Summary
Streptomycetes are soil microorganisms with the potential to produce a broad spectrum of secondary metabolities. The production of antibiotics is accompanied by a decrease in protein synthesis, which raises the question of how these bacteria survived the transition from the primary to the secondary metabolism. Translating ribosomes incapable to properly elongate or terminate polypeptide chain activate bacterial trans‐translation system. Abundance and stability of the tmRNA during growth of Streptomyces collinus and Streptomyces griseus producing kirromycin and streptomycin, respectively, was analysed. The level of tmRNA is mostly proportional to the activity of the translational system. We demonstrate that the addition of sub‐inhibitory concentrations of produced antibiotics to the cultures from the beginning of the exponential phase of growth leads to an increase in tmRNA levels and to an incorporation of amino acids into the tag‐peptides at trans‐translation of stalled ribosomes. These findings suggest that produced antibiotics induce tmRNA that facilitate reactivation of stalled complex of ribosomes and maintain viability. The effect of antibiotics that inhibit the cell‐wall turnover, DNA, RNA or protein synthesis on the level of tmRNA was examined. Antibiotics interfering with ribosomal target sites are more effective at stimulation of the tmRNA level in streptomycetes examined than those affecting the synthesis of DNA, RNA or the cell wall.
Introduction
Ribosomes stalling at the end or within the coding sequence of the mRNA can activate tmRNA function. tmRNA and protein SmpB are main components required for rescueing stalled ribosomes. The defective mRNA is degraded; incomplete polypeptide is labelled for degradation and ribosomal subunits can create a new initiation complex (Roche and Sauer, 1999; Fujihara et al., 2002; Hayes et al., 2002; Li et al., 2006). Production of tmRNA tagged proteins in Escherichia coli has been observed to increase in the presence of both suppressor tRNA (Ueda et al., 2002) and the miscoding drugs kanamycin and streptomycin (Abo et al., 2002) that cause translational read‐through of stop codons. A further study (Luidalepp et al., 2005) showed that cells lacking tmRNA are more sensitive to several inhibitors of protein synthesis and to inhibitors of the cell wall synthesis fosfomycin and ampicillin. Many antibiotics produced by streptomycetes inhibit different steps of protein biosynthesis. Streptomyces griseus is a potent producer of the antibiotic streptomycin that stabilizes aminoacyl‐tRNA binding in the A‐site and alters the rates of GTP hydrolysis by elongation factor Tu on cognate and near‐cognate codons, with similar rates of GTP hydrolysis (Gromadski and Rodnina, 2004). Streptomyces griseus contains the aphE gene encoding aminoglycoside 3′‐phosphotransferase carrying streptomycin resistance (Heinzel et al., 1988). Furthermore, S. griseus possess an ion‐permeable channel with binding site for streptomycin. Negatively charged groups inside the channel are involved in streptomycin binding because this antibiotic is positively charged (Kim et al., 2001). Another mechanism responsible for streptomycin resistance is connected with the loss of specific 16S RNA methyltransferase encoded by gene rsmG (Nishimura et al., 2007). The enzyme catalyses methylation of the residue G518 of 16S RNA in S. coelicolor or G527 of E. coli which interacts directly with streptomycin.
Streptomyces collinus Tü 365 produces antibiotic kirromycin (Wolf and Zahner, 1972). In the presence of kirromycin, complex elongation factor Tu‐GTP can accept aminoacyl‐tRNA and bind to ribosome. Codon‐anticodon recognition is followed by GTP hydrolysis and release of EFTu‐GDP‐kirromycin from ribosomes is blocked, resulting in inhibition of subsequent steps of peptide bond formation (Parmeggiani and Nissen, 2006). In addition, we have found previously that EF‐Tu from S. collinus is sensitive to kirromycin and the rate of polypeptide synthesis in vitro was reduced to 50% by 0.25 µM kirromycin (Mikulík et al., 1982).
Despite the important biological function of tmRNA, its role in streptomycetes producing antibiotics remains unresolved. In this study, the level and stability of tmRNA were examined during the growth and development in two strains of streptomycetes producing antibiotics. We set out to determine how antibiotics with various target sites affect the level of tmRNA and protein synthesis in antibiotics producing strains.
