Abstract
The ubiquitin system regulates virtually all aspects of cellular function. We report a method to target the myriad enzymes that govern ubiquitination of protein substrates. We used massively diverse combinatorial libraries of ubiquitin variants to develop inhibitors of four deubiquitinases (DUBs) and analyzed the DUB-inhibitor complexes with crystallography. We extended the selection strategy to the ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes and found that ubiquitin variants can also enhance enzyme activity. Last, we showed that ubiquitin variants can bind selectively to ubiquitin-binding domains. Ubiquitin variants exhibit selective function in cells and thus enable orthogonal modulation of specific enzymatic steps in the ubiquitin system.
The biological importance of the ubiquitin system for protein posttranslational modification rivals and may exceed that of phosphorylation (1, 2). Consequently, there is great interest in deciphering the role of protein ubiquitination and deubiquitination in both normal and disease states (1). Ubiquitin (Ub) is a small, highly conserved protein that is covalently conjugated to proteins through a cascade of enzyme activities, E1→E2→E3, which can be read by a cohort of Ub-binding domains (UBDs) and reversed by deubiquitinating enzymes (DUBs). Ub monomers and chains of different linkages can be assembled on the substrate, and this complex code can target the substrate for degradation by the 26S proteasome or alter its interactions, localization, or activity (3).
The 58 human ubiquitin-specific proteases (USPs) (4), which represent more than half of the known DUBs, have attracted attention as potential therapeutic targets (5, 6). Numerous USPs have been implicated in cancer and other pathologies, including neurodegenerative, haematological, and infectious diseases (7–13). USPs are cysteine proteases that share a structurally conserved catalytic domain with a well-defined catalytic cleft (14). Thus, it may be possible to develop a general structure-based strategy for inhibiting family members by using similar yet specific molecular entities, as has been the case for kinases that have been targeted with small molecules built on common scaffolds (15). However, only a few weak inhibitors of USPs or other DUBs have been reported (16–18). The paucity of specific inhibitors for DUBs has hampered attempts to understand and manipulate deubiquitination pathways for therapeutic benefit.
We developed a strategy to use Ub as a scaffold to generate highly specific and potent inhibitors of USPs and then extended the approach to target other DUB families, E2 enzymes, E3 enzymes, and UBDs. USP substrates contain a proximal Ub moiety that is conjugated through its C-terminal carboxyl group to a lysine in a target Ub or protein substrate (19, 20). Despite low sequence similarity, USP catalytic domains share a common fold that includes a structurally conserved Ub-binding site for the proximal Ub.
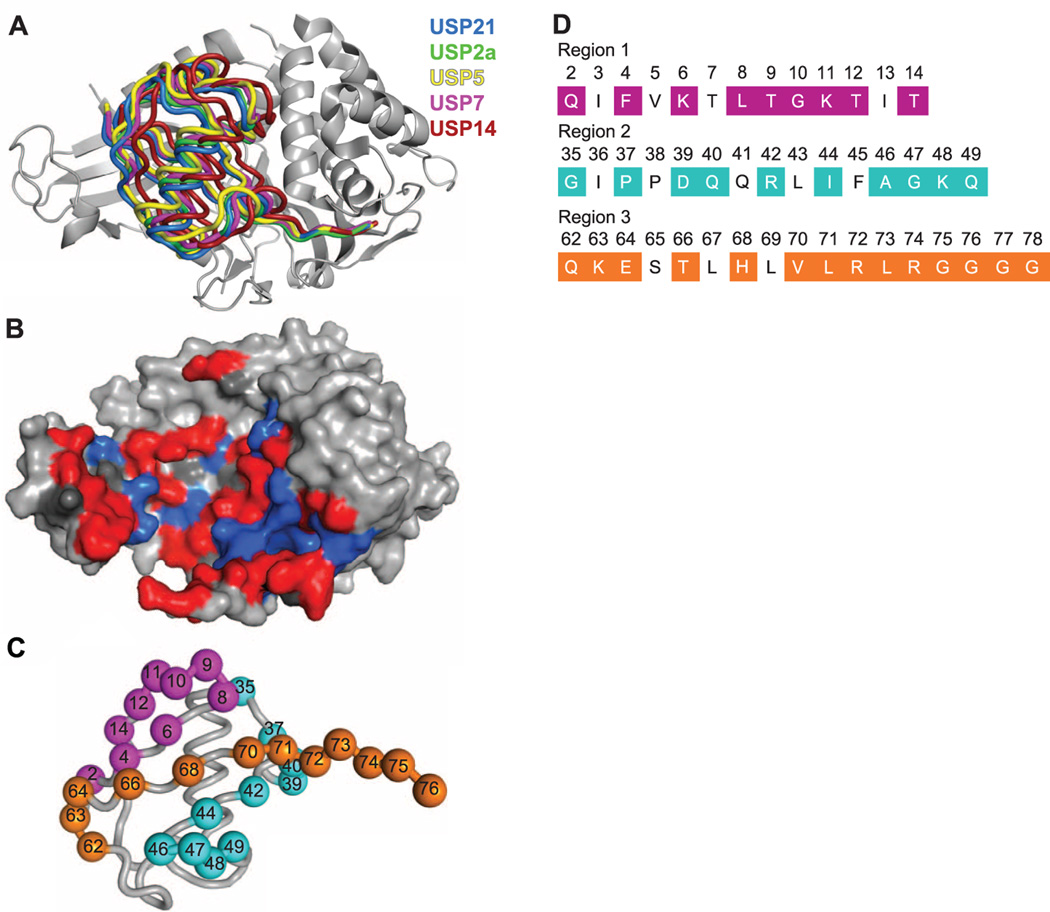
To aid our inhibitor design strategy, we solved the structure of USP21 in complex with a suicide-substrate Ub moiety that forms a covalent bond between its C terminus and the catalytic cysteine of the enzyme (table S1) (21, 22). Superposition of the Ub:USP21 complex structure with structures of four other Ub:USP complexes shows that Ub occupies a similar site on all five enzymes (Fig. 1A). The contact surface between Ub and the USPs is large; for example, 1835 and 1645 Å2 are buried on the surfaces of Ub and USP21, respectively. Approximately 75% of the Ub-binding surface on USP21 is composed of residues that are not conserved within the USP family (Fig. 1B and fig. S1).
