Abstract
Purpose
mTOR pathway hyperactivation occurs in nearly 90% of glioblastomas, but the allosteric mTOR inhibitor rapamycin has failed in the clinic. Here we examine the efficacy of the newly discovered ATP-competitive mTOR kinase inhibitors CC214-1 and CC214-2 in glioblastoma, identifying molecular determinants of response and mechanisms of resistance, and develop a pharmacological strategy to overcome it.
Experimental design
We performed in vitro and in vivo studies in glioblastoma cell lines and an intracranial model to: determine the potential efficacy of the recently reported mTOR kinase inhibitors CC214-1 (in vitro use) and CC214-2 (in vivo use) at inhibiting rapamycin resistant signaling and blocking GBM growth and a novel single cell technology, DNA Encoded Antibody Libraries, was used to identify mechanisms of resistance.
Results
Here we demonstrate that CC214-1 and CC214-2 suppress rapamycin-resistant mTORC1 signaling; block mTORC2 signaling and significantly inhibit the growth of glioblastomas in vitro and in vivo. EGFRvIII expression and PTEN loss enhance sensitivity to CC214 compounds, consistent with enhanced efficacy in strongly mTOR-activated tumors. Importantly, CC214 compounds potently induce autophagy, preventing tumor cell death. Genetic or pharmacologic inhibition of autophagy greatly sensitizes GBM cells and orthotopic xenografts to CC214-1 and CC214-2 induced cell death.
Conclusions
These results identify CC214-1 and CC214-2 as potentially efficacious mTOR kinase inhibitors in GBM and suggest a strategy for identifying patients most likely to benefit from mTOR inhibition. This study also demonstrates a central role for autophagy in preventing mTOR-kinase inhibitor-mediated tumor cell death, and suggests a pharmacological strategy for overcoming it.
Introduction
Glioblastoma (GBM) is the most common malignant primary brain cancer of adults and one of the most lethal of all cancers (1). PI3K signaling is hyperactivated in nearly 90% of GBMs, commonly in association with epidermal growth factor (EGFR) amplification and mutation, and loss of the PTEN tumor suppressor protein (2, 3). The mTOR kinase, which links growth factor receptor signaling through PI3K with protein translation, proliferation and survival, is therefore frequently hyperactivated in many cancer types, including the majority of GBMs (4, 5). mTOR functions through two well-described multiprotein complexes: mTORC1, which promotes many of its cellular activities through phosphorylation of S6 kinase and 4E-BP1 and mTORC2 which phosphorylates Akt to maximize its activity (5). Despite the central importance of mTOR signaling in GBM, the allosteric mTORC1 inhibitor rapamycin has been shown to be ineffective in GBM patients (6, 7). Recent work from our group demonstrates that this is mediated, at least in part, by failure to suppress mTORC2 signaling (6). Further, rapamycin has also been shown to be ineffective at durably suppressing 4E-BP1 phosphorylation in certain cellular contexts, suggesting that rapamycin-resistant mTORC1 signaling may also underlie clinical failure (8, 9).
ATP-competitive mTOR kinase inhibitors, which could potentially suppress rapamycin-resistant mTORC1 and mTORC2 signaling, have the potential to improve the treatment of GBM patients. However to date, their efficacy, molecular determinants of sensitivity, and potential mediators of resistance are not understood. Here, we perform a series of in vitro and in vivo studies in glioblastoma cell lines to: determine the potential efficacy of the recently reported mTOR kinase inhibitors CC214-1 (in vitro use) and CC214-2 (in vivo use) at inhibiting rapamycin resistant signaling and blocking GBM growth (10). We identify molecular determinants of sensitivity and show that autophagy plays a central role in preventing CC214-mediated cell death, which can be reversed by genetic or pharmacologic inhibition of autophagy. These results identify CC214-1 and CC214-2 as potentially effective agents, particularly in combination with lysosomotropic, autophagy-inhibitory compounds.
