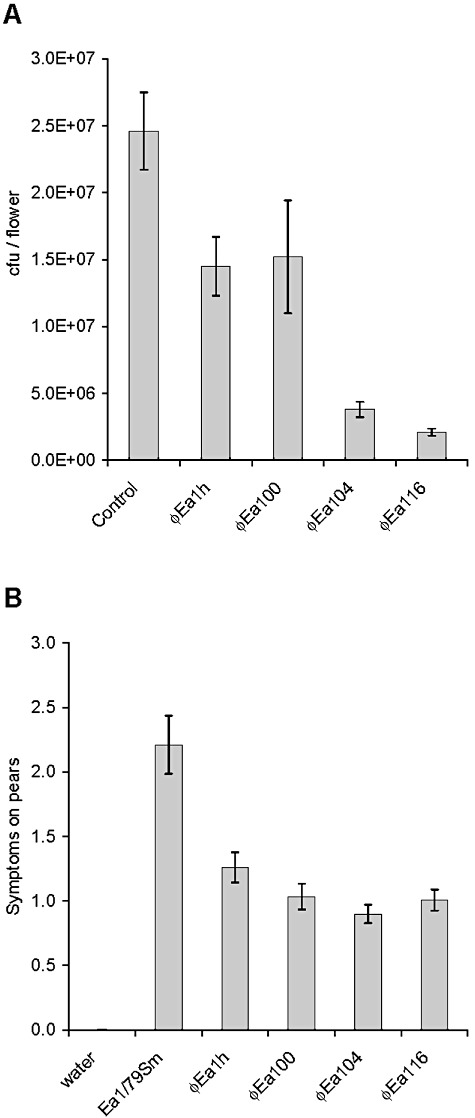
Growth reduction of E. amylovora by phage treatment on apple flowers and immature pears. A. Flower assays with Ea1/79Sm using single bacteriophages. Control flowers were inoculated only with Ea1/79Sm cells. B. Fire blight symptoms on pear slices. Strain Ea1/79Sm applied without and with single bacteriophages.
Symptoms on pear slices treated with a single phage were reduced by half compared with the control without phage treatment. A significant difference among Podoviridae and Myoviridae was not observed for immature pears. Due to high ooze production of E. amylovora on pear slices, the Podoviridae may find a favourable environment for interference with the pathogen (Fig. 5B).
A mixture of Podoviridae and Myoviridae applied with the pathogen for inoculation of apple flowers and pear slices did not significantly change symptom formation compared with treatment with a single myovirus (data not shown).
Partial characterization of new E. amylovora phages
From apple orchards in Germany three E. amylovora phages were isolated in 2008 and were investigated by negative staining for their morphological properties with EM. φEaJ08T and φEaK08T (Fig. 1B and E) belonged to the Podoviridae and resembled φEa1h and φEa100 shown in Fig. 1A and D. The head capsules of φEaJ08T and φEaK08T were 60 nm in diameter (Table 1), comparable to the particle size of φEa1h and φEa100. Their genome length was determined by EM and found to be 40 kb. φEaJ08C was identified as member of the Myoviridae based on its morphology in the EM (see Fig. 1C). Its head capsule was 73 nm in diameter and its contractile tail carrying short tail fibres was 116 nm in length. Its genome size determined by EM was approximately 84 kb, similar to the American Myoviridae (Table 1).
On E. amylovora host strains they formed either turbid (T) or clear plaques (C). In host range assays, φEa1h, φEa100, φEaJ08T and φEaK08T were related as were φEa104, φEa116 and φEaJ08C (Table 3). The lysis pattern of the German strains resembled those of the other Podoviridae and Myoviridae respectively.
PCR primer pairs were designed from the genome sequences of the four American phages and applied to the German phages. The German phages φEaJ08T and φEaK08T showed no or weak signals for primer pairs designed from φEa1h, φEa104 or φEa116. For φEaJ08C partial sequences were recovered, which showed high similarity to phage φEa116 (Table S1) and less to φEa104. Most mismatches were found in two sequences of a putative structural protein.