Fig. 1.
Structural rationale and library design for Ub-based USP inhibitors. (A) Superposition of Ub in complex USP21 (blue; PDB entry 3I3T), USP2a (green; PDB entry 2HD5), USP5 (yellow; PDB entry 3IHP), USP7 (magenta; PDB entry 1NBF), and USP14 (red; PDB entry 2AYO) (22, 45, 46). The superposition was performed with complete coordinates for the Ub:USP complexes, but for clarity, only USP21 is shown (gray). (B) The Ub-binding site of USP21. USP21 is shown as a molecular surface, and residues that form the Ub-binding site are colored red or blue, indicating residues that are <50% or ≥50% conserved, respectively, in the sequences of 48 human USPs (fig. S1). (C) The phage-displayed Ub library design mapped onto the Ub structure (PDB entry 1UBQ). The Ub main chain is shown as a gray tube, and positions that were diversified in the libraries are shown as spheres, colored as follows: region 1, purple; region 2, blue; and region 3, orange. (D) The primary sequence of the regions targeted in the library design. Diversified sequences are shaded and colored as in (C). Two libraries were constructed. Amongst the shaded sequences, library 1 (7 × 1010 distinct variants) did not target six of the positions in region 3 (73 to 78), and library 2 (9 × 109 distinct variants) did not target four of the positions in region 2 (35, 37, 39, and 40).
Because Ub binds to USPs with low affinity but through a large contact area, we reasoned that we should be able to make mutations in Ub that optimize intermolecular contacts and enhance affinity for a particular USP. Because the Ub-binding sites of USPs vary in sequence, Ub variants that bind tightly to one USP should not interact with other USPs. Tight binding Ub variants should act as competitive inhibitors of catalytic activity by blocking the recognition of natural ubiquitinated substrates. To test our hypothesis, we constructed combinatorial, phage-displayed libraries of Ub variants for selection against different USPs and other Ub-associated proteins.
We defined the USP-binding site as ~30 Ub residues that make contact with USP21 in theUb:USP21 complex structure (Fig. 1, C and D). We constructed two phage-displayed libraries using a soft randomization strategy that favored the wild-type (wt) sequence but allowed for diversity across the entire Ub-binding site (23, 24). This approach enabled the selection of variants with mutations that improve affinity for a particular USP without drastically altering the binding interaction.
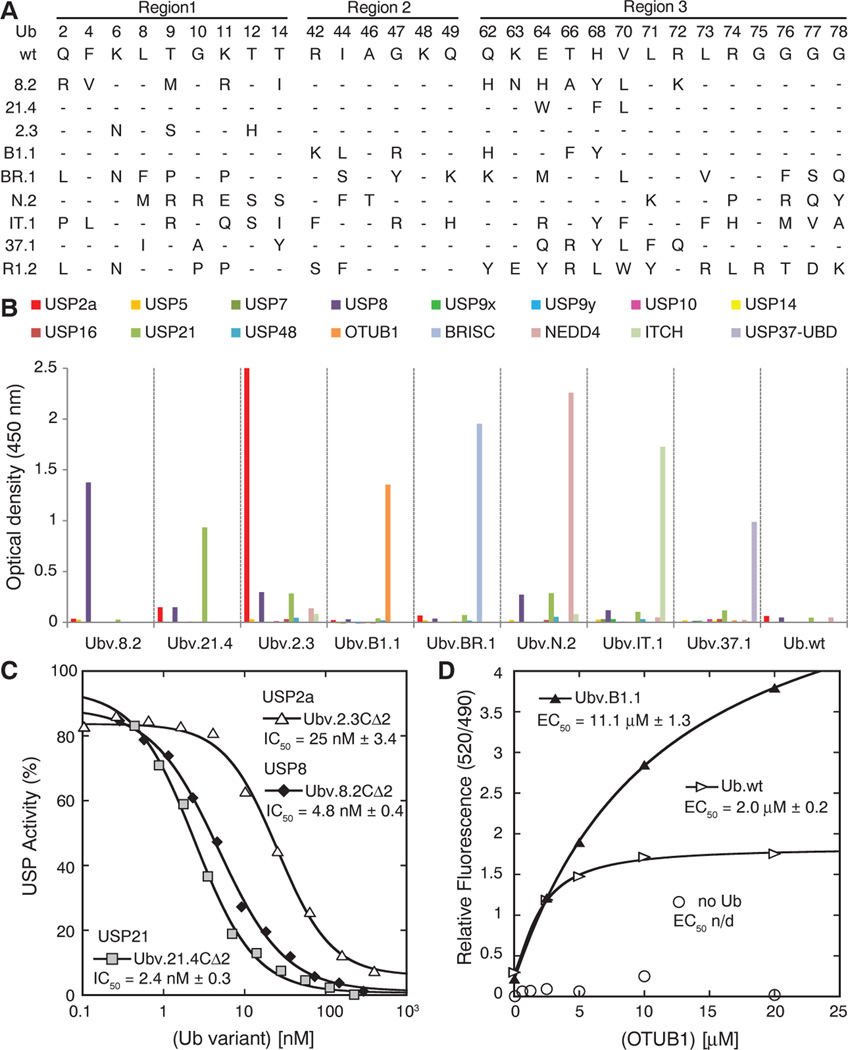
Binding selections yielded variants that bound to either USP8, USP21, or USP2a (Fig. 2A and table S2) but not to 10 other USPs (Fig. 2B).We also isolated variants that bound selectively to members of two other DUB families: the ovarian tumor protease (OTU) family member OTUB1 (25, 26) and the JAB1/MPN/MOV34 metallo-enzyme(JAMM)family member Brcc36-containing isopeptidase complex (BRISC) (27, 28).We also identified variants that bound selectively to the homologous to the E6-AP carboxyl terminus (HECT) domains of the E3 enzymes neural precursor cell expressed developmentally down-regulated protein 4 (NEDD4) or itchy homolog E3 ubiquitin protein ligase (ITCH) (29, 30), to the noncatalytic UBDs of USP37 (USP37-UBD) (31, 32), and to the E2 enzyme Cdc34 (Fig. 2, A and B, and table S2) (33).