Results
tmRNA abundance during development of streptomycetes
Total RNA was isolated from submerse cultures and aliquots (50 µg) were analysed in polyacrylamide‐urea gels. Northern blots were probed with labelled ssrAs and quantified. Growth of S. griseus in medium GYM reaches its maximum between 36 and 38 h of cultivation and production of streptomycin started between 20 and 24 h (Fig. 1A). During the exponential phase of growth (14 h), the highest level of tmRNA was detected and further development was accompanied with decline in the tmRNA level. Growth of S. collinus producing kirromycin reaches its maximum at 36 h cultivation and tmRNA level increases gradually up to 48 h and then decreases near the initial value found in the 14 h cultivation (Fig. 1B). The amount of tmRNA during exponential phase of growth was about four times higher than that found in S. griseus. In parallel experiments that were performed with the same cultures, we followed the amount of 5S RNA to ascertain whether the decrease in the tmRNA is not due to general loss of RNA. No significant changes in the 5S RNA levels were observed during the development of cultures. The decrease in tmRNA during the late exponential phase of growth suggests that the tmRNA was degraded. To test this possibility the half‐life of tmRNA from the exponentially growing cells (14 h cultures) and the cells from stationary phase of growth (40 h and 68 h cultures) were determined by inhibition of transcription with rifampicin and the decay of tmRNA was measured. In cells of S. griseus (Fig. 2A) from exponential phase of growth, the presence of rifampicin (300 µg ml−1) caused complete inhibition of RNA synthesis and about 10% tmRNA was degraded after 60 min cultivation. Significantly lower stability of the tmRNA was found in cultures from stationary phase of growth. In contrast, the level of tmRNA of S. collinus (Fig. 2B) was stable from 14 to 68 h cultivation and the half‐life of the tmRNA in the presence of rifampicin was longer than 50 min. These data show differences in abundance and stability of tmRNA during development of the two streptomycetes strains examined.
Figure 1.
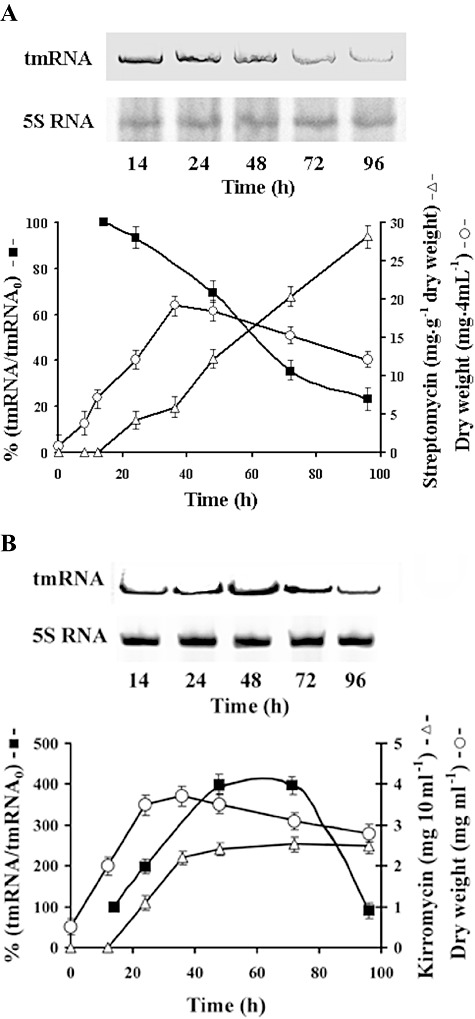
Analysis of tmRNA during development of streptomycetes. Aliquot samples of submerse cultures were taken at 14, 24, 48, 72 and 96 h of cultivation. Development of cultures was followed by examination of dry weight (‐○‐) and production of antibiotics (‐Δ‐). Total RNA was isolated and 50 µg RNA samples were separated by electrophoresis in 7% polyacrylamide‐6M urea gels. Northern blots of tmRNAs from S. griseus (A) and S. collinus (B) were hybridized with Dig‐11‐dUTP labelled ssrA probes and after detection the amount of tmRNA (‐▪‐) was quantified by Bio‐Rad Molecular Imager FX with Quantity One software. The blots were probed for 5S RNA. The amounts of tmRNA were normalized on 5S RNA levels. The values are relative to the average abundance of tmRNA from five separate experiments in 14 h cells. This initial value (tmRNA0) represents 100% independently for strains used.
Figure 2.