Discussion
We have characterized several E. amylovora phages from North America and from Germany for their virion morphology, interaction with host strains and properties of their genomes. They belong to the order Caudovirales and were classified according to their morphotypes C1 and A1 as Podoviridae or Myoviridae respectively (Ackermann, 2001; 2007). The particle size of 60 nm for φEa1h was in agreement with previous data (Ritchie and Klos, 1979). The head capsules of the Myoviridae had a similar size as shown for the related Salmonella phage Felix O1 (Kuhn et al., 2002) and the closely related E. amylovora phage φEa21‐4 (Lehman et al., 2009). The genome structure of the American Myoviridae is completely different from that of the Podoviridae. The Myoviridae genome encodes 24 tRNA genes, one HNH endonuclease gene in contrast to five for the Podoviridae, where also a DNA‐directed RNA polymerase gene was predicted. The Myoviridae lack a small terminase subunit gene, and they do not carry an EPS depolymerase gene. This supports our results that they are not dependent on an amylovoran capsule for host infection, unlike the Podoviridae. Podoviridae and Myoviridae can be classified by their protein sequence similarity with a blastp tool (Lavigne et al., 2008; 2009). Due to the high similarity of our sequenced bacteriophages on nucleotide and protein level to φEra103, φEa1h and φEa100 belong to the newly formed subfamily of Autographivirinae (SP6‐like phages). We observed that φEa1h and φEa100 lack the large terminal repeat described for φEra103. Phages φEa104 and φEa116 are related to φEa21‐4 and belong to the newly suggested genus of Felix O1‐like phages (Lavigne et al., 2009).
The genomes of φEa1h and φEa100 are similar to the genome of φEra103 with 45 455 bp (Vandenbergh and Cole, 1986). HNH endonucleases, encoded in the phage genomes as one or more homologues, can cleave double‐stranded DNA similar to colicin E9 from E. coli (Pommer et al., 2001). In a previous report the genome size of φEa100 and φEa116 was estimated from PFGE analysis to be 34 kb and 75 kb respectively (Schnabel and Jones, 2001). These sizes were corrected by our sequencing data and the EM measurements of the DNA contour lengths.
To test the ability of E. amylovora phages to control fire blight, they were applied to liquid cultures of a pathogenic E. amylovora strain and the decrease of the optical density was measured (Schnabel and Jones, 2001). Similar assays were performed by Gill and colleagues (2003), who also analysed the host range of E. amylovora phages and observed considerable differences. In our experiments the host range of Podoviridae and Myoviridae phages diverged. The former depend on EPS capsules of E. amylovora cells (Billing, 1960; Ritchie and Klos, 1977), as shown for φEa1h (Ayers et al., 1979; Ritchie and Klos, 1979; Bernhard et al., 1996). Erwinia amylovora strains without amylovoran synthesis are not sensitive to the Podoviridae. The EPS depolymerase enables them to degrade the amylovoran capsule (Kim and Geider, 2000), which results in the formation of expanding halos (Ritchie and Klos, 1979; Hartung et al., 1988). The EPS depolymerase genes of φEa1h and φEa100 differ by three bases resulting in three different amino acids. This could explain the lack of halo formation for φEa100.
For fire blight control a combination of bacteriophages and antagonistic bacteria was suggested (Svircev et al., 2006). This approach could not be realized with our potentially antagonistic strains. The two Pantoea strains tested by us did not support phage propagation. Erwinia billingiae Eb661 was weakly lysed without propagation of phages. For the fire blight antagonist E. tasmaniensis Et1/99 no lysis was observed. Cultivation of the Myoviridae on avirulent E. amylovora mutants increased the titre to 1010 pfu per disk. In laboratory and greenhouse trials E. amylovora mutants reduced symptom formation (Tharaud et al., 1997). The use of avirulent E. amylovora mutants for phage propagation is an alternative to the application of epiphytic bacteria, however, meaning the release of genetically modified organisms.
Growth of E. amylovora was well promoted on young flowers (Thomson and Gouk, 2003; Pusey and Smith, 2008) which were also applied in our fire blight control experiments. Phage treatment under lab conditions yielded in significant symptom reduction. Biocontrol of plant pathogens was attempted with Pseudomonas phages to cure bacterial spot on tomato (Iriarte et al., 2007). Bacteriophages were also applied for treatment of human bacterial diseases (Stone, 2002).
Three different phages were isolated during the fire blight season 2008 from blighted plant material in southern Germany. The isolation of bacteriophages from symptomless plant material was not successful, which is not uncommon (Ritchie and Klos, 1977). The soil around infected trees is assumed to be a reservoir for phages (Crosse and Hingorani, 1958; Gill et al., 2003). The Podoviridae or Myoviridae we isolated differed from the American phages in genome size and molecular properties. They may become enriched in E. amylovora populations of infected plants. Their ecology and persistence in nature is difficult to describe. The phages may migrate with spread of fire blight, or they may propagate in E. amylovora as a more sensitive host than other bacteria in their environment.