Fig. 2.
Selective binding of Ub variants to DUBs, Ub ligases, and UBDs, and inhibition of DUB function in vitro. (A) Sequence alignment of Ub.wt and representative Ub variants selected for binding to USP8 (8.2), USP21 (21.4), USP2a (2.3), OTUB1 (B1.1), the BRISC protein complex (BR.1), NEDD4 (N.2), ITCH (IT.1), USP37-UBD (37.1), or Cdc34 (R1.2). The alignment shows only those positions that were diversified in the Ub library, and positions that were conserved as the wt sequence are indicated by dashes. A complete list of selected variants is provided in table S2. (B) Ub variants bind selectively to their cognate targets, as shown by phage enzyme-linked immunosorbent assays for binding to the following immobilized proteins (color coded as indicated in the panel): USP2a, USP5, USP7, USP8, USP9x, USP9y, USP10, USP14, USP16, USP21, USP48, OTUB1, BRISC, NEDD4, ITCH, and USP37-UBD. Bound phage were detected spectrophotometrically (optical density at 450 nm), and background binding to neutravidin was subtracted from the signal. (C) Inhibition of USP2a, USP28, or USP21 shown as dose-response curves for Ub variants. The IC50 value was determined as the concentration of Ub variant that reduced USP activity by 50%. (D) The effects of Ubv.B1.1 or Ub. wt on binding of OTUB1 to Ub~UbcH5B measured as relative fluorescence by means of time-resolved Förster-energy transfer. The median effective concentration (EC50) value is defined as the half-maximal effective concentration of OTUB1.
Ub variants that bound selectively to USP8, USP21, or USP2a were purified as free proteins in a truncated form lacking the last two glycines that are required for conjugation to substrates (19). In vitro proteolysis assays with the substrate Ub-AMC (34) showed that each Ub variant potently inhibited its cognate USP [median inhibitory concentration (IC50) = 4.8 nM, 2.4 nM, or 25 nM for Ubv.8.2CΔ2, Ubv.21.4CΔ2, or Ubv.2.3CΔ2, respectively] (Fig. 2C). In contrast, neither the variants, nor Ub.wt, inhibited non-cognate USPs (IC50 >> 1 µM) (fig. S2). Purified Ubv.B1.1 bound to OTUB1 with a dissociation constant (Kd) of 20 nM (fig. S3A) and efficiently inhibited the cleavage of K48-linked di-Ub (fig. S3B), which is consistent with a substrate-competitive binding mode. OTUB1 binds to the activated E2-conjugating enzyme Ub~UbcH5b, and binding is enhanced by Ub.wt (35). Although Ubv.B1.1 binds to OTUB1 much more tightly than does Ub.wt, it shows a compromised ability to promote complex formation (Fig. 2D), suggesting an alteration in binding mode to OTUB1 relative to Ub.wt.
To elucidate the molecular details of enzyme inhibition,we determined the structures ofUSP21, USP8, USP2a, and OTUB1 in complex with their cognate inhibitors at 2.0, 2.6, 1.85, or 2.5 Å, respectively (Fig. 3 and table S1).
Fig. 3.
