Abstract
A protein corona will be formed on nanoparticles (NPs) entering a biological matrix, which can influence particles’ subsequent behaviors inside the biological systems. For proteins bound stably to the NPs, they can exhibit different association/dissociation rates. The binding kinetics could affect interaction of the NPs with cell surface receptors and possibly contribute to the outcomes of NPs uptake. In the present study, a method to differentiate the corona proteins based on their relative dissociation rates from the NPs was developed, employing flow field-flow fraction (F4) in combination with centrifugation. The proteins bound to the superparamagnetic iron oxide NPs (SPION) present in an IgG/albumin depleted serum were isolated via collection of the SPIONs by either F4 or centrifugation. They were subsequently analyzed by LC-MS/MS and identified. Because the SPION-protein complexes injected to F4 dissociated continuously under the non-equilibrium separation condition, only the proteins with slow enough dissociation rates would be collected with the NPs in the eluent of F4. However, in centrifugation, proteins with good affinity to the SPIONs were collected regardless of the dissociation rates of the complexes. In both cases, the non-binding ones were washed off. Capillary electrophoresis and circular dichroism were employed to verify the binding situations of a few SPION-protein interactions, confirming the effectiveness of our method. Our results support that our method can screen for proteins binding to NPs with fast on-and-off rates, which should be the ones quickly exchanging with the free matrix proteins when the NPs are exposed to a new biological media. Thus, our method will be useful for investigation of the temporal profile of protein corona and its evolution in biological matrices, as well as for high-throughput analysis of the dynamic feature of protein corona related to particle properties.
Keywords: non-equilibrium separation, protein adsorption, nanoparticles, flow field-flow fractionation, binding kinetics, dissociation rate
Introduction
The wide applications of nanoparticles (NPs1–3 have increased their exposure to all living matters, including human beings, calling for a better understanding of the interactions between NPs and biosystems.4,5 Once the NPs enter a biological matrix, their surface would be covered by biomolecules, mainly proteins, which then affect the fate and functions of NPs in the biological system.6–13 Protein corona compositions, protein binding affinities, as well as binding stoichiometries, have been intensely studied to gain more knowledge on the biological roles of protein corona and how its formation could be affected by the properties of NPs and proteins.6,7
Pioneer studies in this field have also pointed out the importance of binding kinetics,14 which affects the temporal development of the corona and its evolution in biological environment. When the NPs are placed in one biological matrix, the rates of proteins binding to and dissociating off the NPs determine the time needed to reach the equilibrium state.15 When the NPs pass onto a new biological environment, the kinetic parameters strongly affect how the corona would evolve:16–18 proteins in the original corona with relatively fast dissociation rates would be exchanged more rapidly by the free proteins in the new environment than those with slower dissociation rates. Still, little is known about how the particle surface and size characteristics would affect the binding kinetics; and how the association/dissociation rates would impact the subsequent interactions between NPs and cell surface receptors.
Several techniques have been employed to analyze the protein corona on NPs. Among them, centrifugation is the simplest, which can bring down a wide variety of NPs together with the adsorbed proteins after they are incubated in biological media. The collected proteins can then be eluted off the NPs and analyzed by SDS-PAGE or LC-MS. It has been applied for identification of corona proteins and for study of corona evolution over time or under different protein concentrations.19,20 For quantification of association/dissociation rates as well as affinity of individual proteins binding to NPs, isothermal calorimetry,21 size exclusion chromatography,14 surface plasmon resonance,22 and capillary electrophoresis,23 have been employed. However, measurement throughput and speed need to be improved to match with the large variety of NPs and the high complexity of biological matrices.
As an open channel separation technique, flow field-flow fractionation (F4) is well suited for analysis of biological interactions, because the absence of column packing significantly reduces disturbance to macromolecular complexes.24,25 F4 separates compounds based on their size and shape, and has been used to isolate antibody-Fc (fragment crystalizable) receptor complexes for determination of binding affinities, for detection of cytochrome C-lipid interactions, and as a cleanup tool to wash away non-specifically-bound proteins from the surface of microbeads employed in affinity pull-down assay.26–28
In the present study, F4 was applied to purify the protein corona formed on the superparamagnetic iron oxide nanoparticles (SPION)s after they were incubated in the Immunoglobulin G (IgG)/albumin-depleted human serum, in parallel with centrifugation. We found out that, while both centrifugation and F4 could isolate proteins with good binding affinity to the SPIONs, F4 could wash off the proteins interacting with NPs with relatively fast association/dissociation rates. The binding affinities of selected proteins were measured by capillary electrophoresis (CE). CE also confirmed that, the proteins being washed off by F4 but remained bound to SPIONs during centrifugation-based complex isolation were those with fast on-and-off rates when binding to proteins, proving the effectiveness of our method. Moreover, circular dichroism (CD) measurement was carried out to show that, the rapidly dissociating SPION-protein interaction would not cast significant disturbance to protein structure. We believe our method can assist with the study of the temporal development of protein corona and its evolution in biological environment with higher throughput than the aforementioned techniques.
Experimental Section
Chemicals
All chemicals for preparation of the buffers used in F4 and protein digestion (sodium phosphate monobasic, sodium phosphate dibasic, FL-70, sodium hydroxide, trifluoroacetic acid, ammonium bicarbonate, and dithiothreitol (DTT)) were purchased from Fisher Scientific (Pittsburgh, PA) except for iodoacetamide (IAA), which was from Sigma Aldrich (St. Louis, MO). Alpha-1-antitrypsin (A1AT), human serum albumin (HSA), apolipoprotein A1 (APOA1), beta-casein, succinyl concanavalin A (ConA), calmodulin, haptoglobin, prealbumin, transferrin, and trypsin were purchased from Sigma Aldrich. Human serum was purchased from Biogenesis (Poole, England).
