Summary
Arabinoxylan‐oligosaccharides (AXOS) are a recently newly discovered class of candidate prebiotics as – depending on their structure – they are fermented in different regions of gastrointestinal tract. This can have an impact on the protein/carbohydrate fermentation balance in the large intestine and, thus, affect the generation of potentially toxic metabolites in the colon originating from proteolytic activity. In this study, we screened different AXOS preparations for their impact on the in vitro intestinal fermentation activity and microbial community structure. Short‐term fermentation experiments with AXOS with an average degree of polymerization (avDP) of 29 allowed part of the oligosaccharides to reach the distal colon, and decreased the concentration of proteolytic markers, whereas AXOS with lower avDP were primarily fermented in the proximal colon. Additionally, prolonged supplementation of AXOS with avDP 29 to the Simulator of Human Intestinal Microbial Ecosystem (SHIME) reactor decreased levels of the toxic proteolytic markers phenol and p‐cresol in the two distal colon compartments and increased concentrations of beneficial short‐chain fatty acids (SCFA) in all colon vessels (25–48%). Denaturant gradient gel electrophoresis (DGGE) analysis indicated that AXOS supplementation only slightly modified the total microbial community, implying that the observed effects on fermentation markers are mainly caused by changes in fermentation activity. Finally, specific quantitative PCR (qPCR) analysis showed that AXOS supplementation significantly increased the amount of health‐promoting lactobacilli as well as of Bacteroides–Prevotella and Clostridium coccoides–Eubacterium rectale groups. These data allow concluding that AXOS are promising candidates to modulate the microbial metabolism in the distal colon.
Introduction
In recent years, many research efforts have focused on the modulation of the colonic microbiota and their fermentation processes with the aim of improving host health (Gibson et al., 1989; Cummings and Macfarlane, 1991; Manning and Gibson, 2004). An approach to achieve this goal is the ingestion of prebiotics, food constituents that are not digestible in the upper gut and that beneficially affect the host by selectively stimulating growth and/or activity of one or of a number of health‐promoting bacteria in the colon (Gibson and Roberfroid, 1995). Most of the currently known or suggested prebiotics in the functional food industry are oligosaccharides such as lactulose, fructo‐oligosaccharides (FOS), galacto‐oligosaccharides (GOS), soybean oligosaccharides (SOS), lactosucrose, isomalto‐oligosaccharides, gluco‐oligosaccharides, xylo‐oligosaccharides (XOS) or palatinose (Manning and Gibson, 2004). Recently, arabinoxylan‐oligosaccharides (AXOS) have been proposed as alternative prebiotics (Kontula et al., 2000; Grootaert et al., 2007; Hughes et al., 2007; Vardakou et al., 2007). AXOS with different average degrees of polymerization (avDP, i.e. the average number of xylose residues in their backbone) and average degrees of substitution (avDS, i.e. the average ratio of arabinose to xylose), which may present different prebiotic properties, can be obtained by enzymatic treatment of arabinoxylans (AX) from cereals (Delcour et al., 1999; Swennen et al., 2005; 2006). Arabinoxylans are made up of β‐1,4‐linked d‐xylopyranosyl residues, substituted at the C(O)‐3 and/or the C(O)‐2 position with monomeric α‐l‐arabinofuranoside. Ferulic acid can be coupled to the C(O)‐5 of arabinose through an ester linkage (Goesaert et al., 2005). These structural elements are also present in AXOS.
Colonic fermentation is an anaerobic process in which carbohydrates and proteins are metabolized by the microbiota of the large intestine. The intestinal microbiome mainly consists of nine bacterial phyla of which Firmicutes, Bacteroides and Actinobacteria are dominant, and plays a key role in obtaining energy from otherwise indigestible compounds (Rajilić‐Stojanovićet al., 2007,Turnbaugh et al., 2007). Whereas fermentation of carbohydrates is generally accepted to be beneficial for the host due to the generation of short‐chain fatty acids (SCFA), fermentation of proteins results in the formation of potentially toxic metabolites such as ammonium ions, indoles, phenols or amines (Gibson et al., 1989; Nowak and Libudzisz, 2006). These compounds can alter the morphology and intermediary metabolism of intestinal epithelial cells, and can increase DNA synthesis and promote tumorigenesis (Kikugawa and Kato, 1986; Macfarlane and Macfarlane, 1997; Ichikawa and Sakata, 1998). The proteolytic metabolism is more evident in the distal part of the colon where proteins are the main substrate available to the microbiota, whereas the proximal colon presents a more saccharolytic environment (Cummings and Macfarlane, 1991). This correlates with the fact that the majority of colonic tumours in humans occur in the distal colon and rectum (Govers et al., 1999).
Many of currently used oligosaccharides are fermented in the proximal colon and the effect on the distal part is minor if not absent (Manning and Gibson, 2004; Van de Wiele et al., 2004; 2007). Therefore, a potential objective for novel prebiotics would be to shift the proteolytic metabolism of the distal colon into a more saccharolytic one. This would also reflect upon the composition of the autochthonous microbial community which can be analysed in terms of Microbial Resource Management (MRM) as a novel approach to characterize the microbial ecology of a given environment (Verstraete, 2007; Marzorati et al., 2008). Previous studies have indicated that prebiotics with higher avDP are more resistant to saccharolytic fermentation so that metabolism takes place more distally in the colon (Hughes and Rowland, 2001).
The aim of the present study was to determine the site of fermentation of AXOS with different avDP and avDS, their impact on the protein/carbohydrate fermentation balance in the distal large intestine, and their influence on the colon microbiome. For this purpose, the Simulator of Human Intestinal Microbial Ecosystem (SHIME) was used. This in vitro system has already been used for several nutrition studies (De Boever et al., 2000; Meddah et al., 2001; Van de Wiele et al., 2004; 2007; Decroos et al., 2006; Ranganathan et al., 2006) and resulted to be a useful tool for predicting in vivo events, in terms of intestinal microbial composition and activity (Molly et al., 1993). For instance, the SHIME results of a synbiotic treatment, using hop and Eubacterium limosum, were recently validated by Possemiers and colleagues (2006; 2008) and Bolca and colleagues (2007) using human microbiota‐associated rats and human trials.
Results
Determination of the fermentation site of AXOS (short‐term SHIME run)
To assess whether AXOS of different avDP and avDS (3‐0.09; 3‐0.25; 8‐0.23; 15‐0.26; 29‐0.30) were fermented by intestinal bacteria and whether inter‐individual variability could affect this metabolic capability, batch fermentation tests in a protein‐rich medium were performed starting from six different faecal inocula. These tests confirmed that, independently from the donor, the colonic microbiota possessed the metabolic capability to hydrolyse these compounds (data not shown). Besides, to confirm AXOS non‐digestibility in the upper gastrointestinal tract (Hopkins et al., 2003), an in vitro digestion batch experiment was performed simulating the stomach and small intestine conditions. Both a simpler AXOS (3‐0.09) and the more complex AXOS (67‐0.58) resulted to be non‐digestible in these conditions, as reported in Table 1. Finally, a test was conducted to assess the main site of AXOS consumption in different colon compartments. Sugar analysis indicated that all AXOS except 15‐0.26 and 29‐0.30 were almost totally consumed after incubation in the transverse colon compartment (Fig. 1A). For the latter two AXOS types, about 30% of the initial amount of sugars remained available for the descending colon where they were completely consumed.
Table 1.