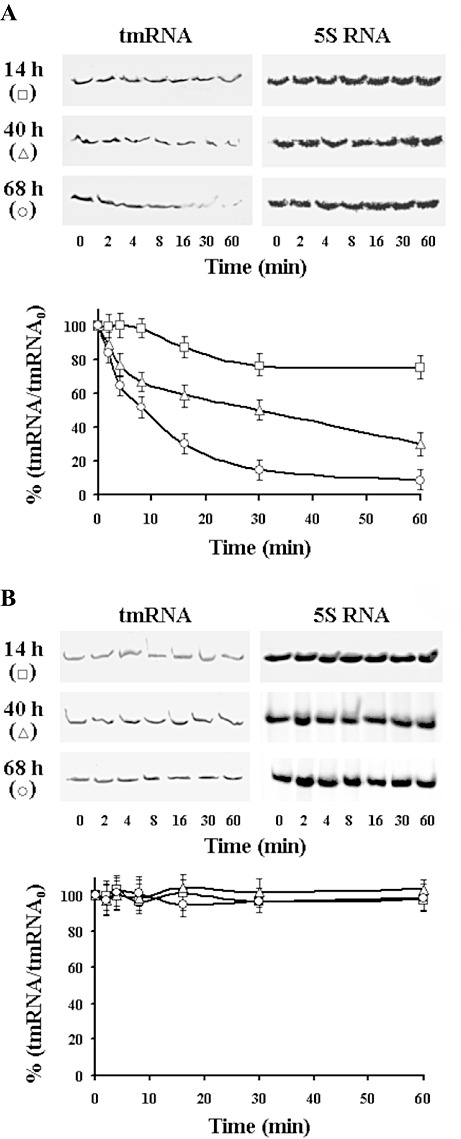
Northern blot analysis of the tmRNA stability from 14 h, 40 h and 68 h cultures of S. griseus (A) and S. collinus (B). Representative blot analyses of the tmRNA abundance. Transcription was inhibited by addition of rifampicin (300 µg ml−1) and aliquots were taken at the indicated times for RNA extraction and Northern analysis. Quantification of the tmRNA was performed as described in Fig. 1, and initial values at time 0 (tmRNA0), before addition or rifampicin, represent 100% separately for each time curve and strain used.
Incorporation of amino acids to stalled complex of ribosomes from streptomycetes
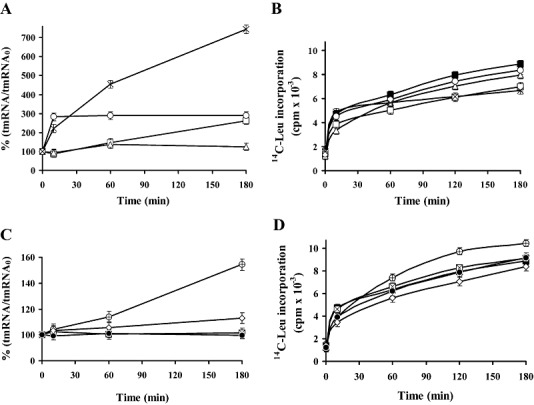
We ask whether the antibiotics produced by streptomycetes influences the level of the tmRNA. To test this possibility, cells from exponential phase of growth (where biosynthesis of antibiotics was not detected) were cultivated with sub‐inhibitory concentrations of streptomycin or kirromycin. Cells of S. griseus were incubated with 3.5 µM streptomycin and cultures of S. collinus with 5 µM kirromycin. At time intervals samples were taken and washed cells were used for preparation of S30 fractions and stalled complexes of ribosomes with poly(U) as previously described (Himeno et al., 1997; Konno et al., 2004; Asano et al., 2005). After 30 min of incubation, the polyphenylalanine incorporation was saturated and a mixture of l‐[U‐14C]‐labelled amino acids was added to the stalled complex of ribosomes, which are constituents of the tag‐peptides of the tmRNAs. Stalled complex of ribosomes from S. griseus (Fig. 3A) and S. collinus (Fig. 3B), which were incubated for 60 min in the presence of antibiotics, were found to incorporate about two times more of l‐[U‐14C]‐amino acids to tag‐peptide than the complexes isolated in absence of antibiotics. Results of these experiments indicate that the sub‐inhibitory concentrations of antibiotics led to an enhancement of tmRNA and trans‐translation in the antibiotic producing strains. Longer incubation of cells with antibiotics up to 240 min had no additional effect on increase of amino acids incorporation to the tag‐peptides. We also examined sensitivity of ribosomes of the S30 fractions from the exponential phase of growth to antibiotics in poly(U)‐dependent synthesis of polyphenylalanine. As shown in Fig. 3C, in vitro system of S. griseus was active in the presence of streptomycin up to 2 µM but higher concentrations of streptomycin inhibited poly(U) translation. In S. collinus, the polyphenylalanine synthesis directed by poly(U) was inhibited by kirromycin at concentrations higher than 0.2 µM (Fig. 3D).
Figure 3.