Experimental procedures
Bacterial strains and bacteriophages
Bacterial strains and bacteriophages used in the experiments are listed in Table 6. The bacteria were grown on StI agar (Merck AG, Darmstadt, Germany), in Luria–Bertani (LB) broth or in MM2C medium (Bereswill et al., 1998). For strains of the genus Pantoea and for E. tasmaniensis, 1% glucose was added to the LB medium.
Table 6.
Bacterial strains and bacteriophages used in the assays.
| Strain | Isolation, properties | Source/reference |
|---|---|---|
| E. amylovora | ||
| CFBP1430 | Crataegus sp.; France, 1972 | CFBP |
| CFBP1430Sm‐amsD | Smr mutant of CFBP1430, Tn5 insertion in amsD | Lab collection |
| PMV6076 | hrp/dsp deletion mutant CFBP1430 | Barny et al. (1990) |
| Ea1/79 | Cotoneaster sp.; Germany, 1979 | Falkenstein et al. (1988) |
| Ea1/79del100 | Cmr, deletion of ams cluster | Bugert and Geider (1995) |
| Ea1/79Sm | Smr spontaneous mutant of Ea1/79 | Bellemann et al. (1994) |
| Ea322A | CFBP1368, avirulent Tn5 mutant of Ea322 | Jock et al. (2000) |
| EaDS05 | Quince; Germany, 2007 | Lab collection |
| EaDS08 | Apple; Germany, 2008 | Lab collection |
| EaOR1/07 | Apple; USA, 2007 | Lab collection |
| EaOR2/07 | Apple; USA, 2007 | Lab collection |
| EaRW1/06 | Cotoneaster floccosus; Germany, 2006 | Lab collection |
| IL6 | Raspberry; USA | Kim et al. (2001) |
| MR1 | Raspberry; USA | Jock and Geider (2004) |
| E. pyrifoliae | ||
| Ep1/96 | Nashi pear; Korea | Kim et al. (1999) |
| Ep2/97 | Nashi pear; Korea, hrpL | Jock et al. (2003) |
| Ejp557 | Nashi pear; Japan | Kim et al. (2001) |
| Ejp617 | Nashi pear; Japan | Kim et al. (2001) |
| E. billingiae | ||
| Eb661T | NCPPB661T, apple tissue; England | Mergaert et al. (1999) |
| E. tasmaniensis | ||
| Et1/99T | Apple flowers; Tasmania | Geider et al. (2006) |
| P. stewartii | ||
| DC283 | Nalr mutant of SS104, Zea mays; Illinois, 1967 | Coplin et al. (2002) |
| P. agglomerans | ||
| C9‐1 | Apple; USA | V. Stockwell |
| MB96 | Corn; Germany, 1996 | Lab collection |
| Bacteriophages | ||
| φEa1h | Blighted Jonathan apple shoots; Michigan, 1975 | Hartung et al. (1988) |
| φEa100 | Soil sample; Michigan, 1996 | Schnabel and Jones (2001) |
| φEa104 | Soil apple orchard; Michigan, 1996 | Schnabel and Jones (2001) |
| φEa116 | Blighted apple tissue; Michigan, 1996 | Schnabel and Jones (2001) |
| φEaJ08C | Blighted apple shoots; Germany, 2008 | This work |
| φEaJ08T | Blighted apple shoots; Germany, 2008 | This work |
| φEaK08T | Blighted Gala apple shoots; Germany, 2008 | This work |
CFBP = Collection Française de Bactéries Phytopathogènes; NCPPB = National Collection of Plant Pathogenic Bacteria (UK).
Isolation of new bacteriophages
Bacteriophages were isolated in two apple orchards in southern Germany. Aerial plant material (approximately 0.5 g of leaves or twigs) was washed in 10 ml of water for 10 min or directly added to 10 ml of LB medium containing 1% sorbitol and E. amylovora cells. The mixtures were grown overnight, a few drops of CHCl3 were added to the culture supernatants and dilutions plated on LB agar with E. amylovora cells in the top agar.