Assessment of non‐digestibility of AXOS 3‐0.09 and AXOS 67‐0.58 in batch experiments simulating stomach and small intestine conditions.
| Incubation | Time (h) | xylose (mg l−1) | arabinose (mg l−1) |
|---|---|---|---|
| AXOS 3 in stomach | 0 | 2.4 ± 0.3 | 1.9 ± 0.4 |
| 2 | 2.0 ± 0.1 | 1.6 ± 0.6 | |
| AXOS 67 in stomach | 0 | 1.5 ± 0.1 | 1.3 ± 0.3 |
| 2 | 1.5 ± 0.1 | 1.2 ± 0.4 | |
| AXOS 3 in small intestine | 0 | 2.2 ± 0.2 | 1.8 ± 0.5 |
| 2 | 1.9 ± 0.1 | 1.6 ± 0.5 | |
| AXOS 67 in small intestine | 0 | 1.9 ± 0.4 | 1.3 ± 0.3 |
| 2 | 1.5 ± 0.0 | 1.2 ± 0.3 |
The amount of released xylose and arabinose are reported as mg l−1. Data are means ± SD. The theoretical yield of sugars from AXOS 3‐0.09 is 49.56 and 4.38 mg l−1 xylose and arabinose respectively; from AXOS 67‐0.58 is 38.82 and 10.38 mg l−1.
Figure 1.
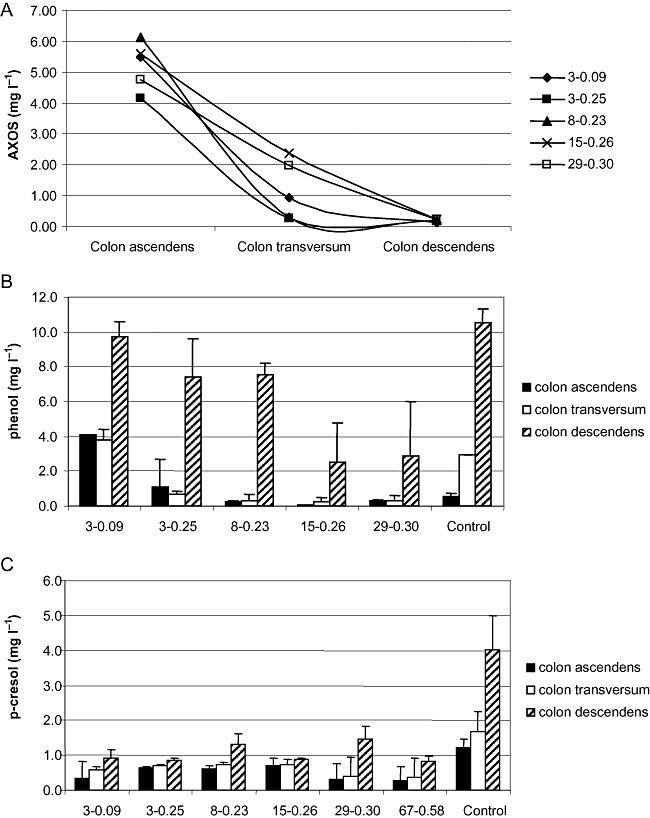
Concentration of AXOS (A), phenol (B) and p‐cresol (C) after successive batch incubations simulating the conditions of the ascending colon (18 h, pH 5.9), transverse colon (30 h, pH 6.2) and descending colon (22 h, pH 6.7) of the SHIME. Concentration of AXOS was determined as (Arabinose + Xylose) × 0.88. Initial AXOS concentration was 6.0 g l−1. AXOS preparations are reported in the legend and below the bars in terms of specific degree of polymerization and degree of substitution [i.e. for the diamond in (A), 3 and 0.09 respectively].
Production of phenol decreased with all supplemented samples after incubation in the transverse and descending colon vessels, except for AXOS 3‐0.09 (Fig. 1B). More interestingly, the levels of phenol after the descending colon incubation were remarkably lower for AXOS 15‐0.26 and 29‐0.30. In comparison with the control samples, a decrease of p‐cresol production was noted for all supplemented AXOS compounds in all colon compartments (Fig. 1C). Ammonium ions and SCFA levels did not show significant variations compared with the control samples, although propionate production seemed to be increased in the transverse colon in all the supplemented samples (data not shown).
Long‐term effect of AXOS 29‐0.30 on the SHIME
Based on the results of the previous tests, AXOS 29‐0.30 was chosen to perform a SHIME run (7 weeks) to assess the long‐term effect of this compound on the colon metabolism and on the indigenous microbial community. The SHIME run consisted of 2 weeks of a basal period during which standard SHIME medium was fed to the reactor, 3 weeks of treatment period during which part of the starch of the standard feeding was replaced by AXOS 29‐0.30, followed by 2 weeks of washout period in which standard SHIME medium was fed again to the reactor.
Table 2 shows that replacement of part of the starch with AXOS 29‐0.30 significantly decreased phenol levels in the descending colon during the treatment period and resulted in an apparent phenol decrease in the transverse colon. The decrease in phenol production continued after that treatment with AXOS had stopped. In the case of p‐cresol, a similar decreasing trend was observed during and after the treatment, although its concentration in all samples was very low and only slightly above the detection limit. The levels of ammonium ions increased between 1.3‐ and 1.5‐foldAXOS treatment period. During the washout period, the concentrations returned to values close to the ones measured during the basal period.
Table 2.
Concentration of SCFA, ammonium ions, phenol, p‐cresol and activity of AXOS‐degrading enzymes measured in the three vessels of the SHIME during the basal period (n ≥ 5), treatment with AXOS 29‐0.30 (n ≥ 9) and washout period (n ≥ 5).a
| Basal period | Treatment | Washout | ||
|---|---|---|---|---|
| Ascending colon | ||||
| SCFA (mmol l−1) | Acetate | 22.5 ± 3.1 | 25.4 ± 3.2 | 21.3 ± 3.2 |
| Propionate | 13.7 ± 2.1 | 17.0 ± 2.1* | 15.9 ± 1.7 | |
| Butyrate | 3.1 ± 1.4 | 5.8 ± 1.3* | 5.1 ± 0.9* | |
| Other SCFAb | 1.1 ± 0.4 | 2.1 ± 0.4* | 1.8 ± 0.1* | |
| Total SCFA | 40.4 ± 7.1 | 50.2 ± 7.0* | 44.1 ± 5.9* | |
| Proteolytic markers (mg l−1) | Ammonium | 183 ± 48 | 292 ± 27* | 237 ± 2* |
| Phenol | 0.41 ± 0.13 | 0.55 ± 0.46 | 0.04 ± 0.01* | |
| p‐Cresol | ND | ND | ND | |
| AXOS‐degrading enzymes (AU) | β‐d‐xylosidase | 3.1 ± 3.0 | 10.2 ± 7.0 | 2.5 ± 2.0 |
| α‐l‐arabinofuranosidase | 5.1 ± 4.2 | 12.4 ± 7.6 | 20.1 ± 4.7* | |
| Endo‐1,4‐β‐xylanase | 0.01 ± 0.01 | 1.52 ± 0.70* | 1.30 ± 0.02* | |
| Transverse colon | ||||
| SCFA (mmol l−1) | Acetate | 26.8 ± 1.0 | 34.8 ± 5.3* | 30.4 ± 2.5* |
| Propionate | 16.7 ± 1.6 | 23.8 ± 2.0* | 22.9 ± 2.5* | |
| Butyrate | 3.5 ± 1.1 | 7.8 ± 1.3* | 7.1 ± 1.8* | |
| Other SCFAb | 1.6 ± 0.2 | 2.9 ± 0.4* | 2.5 ± 0.6* | |
| Total SCFA | 49.5 ± 4.0 | 69.3 ± 9.0* | 62.9 ± 7.3* | |
| Proteolytic markers (mg l−1) | Ammonium | 296 ± 18 | 384 ± 73* | 364 ± 20 |
| Phenol | 6.27 ± 1.33 | 6.0 ± 1.0 | 4.55 ± 1.11 | |
| p‐Cresol | 0.41 ± 0.29 | 0.25 ± 0.21 | 0.03 ± 0.02* | |
| AXOS‐degrading enzymes (AU) | β‐d‐xylosidase | 3.0 ± 3.2 | 25.6 ± 6.1* | 14.6 ± 5.0* |
| α‐l‐arabinofuranosidase | 7.0 ± 5.0 | 58.4 ± 10.0* | 28.4 ± 20.2 | |
| Endo‐1,4‐β‐xylanase | 0.02 ± 0.01 | 2.0 ± 0.5* | 0.4 ± 0.03* | |
| Descending colon | ||||
| SCFA (mmol l−1) | Acetate | 33.1 ± 3.5 | 37.3 ± 2.7 | 33.5 ± 1.9 |
| Propionate | 16.3 ± 2.7 | 22.7 ± 2.7 | 21.8 ± 0.8* | |
| Butyrate | 4.2 ± 1.2 | 8.0 ± 1.3* | 6.4 ± 1.7 | |
| Other SCFAb | 1.8 ± 0.2 | 2.5 ± 1.1* | 3.2 ± 2.1 | |
| Total SCFA | 55.4 ± 7.6 | 70.5 ± 6.9* | 64.9 ± 6.5 | |
| Proteolytic markers (mg l−1) | Ammonium | 321 ± 8 | 431 ± 62* | 398 ± 30* |
| Phenol | 7.87 ± 1.20 | 6.39 ± 0.63* | 4.69 ± 1.27* | |
| p‐Cresol | 0.28 ± 0.10 | 0.21 ± 0.15 | 0.08 ± 0.09* | |
| AXOS‐degrading enzymes (AU) | β‐d‐xylosidase | 4.2 ± 0.9 | 23.3 ± 4* | 8.0 ± 4.1* |
| α‐l‐arabinofuranosidase | 19.1 ± 4.0 | 56.7 ± 6.1* | 31.6 ± 4.8* | |
| Endo‐1,4‐β‐xylanase | 0.02 ± 0.01 | 1.81 ± 0.70* | 0.90 ± 0.31* | |
Data are means ± SD.