Materials and Methods
Cell lines and reagents
The U87, U87EGFRvIII, U87EGFR, U87EGFRvIII/-PTEN cells were obtained as previously described (5); U251, LN229 were cultured in DMEM (Cellgro) supplemented with 10% FBS (vol/vol, Omega Scientific) and 100 U/mL penicillin and streptomycin (Gibco); U373 Tet OFF system were kindly provided by Webster Cavenee group (Ludwig Inst., San Diego, U.S.A.), LN229 Tet ON cell lines were grown as mentioned in Guo et al. (11). GBM39 primary neurospheres were provided by Prof. David James (UCSF, San Francisco, U.S.A.). All cell lines were cultured in a humidified 5% CO2 (vol/vol) incubator, at 37°C. CC214-1 and CC214-2 were provided by Celgene Corporation (San Diego, U.S.A.). The development of the series that led to CC214 compounds (12) and its structure (10) have been described. P-Akt Ser473, P-Akt Thr308, P-NDRG1 Thr346, P-S6 Ser235/236, S6, cleaved PARP, P-4E-BP1 Thr37-46, 4E-BP1, eIF4E, LC3B, Atg-5, Atg-5/12 antibodies were purchased from Cell Signaling Technologies. P-EGFR Tyr1086, P-PRAS40 were from Invitrogen. EGFRvIII was made by Dako (U.S.A.). Actin, p62 and PTEN antibodies were purchased respectively from Novus Biologicals, Progen Biotechnik and Cascade BioScience. Chloroquine was from Sigma.
Immunoblotting
Western blot analysis has been performed using a 10–50 µg range of total protein lysates. Lysates were obtained from cultured cells or snap-frozen tissues using RIPA buffer (Boston BioProducts) and protease plus phosphatase inhibitor cocktail (Thermo Scientific). Mono-dimensional electrophoresis has been applied in 4–12 % gradient gels NuPAGE Bis-Tris Mini Gel (Invitrogen); 10% or 15% acrylamide (vol/vol, National Diagnostics) gels were made and used to improve middle and low MW protein separation. Proteins have then been transferred on nitrocellulose membranes (GE Healthcare), using BioRad transfer chamber, applying 110 Volts for 1 hour. Membranes were subsequently blocked in Tris-buffered saline containing 0.1% Tween20 (vol/vol) and 5% BSA (g/mL, Fischer Scientific) for 1 hour. Primary antibodies incubations were performed overnight, at 4°C. Incubation with secondary HRP conjugated antibodies were done at RT for 1 hour. Detection of the immunoreactivities was obtained with Super Signal West Pico Chemiluminescent Substrate or West Femto Trial kit (Thermo Scientific). Scanned films or digitalized images acquired by Chemidoc (BioRad), Image Lab 4.0.1, were quantified using Image J software (NIH).
Cell proliferation
WST assay was performed with Cell Proliferation Assay Kit (Chemicon). Specifically, cells were seeded at a density of 1×103 cells each well in 1% FBS DMEM (vol/vol), a first reading after adhesion was done, after which drug treatment started and was extended up to 4 days. Each day of reading, plates were incubated for 2 hours with tetrazolium salt WST 1 [2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfo-phenyl)-2H-tetrazolium, monosodium salt] (Chemicon) in the incubator. The absorbance was measured with a microplate reader (BioRad) at 420 to 480 nm.
Viability tests
Fifteen thousand of GBM cells were seeded in 12 well plates and treated, after one night, with CC214-1 0.1 µM, 1 µM, 2 µM, 5 µM and 10 µM. Chloroquine 10 µM was used for combinatory treatment. Cell viability was evaluated after 3 days of treatment and assessed by trypan blue exclusion (Invitrogen).
Flow Cytometry Analysis: Annexin V, PI
Viability test on GBM cell lines was done with Annexin V, PI kit (BD), following the datasheet instructions, after 72 hours of CC214-1 (2 µM) treatment.