Nanoparticles
SPIONs coated with polyacrylic acid (referred below as PAA-SPIONs) were synthesized in our lab using the protocol established by Dr. Yadong Yin’s group.29 In brief, the particles were prepared in a one-step hydrolysis reaction, in which a mixture of sodium hydroxide/diethylene glycol was added to the solution containing PAA and iron(III) chloride at 220 °C. This reaction continued for 12 hours to yield spherical PAA-SPIONs with an average particle diameter of 8.62±2.11 nm, determined by transmission electron microscopy (TEM, Supporting Information Figure S1a). Based on the particle diameter and iron content as determined by ICP-AES, the particle concentration of the stock PAA-SPIONs was found to be 1.2 µM. SPIONs coated with an amphiphilic block copolymer (referred to below as AMP-SPIONs) with an average particle diameter of 10 nm and stock concentration of 4.6 µM (as determined by the manufacturer) were purchased from Ocean Nanotech (Springdale, AK). The hydrodynamic diameters and the zeta potentials for both particles can be found in Figure S1b (Supporting Information).
Flow field-flow fractionation conditions
All F4 collections were conducted in an F1000 symmetric F4 instrument (Postnova, Salt Lake City, UT), coupled to a Shimadzu SP-20A absorbance detector (Columbia, MD). The separation channel was made by clamping a spacer with a thickness of 0.254 mm in between two ceramic blocks; and the bottom of the channel, i.e. the accumulation wall, was made from a 10 kDa MWCO regenerated cellulose membrane and had a surface area of 5900 mm2. This system has a void volume V0 of 1.41 mL. After calibration with SPIONs of similar size to those investigated in this study (Figure S2), the channel dimensions were 20 × 270 × 0.236 mm. The carrier solution was comprised of 10 mM phosphate at pH 7.5 ± 0.1 (prepared from monobasic sodium phosphate monohydrate and anhydrous dibasic sodium phosphate without additional pH adjustment), with 0.025% FL-70 added. Samples containing the PAA-SPIONs were analyzed with the channel and cross flow rates of 1.00 mL/min and 0.75 mL/min, respectively; but both flow rates were at 0.75 mL/min when analyzing the AMP-SPIONs and their protein incubations. Absorbance detection was done at 280 nm.
Incubation of SPIONs with proteins
For all binding experiments, SPIONs were mixed with proteins in 20 µL 10 mM phosphate buffer at pH 7.4, and incubated for two hours at 37 °C prior to isolation of the SPION-protein complex by either centrifugation or F4. Our preliminary study showed that the retention time of the SPIONs did not change after 2 hrs, which means the binding equilibrium was established and stable SPION-protein complexes were formed.
During the test of single protein (HSA, β-casein, calmodulin, and succinyl ConA) binding with the PAA-SPIONs, a protein:particle molar ratio of 12:1 was used. Additionally, protein mixtures with the molar ratios of HSA : β-casein : succinyl ConA at 10:1:1, 5:1:1, 1:1:1, and 0.1:1:1, respectively, were prepared, while still keeping the total protein : particle molar ratio at 12:1.
The study of protein adsorption in the IgG/albumin depleted serum was conducted using the AMP-SPIONs. Preparation of the depleted serum was done via a Cibracon Blue and Protein A-based serum depletion kit (Thermo Fisher), and the obtained serum sample was desalted and buffer-exchanged into 20 mM phosphate using a 7kDa MWCO Zebra Spin Column (Thermo Fisher). Ten µL of the depleted serum was mixed with the AMP-SPIONs in a 20µL volume, giving a final particle concentration of 1.2 µM in 10 mM phosphate buffer at pH 7.4. This mixture was incubated for two hours at 37 °C.
Isolation of SPION-protein complexes by centrifugation and F4
After the SPIONs were incubated with proteins for 2 hrs, the SPION-protein complexes were isolated from the incubated mixture by either centrifugation or F4. Before centrifugation isolation, the incubation sample was diluted from 20 to 400 µL with 10 mM phosphate (pH 7.4). Then, it was centrifuged for 30 minutes at 16.1 krcf, pelleting the SPION-protein complex. The supernatant was removed; the SPIONs were resuspended in the same phosphate buffer. The centrifugation and SPION re-suspension process was repeated once. The SPION-protein complex was then subjected to trypsin digestion and analysis, explained below.
When isolating the PAA-SPIONs by F4, the SPION-protein mixture was injected onto the F4 system immediately following incubation. A 4-mL eluate was collected within the elution window of the SPION-protein complex peak. This collection was split across 4 aliquots, which were then centrifuged for 30 minutes at 16.1 krcf. The bottom 100 µL of each aliquot was transferred to a single, clean tube, and the centrifugation process was repeated. The supernatant was removed, and the SPION-protein pellet was resuspended for analysis. During isolation of the AMP-SPIONs by F4, it turned out to be very difficult to pellet the SPIONs from a 4-mL solution under the same centrifugation condition as used above. Thus, we revised the procedure for this type of SPIONs, and concentrated the eluate using a 4-mL Amicon filter having a MWCO of 10kDa (Millipore).
In both cases, blanks were also prepared, by either centrifuging the depleted serum only under the same conditions as used on the incubation samples, or injecting the depleted serum in F4 and collecting the eluents within the same elution window as the SPION-protein complexes. Proteins identified in the blanks were considered as co-precipitates or co-eluted, unbound proteins, and excluded from the final report of the SPION-interacting proteins.
Protein treatment and identification
In the study of binding between the PAA-SPIONs and the standard proteins, the adsorbed proteins were removed from the SPIONs in the loading buffer, which had a final SDS concentration of 1%, and analyzed by SDS-PAGE, using a 10% polyacrylamide resolving gel and a 4% stacking gel. Protein bands in each sample were visualized using a SYPRO Ruby protein stain (Invitrogen). Gel images were taken on a Typhoon imager, and the presence or absence of each protein was determined.