Sum of valerate, isovalerate and isobutyrate.
Significantly different from basal period (Student's two‐tailed t‐test, P < 0.05).
ND, not detected.
The concentration of SCFA increased in all colon vessels during the treatment period. This increase was higher for butyric (81%, 125% and 91% for ascending, transverse and descending colon respectively) and propionic acids (24%, 43% and 38%) than for acetic acid (13%, 30% and 12%). Total SCFA concentration increased about 25%, 48% and 30% in the ascending, transverse and descending colon, respectively, during the treatment period. Although a decreasing trend was observed during the washout period for all SCFA, the higher concentrations persisted for at least 2 weeks after treatment ceased.
The activity of three enzymes responsible for hydrolysis of AXOS (i.e. α‐l‐arabinofuranosidase, β‐d‐xylosidase and endo‐1,4‐β‐xylanase) increased during the supplementation with AXOS in all vessels indicating an induction effect. This increase was the highest in the transverse and descending colon vessels (Table 2).
AXOS 29‐0.30 effect on the SHIME microbial community
Plate count analysis for total aerobes and anaerobes in the different colon compartments delivered similar concentrations as in previous SHIME runs, yet, AXOS supplementation did not significantly change these levels (Table 3). Looking at more specific groups, no significant changes were observed in the plate counts of coliforms, lactobacilli, enterococci, clostridia, bifidobacteria and staphylococci. However, we need to stress that agar plate techniques have several limitations when dealing with the colon microbiome, which consists of at least 80% of non‐culturable microorganisms (Eckburg et al., 2005). To overcome this problem, a molecular microbial ecology survey based on the molecular fingerprinting technique denaturant gradient gel electrophoresis (DGGE) (on total bacteria) and quantitative PCR (qPCR) (on total bacteria, bifidobacteria, lactobacilli, Bacteroides–Prevotella and Clostridium coccoides–Eubacterium rectale group) was applied.
Table 3.
Total aerobes and anaerobes microbial Log counts in the colon vessels of the SHIME reactor during basal period (n = 4), 3 weeks of treatment (n = 6) and washout period (n = 4).a
| Bacterial group | Period | CA | CT | CD |
|---|---|---|---|---|
| Total aerobes | Basal | 8.2 ± 0.53 | 7.8 ± 0.69 | 7.6 + 0.65 |
| Treatment | 8.4 ± 0.69 | 8.4 ± 0.77 | 8.1 + 0.87 | |
| Washout | 7.7 ± 0.31 | 7.9 ± 0.21 | 7.5 + 0.18 | |
| Total anaerobes | Basal | 8.2 ± 0.34 | 8.0 ± 0.21 | 7.6 ± 0.39 |
| Treatment | 8.4 ± 0.75 | 8.4 ± 0.79 | 8.2 ± 0.86 | |
| Washout | 7.9 ± 0.29 | 8.3 ± 0.76 | 8.3 ± 0.82 |
Data are means ± SD.
CA, colon ascending; CD, colon descending; CT, colon transversum.
DGGE was used to monitor changes in the composition and structure of microbial communities from the three colon compartments, throughout the SHIME run of 7 weeks. Samples were collected at the end of every week from each of the three colon vessels. Clustering of the DGGE lane fingerprints (Fig. 2) showed that all samples from the ascending colon (V1) clustered in a separate group, and that the samples from the transverse (V2) and descending (V3) colon clustered together in another one. Within the transverse/descending colon group, samples from week 5 (end of the treatment) and weeks 6 and 7 (washout) formed separate subclusters (Fig. 2, dashed squares) indicating that the AXOS supplementation had impacted the composition of colonic microbial community in a way that remained stable also after the treatment. Among several small changes in the composition and intensity of the band patterns, a single band appeared to become dominant following AXOS supplementation (Fig. 2, black full square). Sequencing of the DNA eluted from the band identified a bacterial species with a similarity of 95% (on 164 bp) with Selenomonas sp. (AF385576).
Figure 2.
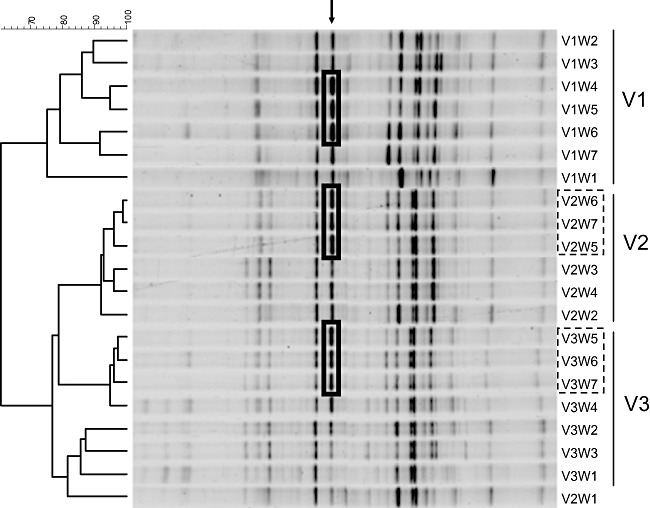
DGGE fingerprint patterns for total bacteria on 45–60% denaturant gradient. Clustering analysis is based on the Pearson product–moment correlation coefficient and dendrograms were created by using UPGMA linkage. V1 refers to ascending colon, V2 to transverse colon and V3 to descending colon. Samples are named as follows: VxWy, where x represents the number of the vessel and y the week of the sampling. Dashed squares indicate subclusters of samples within the same colon compartment, following AXOS supplementation. Black full square and the arrow indicate the band that is enriched following AXOS supplementation.
Range‐weighted richness values, indicative of the carrying capacity of a given environment (Marzorati et al., 2008), increased in all the three colon compartments: from 36 to 51 in the colon ascendens, from 32 to 44 in the colon transversum and from 60 to 68 in the colon descendens. The dynamics of the system, calculated through the weekly rate of change – after the basal period – was of 8.7 ± 2.8%, 3.5 ± 1.6% and 5.9 ± 4.8% for ascending, transverse and descending colon respectively. Finally, Pareto–Lorenz (PL) curve representation of the functional organization and structure of the microbial community was applied to the samples at the end of the basal (W2), treatment (W5) and washout (W7) periods (Fig. S1). During the treatment (Fig. S1, dashed line), the numerical interpretation of the PL curves of the three vessels indicated the presence of structured microbial communities. The values of the y‐axis projection of the respective intercepts with the vertical 20% x‐axis line (Marzorati et al., 2008) were 47%, 45% and 55% for V1, V2 and V3 respectively. In the transverse colon (V2), the effect on the community structure appeared reversible, with the curve for sample V2W7 that returned in the starting position, overlapping the one of sample V2W2. On the contrary, the effect on the structure of the microbial community in the colon descending remained stable even after the treatment had ceased.