In vivo experiments
Flank xenografts: U87EGFRvIII xenografts models were obtained in full compliance with the UCLA-Division of Laboratory Animal Medicine (DLAM) regulation and after approval by the Chancellor’s Animal Research Committee of UCLA. Particularly, U87-EGFRvIII cells were implanted subcutaneously in immunocompromised NOD-SCID gamma null mice. Cells were cultured, trypsinized and resuspended in PBS (Cellgro) plus Matrigel (BD Biosciences), 1:1 solution, at 6×106 cells/ml density. Tumor sizes were monitored daily using automated caliper. CC214-2 treatment, 50 mg/kg, was done by oral gavage, once every day in a suspension containing 0.5% carboxymethylcellulose (g/mL, Sigma), 0.25% teewn-80 (vol/vol, Sigma) in nanopure water. Mice were euthanized when tumors reached 15 mm diameter.
Orthotopic xenografts: U87EGFRvIII-TurboFP635 orthotopic xenografts models were obtained in full compliance with the UCSD-Institutional Animal Core and Use Committee (IACUC). Plasmid and cells: TurboFP635 fluorescent protein was used to tag U87EGFRvIII cells that were injected intracranially. Briefly, the lentiviral LV plasmid (13) was digested with NheI and XhoI. The same restriction enzymes were used to subclone the far-red fluorescent TurboFP635 protein sequence in the vector (Evrogen JSC, Moscow, Russia). The lentivirus was obtained by transient transfection of 293T cells. The U87EGFRvIII cells were then infected with the virus carrying the sequence for the far-red protein TurboFP635 and subjected to two cycles of FACS sorting, to sort and enrich the 5% most fluorescent cell population each cycle.
Intracranial Injection procedure: A total of 1 × 105 U87EGFRvIII cells, tagged with the far-red protein TurboFP635, in a 5 µL volume, were injected intracranially into 4-wk-old athymic nude mice using a stereotactic system according to the protocol described by Lee et al. (14). In vivo tumors were measured using near-infrared quantitative fluorescence imaging by Fluorescence Molecular Tomography (FMT 2500, PerkinElmer, Massachusetts, USA). Tumor fluorescence emission, at 635 nm, was collected the first day of treatment and on the sacrifice day. CC214-2 treatment, 100mg/kg, was done by oral gavage, once every two days in a suspension containing 0.5% carboxymethylcellulose (g/mL, Sigma), 0.25% teewn-80 (vol/vol, Sigma) in nanopure water. Chloroquine, 30mg/kg in nanopure water, was administered intraperitoneally, once every two days. Mice were euthanized in accordance with UCSD institutional guidelines for animal welfare and experimental conduct. Survival until the onset of neurologic sequelae in the vehicle mice was considered for the time of sacrifice.
Immunohistochemistry and immunofluorescence
Paraffin embedded U87EGFRvIII xenografts blocks were sectioned at the UCLA Pathology Histology and Tissue Core Facility and at the UCSD Histology and Immunohistochemistry Shared Resource. Immunohistochemistry was performed as described in Mellinghoff et al. (15). Three images per IHC slide were captured using DP 26 camera mounted on an Olympus BX43 microscope at 40x magnification. Quantitative analysis of the IHC stained slides was performed with the image analyzing software Microsuite Five (Olympus). Immunofluorescence staining was done as previously described in Gini et al. (16).
Cap-binding proteins assay
U87EGFRvIII cells were treated with CC214-1 (2 µM) or Rapamycin (5 nM) for 24 hours. Five hundred of µg of protein lysates from control and treated cells, obtained as described in the Immunoblotting section, were incubated with 7-Methyl GTP Sepharose (GE Healthcare) for 4 hours at 4°C, on rotating shaker. Resin was then centrifuged 90 seconds at 1800 rpm and pellet washed twice with RIPA buffer. Resin was further incubated 5 minutes at 99°C with Loading Buffer 2X. After centrifugation, cap-binding proteins were collected in the supernatant and analyzed by immunoblotting.