In the serum protein binding study, the proteins adsorbed on the SPIONs were digested without removing the particles, because no interference to the activity of trypsin was noticed in our experiments. The samples were first treated with dithiothreitol and iodoacemide for reduction of disulfide bonds and alkylation of the resulting thiol groups, and then underwent a tryptic digest at 37 °C overnight. After digestion, the samples were mixed with a 50% acetonitrile solution to denature any adsorbed peptides. The released peptides were separated from the SPIONs with a 100kDa Amicon filter. The peptides collected in the filtrate were concentrated to dryness, reconstituted in 0.1% trifluoroacetic acid, and desalted and preconcentrated by ZipTip, prior to analysis by 2D MUDPIT LC-nano-MS/MS analysis (Supporting Information).30
Confirmation of AMP-SPION-protein interaction by capillary electrophoresis
All CE experiments were conducted on a Beckman-Coulter P/ACE MDQ Glycoprotein System, using a fused silica capillary (i.d. 75 µm) with an effective length of 40 cm and an overall length of 50 cm. In both CE modes, dimethyl sulfoxide was added as a neutral marker to monitor mobility shift or change in the electroosmotic flow. All separations were conducted at +20 kV and the electropherograms obtained under UV adsorption detection at 200 nm (for neutral marker detection) and 280 nm (for analysis of bound and free nanoparticle) by a photodiode array detector were collected.
The process of affinity measurement using CE was the same as used in our previous work.23 The samples tested by capillary zone electrophoresis (CZE) were prepared by incubating the protein and the AMP-SPIONs at various molar ratios ranging from 0:1 to 10:1 overnight at 37 °C. The running buffer was 10 mM borate at pH 8.5. The peak areas of the complex and the free SPIONs were used to calculate the protein-bound SPION ratios which were plotted against the protein concentrations. The resulting curves were fitted in a Hill plot and the Kd values were calculated. In the case of affinity capillary electrophoresis (ACE), the running buffer was 15 mM phosphate (pH 7.5), to maintain an appropriate binding environment inside the capillary. The protein was added to this running buffer at various protein:nanoparticle ratios (from 0:1 to 50:1). The mobility shift of the nanoparticle peak was plotted versus protein concentration, and a Hill equation was used to calculate the Kd value from the resulting curve.
Circular dichroism (CD) measurements
All CD measurements were taken on a Jasco J-815 circular dichroism instrument, using a quartz cuvette with a 1-mm pathlength. Single protein and the AMP-SPIONs were incubated at a 25:1 molar ratio in 10 mM phosphate for 3 hrs at 37 °C. The samples were diluted 3 fold prior to CD measurements. CD spectra were obtained across a wavelength range of 190–260 nm, with 1 nm scan increment. Background subtraction (either a phosphate blank for protein-only measurements, or a SPIONs-only sample for protein incubated particle measurements) was done on the raw spectra. Once the CD data was obtained, the percentage of each secondary structure (helix, sheet, random coil) was estimated via the concentration-independent method designed by Raussens et al.31 The changes in the secondary structure fraction before and after incubation with SPIONs were then compared.
Results and Discussion
Washing off proteins with fast dissociation rates by F4
When the equilibrium mixture of the SPIONs and the proteins was injected as a short plug into the F4 column, the SPION-protein complexes were subject to the non-equilibrium separation condition since no proteins were present in the carrier solution of F4.32–37 Thus, the complex started to dissociate once the free proteins in the sample diffused out of the sample zone (Figure 1). Re-association could be ignored if the free, dissociated proteins are well separated from the complexes. Then, the proteins dissociating off the SPIONs with relatively fast dissociation rates would be found in the eluent at a much lower or even not-detectible content than those with slower dissociation rates, if the initial complex concentration was the same (Fig. 1). Similarly, the non-binding proteins would be washed away due to their fast dissociation rates (Fig. 1).
Figure 1.
Dissociation of proteins from the NPs in non-equilibrium F4 allows for differentiation between the slowly and rapidly dissociating NP-protein complexes. Grey circles – NPs. Black circles – proteins.
To get an idea of how soon complex dissociation would occur on column, we estimated how quickly the representative protein, HSA, would diffuse out of the zone containing the PAA-SPION-HSA complex, using the simple, one-dimensional diffusion of HSA towards the channel center in the direction perpendicular to the membrane surface. Under this condition, the time (t) needed for the protein to diffuse a mean distance can be calculated by the relationship of (diffusion distance2 = 2Dt, with D being the diffusion coefficient of HSA.
In F4, the mean height of the sample zone on top of the membrane, l, can be calculated using the equation shown below:
| (1) |
in which t0 is the void time; tr is the retention time; and w is the effective channel thickness.38 In our system, t0 was 1.19 ± 0.02 min and w was 0.236 mm (Experimental Section). With an average tr of 7.02 ± 0.16 min (Figure 2), the l of the PAA-SPION-HSA complex was then calculated to be 6.67 ± 0.15 µm. The D of HSA in water has been reported to be 6.1×10−7 cm2/sec in water.39,40 Thus, for HSA to diffuse out of the complex zone with a gravity center at 6.67 µm above the accumulation wall, it would take only 0.36 sec. If the time scale for HSA binding to the PAA-SPIONs is shorter than 0.36 sec, the complex would not be able to reform due to protein diffusing out of the sample zone. The low possibility of complex reformation can also be seen from the large retention time difference between HSA and the PAA-SPION-HSA complex: HSA had a tr of 1.78 ± 0.20 min under the flow rates used in the study (data not shown), and would be well resolved from the complex peak centered at 7.02 ± 0.16 min.