Propagation of phages on nitrocellulose filters
Erwinia amylovora, E. billingiae or P. stewartii cells (100 µl, 1 × 108 cfu) were mixed with phages (1 × 104 pfu) and incubated overnight on nitrocellulose filters (25 mm in diameter, Whatman) placed on MM2C agar or LB agar with 1% glucose. Bacteria and phages were resuspended in 5 ml of MM2C medium by vortexing. The OD600 and EPS production were measured, and the phage titre was estimated by soft agar drop tests of dilutions.
Electron microscopy of bacteriophages and their DNA
Prior to EM assays, the bacteriophages were purified according to Boulanger (2009). Electron microscopic pictures were taken after negative staining with 2% uranyl acetate according to Steven and colleagues (1988). For DNA purification the phages were concentrated using a Beckman Airfuge followed by phenol extraction. The genome length of the phage DNAs was measured with the droplet method as described before (Spiess and Lurz, 1988), and circular DNA of plasmid RSF1010 (8684 bp) was used as an internal standard.
MALDI‐TOF MS analysis of viral proteins
Bacteriophage protein solutions were precipitated with 10% TCA, desalted and the proteins dissolved by heating in SDS loading buffer. The denatured proteins were separated on a 12% SDS‐PAGE for 120 min and stained with Coomassie Brilliant Blue G‐250 (PageBlue Protein Staining Solution; Fermentas, St. Leon‐Rot, Germany). As molecular marker the PageRuler Prestained Protein Ladder from Fermentas was used. Protein bands were excised and digested by trypsin. After tryptic digests of gel‐separated proteins (Gobom et al., 2001) mass spectra were recorded on a Bruker Scout 384 Reflex II instrument (Bruker Daltonik). Protein identification by MALDI‐TOF MS peptide mapping was accomplished with the search engine MASCOT (Matrix Science, UK).
DNA manipulation and sequencing
Purified bacteriophages were lysed with SDS and the solutions extracted with phenol : chloroform : isoamyl alcohol (25:24:1). After a final treatment with chloroform and ethanol precipitation, the DNA was digested with the fast‐digest versions of enzymes BamHI, BglII, EcoRI and HindIII (Fermentas, St. Leon‐Rot, Germany). Other restriction enzymes were from AGS/Hybaid, Heidelberg, Germany.
The partial sequence of phage φEa116 was determined by whole‐genome shotgun sequencing. Two plasmid libraries with an average insert size of 1 and 2 kb were generated, as previously described (Kube et al., 2005). Sequencing was performed by using dye terminator sequencing on ABI3730XL capillary sequencers. Shotgun sequences were assembled using PHRAP (http://www.phrap.org) and GAP4 (http://www.gap‐system.org). ORFs were identified by GLIMMER3 [Gene Locator and Interpolated Markov ModelER according to Delcher et al. (1999)].
After alignment of φEa116 sequences to the genomes of φEa104 and φEa21‐4, missing or ambiguous DNA regions were sequenced from PCR fragments, applying freshly designed primers and primer walking.
The sequences for the contigs and their annotation of E. amylovora phage φEa116 have been submitted in the EMBL database under Accession No. FQ857195. Their corresponding positions in the genome of φEa104 (FQ482083) are given in Table 2.
Amylovoran and stewartan synthesis
The amount of EPS was measured with the CPC‐turbidity assay (Bellemann et al., 1994).
Plant assays
Fully expanded apple flowers from greenhouse trees were placed on top of water‐filled Eppendorf tubes. The stigmata of the flowers were inoculated with 5000 cfu of the streptomycin‐resistant E. amylovora strain Ea1/79Sm and with 5 × 107 to 1 × 108 pfu of single or mixed E. amylovora phages in water. The flowers were incubated in a climate chamber with 24°C for day and 21°C for night conditions. After 5 days petals and stems of the flowers were removed, and the flowers were extracted in water for 10 min. From the extracts, 100 µl of a 10−4 dilution was plated on StI agar containing streptomycin (500 µg ml−1) and cycloheximide (50 µg ml−1).
Immature pears were cut in slices and soaked in a suspension of 5 × 108 pfu ml−1 of an E. amylovora phage or in water as a control. The briefly dried slices were inoculated with 500 cfu of the E. amylovora strain Ea1/79Sm and incubated in sealed Petri dishes for 5 days at 28°C. The pears were rated for symptoms from 0 (no symptoms) to 3 (browning and large drops of ooze).