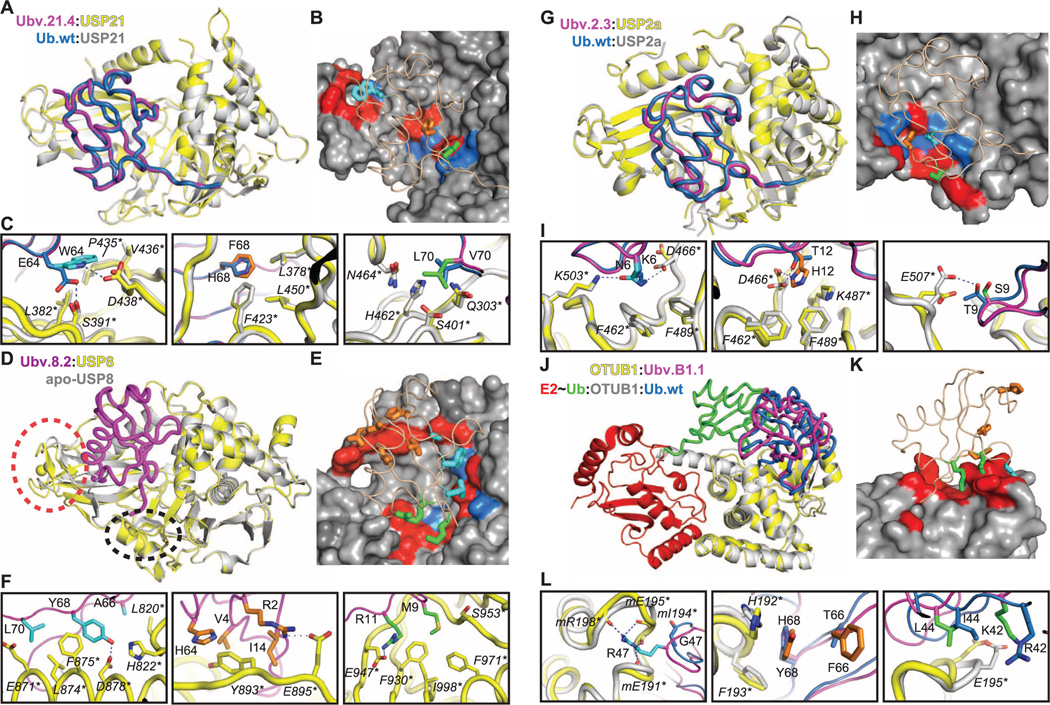
Molecular basis for DUB inhibition by Ub variants. (A) Superposition of USP21 (yellow) in complex with Ubv.21.4 (purple) and USP21 (gray) in complex with Ub.wt (blue). (B) Surface representation of the Ub-binding site of USP21 in complex with Ubv.21.4 (wheat tube). Residues in Ubv.21.4 that are mutated relative to Ub.wt are shown as colored sticks, and residues on the USP21 surface that make contact with these residues are colored red or blue if they are <50% or ≥50% conserved in the sequences of 48 human USPs, respectively. (C) Details of the superposition of USP21 in complex Ubv.21.4 or Ub.wt, showing the changes in molecular interactions caused by the mutations at positions 64 (left), 68 (center), and 70 (right). The Ubv.21.4 side chains are colored as in (B), and Ub.wt side chains are colored blue. Main chains and USP21 side chains are colored as in (A). Residues are numbered according to the PDB files, and asterisks indicate USP residues. (D) Superposition of USP8 (yellow) in complex with Ubv.8.2 (purple) and apo-USP8 (gray). Dashed ovals demarcate structural changes in the zinc finger region (red) or the BL2 loop (black) of USP8. (E) Surface representation of the Ub-binding site of USP8 in complex with Ubv.8.2 (wheat tube). Residues in Ubv.8.2 that are mutated relative to Ub.wt and make contact with USP8 are shown as colored sticks, and residues on the USP8 surface that make contact with these residues are colored red or blue, as in (B). (F) Details of USP8 in complex with Ubv.8.2, showing molecular interactions involving residues in Ubv.8.2 that are mutated relative to Ub.wt and are in contact with USP8. The Ubv.8.2 side chains are colored as in (E). Main chains and USP8 side chains are colored as in (D). (G) Superposition of USP2a (yellow) in complex with Ubv.2.3 (purple) and USP2a (gray) in complex with Ub.wt (blue). (H) Surface representation of the Ub-binding site of USP2a in complex with Ubv.2.3 (wheat tube). Residues in Ubv.2.3 that are mutated relative to Ub.wt are shown as colored sticks, and residues on the USP2a surface that make contact with these residues are colored red or blue, as in (B). (I) Details of the superposition of USP2a in complex with Ubv.2.3 or Ub.wt, showing changes in molecular interactions caused by the mutations at positions 6 (left), 12 (center), and 9 (right). The Ubv.2.3 side chains are colored as in (H). Main chains and USP2a side chains are colored as in (G). (J) Superposition of OTUB1 (yellow) in complex with Ubv. B1.1 (purple) and OTUB1 (gray) in complex with distal Ub.wt (blue) and an E2-Ub covalent conjugate consisting of E2 conjugating enzyme UbcH5b (red) and Ub.wt (green). (K) Surface representation of the Ub-binding site of OTUB1 in complex with Ubv.B1.1 (wheat tube). Residues in Ubv.B1.1 that are mutated relative to Ub.wt are shown as colored sticks, and residues on the OTUB1 surface that are in contact with these residues are colored red. (L) Details of the superposition of OTUB1 in complex with Ubv.B1.1 or Ub.wt, showing changes in molecular interactions caused by the mutations at positions 47 (left), 66 and 68 (center), and 42 and 44 (right). The Ubv.B1.1 side chains are colored as in (K). Main chains and OTUB1 side chains are colored as in (J).
The structures of USP21 in complex with Ubv.21.4 or Ub.wt were highly similar; the Cα atoms of the USP and Ub moieties superim-pose with root-mean-square deviation (RMSD) of less than 0.7 or 0.4 Å2, respectively (Fig. 3A). Ubv.21.4 was purified with a C-terminal extension, but mass spectrometry of protein sample before crystallization showed a mass consistent with cleavage of the tail after Gly76, the last residue of natural Ub (fig. S4 and table S3). These results indicate that Ubv.21.4 is a substrate for USP21 but is not released after cleavage. This result is consistent with the crystal structure, which shows excellent superposition between the tails of Ubv.21.4 and Ub.wt in the catalytic cleft of the enzyme.
The three mutated residues in Ubv.21.4 contact residues of USP21 that are not conserved in the USP family (Fig. 3B), and this provides a structural explanation for the selectivity of Ubv.21.4 for USP21 (fig. S5). Compared with Ub.wt, two of the three substitutions in Ubv.21.4 serve to remove repulsive interactions and optimize hydrophobic packing with USP21. The Glu64 side chain in Ub.wt is located close to Asp438* of USP21 (USP residues are marked with asterisks), but in Ubv.21.4, this unfavorable charge interaction is abrogated by substitution with Trp64, which makes a hydrogen bond with Asp438* and hydrophobic contacts with the side chains of Pro435* and Val436*. (Fig. 3C, left). The Ub side chain at position 68 packs against three hydrophobic side chains of USP21, and consequently, the hydrophobic Phe68 of Ubv.21.4 appears to make more favorable interactions than those of the polar His68 side chain of Ub.wt (Fig. 3C, center).