Effect of antibiotics on in vitro incorporation of amino acids by stalled complex of ribosomes from S. griseus and S. collinus. Cells from the exponential phase of growth were incubated in absence (▪) or presence (□) of sub‐inhibitory concentrations of produced antibiotics. In S. griseus with 3.5 µM streptomycin (A) and S. collinus with 5 µM kirromycin (B). At indicated time intervals samples were taken for preparation of S30 fractions. Pre‐incubated S30 fractions (2 A260 units) were used for translation of poly(U) in reaction mixtures (100 µl) containing 50 mM Tris‐HCl pH 7.6, 10 mM MgCl2, 80 mM NH4Cl, 1 mM dithiothreitol, 5 mM phosphoenol pyruvate, 1 mM ATP, 0.5 mM GTP, 15 mM phenylalanine and 100 µg poly(U). After 15 min of incubation, phenylalanine incorporation was saturated. To the stalled complex of ribosomes was added 20 µM each of U14C‐l‐alanine, U14C‐l‐arginine, U14C‐l‐leucine, U14C‐l‐lysine and 50 µM each of remaining unlabelled amino acids. These mixtures were incubated at 32°C for 15 min. Reactions were terminated by trichloroacetic acid, boiled at 95°C for 20 min and filtered over Whatmam GF/C filters and assayed for radioactivity. The sensitivity of ribosomes of the 10 h cultures to the presence of produced antibiotics was examined at the translation of poly(U). Dialysed and pre‐incubated S30 fractions of S. griseus (C) and S. collinus (D) were incubated in the reaction mixtures with U14C‐l‐phenylalanine (0.2 µCi) and antibiotics as indicated. Radioactivity in the hot trichloroacetic acid insoluble fractions was measured by liquid scintillation counter. The values are average from four independent experiments.
The effect of antibiotics on the tmRNA abundance
Previously, it was shown that sub‐inhibitory concentrations of several protein synthesis inhibitors induced an increase of the tmRNA in Streptomyces aureofaciens producing tetracycline (Palečkováet al., 2006). We examined the abundance of tmRNA in cells from the exponential phase of growth of S. griseus and S. collinus incubated in the presence of antibiotics that inhibit protein synthesis or other cellular functions as synthesis of DNA, RNA or the cell wall. When the cells of S. griseus (Fig. 4A) were incubated in the presence of streptomycin, the level of tmRNA rapidly increased during the first 10 min and remained unchanged for up to 180 min. At the presence of tetracycline, protein synthesis decreased to 75% of control value without tetracycline (Fig. 4B) and tmRNA synthesis increased more than seven times. Presence of kirromycin led to a gradual increase in tmRNA and after 3 h cultivation the level was three times higher than that of the control value. No significant changes in the level of tmRNA and protein synthesis were observed in the cells incubated with sub‐inhibitory concentration of chloramphenicol. Out of the antibiotics that interfere with other cellular activities than protein synthesis, only nalidixin slightly stimulated the elevation of the tmRNA level (Fig. 4C) and protein synthesis (Fig. 4D). Low stimulative effect on the level of tmRNA also caused vancomycin. Sub‐inhibitory concentrations of rifampicin and ampicillin had no effect on the tmRNA level and protein synthesis. In experiments with S. collinus (Fig. 5A), the presence of tetracycline gave rise to an increase in tmRNA more than five times and the rate of protein synthesis remained the same as in the control experiment without antibiotics (Fig. 5B). The presence of streptomycin had no effect on the tmRNA abundance, but protein synthesis decreased to 40% of control without the drug. The addition of kirromycin to cells from the exponential phase of growth induced an increase of the tmRNA during the first 10 min and then remained unchanged. Chloramphenicol caused a significant increase in the tmRNA after 3 h of cultivation, while protein synthesis decreased to 50% of control value without antibiotics. The presence of sub‐inhibitory concentrations of antibiotics interfering with synthesis of RNA and DNA (Fig. 5C) had no valuable effect on tmRNA abundance. The low level of rifampicin had no effect on tmRNA synthesis, but exerted an influence on the protein synthesis which was reduced by about 40% compared with that of control without antibiotics (Fig. 5D). Slight stimulation effect on tmRNA abundance exhibited ampicillin during the first 60 min of cultivation.
Figure 4.
Effect of antibiotics on the level of tmRNA and incorporation of U14C‐leucine to proteins in S. griseus. A. To cultures from the exponential phase of growth (14 h) the antibiotics (□) kirromycin (5 µg ml−1), (Δ) chloramphenicol (8 µg ml−1), (○) streptomycin (5 µg ml−1) or (×) tetracycline (1 µg ml−1) interfering with function of ribosomes were added. At the time intervals, total RNA was isolated and aliquot parts were analysed by electrophoresis in polyacrylamide‐urea gels. Northern blots were hybridized with Dig‐labelled probes. The results represent average from three independent experiments. The amounts of tmRNA were normalized on 5S RNA levels and on parallel control experiments. The level of tmRNA in cells from exponential phase of growth without addition of antibiotics represents 100%. B. At the same time cultures were labelled with U14C‐leucine (0.5 µCi ml−1), two 1 ml samples were taken at indicated time intervals and incorporation was stopped with 1 ml 10% trichloroacetic acid (TCA). Samples were boiled for 20 min at 95°C and filtered using Whatman membrane filters (0.4 µm). Washed samples with 5% TCA were dried and radioactivity measured. The data were obtained from three independent experiments. The designations are as in (A) and (▪) represents control without addition of antibiotics. C. Cultures were incubated in the presence or absence of antibiotics affecting other targets: (⊠) ampicillin (10 µg ml−1), (●) rifampicin (1 µg ml−1), (⊕) nalidixic acid (20 µg ml−1) or (◊) vankomycin (1 µg ml−1); and tmRNA was analysed as above. D. Incorporation of U14C‐leucine (0.5 µCi ml−1) to proteins in presence or absence (▪) of antibiotics was examined.