Acknowledgments
We thank M. Gernold for excellent technical support and B. Schneider for comments on the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Partial sequence analysis of the German isolate φEaJ08C amplified with primers from φEa104 and φEa116. The positions refer to the partial sequence of φEa116 (Accession No. FQ85719).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ackermann H.‐W. Frequency of morphological phage descriptions in the year 2000. Arch Virol. 2001;146:843–857. doi: 10.1007/s007050170120. [DOI] [PubMed] [Google Scholar]
- Ackermann H.‐W. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- Ayers A.R., Ayers S.B., Goodman R.N. Extracellular polysaccharide of Erwinia amylovora: a correlation with virulence. Appl Environ Microbiol. 1979;38:659–666. doi: 10.1128/aem.38.4.659-666.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barny M.A., Guinebretière M.H., Marcais B., Coissac E., Paulin J.P., Laurent J. Cloning of a large gene‐cluster involved in Erwinia amylovora CFBP1430 virulence. Mol Microbiol. 1990;4:777–786. doi: 10.1111/j.1365-2958.1990.tb00648.x. [DOI] [PubMed] [Google Scholar]
- Bellemann P., Bereswill S., Berger S., Geider K. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int J Biol Macromol. 1994;16:290–296. doi: 10.1016/0141-8130(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Bereswill S., Jock S., Bellemann P., Geider K. Identification of Erwinia amylovora by growth morphology on agar containing copper sulfate and by capsule staining with lectin. Plant Dis. 1998;82:158–164. doi: 10.1094/PDIS.1998.82.2.158. [DOI] [PubMed] [Google Scholar]
- Bernhard F., Schullerus D., Bellemann P., Nimtz M., Coplin D.L., Geider K. Genetic transfer of amylovoran and stewartan synthesis between Erwinia amylovora and Erwinia stewartii. Microbiology. 1996;142:1087–1096. doi: 10.1099/13500872-142-5-1087. [DOI] [PubMed] [Google Scholar]
- Billing E. An association between capsulation and phage sensitivity in Erwinia amylovora. Nature. 1960;186:819–820. doi: 10.1038/186819a0. [DOI] [PubMed] [Google Scholar]
- Boulanger P. Purification of bacteriophages and SDS‐PAGE analysis of phage structural proteins from ghost particles. In: Clokie M.R.J., Kropinski A.M., editors. Humana Press; 2009. pp. 227–238. [DOI] [PubMed] [Google Scholar]
- Bugert P., Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- Coplin D.L., Majerczak D.R., Zhang Y., Kim W.‐S., Jock S., Geider K. Identification of Pantoea stewartii subsP. stewartii by PCR and strain differentiation by PFGE. Plant Dis. 2002;86:304–311. doi: 10.1094/PDIS.2002.86.3.304. [DOI] [PubMed] [Google Scholar]
- Crosse J.E., Hingorani M.K. A method for isolating Pseudomonas morsprunorum phages from soil. Nature. 1958;181:60–61. [Google Scholar]
- Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erskine J.M. Characteristics of Erwinia amylovora bacteriophage and its possible role in the epidemiology of fire blight. Can J Microbiol. 1973;19:837–845. doi: 10.1139/m73-134. [DOI] [PubMed] [Google Scholar]
- Falkenstein H., Bellemann P., Walter S., Zeller W., Geider K. Identification of Erwinia amylovora, the fireblight pathogen, by colony hybridization with DNA from plasmid pEA29. Appl Environ Microbiol. 1988;54:2798–2802. doi: 10.1128/aem.54.11.2798-2802.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachowsky H., Richter K., Kim W.S., Geider K., Hanke M.V. Transgenic expression of a viral EPS‐depolymerase is potentially useful to induce fire blight resistance in apple. Ann Appl Biol. 2008;153:345–355. [Google Scholar]
- Geider K., Auling G., Du Z., Jakovljevic V., Jock S., Völksch B. Erwinia tasmaniensis sp. nov., a non‐phytopathogenic bacterium from apple and pear trees. Int J Syst Evol Microbiol. 2006;56:2937–2943. doi: 10.1099/ijs.0.64032-0. [DOI] [PubMed] [Google Scholar]