We could not solve the structure of USP8 in complex with Ub.wt, but we compared the structure of USP8 in complex with Ubv.8.2 with a previous structure of the USP8 catalytic domain alone (apo-USP8) (36). An auto-inhibited or inactive state was ascribed to apo-USP8 because the Ub-binding site is partially occluded by a zinc finger domain, implying an induced fit mechanism for substrate binding. Overall, the structures of apo-USP8 and USP8 in complex with Ubv.8.2 are similar, and the Cα atoms superimpose with a RMSD of 1.4 Å2 (Fig. 3D). However, there are substantial changes in the zinc finger region, which is opened by ~8 Å (measured by the change in position of Asn980*) in the inhibitor-bound form. Additionally, the BL2 loop adjacent to the active site of the protein is shifted slightly outward by ~3 Å (measured by the change in position of Gly1018*). Surprisingly, Ubv.8.2 is rotated by ~85° and shifted by ~5 Å relative to the positions of the Ub moieties in the complexes with USP21 and other USPs (Figs. 1 and 3). Consequently, the tail of Ubv.8.2 does not extend into the active site cleft, and consistently, mass spectrometry showed no cleavage of the C-terminal extension that was added downstream of Gly76 (fig. S2 and table S3). Thus, Ubv.8.2 binds to a conformation of USP8 that resembles apo-USP8 but in an orientation that differs drastically from known structures of Ub.wt bound to active USPs. Despite these unusual features, Ubv.8.2 occludes the Ub-binding site and potently inhibits USP8 activity.
Nine of the 12 mutated residues in Ubv.8.2 make contacts with USP8, and 70% of these contacts involve nonconserved residues in the USP family (Fig. 3E), likely accounting for the high specificity of Ubv.8.2 for USP8 (Fig. 2B). The tail of Ubv.8.2 contains three mutated contact residues (Ala66, Tyr68, and Leu70) and interacts with an α-helix formed by residues 870* to 882* of USP8 (Fig. 3F, left). A second cluster of contacts involving mutated residues is formed by the interactions of Arg2, Val4, Ile14, and His64 with residues 891* to 895* (Fig. 3F, center). The remaining two mutated contact residues (Met9 and Arg11) are in close proximity (Fig. 3F, right).
The structures of USP2a in complex with Ub.wt or Ubv.2.3 are highly similar (Fig. 3G) because the Cα atoms of the USP and Ub moieties superimpose with a RMSD of 1.0 or 0.3 Å, respectively. The three mutated residues in Ubv.2.3 make contacts with residues that are not conserved in the USP family (Fig. 3H), again explaining the selectivity. Relative to Ub.wt, the three substitutions alter hydrogen-bonding patterns (Fig. 3I). Whereas Lys6 of Ub.wt formed a hydrogen bond with Asp466*, Asn6 of Ubv.2.3 forms a hydrogen bond with Lys503* (Fig. 3I, left). Consequently, Asp466* reorients and forms a hydrogen bond with His12 of Ubv.2.3 (Fig. 3I, middle).
The structure of the OTUB1 OTU domain (OTUB145–271) in complex with Ubv.B1.1 (table S1) reveals that Ubv.B1.1 binds to the distal Ub-binding site of OTUB1 (Fig. 3J) and enables a rationalization of its altered binding and allosteric properties. Compared with a previously published Ub~UbcH5b-OTUB1-Ub complex structure (35), Ubv.B1.1 is rotated by 22° relative to the distal site Ub.wt (Fig. 3J). This deviation in binding mode may account for the compromised ability to allosterically promote binding of Ub~UbcH5b to OTUB1. Four of the six mutated residues in Ubv.B1.1 are in contact with OTUB1 (Fig. 3K) and bind to a turn motif connecting α8 and α9 of OTUB1. Most prominent in this interaction are Leu44, Arg47, and Tyr68, which improve polar and hydrophobic contacts. Arg47 establishes hydrogen bonds to four main chain carbonyl groups in the turn motif (Fig. 3L, left), whereas Tyr68 makes favorable packing interactions with His192* and Phe193* (Fig. 3L, middle). Leu44 establishes van der Waals contacts with the peptide bonds of the α8:α9 loop (Fig. 3L, right). These results demonstrate that high-affinity Ub variants can be developed against specific UBDs.
We investigated the in vivo effects of different variants expressed as C-terminal fusions with Flag-tagged green fluorescent protein in mammalian cells. Mass spectrometric analysis of associated proteins showed that each inhibitor interacted specifically with its cognate USP (table S4).