Figure 5.

The tmRNA abundance and incorporation of U14C‐leucine to proteins in the presence or absence of antibiotics in S. collinus. A. To cultures from the exponential phase of growth (14 h) the antibiotics (□) kirromycin (5 µg ml−1), (Δ) chloramphenicol (8 µg ml−1), (○) streptomycin (5 µg ml−1) or (×) tetracycline (1 µg ml−1) were added. At the time intervals tmRNA was analysed as in Fig. 4. B. At the same time incorporation of U14C‐leucine (0.5 µCi ml−1) was determined as above. C. Cultures were incubated in the presence or absence of (⊠) ampicillin (10 µg ml−1), (●) rifampicin (1 µg ml−1), (⊕) nalidixic acid (20 µg ml−1) or (◊) vankomycin (1 µg ml−1), and tmRNA was examined as in Fig. 4. D. Incorporation of U14C‐leucine (0.5 µCi ml−1) to proteins in the presence or absence (▪) of antibiotics was measured. The figure represents average data from three independent experiments.
These experiments demonstrate differences between streptomycetes in sensitivity to the presence of antibiotics and in the responses leading to changes in the tmRNA abundance.
Discussion
The translation of genetic information consumes a significant amount of cell energy. Altered translation rates occur under conditions such as the exhausting of the nutrient limitation, stress, during transition from active growth to antibiotic production phase (secondary metabolism) and cell differentiation. Antibiotics that cause ribosome to stall or pause could increase mRNA within or in adjacent to the A‐site codon. The A‐site mRNA cleavage pathway accompanied with pausing ribosomes and induction of the tmRNA system can reduce translation errors and production of aberrant polypeptides. In the present study and previous work with S. aureofaciens (Palečkováet al., 2006) we have found that sub‐inhibitory concentrations of antibiotics produced or exogenously added to the cultures at the beginning of the exponential phase of growth led to an enhancement of tmRNA level and the incorporation of labelled amino acids to the tag‐peptides. The translation of ORF from tmRNA and normal termination releases the tagged polypeptides for degradation and permits disassembly and recycling of ribosomal subunits for the new rounds of polypeptide synthesis (Keiler et al., 1996; Hayes and Sauer, 2003). Streptomycetes differ in amount of tmRNA and in the stability during development. The highest level of the tmRNA was found in cells from the exponential phase of growth. During antibiotic biosynthesis both protein synthesis and tmRNA level decreases. Our data indicate that abundance of the tmRNA in the S. griseus, S. collinus is correlated mostly with the activity of the translational system while in S. lividans tmRNA was constitutively expressed during cultivation (Braud et al., 2005). To support the suggestion demonstrating differences in tmRNA abundance during development, the stability of tmRNA in the two streptomycetes strains was investigated. The decay of tmRNA was examined at a high concentration of rifampicin (300 µg ml−1) because at a low concentration (25 µg ml−1) the drug stimulates transcription from σR‐dependent promoters (Newell et al., 2006). Ribosomes, having a highly cooperative structure, are also a potential target for control mechanisms that generate signals and activate adaptive regulons. The function of tmRNA in trans‐translation is dependent on the presence of several protein ligands: protein synthesis elongation factor Tu (Rudinger‐Thirion et al., 1999), SmpB (small protein B) (Karzai et al., 1999) and ribosomal protein S1 (Wower et al., 2000). RNase R was identified as the nuclease responsible for degradation of the tmRNA. It is known that SmpB and EF‐Tu binds to aminoacyl moiety of alanyl‐tmRNA (Shimizu and Ueda, 2006). It is likely that in the cells from the exponential phase of growth, tmRNA is stabilized by interaction with SmpB and its stability could be regulated by exposing or protecting the 3′ end of the tmRNA (Hong et al., 2005). In experiments with S. aureofaciens, we found that the highest level of SmpB was in the cells from exponential phase of growth and a descending amount in the cells from stationary phase (Mikulík et al., 2008), which indicates that availability of the SmpB can be involved in regulation of tmRNA stability.
A previous study (Ueda et al., 2002) demonstrated that antibiotics causing amino acid misreading (kanamycin, streptomycin, hygromycin B, paromomycin and gentamicin) induce read‐through of stop codons and enhance the tmRNA mediated protein tagging. Differences among streptomycetes in response to the presence of antibiotic led to the analysis of ssrA genes. The most significant differences between ssrA genes of S. aureofaciens, S. collinus and S. griseus are in the central regions of the tag‐encoding parts and in the sequences about 20 nt following termination codons. These differences in the tag‐encoding domains are most probably involved in recognition signals of specific proteolysis system (Mikulík et al., 2008).