The qPCR data on total bacteria (data not shown) confirmed the general increase in cell numbers observed through plate counts. AXOS supplementation induced an increase of the amount of both bifidobacteria and lactobacilli (Fig. 3). For bifidobacteria, a significant 2.2 times increase of the 16S rRNA gene copies per microlitre in the ascending colon was observed (P < 0.05), while, for lactobacilli, the increase was of four times in the ascending colon and of 1.3 and 1.8 times in the transverse and descending colon respectively. The increase in bifidobacteria concentrations did not persist after the end of the treatment period, while the lactobacilli concentration in the transverse colon kept increasing also when the AXOS supplementation ceased (P < 0.05). Moreover, as reported by other studies (Kabel et al., 2002; Chassard et al., 2007; Hughes et al., 2007) also bacteroides and clostridia groups took advantage of AXOS supplementation. The amount of microorganisms associated to C. coccoides–E. rectale group increased of 1.2 Log units after the treatment in the descending colon and also significantly slightly start to increase both in the ascending and in the descending colon during the treatment period (Fig. 3C). Bacteroides concentration significantly increased in all the three colon compartments becoming 65% of the total microbial community both in the transverse and in the descending colon during the washout period (Fig. 3D).
Figure 3.
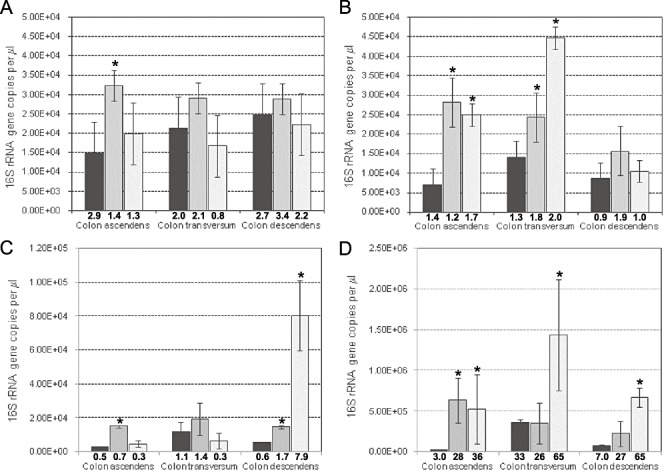
Concentrations of bifidobacteria (A), lactobacilli (B), Clostridium coccoides–Eubacterium rectale group (C) and Bacteroides–Prevotella group (D) in the intestinal suspension from the three colon vessels, derived from quantitative PCR. Black bars refer to the basal period; dark grey bars to the treatment period; light grey bars to the washout period. Below each bar is indicated a number, the percentage of the analysed group on the total of the microbial community. Asterisks indicate those bars that are significantly different from the basal period (P < 0.05).
Discussion
This work demonstrates the capacity of AXOS to modulate the protein/carbohydrate fermentation balance by the microbiota of the distal colon, which might have health‐related effects. In this context, the presence of AXOS in a protein‐rich environment, which is the case for the distal colon, led to a decrease in the production of phenol and p‐cresol. Phenol and p‐cresol, the main products of tyrosine fermentation by anaerobic bacteria, can contribute to colon carcinogenesis (Kikugawa and Kato, 1986; Macfarlane and Macfarlane, 1997; Smith and Macfarlane, 1997a). The decrease in proteolytic markers was also described by Smith and Macfarlane (1998) , who observed that the dissimilatory metabolism of proteins and amino acids was decreased in the presence of fermentable carbohydrates such as starch. AXOS supplementation also increased SCFA production, which is considered beneficial. SCFA contribute to normal large bowel function and prevent pathology through their actions in the lumen, as described previously (Demigne et al., 1995; Jan et al., 2002), and on the colonic musculature and vasculature as well as through their metabolism by colonocytes (Topping and Clifton, 2001).
The starting batch fermentation tests in a protein‐rich medium confirmed that the colonic microbiota possessed the metabolic capability to hydrolyse AXOS. Besides, this initial test conducted with the microbial communities deriving from faecal samples of six different donors clearly showed that the metabolic capability of fermenting AXOS was not limited by inter‐individual variability. The latter is a factor that must be always taken into account in the evaluation of results of specific in vitro and in vivo studies. However, this preliminary experiment did not take into consideration that, moving towards the distal part of the colon, the availability of fermentable carbohydrates normally decreases due to their consumption by saccharolytic bacteria. Apart from this, fermentable carbohydrate availability may also be dependent on the structure of the oligomers. For instance, fructans of longer avDP are more resistant to saccharolytic fermentation and, as a consequence, their metabolism takes place more distally in the colon (Van de Wiele et al., 2007). For this reason, a simulated short‐term SHIME run was performed to determine the principal site of fermentation of AXOS with different avDP and avDS (ascending, transverse or descending colon) and their contribution to the proteolytic metabolism of the different segments of the large intestine. All the AXOS preparations, with the exception of 3‐0.09, resulted in a decrease in the production of the proteolytic markers phenol and p‐cresol in both the transverse and descending colon. The effect in the distal colon was higher for the two AXOS samples with the highest avDP (i.e. AXOS 15‐0.26 and 29‐0.30) as observed for the lower levels of phenol. This was in agreement with the results obtained from the sugar analysis, which indicated a slower fermentation rate of these two AXOS preparations along the ‘intestinal transit’. These results suggested that a higher avDP was associated with more distal fermentation and an accompanying effect on proteolytic metabolism in the distal colon. Results for the proteolytic marker ammonium ion were less clear‐cut than those of phenol and p‐cresol and indicated only slight differences between AXOS and control samples. Based on the results obtained in the short‐term SHIME run, AXOS 29‐0.30 was selected as an appropriate candidate to study its long‐term effect in the SHIME reactor.
A 3‐week supplementation period resulted in a decrease of the proteolytic markers phenol and p‐cresol. The concentration of these two compounds continued decreasing in the transverse and descending colon also after the treatment had ceased. This would suggest a long‐term effect on the microbial population. Unlike the other proteolytic markers, ammonium ion concentration increased in all colon vessels during treatment. A similar effect was observed by Propst and colleagues (2003) in an in vivo trial using dogs fed with oligofructose and inulin. In both cases, the faecal concentration of ammonium ions increased with respect to the control diet, while the level of other proteolytic markers decreased and that of SCFA increased. Smith and Macfarlane (1997b) observed that the expected production of ammonium ions by faecal microbiota, based on amino acid depletion, was rarely observed suggesting that ammonium ions formed in deamination reactions were subsequently assimilated and incorporated into cellular material. However, it is well established that ammonium ions are commonly absorbed and transported from the colonic lumen to the portal circulation (Singh et al., 1995). Such absorption is not possible in an in vitro system like the SHIME, and this drawback becomes more prominent in case of microbial communities with high proteolytic activity. These observations suggest that the level of ammonium ions must be carefully evaluated as a marker for proteolytic metabolism in long‐term in vitro experiments, due to the continuous equilibrium between protein digestion and microbial uptake.
Apart from this, long‐term supplementation with AXOS increased the concentration of beneficial SCFA such as butyric and propionic acids in all colon vessels. The increase in total SCFA in the ascending colon compartment (25%) was lower than that observed in the last two vessels (48% and 30% respectively). In a similar study (Van de Wiele et al., 2004), supplementation of the SHIME with inulin resulted in the highest increase in total SCFA (44%) levels in the ascending colon, whereas this was lower in the transverse and descending colon (23% and 33% respectively). In our case, the difference can be explained by the lower activity of the AXOS‐degrading enzymes (i.e. xylanases, arabinofuranosidases and xylosidases) in the first colon vessel. This may well be due to partial repression caused by the presence of glucose (Kulkarni et al., 1999) which is a component of the SHIME feed. Overall, these results indicate that long‐term supplementation with AXOS promoted a beneficial metabolic shift in the SHIME reactor towards more saccharolytic and less proteolytic conditions.