Figure 2.
(a) F4 fractograms of the PAA-SPIONs incubated with various single proteins, with an increase in the retention time indicating an increase in the hydrodynamic diameter due to protein adsorption. (b) SDS-PAGE of single protein incubations with the PAA-SPIONs collected by F4 and centrifugation, showing F4’s ability to isolate only proteins with slow interaction kinetics. H – human serum albumin. B – β-casein. S – succinyl concanavalin A.
The rapid diffusion of free proteins from the complex zone causes continuous dissociation of the SPION-protein complexes. Thus, if the initial particle-bound protein amounts are comparable, the proteins that dissociate from the SPIONs more rapidly will be collected at much lower amounts, or even not-detectible, in the eluent containing the SPION-protein complexes than those with much slower dissociation rates.
Compared to the <10 minutes separation in F4, centrifugation, took much longer time, ~ 30 min, to precipitate the SPIONs used in our study. It may not provide quick enough separation of the free proteins and the complex, and re-association of the proteins and the SPIONs could occur during the long isolation. Within this long period, re-equilibrium may even be reached if the interacting proteins have fast enough association/dissociation rates, although the complex concentrations were reduced as the result of sample dilution by the wash solution. Those proteins could be collected together with the NPs in the pellet. The non-binding ones, due to their slow association/fast dissociation rate profile, would still be washed off during multiple rounds of washes.
Single protein adsorption on SPIONs
To investigate the capability of F4 in removing the proteins with fast dissociation rates from the particles, the PAA-SPIONs were incubated with several single proteins, the binding of which was initially probed by CE. Our group has developed a quick, simple CE method of measuring the binding between individual protein and NPs.23 Using this CE method, we found that, calmodulin and succinyl ConA did not bind to the PAA-SPIONs; β-casein bound with a fast on-and-off rate; and the complex formed between HSA and the particles did not dissociate to a noticeable level within the time scale of CE separation (Supporting Information Figure S3).
Upon injection into the F4 system, only those proteins capable of binding to the PAA-SPIONs showed migration time shift in the F4 fractogram (Figure 2a), which indicated change in particle diameter upon protein binding. Beta-casein and HSA increased the hydrodynamic diameter of the particles by 9% (from 17.4 to 19.0 nm) and 18% (to 20.5 nm), respectively (Table 1). Calmodulin and ConA, being non-interacting, did not induce statistically significant size change to the PAA-SPIONs.
Table 1.
Diameter increases of AMP-SPION incubated with single proteins measured by F4.
| Analyte | RT (min) | Diameter (nm) | Diameter increase (nm) (% increase) |
|---|---|---|---|
| PAA-SPION | 5.95 ± 0.09 | 17.4 ± 0.3 | N/A |
| Inc. with ConA | 5.85 ± 0.21 | 17.1 ± 0.6 | −0.3 (−2%) |
| Inc. with Calm | 6.07 ± 0.24 | 17.7 ± 0.7 | 0.3 (2%) |
| Inc. with β-Cas | 6.48 ± 0.12 | 19.0 ± 0.4 | 1.6 (9.2%) |
| Inc. with HAS | 7.02 ± 0.16 | 20.5 ± 0.5 | 3.1 (18%) |
The SPION-protein complexes isolated by either centrifugation or F4 were analyzed by SDS-PAGE to reveal the presence of the proteins in the complex collection (Figure 2b). While both SPION-binding proteins were found in the centrifugation collection, only HSA was collected along with the SPIONs in F4. However, the band intensity of HSA collected in F4 reduced by more than 3 fold when quantified by Image J (data not shown). Several reasons can be accounted for the reduction in protein collection. The recovery of PAA-SPION in F4 was only 40%, lower than the 80% recovery by centrifugation (measured by both UV-Vis absorbance and inductively coupled plasma – atomic emission spectroscopy). The additional HSA loss could be attributed to the non-equilibrium F4 process: although the protein has a slow-dissociation rate, some of the dissociated HSA would be removed from the SPIONs and not collected at the end of F4.
Adsorption on PAA-SPION when incubated with a protein mixture
To further confirm F4’s ability to distinguish proteins dissociating from the particles with relatively slow or fast rates, the PAA-SPIONs were incubated with a mixture of HSA, β-casein, and ConA. The ratios of HSA:β-casein:ConA in the mixture were 10:1:1, 5:1:1, 1:1:1, and 0.1:1:1, with the amount of the protein with a slower dissociation rate, i.e. HSA, decreasing from 10-fold higher to 10-fold lower than others. Judged from the fractograms, HSA was the deciding factor for the final size of the SPION-protein complex: the hydrodynamic diameter calculated from the elution time in F4 increased linearly with the molar fraction of HSA in the protein mixture (Supporting Information Figure S4a). In the mixture with the highest HSA concentration, a 17% size increase (from 12.9 to 15.0 nm) was gained by the SPIONs.
The collected samples were analyzed by SDS-PAGE (Supporting Information Figure S4b). Similar to the single protein incubations, the centrifugation collections possessed both β-casein and HSA; and the F4 collections contained only HSA. This study proved that even in a more complex protein sample, only the proteins dissociating from the particles with relatively slow rates were successfully isolated by F4. The rapidly dissociating proteins, such as β-casein, were not collected by F4 even when the starting concentration was 10 fold higher than HSA.