- Gill J.J., Svircev A.M., Smith R., Castle A.J. Bacteriophages of Erwinia amylovora. Appl Environ Microbiol. 2003;69:2133–2138. doi: 10.1128/AEM.69.4.2133-2138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobom J., Schürenberg M., Müller M., Theiss D., Lehrach H., Nordhoff E. a‐Cyano‐4‐hydroxycinnamic acid affinity sample preparation. A protocol for MALDI‐MS peptide analysis in proteomics. Anal Chem. 2001;3:434–438. doi: 10.1021/ac001241s. [DOI] [PubMed] [Google Scholar]
- Hartung J.S., Fulbright D.W., Klos E.J. Cloning of a bacteriophage polysaccharide depolymerase gene and its expression in Erwinia amylovora. Mol Plant Microbe Interact. 1988;1:87–93. [Google Scholar]
- Iriarte F.B., Balogh B., Momol M.T., Smith L.M., Wilson M., Jones J.B. Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl Environ Microbiol. 2007;73:1704–1711. doi: 10.1128/AEM.02118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jock S., Geider K. Molecular differentiation of Erwinia amylovora strains from North America and of two Asian pear pathogens by analyses of PFGE patterns and hrpN genes. Environ Microbiol. 2004;6:480–490. doi: 10.1111/j.1462-2920.2004.00583.x. [DOI] [PubMed] [Google Scholar]
- Jock S., Kim W.‐K., Barny M.‐A., Geider K. Molecular characterization of natural Erwinia pyrifoliae strains deficient in the hypersensitive response. Appl Env Microbiol. 2003;69:679–682. doi: 10.1128/AEM.69.1.679-682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jock S., Rodoni B., Gillings M., Kim W.‐S., Copes C., Merriman P., Geider K. Screening of ornamental plants from the Botanic Gardens of Melbourne and Adelaide for the occurrence of Erwinia amylovora. Australas Plant Pathol. 2000;29:120–128. [Google Scholar]
- Jones J., Jackson L., Balogh B., Obradovic A., Iriarte F., Momol M. Bacteriophages for plant disease control. Annu Rev Phytopathol. 2007;45:245–262. doi: 10.1146/annurev.phyto.45.062806.094411. [DOI] [PubMed] [Google Scholar]
- Kim W.‐S., Geider K. Characterization of a viral EPS‐depolymerase, a potential tool for control of fire blight. Phytopathology. 2000;90:1263–1268. doi: 10.1094/PHYTO.2000.90.11.1263. [DOI] [PubMed] [Google Scholar]
- Kim W.‐S., Gardan L., Rhim S.‐L., Geider K. Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia Nakai) Int J Syst Bacteriol. 1999;49:899–906. doi: 10.1099/00207713-49-2-899. [DOI] [PubMed] [Google Scholar]
- Kim W.‐S., Hildebrand M., Jock S., Geider K. Molecular comparison of pathogenic bacteria from pear trees in Japan and the fire blight pathogen Erwinia amylovora. Microbiology-SGM. 2001;147:2951–2959. doi: 10.1099/00221287-147-11-2951. [DOI] [PubMed] [Google Scholar]
- Kube M., Beck A., Zinder S.H., Kuhl H., Reinhardt R., Adrian L. Genome sequence of the chlorinated compound‐respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23:1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- Kuhn J.M.S., Chiswell D., Azriel A., Berman B., Shahar D. A bacteriophage reagent for Salmonella: molecular studies on Felix 01. Int J Food Microbiol. 2002;74:217–227. doi: 10.1016/s0168-1605(01)00682-1. et al. [DOI] [PubMed] [Google Scholar]
- Lang J.M., Gent D.H., Schwartz H.F. Management of Xanthomonas leaf blight of onion with bacteriophages and a plant activator. Plant Dis. 2007;91:871–878. doi: 10.1094/PDIS-91-7-0871. [DOI] [PubMed] [Google Scholar]
- Lavigne R., Seto D., Mahadevan P., Ackermann H.W., Kropinski A.M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using blastp‐based tools. Res Microbiol. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Lavigne R., Darius P., Summer E.J., Seto D., Mahadevan P., Nilsson A.S. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009;9:244. doi: 10.1186/1471-2180-9-224. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman S.M., Kropinski A.M., Castle A.J., Svircev A.M. Complete genome of the broad‐host‐range Erwinia amylovora phage φEa21‐4 and its relationship to Salmonella phage Felix O1. Appl Environ Microbiol. 2009;75:2139–2147. doi: 10.1128/AEM.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna F., El‐Tarabily K.A., Hardy K.A., Hardy G.E.S.J., Dell B. Novel in vivo use of a polyvalent Streptomyces phage to disinfest Streptomyces scabies‐infected seed potatoes. Plant Pathol. 2001;50:666–675. [Google Scholar]