All these data suggested a long‐term effect of AXOS on the colonic microbial community, even if this hypothesis was not confirmed by the results of the microbial counts. Yet, a culture‐independent technique such as DGGE showed that, within a colon compartment, samples collected after AXOS supplementation subclustered together. Within the transverse/descending colon group samples from week 5 (end of the treatment) and weeks 6 and 7 (washout) formed separate subclusters (Fig. 2, dashed squares), indicating that the AXOS supplementation impacted the composition of colonic microbial community that remained stable also after the treatment. In any case, as already reported by Van de Wiele and colleagues (2004), the dominant factor for clustering was the colon compartment itself from which the samples were taken.
Calculation of the range‐weighted richness (a correlation between the number of bands and the GC content diversity in a given fingerprint) and of the weekly rate of change (based on the moving window analysis) of the microbial community in each colon compartment (Marzorati et al., 2008) indicated that AXOS supplementation did not cause an imbalance in the total microbial community. The microbial communities had similar balanced structures, as shown by PL curves, with a low weekly rate of change also in the washout period. During the treatment (Fig. S1, dashed line), PL curves also showed in all the three vessels balanced microbial communities characterized by a medium ‘functional organization’, where some species were dominant and many others resilient, thus normally preserving their functionality (Marzorati et al., 2008).
In all the three vessels a common band, phylogenetically related to Selenomonas, increased during and after treatment with AXOS. Selenomonas are common inhabitants of the rumen of animals, where they play an important role in the fate of xylan from the diet (Cotta and Zeltwanger, 1995; Cotta and Whitehead, 1998). They have also been isolated from the human oral cavity and faeces (Tyrrel et al., 2003; Duncan et al., 2004). Although the ability of Selenomonas to degrade high avDP polysaccharides such as xylan is low, some species have been described as effective users of low‐avDP oligosaccharides. These oligosaccharides normally arise during hydrolysis of xylan by other xylanolytic bacteria such as Butyrivibrio, Fibrobacter, Eubacterium, Prevotella, Roseburia or Bacteroides (Cotta and Zeltwanger, 1995; Chassard et al., 2007). Selenomonas can also utilize lactate and convert it to acetate and propionate (Duncan et al., 2004), which would result in positive effects for the host's health. Finally, qPCR showed a significant lactobacillogenic effect on AXOS supplementation. AXOS promote the growth of some lactobacilli (Kontula et al., 2000) as they possess the needed set of enzymes to hydrolyse the AXOS structure, i.e. α‐l‐arabinofuranosidase, β‐d‐xylosidase and endo‐1,4‐β‐xylanase (Grootaert et al., 2007). The lactobacillogenic effect remained stable also in the washout period, especially in the transverse colon where the AXOS were mainly consumed. Moreover, AXOS supplementation also induced a positive effect on the concentration of clostridia and bacteroides in the distal colon. Within these two groups there are known xylanolytic bacteria such as Bacteroides intestinalis, Bacteroides dorei or Roseburia (Duncan et al., 2002; Chassard et al., 2007; 2008). Finally, the presence of clostridia and bacteroides may well explain the production of propionate and butyrate. Bacteroides have a wide metabolic capability and can produce proprionate from xylose fermentation (Chassard et al., 2007; 2008). Clostridia (i.e. Roseburia) can directly produce butyrate from xylan or this SCFA can be obtained by clostridia starting from acetate which originates from AXOS fermentation by other intestinal microbial groups (Kabel et al., 2002; Louis et al., 2004; Chassard et al., 2007; 2008; Lin et al., 2008; Seedorf et al., 2008).
In conclusion, this work indicates that AXOS of different avDP and avDS can be fermented by intestinal bacteria in a way that brings about positive effects for human health. To exert this effect in the distal part of the colon, AXOS preparations with higher avDP, such as AXOS 29‐0.30, must be used in order to prevent total fermentation during transit through previous compartments. Prolonged supplementation of the SHIME with AXOS 29‐0.30 resulted in: (i) a decreased production of some potentially toxic compounds derived from microbial degradation of proteins, which may help to decrease the risk of alterations of the colon epithelium, (ii) an increase in levels of beneficial SCFA, especially butyrate and propionate, and (iii) a long‐term effect on the colonic microbial community. The obtained in vitro results indicated that AXOS, in terms of MRM are promising candidates to influence the potential of the microbiota normally associated to the host to modulate a positive response for the human health. Further studies must be conducted in placebo‐controlled dietary intervention trials in humans to obtain the final proof that these new candidate prebiotic compounds can be really classified as prebiotic.
Experimental procedures
AXOS preparation
In this work, the different AXOS structures were identified with two numbers in the form avDP–avDS, where avDP indicated the average degree of polymerization and avDS the average degree of substitution (ratio arabinose/xylose).
AXOS 3‐0.09 was the commercial oligosaccharide preparation Xylooligo‐95P (Suntory, Tokyo, Japan). This product consists predominantly of xylobiose, xylotriose and xylotetraose.
AXOS 3‐0.25 was prepared from commercial wheat bran (Dossche Mills & Bakery, Deinze, Belgium) by endoxylanase‐mediated extraction from de‐starched and de‐proteinized bran as described by Swennen and colleagues (2006), except that the endoxylanase extraction was performed for 10 h at 50°C with a GHF11 endoxylanase from Bacillus subtilis (Grindamyl H640, Danisco, Brabrand, Denmark) at 1.2 units per gram of de‐starched and de‐proteinized wheat bran, and for another 10 h at 50°C after addition of an endoxylanase from Aspergillus aculeatus (Shearzyme 500L, Novozymes, Bagsvaerd, Denmark) at 21 units per gram of de‐starched and de‐proteinized wheat bran.
AXOS 8‐0.23 was prepared by incubating a solution (1:10 w/v) of AXOS 15‐0.26 at 30°C during 60 min with the above GHF10 endoxylanase from A. aculeatus at 75 units per gram of AXOS 15‐0.26. After inactivation of the enzyme by boiling (30 min), the solution was lyophilized and the obtained material was homogenized and sieved through a 250 µm sieve.
AXOS 15‐0.26 and AXOS 29‐0.30 were prepared from the above wheat bran by endoxylanase‐mediated extraction from de‐starched and de‐proteinized bran as described by Swennen and colleagues (2006). However, for the latter, the endoxylanase extraction was performed at 52°C with the above GHF11 endoxylanase from B. subtilis at 1.2 units per gram of de‐starched and de‐proteinized wheat bran.
Analytical techniques
SCFA and ammonium ion levels were determined as described by Van de Wiele and colleagues (2004). Analysis of total sugar and reducing end sugars were performed as described by Swennen and colleagues (2005). Phenol and p‐cresol levels were determined as follows: 20 µl of internal standard solution (4‐ethylphenol, 0.1 g l−1) was added to 50 µl of sample which was then acidified with 50 µl of HCl 1.0 M, saturated with 0.2 g of NaCl and extracted by shaking for 5 min with 0.6 ml of ethyl acetate. After centrifugation at 3000 g for 3 min, 20 µl supernatant was used for HPLC analysis. Calibration curves were obtained by analysing standard solutions of phenol and p‐cresol in demineralized water, treated in a similar way as the samples. HPLC was performed using a Zorbax SB‐C18 column [5 µm, 4.6 mm (inside diameter ) × 150 mm] (Agilent Technologies, Foster City, CA, USA). Solvents were acetonitrile (A) and 2.0% acetic acid (B). The solvent programme was: 0–22 min, gradient from 85% to 50% B (15% to 50% A); 22–25 min, gradient from 50% to 0% B; 25–30 min, 0% B; 30–35 min, gradient from 0% to 85% B. The flow rate was 1.0 ml min−1 and the temperature was set at 25°C. The elution was monitored by detection of fluorescence (excitation at 260 nm and emission at 305 nm). Data were collected and peaks integrated using the Chromeleon chromatography manager software (Dionex, Tienen, Belgium).