Protein adsorption on SPIONs in depleted serum
Next, we tested the capability of F4 to screen for NP-protein interactions based on their relative dissociation rates from a complex biological matrix. AMP-SPIONs were selected for this step of the study. They are commercially available and supplied with high-quality and detailed characterization documents, and thus will be more widely used in research labs than the synthesized-in-lab PAA-SPIONs. Similar to the PAA-SPIONs, these particles were coated with polymers having high density of carboxyl groups that rendered high particle stability in aqueous and salty solutions. The SPIONs were incubated with the serum that had been depleted of albumin and IgG, two of the most abundant components in human serum. Removing the highly abundant components gives more comparable concentrations of the remaining proteins. Since the final amount of the recovered protein depends both on the dissociation rate constant as well as the initial protein amount in the mixture, leveraging the initial amounts of proteins could make the final recovered protein amounts more closely reflecting the difference in dissociation rate constants. Additionally, using the depleted serum could be beneficial for discovery of the proteins present at lower abundance but bound with higher affinities than HSA and IgG; the binding of these low abundance proteins to SPIONs have been commonly found in other studies.
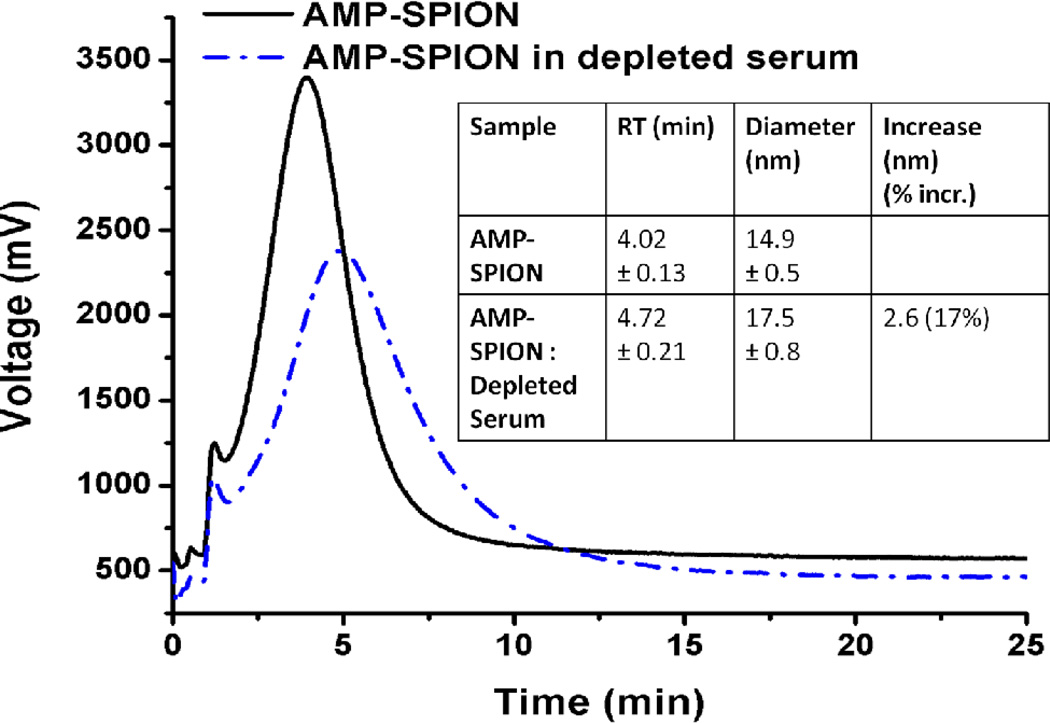
The hydrodynamic diameter of the SPIONs after incubation with the depleted serum increased by 17%, from 14.9 ± 0.5 nm to 17.5 ± 0.8 nm, based on the migration times in F4 (Figure 3). After incubation and collection by both F4 and centrifugation, the proteins co-isolated with the AMP-SPIONs were digested and subjected to 2D nano-LC-MS/MS analysis for protein identification.
Figure 3.
F4 fractograms of AMP-SPION before and after incubation with the serum sample depleted of IgG and HSA. Inset – Diameter increase of the AMP-SPIONs after protein binding determined by F4.
A total of 53 non-homologous protein hits were found in the collections. The protein list was then narrowed down through additional manual removal of the homologous proteins, as well as elimination of proteins not found in human serum (determined through searching in the Plasma Proteome Database) (Supporting Information Table S1).41 Of the 20 serum proteins found to bind to the AMP-SPIONs, 11 were believed to be proteins with relatively slow rates dissociating off the SPIONs, based on their presence in both the centrifugation and F4 collections. The remaining proteins were believed to bind to the AMP-SPIONs with relatively fast on-and-off rates, because they were absent from the F4 collections.
Binding confirmation using CE and CD spectroscopy
In order to confirm the dissociation rate situation observed in the F4-centrifugation study, CE was used to assess the binding between the AMP-SPIONs and a few selected serum proteins. Alpha-1-antitrypsin (A1AT), haptoglobin, and transferrin were chosen to represent the proteins dissociating from the SPIONs with relatively slow rates; and apolipoprotein A1 (APOA1) was the representative for the proteins rapidly dissociating off the SPIONs. Pre-albumin was not found in either centrifugation or F4-based collections of the protein-bound 10-AMP particles, and thus acted as a non-binding model protein.