A sub‐inhibitory concentration of antibiotics induces specific cell responses at the level of gene expression (Goh et al., 2002). This response is drug‐specific and need not to be directly linked to the mode of antibiotics action (Tsui et al., 2004). As trans‐translation is involved in various steps of bacterial physiology, it is assumed that the inactivation of the ssrA could induce changes in sensitivity to antibiotics that affect other target sites than the ribosome (Luidalepp et al., 2005). We examined sensitivity of S. griseus and S. collinus to the presence of antibiotics interfering with different target sites. The enhancement of the tmRNA level stimulated by the presence of antibiotic that is naturally produced by the strain is instantaneous. Similar results were obtained in experiments with S. collinus.
The outcome of these experiments indicates that the trans‐translation system responds to the presence of produced antibiotics and contributes to the reactivation of stalled complexes of ribosomes and thereby plays a significant role in the survival of streptomycetes under adverse conditions. This activity is limited by drug produced intracellular concentration and the sensitivity of ribosomes. Overexpression of antibiotics is connected with a decrease in activity of transcription and translation system and degradation of cell macromolecules, which are used as building blocks for biosynthesis of secondary metabolites (Voigt et al., 2002; Ostash et al., 2007). The system activates efficient resistance mechanisms, e.g. active efflux of antibiotics from cells, changes in the composition of the cell wall or it induces the drug modification.
Antibiotics that interfere with RNA polymerase, DNA and the cell wall were less effective in stimulation of tmRNA abundance than drugs targeting the ribosome itself. From antibiotics having other target sites than ribosomes, a small increase in tmRNA was observed in the presence of nalidixic acid and vancomycin in S. griseus. Inhibitory action of nalidixic acid involves trapping a gyrase–DNA complex in which DNA is broken. During the gyrase poisoning hydroxyl radicals are generated, which play an important role in cell killing (Dwyer et al., 2007). Hydroxyl radical cleaves RNA independently of base sequence and secondary structure. Significant increase in tmRNA was observed in the presence of ampicillin in S. collinus. It remains to be seen whether the tmRNA is induced by ampicillin as a response to oxidative stress. It was recently showed (Kohanski et al., 2007) that bactericidal antibiotics, regardless of drug‐target interaction, stimulate the production of toxic hydroxyl radicals that contribute to cell death.
Experimental procedures
Strains and cultivation
Streptomyces collinus Tü 365 was cultivated in medium (g l−1) peptone 4.0, yeast extract 4.0, malt extract 2.0, MgSO4 0.5, K2HPO4 2.0, KH2PO4 2.0, glucose 8.0 (sterilized separately), pH 7.2.
Streptomyces griseus MBU was cultivated in medium GYM (g l−1) glucose 4.0, yeast extract 4.0, malt extract 10.0, FeSO4.7H2O 7.5 mg, CuSO4.5H2O 5.0 mg, MnSO4.5H2O 4.0 mg, CaCl2.2H2O 15.0 mg, ZnSO4.7H2O 9.0 mg, pH 7.3.
Both strains were cultivated in 500 ml flasks with 80 ml media at 28°C on a reciprocal shaker at 125 r.p.m. Cells from different stages of development were harvested by centrifugation and washed with the standard buffer 20 mM Tris‐HCl, pH 7.6, 40 mM NH4Cl, 10 mM MgCl2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride.
Isolation of RNA
Vegetative cells (1.0 g wet material) were mixed with RNA Blue containing guanidium thiocyanate, sodium citrate, 2‐mercaptoethanol, Na3VO4 (Top‐Bio) and disrupted with glass beads in FastPrep homogenizer (Qbiogene). Homogenate (1 ml) was mixed with 50 µl of 10% sarkosyl and incubated for 5 min at room temperature. The sample was shaken with 0.2 ml of chloroform and then centrifuged at 14 500 g for 10 min at 4°C. The water phase containing RNA was taken off and precipitated by 0.1 vol 3 M CH3COONa, pH 5.5:1 vol isopropanol. RNA was sediment at 14 500 g for 10 min and 4°C and washed with 1 ml of 75% ethanol. The final sediment was solubilized in a sterile 10 mM Tris‐HCl, pH 8.0. RNA was analysed in 7% polyacrylamide‐6M urea sequencing gels and visualized with ethidium bromide.
Analysis of tmRNA by Northern hybridization
Aliquots of the total RNA were separated by electrophoresis in a 7% polyacrylamide‐6M urea gels. A nylon membrane was blotted with separated RNAs using a vacuum blotter Hybaid. Dig‐11‐dUTP‐labelled DNA‐probe was prepared by PCR amplification of corresponding DNAs using upstream primer 5′‐GGGGATGATCGGTTTCGACAG‐3′ and downstream primer 5′‐TGGTGGAGATGGCGGGAATC‐3′. After hybridization at 68°C for 16 h, tmRNA was detected by anti‐digoxigenin‐AP‐conjugate and colorimetric substrates. The relative amounts of tmRNA were quantified using Bio‐Rad Molecular Imager FX and Aida Image analyser. The ssrA genes from S. collinus and S. griseus have been deposited in the GenBank (accession numbers AY485228 and DQ471924 respectively).