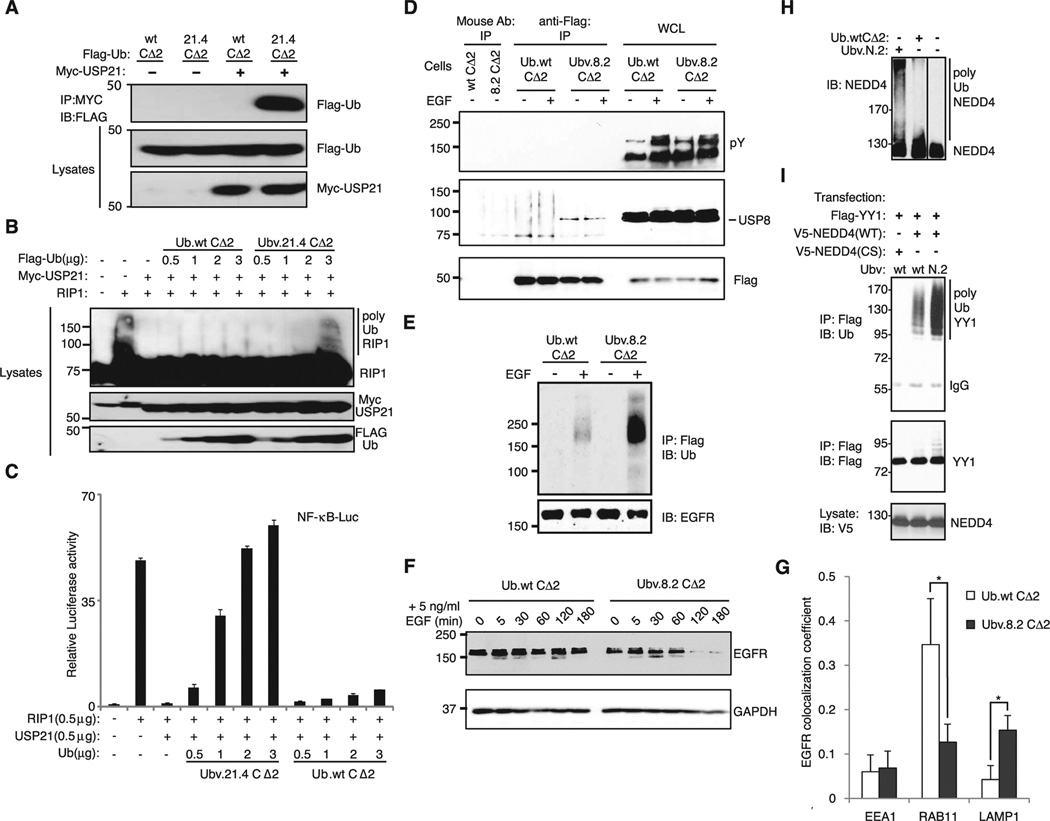
Ubiquitinated receptor-interacting protein 1 (RIP1) is a positive regulator of nuclear factor κB (NF-κB) activation induced by tissue necrosis factor–α (TNFα), and in turn, USP21 has been shown to down-regulate TNFα-induced NF-κB activation by deubiquitinating RIP1 (37). Ubv.21.4CΔ2 coimmunoprecipitated with USP21 in cotrans-fected human embryonic kidney (HEK) 293T cells (Fig. 4A), blocked the deubiquitination of RIP1 by USP21 (Fig. 4B), and restored NF-κB activation (Fig. 4C). No such effects were observed when Ub.wt was cotransfected rather than Ubv.21.4CΔ2. These results show that Ubv.21.4CΔ2 binds and inhibits USP21 in cells and establish that Ub variants can act as orthogonal genetic probes of USP function.
Fig. 4.
Modulation of enzyme function in cells by Ub variants. (A) Flag-tagged Ubv.21.4CΔ2, but not Ub.wtCΔ2, interacts with Myc-tagged USP21, as evidenced by immunoprecipitations with anti-Myc followed by immunoblotting with anti-Flag. (B) Transfection of Ubv.21.4CΔ2, but not Ub.wtCΔ2, causes increased RIP1 ubiquitination. The blot is overexposed so as to reveal the ubiquitination state of RIP1. (C) NF-κB–mediated luciferase activity, which is activated by RIP1 transfection and down-regulated by USP21 cotransfection, is restored by cotransfection of Ubv.21.4CΔ2 but not by cotransfection of Ub.wt CΔ2. (D) Ubv.8.2CΔ2, but not Ub.wtCΔ2, binds to USP8 in an EGFstimulation– independent manner, as shown by means of immunoprecipitation of Flag-tagged Ub variants, followed by immunoblotting with antibodies to USP8. Immunoblotting of whole-cell lysates (WCLs) with anti-phosphotyrosine (pY) and antibodies to USP8 indicates that EGFR phosphorylation and endogenous USP8 concentrations are not affected by the expression of Ub.wtCΔ2 or Ubv.8.2CΔ2. An irrelevant mouse antibody was included as negative control (first two lanes). (E) Transient expression of Ubv.8.2CΔ2, but not Ub.wtCΔ2, leads to increased ubiquitination of EGFR in response to stimulation with EGF (10 ng/ml, 10 min) in HEK293 cells stably expressing Flag-tagged EGFR, as shown by means of immunoprecipitation with anti-Flag antibody from denatured lysates, followed by immunoblotting with anti-Ub antibody. (F) Expression of Ubv.8.2CΔ2, but not Ub.wtCΔ2, leads to decreased levels of endogenous EGFR in response to stimulation with EGF, as shown through analysis of total cellular EGFR levels by immunoblotting with antibody to EGFR. Immunoblotting of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) indicates that similar amounts of sample were probed. (G) Quantification of colocalization of EGFR with EEA1, RAB11, or LAMP observed in immunoflourescence microscopy images. The mean colocalization coefficients, averaged from 10 independent single-cell images, represent pixel overlap between EGFR and EEA1, Rab11, or LAMP. The coefficients vary from 0 to 1, with 0 corresponding to nonoverlapping images and 1 corresponding to 100% colocalization. All error bars represent SD *P < 0.001. (H) Auto-ubiquitination of NEDD4 in vitro. Recombinant full-length NEDD4 was incubated for 3 hours with E1, E2 (UbcH7), adenosine 5´-triphosphate (ATP), Ub, and Ub.wtCΔ2 or Ubv.N.2, and the reaction mixture was immunoblotted with antibody to NEDD4. Ubv.N.2 is not a substrate for the E1–E2–E3 enzyme ubiquitination cascade because its C terminus does not contain a di-glycine motif (Fig. 2A). (I) Ubiquitination of the NEDD4 substrate YY1 in cells. HEK293T cells were transfected with Flag-tagged YY1, V5-tagged NEDD4 (wt or catalytically inactive CS mutant), and Ub.wtCΔ2 or Ubv.N.2. YY1 was immunoprecipitated with antibody to Flag, and gels were immunoblotted for Ub (top) or YY1 (middle). (Bottom) The amount of NEDD4 in the lysates.