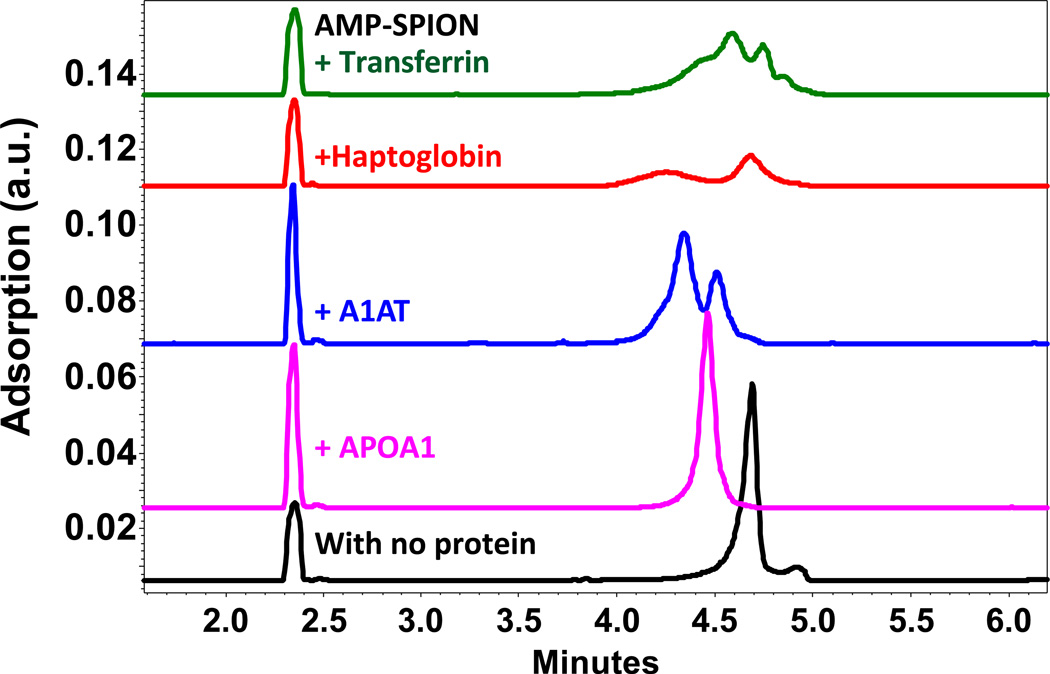
The samples were incubated in a 3:1 protein:SPION molar ratio overnight before being analyzed by CZE (Figure 4), in which the SPION-protein complex with slow enough dissociation rates, i.e. longer half-life than the separation time, should form a complex peak separable from the SPIONs peak and the proteins binding to the SPIONs with fast on-and-off rates would show a migration time shift of the particle peak. Of the five proteins, A1AT, haptoglobin and transferrin formed observable complex peaks with the AMP-SPIONs; APOA1 caused migration time shift of the SPIONs; and pre-albumin did not induce any change in the separation profile. The Kd values of the interacting proteins measured by CZE or ACE were displayed in Table 2. It is worth of notice that, APOA1, although dissociating more rapidly from the SPIONs than other proteins, had the strongest binding affinity to the SPIONs.
Figure 4.
Electropherograms of the SPION-protein complexes detected by CZE in 10 mM borate (pH 8.5), with a 3:1 molar ratio of protein to nanoparticles. The complex dissociating slowly (those with transferring, haptoglobin, or A1AT) showed a separable peak from the particle itself; and the complex with a fast dissociation rate (with APOA1) was detected as a shift of the peak for the SPIONs.
Table 2.
Confirmation of SPION-protein binding situations revealed by the F4-centrifugation method, using CE.
| Protein | Centri- fugation? |
F4 Isolation? |
Binding situation detected by CE |
CE Method | Kd(µM) |
|---|---|---|---|---|---|
| Antitrypsin | X | X | Slow dissociation | CZE | 1.01 ± 0.10 |
| Transferrin | X | X | Slow dissociation | CZE | 0.233 ± 0.011 |
| Haptoglobin | X | X | Slow dissociation | CZE | 0.612 ± 0.074 |
| Apolipoprotein Al | X | Fast dissociation | ACE | 0.155 ± 0.001 | |
| Prealbumin | Non-binding | CZE, ACE | Non-binding |
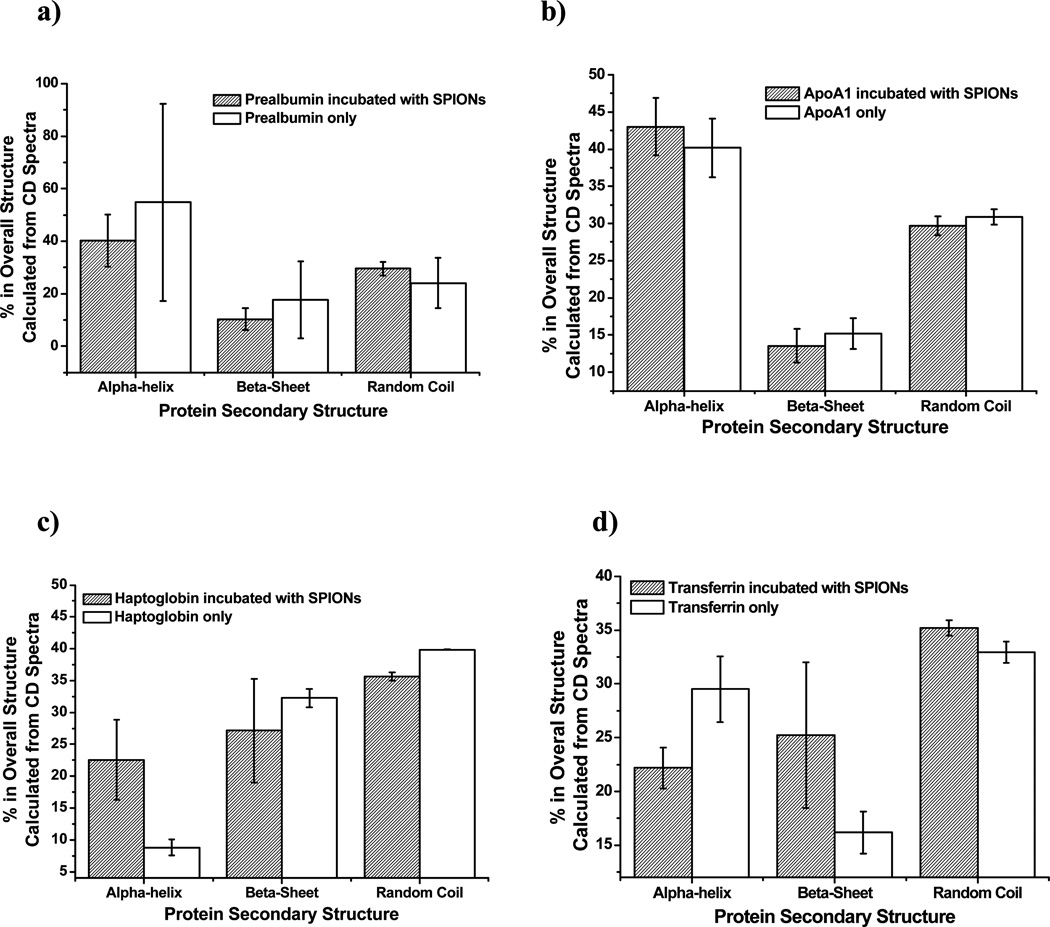
In addition to CE, CD measurements of prealbumin, APOA1, transferrin and haptoglobin incubated with the AMP-SPIONs was conducted (Figure 5). We hypothesized that the proteins with fast on-and-off rates when forming complexes with SPIONs would experience less change in their secondary structure compared to those with relatively slower dissociation rates, because they were constantly changing in between SPION-bound and -unbound states. Indeed, in the case of the prealbumin (non-binding) (Fig. 5a) and APOA1 (fast dissociation) (Fig. 5b), there were no statistically significant changes in the secondary structure of the protein after incubation with the AMP-SPIONs. However, the two proteins dissociating relatively slowly from the SPIONs (haptoglobin (Fig. 5c) and transferrin (Fig. 5d) showed noticeable changes in their secondary structure after incubation. A decrease in the helical structure, as well as an increase in the random structure, was observed in haptoglobin. Transferrin displayed an increase in the helical structure and a decrease in both the beta-sheet and random coil structure (Supporting Information Figure S5). CD, although able to recognize NP-protein interactions with slow dissociation rates, is unable to detect the interactions with fast dissociation rates, which could actually have strong affinity. This perspective of CD is similar to F4, but CD can only measure the binding of one protein, and consumes larger amounts of material to overcome its low signal intensity and the high background noise resulted from the SPIONs.