- Malnoy M., Faize M., Venisse J.S., Geider K., Chevreau E. Expression of viral EPS‐depolymerase reduces fire blight susceptibility in transgenic pear. Plant Cell Rep. 2005;23:632–638. doi: 10.1007/s00299-004-0855-2. [DOI] [PubMed] [Google Scholar]
- Mergaert J., Hauben L., Cnockaert M.C., Swings J. Reclassification of non‐pigmented Erwinia herbicola strains from trees as Erwinia billingiae sp. nov. Int J Syst Bacteriol. 1999;49:377–383. doi: 10.1099/00207713-49-2-377. [DOI] [PubMed] [Google Scholar]
- Momol M.T., Aldwinckle H.S. Genetic diversity and host range of Erwinia amylovora. In: Vanneste J.L., editor. CABI Publishing; 2000. pp. 55–72. , and . In Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora (ed.). Wallingford, UK: , pp. [Google Scholar]
- Müller I., Kube M., Reinhardt R., Jelkmann W., Geider K. The complete genome sequences of three Erwinia amylovora phages isolated in North America and a bacteriophage induced from an Erwinia tasmaniensis strain. J Bacteriol. 2011;193:795–796. doi: 10.1128/JB.01293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic A., Jones J.B., Momol M.T., Balogh B., Olson S.M. Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Dis. 2004;88:736–740. doi: 10.1094/PDIS.2004.88.7.736. [DOI] [PubMed] [Google Scholar]
- Pommer A.J., Cal S., Keeble A.H., Walker D., Evans S.J., Kühlmann U.C. Mechanism and cleavage specificity of the H‐N‐H endonuclease colicin E9. J Mol Biol. 2001;314:735–749. doi: 10.1006/jmbi.2001.5189. et al. [DOI] [PubMed] [Google Scholar]
- Pusey P.L., Smith T.J. Relation of apple flower age to infection of hypanthium by Erwinia amylovora. Plant Dis. 2008;92:137–142. doi: 10.1094/PDIS-92-1-0137. [DOI] [PubMed] [Google Scholar]
- Ritchie D.E., Klos E.J. Isolation of Erwinia amylovora bacteriophage from aerial parts of apple trees. Phytopathology. 1977;67:101–104. [Google Scholar]
- Ritchie D.E., Klos E.J. Some properties of Erwinia amylovora bacteriophages. Phytopathology. 1979;69:1078–1083. [Google Scholar]
- Schnabel E.L., Jones A.L. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl Environ Microbiol. 2001;67:59–64. doi: 10.1128/AEM.67.1.59-64.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess E., Lurz R. Electron microscopic analysis of nucleic acids and nucleic acid–protein complexes. Methods Microbiol. 1988;20:293–323. [Google Scholar]
- Steven A.C., Trus B.L., Maizel J.V., Unser M., Parry D.A.D., Wall J.S. Molecular substructure of a viral receptor‐recognition protein. The gp17 tail‐fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. et al. [DOI] [PubMed] [Google Scholar]
- Stone R. Bacteriophage therapy: Stalin's forgotten cure. Science. 2002;298:728–731. doi: 10.1126/science.298.5594.728. [DOI] [PubMed] [Google Scholar]
- Svircev A.M., Sholberg P.L., Castle A.J. Bacteriophages: fighting fire with a twist. Biocontrol Files/Les Dossiers Biocontrôle. 2006;7:5. [Google Scholar]
- Tharaud M., Laurent J., Faize M., Paulin J.‐P. Fire blight protection with avirulent mutants of Erwinia amylovora. Microbiology/SGM. 1997;143:625–632. doi: 10.1099/00221287-143-2-625. [DOI] [PubMed] [Google Scholar]
- Thomson S.V., Gouk S.C. Influence of age of apple flowers on growth of Erwinia amylovora and biological control agents. Plant Dis. 2003;87:502–509. doi: 10.1094/PDIS.2003.87.5.502. [DOI] [PubMed] [Google Scholar]
- Vandenbergh P.A., Cole R.L. Cloning and expression in Escherichia coli of the polysaccharide depolymerase associated with bacteriophage‐infected Erwinia amylovora. Appl Environ Microbiol. 1986;51:862–864. doi: 10.1128/aem.51.4.862-864.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Partial sequence analysis of the German isolate φEaJ08C amplified with primers from φEa104 and φEa116. The positions refer to the partial sequence of φEa116 (Accession No. FQ85719).