Capture of EGFR, EGFRvIII positive cells by DNA Encoded Antibody Library (DEAL)
Capture of EGFR, EGFRvIII positive cells from GBM39 neurospheres has been performed by DEAL as described in Bailey et al. (17). Briefly, DEAL array (CalTech) were blocked with 1% BSA solution (g/mL) for 30 minutes, washed in PBS and deionized water and then incubated with oligo-Cetuximab (Bristol-Myers) conjugate for 30 minutes, at 37°C. The array was washed with 0.1% BSA in PBS (g/mL) and single cell suspension of GBM39 cells were applied on it for 40 minutes, in ice. After washing, arrays of cells were used for immunofluorescence.
sh-RNA assay
E.coli carrying sh-Atg-5 pLKO.1 plasmids (SIGMA) were expanded in LB medium (Fisher Scientific) and then the plasmids were extracted using MidiPrep kit (Qiagen). For lentiviral production, T293 cells were transfected with sh-Atg-5 pLKO.1 or sh-scramble pLKO.1, together with packaging, envelope plasmids and polybrene (Sigma). Viral harvesting was achieved after 2 days from transfection, in high BSA growth medium. Sixty-thousand U87EGFRvIII cells were seeded in 6 cm plates and infected over night with lentivirus carrying sh-Atg5 or sh-scramble plasmids. Selection of sh-positive cells was achieved after 10 days of growth in medium containing puromycin (2 ug/mL, Sigma).
Crystal violet analysis
Twenty thousand of U87EGFRvIII sh-scramble and U87EGFRvIII sh-Atg5 cells were seeded in 6 well plates under puromycin selection. After 3 days of incubation in CC214-1 2 µM, cells were shortly air dried, fixed 5 minutes in PFA 4% (vol/vol, Santa Cruz Biotech.) and stained in 0.05% crystal violet (g/mL, Sigma) solution for 30 minutes. Plates were then washed twice with tap water and air dried for 2 min. Pictures of the plates were taken using Nikon Eclipse TS100 scope equipped with Canon S51S camera.
Results
CC214-1 inhibits rapamycin-resistant mTORC1 and mTORC2 signaling, limits protein translation and blocks GBM cell proliferation
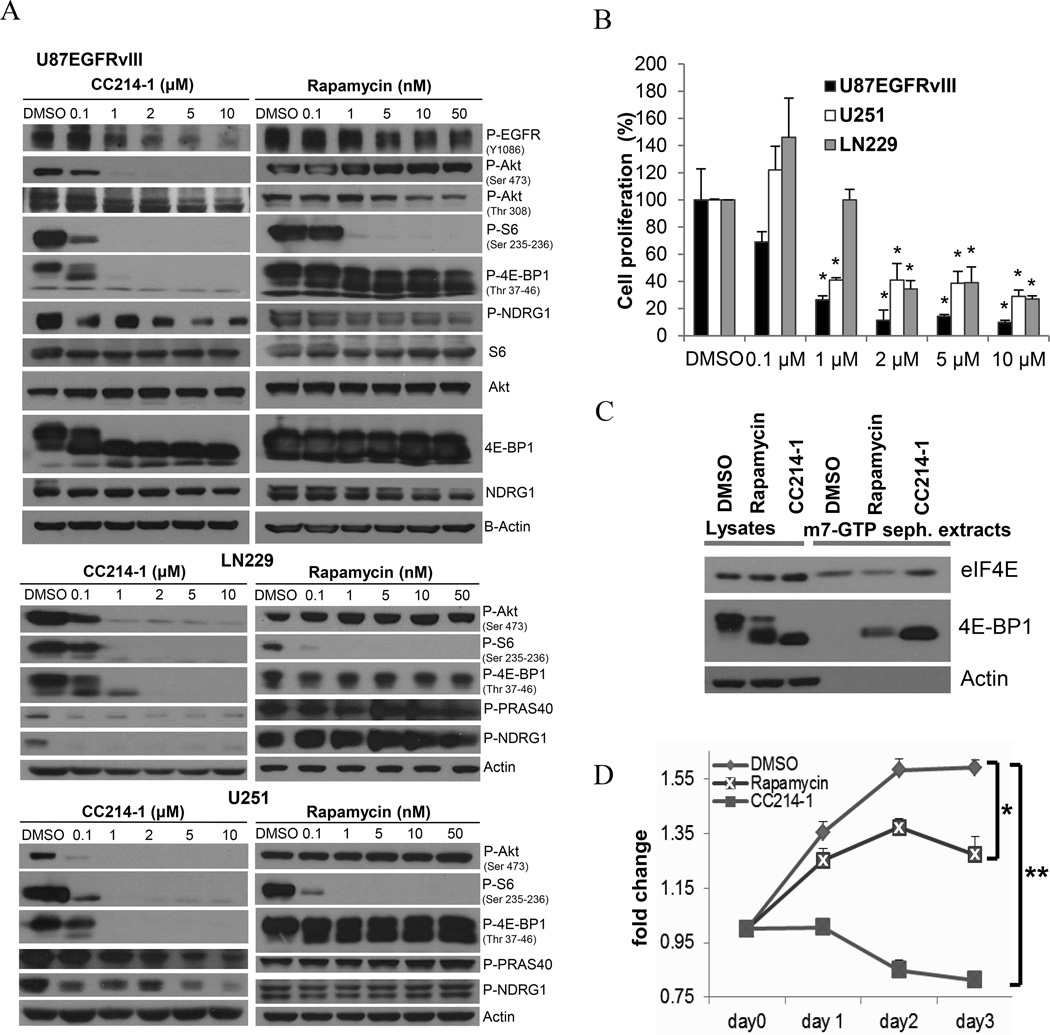