Figure 5.
Percent changes in the protein secondary structures upon binding with the SPIONs calculated from CD spectra for a) prealbumin; b) ApoA1; c) haptoglobin; and d) transferrin.
Conclusions
In this study, it was determined that F4 can be used as an effective tool for screening the NP-protein interactions based on their relative dissociation rates, when used in parallel with centrifugation. Because proteins with relatively slow dissociation rates off the NPs are rapidly exchanging between the particle-bound or –unbound status, they could reach binding equilibrium faster than others when the NPs are exposed to a new biological environment, and be replaced more quickly with the free matrix proteins if the NPs are passing in between matrices. They should be the main contributors to the dynamic feature of the protein corona of NPs. Thus, our screening method can assist with the evolution study of protein corona in different biological matrices. The higher speed of F4 than gel filtration chromatography and its higher sample loading than CE make it an approach of higher throughput in study of the interaction between proteins and NPs. It will be helpful for rapid assessment of how the interaction kinetics would be altered by the properties of NPs and proteins.
There is no doubt that further investigation is needed to explore the full potential of this method in probing the kinetic feature of NP-protein interactions from a highly complex protein mixture. Because the final recovered protein amount depends on both the initial bound protein amount as well as the dissociation rate constant, quantification of protein abundances in the original matrix and in both the centrifugation and F4 collections will be extremely valuable for more accurate interpretation of the collection profile in F4: is the protein recovered in F4 because of a slow dissociation rate or due to a much higher abundance than others? Coupled with the appropriate protein quantification techniques, this method will also be used for measurement of NP-protein dissociation rate constants in a high-throughput manner when mixing the proteins in interest at the same concentrations and allowing the binding to reach equilibria. Application of this method to investigate other interactive systems is also possible.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Environmental Health Sciences Grant No. 1R21ES017870-01A1. Jonathan Ashby was supported by the National Science Foundation Graduate Research Fellowship under Grant Number DGE-0813967. The authors are also very grateful for the support of Dr. Yadong Yin on SPION synthesis, as well as UCR’s Institute for Integrative Genome Biology for the usage of the Typhoon imager and MUDPIT analysis.