Enzymatic activities
α‐l‐arabinofuranosidase, β‐d‐xylosidase and endo‐1,4‐β‐xylanase activities were determined with the substrates p‐nitrophenyl‐α‐l‐arabinofuranoside (Sigma‐Aldrich, Bornem, Belgium), p‐nitrophenyl‐β‐d‐xylopyranoside (Sigma‐Aldrich) and Azo‐wheat‐arabinoxylan (Megazyme, Wicklow, Ireland) respectively. For the α‐l‐arabinofuranosidase and β‐d‐xylosidase activities, 100 µl of the sample (10 000 g centrifugated) was pipetted into a 96‐well plate, with 100 µl of a 5.0 mmol l−1 solution of substrate, prepared in a 0.1 mmol l−1 potassium phosphate buffer (pH 6.5). The plates were incubated at 37°C and the absorbance at 405 nm was read after 30 min with a Sunrise multi‐well spectrophotometer (Tecan, Mechelen, Belgium). The p‐nitrophenol released was quantified based on a p‐nitrophenol standard curve. The results were expressed in µmol p‐nitrophenol released l−1 min−1 (AU ). To determine the endo‐1,4‐β‐xylanase activity, 200 µl of the sample was added to 50 µl of sodium acetate buffer (25 mM, pH 5.0) and 250 µl of Azo‐wheat solution as previously described by De Schryver and colleagues (2008).
In batch studies to assess AXOS resistance in the upper gut
The stomach solution, consisting of 14 ml of SHIME feed at pH 2 (per litre: 1 g of arabinogalactan, 2 g of pectin, 1 g of xylan, 3 g of starch, 0.4 g of glucose, 3 g of yeast extract, 3 g of proteose peptone, 1 g of mucin and 0.5 of l‐cystein), pepsin (10 mg l−1) and AXOS 3‐0.09 (XOS) or AXOS 67‐0.58 (WPC) (6 g l−1after, 6 ml of pancreatic solution (per litre: 12.5 g of NaHCO3, 6 g of Oxgal and 1.9 g of pancreatin) was added to simulate small intestinal conditions, and this solution was incubated for other 2 h at 37°C. Samples for sugar analysis were taken before and after 2 (stomach) and 4 h (small intestine) of incubation. Xylose and arabinose concentrations were measured with a DX‐500 BioLC system (Dionex, Antwerpen, Belgium) containing a GP 50 gradient pump, an ED 50 electrochemical detector, a CarboPac PA10 column and an AS 50 autosampler. The sample was filter‐sterilized and 10 times diluted and a volume of 250 µl was injected. The flow was set on 1 ml min−1. Milli‐Q water (solution A) and NaOH solutions of 200 mM (solution B) and 1M (solution C) were used to generate a gradient in the eluent. The gradient programme was as followed: 8% B from 0 to 10 min, 70% B from 10 to 40 min and 100% C from 40 to 45 min.
Determination of the fermentation site of AXOS (short‐term SHIME run)
In order to study different AXOS simultaneously, a simulation of a short‐term SHIME run was performed. This consisted of three successive batch incubations simulating the conditions of the three SHIME vessels, with a total incubation time of 72 h (the retention time of the SHIME reactor). Samples (30 ml) taken from the ascending colon vessel of the SHIME were buffered with KH2PO4 (5.3 g l−1) and Na2HPO4 (1.4 g l−1) and supplemented with 6.0 g l−1 of the corresponding AXOS preparation. After anaerobic incubation for 18 h (ascending colon incubation), 10 ml was taken for analysis and the remaining volume was used to re‐suspend the pellet obtained after centrifugation (7000 g, 10 min) of an equivalent volume of sample taken from the transverse colon vessel of the SHIME reactor. The pH was adjusted to 6.2 and samples were incubated for 30 h (transverse colon incubation). The same procedure was repeated using pellet from the descending colon vessel of the SHIME. The pH was adjusted to 6.7 and samples were incubated for 22 h (descending colon incubation). After each incubation, samples were analysed for AXOS consumption, proteolysis markers and SCFA production.
Long‐term SHIME run
The SHIME is a dynamic model of the human gastrointestinal tract. It consists of five double‐jacketed vessels maintained at a temperature of 37°C, simulating the stomach, small intestine, ascending colon, transverse colon and descending colon, respectively, with a total retention time of 76 h. The colon vessels harbour a mixed microbial community and pH controllers (pH controller R301; Consort, Turnhout, Belgium) maintain the pH in the range 5.6–5.9, 6.2–6.5 and 6.6–6.9 in the ascending, transverse and descending colon simulations respectively. There is no gas exchange between the different vessels and the headspace of the culture system was flushed twice a day for 15 min with N2 to ensure anaerobic conditions.
The three colon vessels of the SHIME reactor were inoculated with bacteria from the same faecal sample of a healthy adult volunteer who had no history of antibiotic treatment in the 6 months prior to the faecal sample collection for the study. A stabilization period of 2 weeks after the inoculation was considered before beginning any assay. The experimental set‐up of the SHIME run was performed as previously described (Van de Wiele et al., 2004). This included 2 weeks of a basal period, 3 weeks of treatment period and 2 weeks of washout period. The composition of the SHIME feed used during the basal and the washout period was starch (4.0 g l−1), arabinogalactan (1.0 g l−1), pectin (2.0 g l−1), xylan (1.0 g l−1), glucose (0.4 g l−1), yeast extract (3.0 g l−1), peptone (1.0 g l−1), mucin (4.0 g l−1), cysteine (0.5 g l−1) (Sigma‐Aldrich). During the treatment period, 3.0 g l−1 starch was replaced by 3.0 g l−1 AXOS 29‐0.30 .
Classical microbiological analysis
Decimal dilutions of the samples were plated and incubated at 37°C. Counts were performed on McConkey‐agar (Oxoid, Basingstoke, UK) for coliforms, MRS‐agar (Oxoid) for lactobacilli, Enterococcus‐agar (Difco, Sparks, MD, USA) for enterococci, TSC‐agar (Merck, Darmstadt, Germany) for clostridia, RB‐agar (Hartemink et al., 1996) for bifidobacteria, MSA‐agar (Oxoid) for staphylococci and BHI‐agar (Oxoid) for total aerobes and anaerobes.
Molecular microbiological analyses
DGGE on total bacteria and qPCR on total bacteria, bifidobacteria and lactobacilli were performed to study the effect of AXOS on the structure and composition of the colonic microbial community. Metagenomic DNA was extracted from samples collected every week for a total of 7 weeks from each vessel, using QIAamp DNA stool mini kit (Qiagen, Venlo, the Netherlands) according to the manufacturer's instructions. Samples were marked as VxWy, where x represents the number of the vessel and y the week of the sampling. DGGE with a 45–60% denaturant gradient and with primers 338F‐GC and 518R was performed as reported in Van de Wiele and colleagues (2004). The obtained DGGE patterns were subsequently analysed using Bionumerics software version 2.0 (Applied Maths, Sint‐Martens‐Latem, Belgium). A matrix of similarities for the densiometric curves of the band patterns was calculated based on the Pearson product–moment correlation coefficient and dendrograms were created by using UPGMA linkage. MRM interpretation (range‐weighted richness, dynamics and PL curve) was conducted as suggested by Marzorati and colleagues (2008). The distribution of bands in the DGGE pattern can be correlated with the percentage on denaturant gradient of the gel needed to represent the sample's total diversity (within the limit of the technique). The more habitable the environment, the higher the probability it can host a high number of bands with a wide GC variability (in terms of both percentage and positioning of the GC stretches within the 16S rRNA gene). This concept can be mathematically expressed by defining an index for the range‐weighted richness (Rr) = (N2 × Dg), where N represents the total number of bands in the pattern, and Dg the denaturant gradient comprised between the first and the last band of the pattern. Rr < 10 are characteristic of environments with a low carrying capacity; values higher than 30, of environments with a high carrying capacity (Marzorati et al., 2008). The moving window analysis and the rate of change (dynamics) are measures to estimate the number of species that, on average, come to significant dominance (above the detection limit of the technique) at a given habitat, during a defined time interval. The rate of change parameter averages the degree of change between consecutive DGGE profiles of the same community over a fixed time interval (Wittebolle et al., 2005). Pareto–Lorenz evenness distribution curves can be constructed based on the DGGE profiles as previously described (Mertens et al., 2005; Marzorati et al., 2008; Wittebolle et al., 2008). Quantitative PCR for total bacteria using primers PRBA338f and P518r, for bifidobacteria and for lactobacilli was performed as reported by Possemiers and colleagues (2006), while qPCRs for C. coccoides–E. rectale group (also detecting the recently detected xylan‐degrader Roseburia) and Bacteroides–Prevotella group have been conducted according to Rinttilä and colleagues (2004).