We examined the biochemical and anti-proliferative effect of CC214-1 on a panel of GBM cell lines (Fig. 1A-B, Supplementary Fig. S1). CC214-1 dose-dependently inhibited mTORC1 signaling in all GBM cell lines tested, potently suppressing rapamycin-resistant 4E-BP1 and mTORC2 signaling, as demonstrated by effects on 4E-BP1 (Thr 37-46), and Akt (Ser 473), PRAS40 and NDRG-1 phosphorylation, respectively (Fig. 1A, Supplementary Fig. S1). This was confirmed even in presence of a rapamycin concentration 10 times higher (50 nM) than the minimal effective dose (5 nM) that inhibits mTORC1 (Fig. 1A). Consistent with the enhanced inhibition of 4E-BP1 phosphorylation, CC214-1 demonstrated markedly suppressed m7G cap-dependent translation relative to rapamycin, indicating that the dual mTOR kinase inhibitor was significantly more efficacious at blocking mTOR-dependent protein translation (Fig. 1C). Further, CC214-1 was significantly more effective than rapamycin at blocking GBM cell proliferation in culture (Fig. 1D). We have recently shown that ATP-competitive mTOR kinase inhibitors synergize with rapalogs (18). Consistent with these findings, CC214 synergized with rapamycin, inhibiting mTORC1 signaling and tumor cell proliferation at sub-therapeutic doses of each compound (Supplementary Fig. S2A).
Figure 1. CC214-1 induces potent mTORC1/mTORC2 inhibition and tumor growth reduction in GBM cell lines in vitro.
Immunoblotting analysis of mTORC1/2 effectors activities, following 8 hours treatment with CC214-1 or Rapamycin at increasing concentrations, in U87EGFRvIII, LN229 and U251 GBM cell lines (A). Cellular proliferation in three GBM cell lines as assessed by trypan blue exclusion cell count and normalized to DMSO control (B). U87EGFRvIII proved to be the most sensitive GBM cell line assessed, with IC50 in the 0.5 µM range (B). Inhibition of cap-dependent translation by CC214-1 (2 µM) or Rapamycin (5 nM), was assessed by the level of 4E-BP1 bound to the 5′-cap in U87EGFRvIII cell lysates after 24 hours of treatment (C). WST proliferation experiment in U87EGFRvIII cells comparing DMSO, CC214-1 (2 µM) and Rapamycin (5 nM) treatments (D). (P-S6, P-4E-BP1 have been chosen as mTORC1 biomarker's activity; P-Akt-S473, P-PRAS40, P-NDRG1 were the selected mTORC2 effectors). * indicates P<0.05; ** indicates P<0.0001 (Student’s t-Test). Data represent average of three independent experiments.