USP8 regulates ligand-mediated endocytosis of several receptors (38–40), including the epidermal growth factor receptor (EGFR), de ubiquitination of which protects it from degradation in lysosomes (38). Immunoprecipitated Ubv.8.2CΔ2, but not Ub.wtCΔ2, was associated with endogenous USP8 (Fig. 4D). EGF-stimulated ubiquitination of EGFR in transfected HEK293 cells that express large amounts of Flag-tagged EGFR (~106/cell) (41) was greater in cells expressing Ubv. 8.2CΔ2 than in cells expressing Ub.wtCΔ2 (Fig. 4E). Expression of Ubv.8.2CΔ2 in HEK293 cells, but not Ub.wtCΔ2, resulted in more rapid down-regulation of endogenous EGFR in response to a low dose (5 ng/ml) of EGF (Fig. 4F), which is the expected outcome of USP8 inhibition (38, 42). These data illustrate that Ubv.8.2CΔ2 can modulate EGFR ubiquitination over a broad range of receptor densities.
Confocal microscopy of cells expressing Ubv.8.2CΔ2 or Ub.wtCΔ2 (fig. S6) showed no difference in the colocalization of EGFR with the early endosomal antigen 1 (EEA1), indicating that endosomal uptake of EGFR in response to EGF stimulation was not affected by the Ub variant (Fig. 4G). However, Ubv.8.2CΔ2, but not Ub.wtCΔ2, resulted in greater colocalization of EGFR with the lysosome-associated protein 1 (LAMP1) and correspondingly less colocalization with the endosomal recycling marker Ras-related protein RAB11, indicating that Ubv.8.2CΔ2 enhances transfer of EGFR from endosomes to lysosomes (Fig. 4G and fig. S6). These results show that Ubv.8.2CΔ2 interacts with and inhibits USP8 in cells, and this results in increased ubiquitination and accelerated degradation of activated EGFR in lysosomes.
Last, we explored the effects of Ubv.N.2 on the activity of the NEDD4 ligase. Purified Ubv. N.2 bound to the HECT domain of NEDD4 more avidly than did Ub.wt (fig. S7). Unexpectedly, Ubv.N.2 enhanced the auto-ubiquitination of recombinant full-length NEDD4 in vitro (Fig. 4H). We tested the effect of Ubv.N.2 on NEDD4 in cells by monitoring a NEDD4 substrate, the transcription factor Ying-Yang1 (YY1) (43). No ubiquitination of YY1 was detected in cells transfected with catalytically inactive NEDD4, whereas YY1 was ubiquitinated in cells transfected with wt NEDD4, and this ubiquitination was strongly enhanced by the presence of Ubv.N.2 as compared with Ub.wtCΔ2 (Fig. 4I). NEDD4 is auto-inhibited by intramolecular protein-protein interactions (44), and it is likely that Ubv.N.2 alleviates this inhibition.
Through engineered optimization of the low-affinity interactions between Ub and enzymes of the ubiquitin system, we were able to develop potent and selective modulators of ubiquitin system activity. Structural analyses explained the higher affinity of the Ub variants and underscored the plasticity of the Ub fold as a recognition scaffold. Ub variants cause target-specific effects on enzyme activity and can act as simple competitive inhibitors in the case of some USPs, as both a competitive inhibitor of catalytic function and an allosteric attenuator of Ub~UbcH5b docking in the case of OTUB1, or as enhancers of ubiquitin ligase activity in the case of NEDD4.
Collectively, these results show that the dynamic interactions of Ub can be harnessed to develop selective modulators of major enzyme classes in the ubiquitin system. Our findings illustrate a previously unappreciated feature of ubiquitin itself, namely that the extreme sequence conservation of ubiquitin is likely dictated by the constraints of low-affinity interactions with hundreds of partners, few if any of which can tolerate mutational drift toward higher-affinity interaction. As we illustrate here, Ub variants will serve as useful genetic probes to assess and modulate ubiquitin system function in vivo. Applications of Ub variants in drug discovery will include target validation, competition-based screening assays, and structure-guided design of small-molecule mimetics of the stabilized Ubtarget interface.