Incorporation of l ‐[U‐14C]‐leucine to proteins
Cells from 14 h cultures were labelled with l‐[U‐14C]‐leucine (4 Mbq mol−1) in the absence or presence of streptomycin (5 µg ml−1), kirromycin (5 µg ml−1), tetracycline (2.5 µg ml−1), chloramphenicol (10 µg ml−1), rifampicin (2.5 µg ml−1), nalidixic acid (10 µg ml−1), ampicillin (10 µg ml−1) or vancomycin (1 µg ml−1). At the indicated time intervals three 1 ml samples were taken into 1 ml of 10% TCA containing 1 mg ml−1 of cold l‐leucine. Samples were boiled for 20 min at 95–98°C. Chilled samples were collected and washed on Synpor 0.4 µm nitrocellulose filters. Washed samples were dried and radioactivity measured in liquid scintillation counter.
Preparation of S30 fractions
Cells from 14‐, 24‐, 48‐ and 72‐h‐old cultures were homogenized with glass beads in FastPrep homogenizer and extracted with standard buffer containing 20 mM Tris‐HCl, pH 7.6, 80 mM NH4Cl, 10 mM magnesium acetate, 1 mM DTT (dithiothreitol) and 1 µg ml−1 DNase RQ1 (RNase free). Extracts were centrifuged at 10 000 g for 10 min and supernatant solutions were again centrifuged at 30 000 g for 30 min. Supernatant S30 fractions so prepared were incubated for 40 min in mixture of 20 amino acids (0.2 mM each), 5 mM ATP, 5 mM phosphoenol‐pyruvate, 0.2 mM GTP. Dialysed S30 fractions were frozen in aliquots and stored at −70°C.
In vitro trans‐translation using the stalled complex of ribosomes
The composition of the reaction mixture was similar to that as described previously (Himeno et al., 1997; Konno et al., 2004; Asano et al., 2005). Polyphenylalanine was synthesized in 100 µl reaction mixtures containing 50 mM Tris‐HCl pH 7.6, 10 mM magnesium acetate, 80 mM NH4Cl, 1.0 mM DTT, 5 mM phosphoenol‐pyruvate, 1 mM ATP, 0.5 mM GTP, 15 pmol l‐phenylalanine, 2 A260 units of S30 fraction and 100 µg poly(U). In parallel experiment, time‐dependent incorporation of l‐[3H]‐phenylalanine (15 pmol) into polyphenylalanine was examined. After 15 min incubation at 32°C, phenylalanine incorporation was saturated (not shown). To the stalled complex of ribosomes was added 20 µM each of l‐[U‐14C]‐alanine, l‐[U‐14C]‐arginine, l‐[U‐14C]‐leucine, l‐[U‐14C]‐lysine and 50 µM each of remaining unlabelled amino acids. After incubation at 32°C for 15 min, the reaction was terminated by trichloroacetic acid to final concentration of 5%. Material insoluble in hot TCA was filtered over Whatman GF/C filters and assayed for radioactivity.
Acknowledgments
This work was supported by a grant from the Grant agency of the Czech Republic to K.M. 203/05/0106, 310/07/1009, EC Integrated Project ActinoGEN LSHM‐2004‐005224 and Research Concept AVOZ 500510.