EGFRvIII expression and PTEN loss sensitize tumor cells to CC214-1
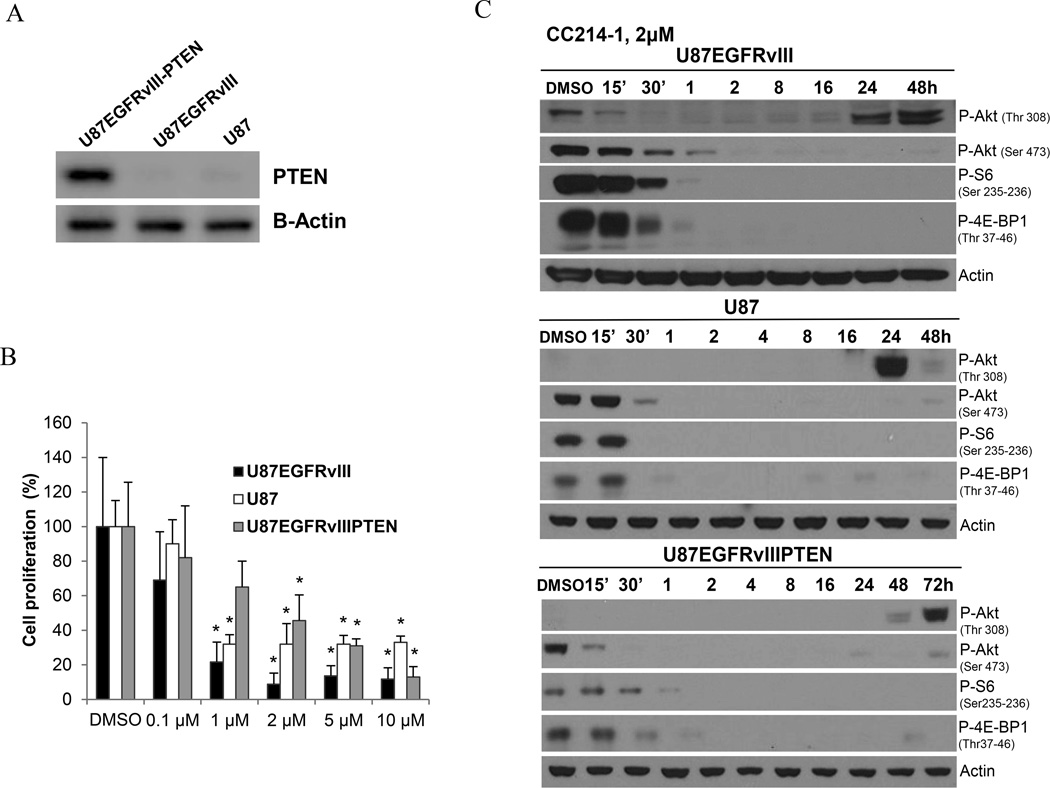
To test the hypothesis that EGFR activation promotes sensitivity to CC214-1, we examined the impact of EGFRvIII or EGF-stimulation of wild type EGFR, in isogenic GBM cell lines. Biochemical analyses confirmed that CC214-1 successfully inhibited mTORC1 dependent 4E-BP1 and S6 phosphorylation in EGFRvIII-expressing GBM cells (Fig. 2A-C and Supplementary Fig. S2B, S3A), as well as blocked EGF-induced 4E-BP1 and S6 phosphorylation in GBM cells overexpressing wild type EGFR (Supplementary Fig. S3B). Further, CC214-1 showed superior inhibition of P-Akt (Ser 473), P-S6 (Ser 235-236) and P-4E-BP1 (Thr 37-46) phosphorylation relative to any of the other EGFR, PI3K, or mTOR inhibitors tested (Supplementary Fig. S3B). Importantly, EGFRvIII sensitized U87 GBM cells to CC214-1-mediated growth inhibition (Fig. 2C), an observation that was validated in another isogenic cell system, U373 GBM cells, in which EGFRvIII was under the control of a tetracycline-regulatable promoter (Supplementary Fig. S4A). PTEN reconstitution rendered GBM cells less sensitive, despite similar levels of pathway inhibition (Fig. 2C). We used a novel technology, DEAL (DNA-encoded antibody library) (19), to capture and analyze individual EGFRvIII expressing tumor cells from the heterogeneous-patient-derived GBM model, GBM39, in which individual tumor cells varied in EGFRvIII and PTEN expression (20). GBM39 cells were treated with CC214-1 in neurosphere cultures, after which the EGFR and EGFRvIII positive tumor cells were sorted using Cetuximab-conjugated oligonucleotides and captured on a glass slide arrayed with their complementary oligonucleotide barcodes (Supplementary Fig. S2C). Immunofluorescent stain for PTEN was then performed (Supplementary Fig. S2C). Consistently, in this patient-cell model, in which PTEN is heterogeneously expressed, a shift in the ratio of PTEN positive/PTEN negative cells was observed, with a significant increase in the percentage of PTEN positive tumor cells in response to CC214-1 treatment relative to control. These data suggest a preferential effect of CC214-1 on the PTEN deficient tumors, consistent with our studies in the isogenic U87-GBM cell lines. Taken together, these data suggest that EGFRvIII expression and PTEN loss sensitizes GBM cells to CC214-1-mediated cell growth arrest.