Acknowledgments
We thank Ellen Van Gysegem for technical assistance. We also thank the IWT (Instituut voor de aanmoediging van innovatie door Wetenschap en Technologie) for funding the SBO project Impaxos (coordinated by KU Leuven). J.I. Sanchez was supported by a postdoctoral grant from the Gobierno del Principado de Asturias (Plan I+D+I), and M. Marzorati by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT‐Vlaanderen) by SBO project No. 40073 ‘Impaxos’. T. Van de Wiele is a Postdoctoral Fellow of the Fund for Scientific Research of Flanders (Belgium).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Pareto-Lorenz curves derived from the analysis of samples in ascending (V1), transverse (V2), and descending V3) colon. Full lines refer to samples at the end of the basal period (W2); dashed lines to samples at the end of the treatment (W5); dotted lines to samples at the end of the washout period (W7). The interpretation of the curves is based on the axis projection of their respective intercepts with the 20% x-axis line (grey dash-dotted line), and refers to a low, medium and high functional strength (M. Marzorati et al., 2008). The cumulative proportion of species is used as x-axis, and their respective cumulative proportions of abundances represent the y-axis. The 45° diagonal represents the perfect evenness of a community (grey full line).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bolca S., Possemiers S., Herregat A., Huybrechts I., Heyerick A., De Vniese S. Microbial and dietary factors are associated with the equol producer phenotype in healthy postmenopausal women. J Nutr. 2007;137:2242–2246. doi: 10.1093/jn/137.10.2242. et al. [DOI] [PubMed] [Google Scholar]
- Chassard C., Goumy V., Leclerc M., Del'homme C., Bernalier‐Donadille A. Characterization of the xylan‐degrading microbial community from human faeces. FEMS Microbiol Ecol. 2007;61:121–131. doi: 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- Chassard C., Delmas E., Lawson P.A., Bernalier‐Donadille A. Bacteroides xylanisolvens sp. nov., a xylan‐degrading bacterium isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:1008–1013. doi: 10.1099/ijs.0.65504-0. [DOI] [PubMed] [Google Scholar]
- Cotta M.A., Whitehead T.R. Xylooligosaccharide utilization by the ruminal anaerobic bacterium Selenomonas ruminantium. Curr Microbiol. 1998;36:183–189. doi: 10.1007/s002849900291. [DOI] [PubMed] [Google Scholar]
- Cotta M.A., Zeltwanger R.L. Degradation and utilization of xylan by the ruminal bacteria Butyrivibrio fibrisolvens and Selenomonas ruminantium. Appl Environ Microbiol. 1995;61:4396–4402. doi: 10.1128/aem.61.12.4396-4402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J.H., Macfarlane G.T. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- De Boever P., Deplancke B., Verstraete W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J Nutr. 2000;130:2599–2606. doi: 10.1093/jn/130.10.2599. [DOI] [PubMed] [Google Scholar]
- De Schryver P., Sesena S., Decaigny B., Van De Wiele T., Verstraete W., Boon N. Xylanases from microbial origin induce syrup formation in dough. J. Cereal Sci. 2008;47:18–28. [Google Scholar]
- Decroos K., Eeckhaut E., Possemiers S., Verstraete W. Administration of equol‐producing bacteria alters the equol production status in the Simulator of the Gastrointestinal Microbial Ecosystem (SHIME) J Nutr. 2006;136:946–952. doi: 10.1093/jn/136.4.946. [DOI] [PubMed] [Google Scholar]
- Delcour J.A., Van Win H., Grobet P.J. Distribution and structural variation of arabinoxylans in common wheat mill streams. J Agric Food Chem. 1999;47:271–275. doi: 10.1021/jf9805294. [DOI] [PubMed] [Google Scholar]
- Demigne C., Morand C., Levrat M.A., Besson C., Moundras C., Remesy C. Effect of propionate on fatty acid and cholesterol synthesis and on acetate metabolism in isolated rat hepatocytes. Br J Nutr. 1995;74:209–219. doi: 10.1079/bjn19950124. [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Hold G.L., Barcenilla A., Stewart C.S., Flint H.J. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrateproducing bacterium from the human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Flint H.J. Lactate‐utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G.R., Roberfroid M.B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- Gibson S.A., Macfarlane C., Hay S., Macfarlane G.T. Significance of microflora in proteolysis in the colon. Appl Environ Microbiol. 1989;55:679–683. doi: 10.1128/aem.55.3.679-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goesaert H., Brijs K., Veraverbeke W.S., Courtin C.M., Gebruers K., Delcour J.A. Wheat flour constituents: how they impact bread quality, and how to impact their functionality. Trends Food Sci Technol. 2005;16:12–30. [Google Scholar]
- Govers M.J.A.P., Gannon N.J., Dunshea F.R., Gibson P.R., Muir J.G. Wheat bran affects the site of fermentation of resistant starch and luminal indexes related to colon cancer risks. Gut. 1999;45:840–847. doi: 10.1136/gut.45.6.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootaert C., Delcour J.A., Courtin C.M., Broekaert W.F., Verstraete W., Van de Wiele T. Microbial metabolism and prebiotic potency of Arabinoxylan oligosaccharides in the human intestine. Trends Food Sci Technol. 2007;18:64–71. [Google Scholar]
- Hartemink R., Kok B.J., Weenk G.H., Rombouts F.M. Raffinose‐Bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J Microbiol Methods. 1996;27:33–43. [Google Scholar]
- Hopkins M.J., Englyst H.N., Macfarlane S., Furrie E., Macfarlane G.T., McBain A.J. Degradation of cross‐linked and non‐cross‐linked arabinoxylans by the intestinal microbiota in children. Appl Environ Microbiol. 2003;69:6354–6360. doi: 10.1128/AEM.69.11.6354-6360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R., Rowland I.R. Stimulation of apoptosis by two prebiotic chicory fructans in the rat colon. Carcinogenesis. 2001;22:43–47. doi: 10.1093/carcin/22.1.43. [DOI] [PubMed] [Google Scholar]
- Hughes S.A., Shewry P.R., Li L., Gibson G.R., Sanz M.L., Rastall R.A. In vitro fermentation by human fecal microflora of wheat arabinoxylans. J Agric Food Chem. 2007;55:4589–4595. doi: 10.1021/jf070293g. [DOI] [PubMed] [Google Scholar]
- Ichikawa H., Sakata T. Stimulation of epithelial cell proliferation of isolated distal colon of rats by continuous colonic infusion of ammonia or short‐chain fatty acids is nonadditive. J Nutr. 1998;128:843–847. doi: 10.1093/jn/128.5.843. [DOI] [PubMed] [Google Scholar]
- Jan G., Belzacq A.S., Haouzi D., Rouault A., Metivier D., Kroemer G., Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short‐chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- Kabel M.A., Kortenoeven L., Schols H.A., Voragen A.G. In vitro fermentability of differently substituted xylo‐oligosaccharides. J Agric Food Chem. 2002;21:6205–6210. doi: 10.1021/jf020220r. [DOI] [PubMed] [Google Scholar]
- Kikugawa K., Kato T. Formation of a mutagenic diazoquinone by interaction of phenol with nitrite. Food Chem Toxicol. 1986;26:209–214. doi: 10.1016/0278-6915(88)90121-4. [DOI] [PubMed] [Google Scholar]
- Kontula P., Suihko M.‐L, Suortti T., Tenkanen M., Mattila‐Sandholmand T., Von Wright A. The isolation of lactic acid bacteria from human colonic biopsies after enrichment on lactose derivatives and rye arabinoxylo‐oligosaccharides. Food Microbiol. 2000;17:13–22. [Google Scholar]