Footnotes
Assorted Content:
Supporting Information. Methods used for protein analysis, SPION physical parameters, measurement of the effective channel thickness, CE of proof-of-concept proteins, simple protein mixture F4 analysis, identification of SPION-protein corona, circular dichroism spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, Tamanoi F, Zink JI. ACS Nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuberger T, Schopf B, Hofmann H, Hofmann M, von Rechenberg B. J. Magn. Magn. Mater. 2005;293:483–496. [Google Scholar]
- 3.Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 4.Schulze C, Kroll A, Lehr CM, Schafer UF, Becker K, Schnekenburger J, Isfort CS, Landsiedel R, Wohlleben W. Nanotoxicology. 2008;2:51–U17. [Google Scholar]
- 5.Montes-Burgos I, Walczyk D, Hole P, Smith J, Lynch I, Dawson K. J. Nanopart. Res. 2010;12:47–53. [Google Scholar]
- 6.Lynch I, Dawson KA. Nano Today. 2008;3:40–47. [Google Scholar]
- 7.Casals E, Puntes VF. Nanomedicine. 2012;7:1917–1930. doi: 10.2217/nnm.12.169. [DOI] [PubMed] [Google Scholar]
- 8.Monopoli MP, Aberg C, Salvati A, Dawson KA. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 9.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Weise S, Hafner M, Rocker C, Zhang F, Parak WJ, Nienhaus GU. J. R. Soc. Interface. 2010;7:S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keck CM, Jansch M, Mueller RH. Pharmaceutics. 2013;5:36–68. doi: 10.3390/pharmaceutics5010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreuter J, Alyautdin RN, Kharkevich DA, Ivanov AA. Brain Res. 1995;674:171–174. doi: 10.1016/0006-8993(95)00023-j. [DOI] [PubMed] [Google Scholar]
- 13.Wohlfart S, Gelperina S, Kreuter J. J. Controlled Release. 2012;161:264–273. doi: 10.1016/j.jconrel.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dell'Orco D, Lundqvist M, Celilia O, Cedervall T, Linse S. PloS One. 2010;5:e10949. doi: 10.1371/journal.pone.0010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Foglia P, Amenitsch H, Lagana A. Langmuir. 2011;27:15048–15053. doi: 10.1021/la202912f. [DOI] [PubMed] [Google Scholar]
- 17.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 18.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes VF. Small. 2011;7:3479–3486. doi: 10.1002/smll.201101511. [DOI] [PubMed] [Google Scholar]
- 19.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 21.Li LW, Mu QX, Zhang B, Yan B. Analyst. 2010;135:1519–1530. doi: 10.1039/c0an00075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagner JE, Qian X, Lopez MM, Dordick JS, Siegel RW. Biomaterials. 2012;33:8503–8516. doi: 10.1016/j.biomaterials.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Li N, Zeng S, He L, Zhong W. Anal. Chem. 2010;82:7460–7466. doi: 10.1021/ac101627p. [DOI] [PubMed] [Google Scholar]
- 24.Giddings JC. Science. 1993;260:1456–1465. doi: 10.1126/science.8502990. [DOI] [PubMed] [Google Scholar]
- 25.Rambaldi DC, Reschiglian P, Zattoni A. Anal. Bioanal. Chem. 2011;399:1439–1447. doi: 10.1007/s00216-010-4312-5. [DOI] [PubMed] [Google Scholar]
- 26.Li JS, Ge JP, Yin YD, Zhong WW. Anal. Chem. 2008;80:7068–7074. doi: 10.1021/ac801251y. [DOI] [PubMed] [Google Scholar]
- 27.Pollastrini J, Dillon TM, Bondarenko P, Chou RYT. Anal. Biochem. 2011;414:88–98. doi: 10.1016/j.ab.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Yohannes G, Wiedmer SK, Tuominen EKJ, Kinnunen PKJ, Riekkola ML. Anal. Bioanal. Chem. 2004;380:757–766. doi: 10.1007/s00216-004-2842-4. [DOI] [PubMed] [Google Scholar]
- 29.Ge JP, Hu YX, Biasini M, Dong CL, Guo JH, Beyermann WP, Yin YD. Chem-a. Eur. J. 2007;13:7153–7161. doi: 10.1002/chem.200700375. [DOI] [PubMed] [Google Scholar]
- 30.Wolters DA, Washburn MP, Yates JR. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 31.Raussens V, Ruysschaert JM, Goormaghtigh E. Anal. Biochem. 2003;319:114–121. doi: 10.1016/s0003-2697(03)00285-9. [DOI] [PubMed] [Google Scholar]
- 32.Berezovski M, Krylov SN. J. Am. Chem. Soc. 2002;124:13674–13675. doi: 10.1021/ja028212e. [DOI] [PubMed] [Google Scholar]
- 33.Berezovski MV, Okhonin V, Petrov A, Krylov SN. Proc. SPIE- Int. Soc. Opt. Eng. 2005;5969:59690Y/59691-59690Y/59613. [Google Scholar]
- 34.Krylov SN. J. Biomolecular Screening. 2006;11:115–122. doi: 10.1177/1087057105284339. [DOI] [PubMed] [Google Scholar]
- 35.Krylov SN. Electrophoresis. 2007;28:69–88. doi: 10.1002/elps.200600577. [DOI] [PubMed] [Google Scholar]
- 36.Krylov SN, Berezovski M. Analyst. 2003;128:571–575. doi: 10.1039/b212913b. [DOI] [PubMed] [Google Scholar]
- 37.Krylova SM, Dove PM, Kanoatov M, Krylov SN. Anal. Chem. 2011;83:7582–7585. doi: 10.1021/ac2018876. [DOI] [PubMed] [Google Scholar]
- 38.Wahlund KG, Nilsson L. In: Field-Flow Fractionation in Biopolymer Analysis. Williams SKR, Caldwall KD, editors. Springer-Verlag/Wien; 2012. pp. 1–21. [Google Scholar]
- 39. http://www.sigmaaldrich.com/content/dam/sigmaaldrich/docs/Sigma/Product_Information_Sheet/a9511.pis.pdf.
- 40.Putnam FW, editor. The Plasma Proteins. Vol. 1. New York: Academic Press; 1975. pp. 133–181. [Google Scholar]
- 41.Muthusamy B, Hanumanthu G, Suresh S, Rekha B, Srinivas D, Karthick L, Vrushabendra BM, Sharma S, Mishra G, Chatterjee P, Mangala KS, Shivashankar HN, Chandrika KN, Deshpande N, Suresh M, Kannabiran N, Niranjan V, Nalli A, Prasad TSK, Arun KS, Reddy R, Chandran S, Jadhav T, Julie D, Mahesh M, John SL, Palvankar K, Sudhir D, Bala P, Rashmi NS, Vishnupriya G, Dhar K, Reshma S, Chaerkady R, Gandhi TKB, Harsha HC, Mohan SS, Deshpande KS, Sarker M, Pandey A. Proteomics. 2005;5:3531–3536. doi: 10.1002/pmic.200401335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.