References
- Abo T., Ueda K., Sunohara T., Ogawa K., Aiba H. Ssra‐mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells. 2002;7:629–638. doi: 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
- Asano K., Kurita D., Takada K., Konno T., Muto A., Himeno H. Competition between trans‐translation and termination or elongation of translation. Nucl Acids Res. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud S., Lavire C., Bellier A., Mazodier P. Effect of SsrA (tmRNA) tagging system on translational regulation in Streptomyces. Arch Microbiol. 2005;184:343–352. doi: 10.1007/s00203-005-0051-y. [DOI] [PubMed] [Google Scholar]
- Dwyer D.J., Kohanski M.A., Hayete B., Collins J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara A., Tomatsu H., Inagaki S., Tadaki T., Ushida C., Himeno H., Muto A. Detection of tmRNA‐mediated trans‐translation products in Bacillus subtilis. Genes Cells. 2002;7:343–350. doi: 10.1046/j.1365-2443.2002.00523.x. [DOI] [PubMed] [Google Scholar]
- Goh E.B., Yim G., Tsui W., McClure J.E., Surette M.G., Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski K.B., Rodnina M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat Struct Mol Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Hayes C.S., Bose B., Sauer R.T. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C.S., Sauer R.T. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translation quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- Heinzel P., Werbitzky O., Distler J., Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin‐3‐phosphotransferase. Relationships between antibiotic and protein kinases. Arch Microbiol. 1988;150:184–192. doi: 10.1007/BF00425160. [DOI] [PubMed] [Google Scholar]
- Himeno H., Sato M., Tadaki T., Fukushima M., Ushida C., Muto A. In vitro Trans‐translation mediated by alanine‐charged 10Sa RNA. J Mol Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- Hong S‐J., Tran Q‐A., Keiler K.C. Cell cycle regulated degradation of tmRNA is controlled by RNaseR and SmpB. Mol Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A.W., Susskind M.M., Sauer R.T. SmpB, a unique RNA‐binding protein essential for the peptide‐tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged mRNA. Science. 1996;271:955–956. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kim B‐H., Andersen C., Benz R. Identification of a cell wall channel of Streptomyces griseus: the channel contains a binding site for streptomycin. Mol Microbiol. 2001;41:665–673. doi: 10.1046/j.1365-2958.2001.02544.x. [DOI] [PubMed] [Google Scholar]
- Kohanski M.A., Dwyer D., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Konno T., Takahashi T., Kurita D., Muto A., Himeno H. A minimum structure of aminoglycosides that causes an initiation shift of trans‐translation. Nucl Acids Res. 2004;32:4119–4126. doi: 10.1093/nar/gkh750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hirano R., Tagami H., Aiba H. Protein tagging at rare codons is caused by tmRNA action at 3′ end of nonstop mRNA generated in response to ribosome stalling. RNA. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H., Hallier M., Felden B., Tenson T. tmRNA decreases the bactericidal activity of aminoglycosides and succeptibility to inhibitors of cell wall synthesis. RNA Biol. 2005;2:70–74. doi: 10.4161/rna.2.2.2020. [DOI] [PubMed] [Google Scholar]
- Mikulík K., Weiser J., Hašková D. Protein synthesis elongation factor EF‐Tu from Streptomyces collinus producing kirromycin. Biochem Biophys Res Commun. 1982;108:861–867. doi: 10.1016/0006-291x(82)90910-x. [DOI] [PubMed] [Google Scholar]
- Mikulík K., Palečková P., Felsberg J., Bobek J., Zídková J., Halada P. SsrA genes of streptomycetes and association of proteins to the tmRNA during development and cellular differentiation. Proteomics. 2008;8:1429–1441. doi: 10.1002/pmic.200700560. [DOI] [PubMed] [Google Scholar]
- Newell K.V., Thomas D.P., Brekasis D., Paget M.S.B. The RNA polymerase‐binding protein RbpA confers basal levels of rifampicin resistance of Streptomyces coelicolor. Mol Microbiol. 2006;60:687–696. doi: 10.1111/j.1365-2958.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Hosaka T., Tokoyama S., Okamoto S., Ochi K. Mutations in rsmG encoding a 16S rRNA methyltransferase result in low‐level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2) J Bacteriol. 2007;189:3876–3883. doi: 10.1128/JB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostash B., Saghatelian A., Walker S. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palečková P., Bobek J., Felsberg J., Mikulík K. Activity of translation system and abudance of tmRNA during development of Streptomyces aureofaciens producing tetracycline. Fol Microbiol. 2006;51:517–524. doi: 10.1007/BF02931615. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A., Nissen P. Elongation factor Tu‐targeted antibiotics: Four different structures, two mechanisms of action. FEBS Lett. 2006;580:4576–4581. doi: 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Roche E.D., Sauer R.T. SsrA‐mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger‐Thirion J., Giege R., Felden B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Ueda T. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon‐anticodon interaction during trans‐translation initiation. J Biol Chem. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- Tsui W.H., Yim G., Wang H.H., McClure J.E., Surrete M.G., Davies J. Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem Biol. 2004;11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Ueda K., Yamamoto Y., Ogawa K., Abo T., Inokuchi H., Aiba H. Bacterial SsrA system plays a role in coping with unwanted translational read through caused by suppressor tRNAs. Genes Cells. 2002;7:509–519. doi: 10.1046/j.1365-2443.2002.00537.x. [DOI] [PubMed] [Google Scholar]
- Voigt C.A., Martinez C., Wang Z.G., Mayo S.L., Arnold F.H. Protein building blocks preserved by recombination. Nat Struct Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- Wolf H., Zahner H. Metabolic products of microorganisms. Kirromycin. Arch Microbiol. 1972;83:147–154. [PubMed] [Google Scholar]
- Wower I.K., Zwieb C.W., Guven S.A., Wower J. Binding and cross‐linking of tmRNA to ribosomal protein S1, on and off the Eschrichia coli ribosome. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]