- Kulkarni N., Shendye A., Rao M. Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev. 1999;23:411–456. doi: 10.1111/j.1574-6976.1999.tb00407.x. [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Wu C.C., Hung C.H. Temperature effects on fermentative hydrogen production from xylose using mixed anaerobic cultures. Int J Hydrogen Energy. 2008;33:43–50. [Google Scholar]
- Louis P., Duncan S.H., McCrae S.I., Millar J., Jackson M.S., Flint H.J. Restricted distribution of the butyrate kinase pathway among butyrate‐producing bacteria from the human colon. J. Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane G.T., Macfarlane S. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scand J Gastroenterol. 1997;32:3–9. doi: 10.1080/00365521.1997.11720708. [DOI] [PubMed] [Google Scholar]
- Manning T.S., Gibson G.R. Microbial‐gut interactions in health and disease: prebiotics. Best Pract Res Clin Gastroenterol. 2004;18:287–298. doi: 10.1016/j.bpg.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Marzorati M., Wittebolle L., Boon N., Daffonchio D., Verstraete W. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol. 2008;10:1571–1581. doi: 10.1111/j.1462-2920.2008.01572.x. [DOI] [PubMed] [Google Scholar]
- Meddah A.T., Yazourh A., Desmet I., Risbourg B., Verstraete W., Romond M.B. The regulatory effects of whey retentate from bifidobacteria fermented milk on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) J Appl Microbiol. 2001;91:1110–1117. doi: 10.1046/j.1365-2672.2001.01482.x. [DOI] [PubMed] [Google Scholar]
- Mertens B., Boon N., Verstraete W. Stereospecific effect of hexachlorocyclohexane on activity and structure of soil methanotrophic communities. Environ Microbiol. 2005;7:660–669. doi: 10.1111/j.1462-2920.2005.00735.x. [DOI] [PubMed] [Google Scholar]
- Molly K., Vande Woestyne M., Verstraete W. Development of a 5‐step multi‐chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- Nowak A., Libudzisz Z. Influence of phenol, p‐cresol and indole on growth and survival of intestinal lactic acid bacteria. Anaerobe. 2006;12:80–84. doi: 10.1016/j.anaerobe.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Possemiers S., Bolca S., Grootaert C., Heyerick A., Decroos K., Dhooge W. The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8‐prenylnaringenin in vitro and in the human intestine. J Nutr. 2006;136:1862–1867. doi: 10.1093/jn/136.7.1862. et al. [DOI] [PubMed] [Google Scholar]
- Possemiers S., Rabot S., Espín J.C., Bruneau A., Philippe C., Gonzales‐Sarrias A. Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8‐prenylnaringenin in vitro and in rat intestine. J Nutr. 2008;138:1310–1316. doi: 10.1093/jn/138.7.1310. et al. [DOI] [PubMed] [Google Scholar]
- Propst E.L., Flickinger E.A., Bauer L.L., Merchen N.R., Fahey G.C. A dose–response experiment evaluating the effects of oligofructose and inulin on nutrient digestibility, stool quality, and fecal protein catabolites in healthy adult dogs. J Anim Sci. 2003;81:3057–3066. doi: 10.2527/2003.81123057x. [DOI] [PubMed] [Google Scholar]
- Rajilić‐Stojanović M., Smidt H., De Vos W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Ranganathan N., Patel B.G., Ranganathan P., Marczely J., Dheer R., Pechenyak B. In vitro and in vivo assessment of intraintestinal bacteriotherapy in chronic kidney disease. ASAIO J. 2006;52:70–79. doi: 10.1097/01.mat.0000191345.45735.00. et al. [DOI] [PubMed] [Google Scholar]
- Rinttilä T., Kassinen A., Malinen E., Krogius L., Palva A. Development of an extensive set of 16S rDNA‐targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real‐time PCR. J Appl Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- Seedorf H., Fricke W.F., Veith B., Brüggemann H., Liesegang H., Strittmatter A. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci USA. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Binder H.J., Geibel J.P., Boron W.F. An apical permeability barrier to NH3/NH4+ in isolated, perfused colonic crypts. Proc Natl Acad Sci USA. 1995;92:11573–11577. doi: 10.1073/pnas.92.25.11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.A., Macfarlane G.T. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol. 1997a;33:180–188. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- Smith E.A., Macfarlane G.T. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe. 1997b;3:327–337. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- Smith E.A., Macfarlane G.T. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998;366:355–368. [Google Scholar]
- Swennen K., Courtin C.M., Van der Bruggen B., Vandecasteele C., Delcour J.A. Ultrafiltration and ethanol precipitation for isolation of arabinoxylooligosaccharides with different structures. Carbohydr Polym. 2005;62:283–292. [Google Scholar]
- Swennen K., Courtin C.M., Lindemans G.C.J.E., Delcour J.A. Large scale production and characterisation of wheat bran arabinoxylooligosaccharides. J Sci Food Agric. 2006;86:1722–1731. [Google Scholar]
- Topping D.L., Clifton P.M. Short‐chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Hamady M., Fraser‐Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell K.L., Citron D.M., Warren Y.A., Nachnani S., Goldstein E.J. Anaerobic bacteria cultured from the tongue dorsum of subjects with oral malodor. Anaerobe. 2003;9:243–246. doi: 10.1016/S1075-9964(03)00109-4. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T., Boon N., Possemiers S., Jacobs H., Verstraete W. Prebiotic effects of chicory inulin in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004;51:143–153. doi: 10.1016/j.femsec.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T., Boon N., Possemiers S., Jacobs H., Verstraete W. Inulin‐type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J Appl Microbiol. 2007;102:452–460. doi: 10.1111/j.1365-2672.2006.03084.x. [DOI] [PubMed] [Google Scholar]
- Vardakou M., Nueno‐Palop C., Gasson M., Narbad A., Christakopoulos P. In vitro three‐stage continuous fermentation of wheat arabinoxylan fractions and induction of hydrolase activity by the gut microflora. Int J Biol Macromol. 2007;41:584–589. doi: 10.1016/j.ijbiomac.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Verstraete W. Microbial ecology and environmental biotechnology. ISME J. 2007;1:4–8. doi: 10.1038/ismej.2007.7. [DOI] [PubMed] [Google Scholar]
- Wittebolle L., Boon N., Vanparys B., Heylen K., De Vos P., Verstraete W. Failure of the ammonia oxidation process in two pharmaceutical wastewater treatment plants is linked to shifts in the bacterial communities. J Appl Microbiol. 2005;99:997–1006. doi: 10.1111/j.1365-2672.2005.02731.x. [DOI] [PubMed] [Google Scholar]
- Wittebolle L., Vervaeren H., Verstraete W., Boon N. Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl Environ Microbiol. 2008;74:286–293. doi: 10.1128/AEM.01006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pareto-Lorenz curves derived from the analysis of samples in ascending (V1), transverse (V2), and descending V3) colon. Full lines refer to samples at the end of the basal period (W2); dashed lines to samples at the end of the treatment (W5); dotted lines to samples at the end of the washout period (W7). The interpretation of the curves is based on the axis projection of their respective intercepts with the 20% x-axis line (grey dash-dotted line), and refers to a low, medium and high functional strength (M. Marzorati et al., 2008). The cumulative proportion of species is used as x-axis, and their respective cumulative proportions of abundances represent the y-axis. The 45° diagonal represents the perfect evenness of a community (grey full line).