Supplementary Material
Acknowledgments
The authors thank I. Pot and J.J. Adams for helpful comments on the manuscript and Y. Li for cloning protease constructs. This project was supported by operating grants from the Canadian Institutes for Health Research to S.S.S. (MOP-111149), D.R. (MOP-13494), F.S., M.T. (MOP-57795), and M.F.M. (MOP-102536). J.Y. receives funding from a NIH–National Institute of Neurological Disorders and Stroke grant (1R01NS072420-01). F.S., D.R., M.T., and M.F.M. hold Canada Research Chairs (Tier 1) in Structural Principles of Signal Transduction, Canadian Foundation for Innovation, Systems and Synthetic Biology, and Molecular Signatures, respectively. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. A.E., G.A., S.D., and S.S.S. designed the study. A.E., A.V., P.G., and L.B. performed phage display selections, protein purification, and in vitro inhibition assays. G.A., A.-M.N., D.N., J.W., Y.-C.J., and S.D. purified proteins used in this study and solved all the crystal structures. J.T. and M.F.M. performed intracellular USP8 inhibition experiments. Y.F., Y.Z., and J.Y. performed intracellular USP21 inhibition experiments. Y.-C.J., M.-C.L., C.Y., E.Z, F.S., L.P., M.T., and D.D. purified proteins used as targets for phage display selections. K.K. and J.M. performed initial intracellular characterizations of Ub variants. A.A.-H. and M.V. performed in vitro enzymatic assays. P.-K.C. and B.R. provided Ub variants expressing stable cell lines and performed mass spectrometry experiments. P.A., A.P., and D.R. performed NEDD4 ubiquitination experiments; A.E. and S.S.S. analyzed data, prepared figures, and wrote the paper. S.S.S., L.B., and A.E. are inventors on a pending patent, “Specific Active Site Inhibitors of Enzymes or Substrate Binding Partners and Methods of Producing the Same” (PCT/US11/39601).
Footnotes
Supplementary Materials
www.sciencemag.org/cgi/content/full/science.1230161/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S7
Tables S1 to S5
References (47–69)
References and Notes
- 1.Cohen P, Tcherpakov M. Cell. 2010;143:686. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Komander D. Biochem. Soc. Trans. 2009;37:937. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M. Annu. Rev. Biochem. 2012;81:203. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Nijman SMB, et al. Cell. 2005;123:773. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Yang YL, Kitagaki J, Wang H, Hou DX, Perantoni AO. Cancer Sci. 2009;100:24. doi: 10.1111/j.1349-7006.2008.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daviet L, Colland F. Biochimie. 2008;90:270. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Edelmann MJ, Kessler BM. Biochim. Biophys. Acta. 2008;1782:809. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain S, Zhang Y, Galardy PJ. Cell Cycle. 2009;8:1688. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 9.Grunda JM, et al. J. Neurooncol. 2006;80:261. [Google Scholar]
- 10.Zhang X, Berger FG, Yang J, Lu X. EMBO J. 2011;30:2177. doi: 10.1038/emboj.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummins JM, Vogelstein B. Cell Cycle. 2004;3:687. [PubMed] [Google Scholar]
- 12.Li MY, Brooks CL, Kon N, Gu W. Mol. Cell. 2004;13:879. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 13.Priolo C, et al. Cancer Res. 2006;66:8625. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 14.Komander D, Clague MJ, Urbé S. Nat. Rev. Mol. Cell Biol. 2009;10:550. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 15.Fedorov O, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20523. [Google Scholar]
- 16.Colland F. Biochem. Soc. Trans. 2010;38:137. doi: 10.1042/BST0380137. [DOI] [PubMed] [Google Scholar]
- 17.Lee BH, et al. Nature. 2010;467:179. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapuria V, et al. Cancer Res. 2010;70:9265. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 19.Pickart CM, Eddins MJ. Biochim. Biophys. Acta. 2004;1695:55. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Ye Y. Cell. Mol. Life Sci. 2008;65:2397. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson KD, Gan-Erdene T, Kolli N. Methods Enzymol. 2005;399:37. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, et al. Cell. 2002;111:1041. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 23.Fellouse FA, Pal G. In: Phage Display in Biotechnology and Drug Discovery. Sidhu SS, editor. vol. 3. Boca Raton, FL: Taylor and Francis Group; 2005. pp. 111–142. [Google Scholar]
- 24.Sidhu SS, et al. Methods in Enzymology. vol. 328. Academic Press; 2000. p. 333–IN5. [Google Scholar]
- 25.Edelmann MJ, et al. Biochem. J. 2009;418:379. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 26.Nakada S, et al. Nature. 2010;466:941. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 27.Cooper EM, Boeke JD, Cohen RE. J. Biol. Chem. 2010;285:10344. doi: 10.1074/jbc.M109.059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper EM, et al. EMBO J. 2009;28:621. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry WL, et al. Nat. Genet. 1998;18:143. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 30.Melino G, et al. Cell Death Differ. 2008;15:1103. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 31.Hurley JH, Lee S, Prag G. Biochem. J. 2006;399:361. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, et al. Mol. Cell. 2011;42:511. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 33.Wenzel DM, Stoll KE, Klevit RE. Biochem. J. 2010;433:31. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang LC, Melandri FD, Stein RL. Biochemistry. 1998;37:1868. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 35.Juang Y-C, et al. Mol. Cell. 2012;45:384. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avvakumov GV, et al. J. Biol. Chem. 2006;281:38061. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 37.Xu GF, et al. J. Biol. Chem. 2010;285:969. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berlin I, Schwartz H, Nash PD. J. Biol. Chem. 2010;285:34909. doi: 10.1074/jbc.M109.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balut CM, Loch CM, Devor DC. FASEB J. 2011;25:3938. doi: 10.1096/fj.11-187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer IMJ, van Leeuwen JEM. Cell. Signal. 2011;23:458. doi: 10.1016/j.cellsig.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 41.Tong J, Taylor P, Peterman SM, Prakash A, Moran MF. Mol. Cell. Proteomics. 2009;8:2131. doi: 10.1074/mcp.M900148-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niendorf S, et al. Mol. Cell. Biol. 2007;27:5029. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persaud A, et al. Mol. Syst. Biol. 2009;5:333. doi: 10.1038/msb.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiesner S, et al. Cell. 2007;130:651. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 45.Renatus M, et al. Structure. 2006;14:1293. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu M, et al. EMBO J. 2005;24:3747. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.