Figure 2. EGFRvIII expression and PTEN loss sensitize GBM cells to CC214-1 dependent inhibition of proliferation.
CC214-1 efficacy has been evaluated in a panel of isogenic U87 cell lines differentially expressing EGFRvIII and PTEN (A). Reduction of mTORC1 and mTORC2 biomarkers followed CC214-1 treatment (2 µM) was observed in all of the three cell lines by Western blot (B). Cellular proliferation in three isogenic U87 cell lines as assessed by trypan blue exclusion cell count after treatment with CC214-1 and normalization to DMSO control (C). The EGFRvIII positive and PTEN negative U87 cell line (U87EGFRvIII) proved to be the most sensitive, when compared to EGFRvIII negative, PTEN negative cells (U87) or EGFRvIII positive, PTEN positive cells (U87EGFRvIIIPTEN) (C). Data represent average of three independent experiments. * indicates P<0.05 (Student’s t-Test).
CC214 potently blocks tumor growth in vivo
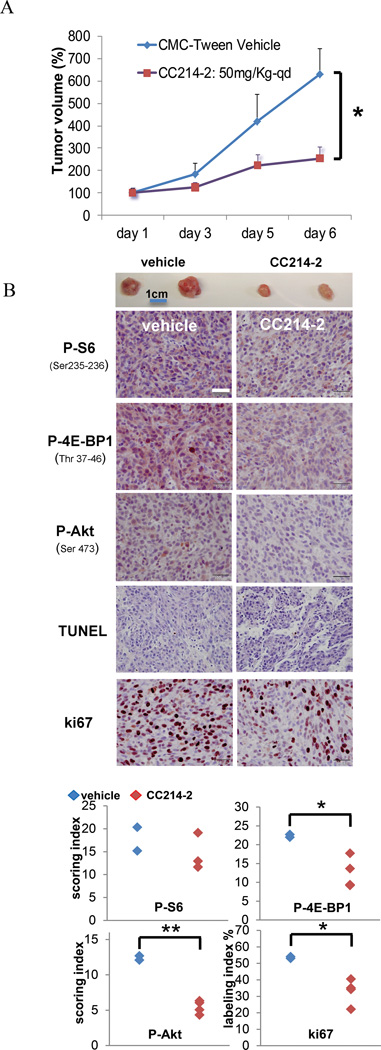
We then assessed the potential of CC214-2, an analog related to CC214-1 with PK properties suitable for in vivo application (10), to inhibit the growth of U87EGFRvIII cells in vivo (Fig. 3). CC214-2 significantly reduced the growth of U87EGFRvIII flank xenografts (Fig. 3A) by more than 50%, following 1 week of treatment via oral gavage. Immunohistochemical analysis demonstrated mTORC1 and mTORC2 inhibition, as measured by diminished P-S6, P-4E-BP1 and P-Akt cytoplasmic staining (Fig 3B). CC214-2 treatment resulted in markedly diminished ki67 staining (Fig. 3B). In contrast, no change in TUNEL staining was detected (Fig. 3B), indicating that CC214-2-mediated growth inhibition was due to its cytostatic effect, rather than a consequence of tumor cell death.
Figure 3. In vivo CC214-2 treatment of U87EGFRvIII flank xenografts down-regulates mTORC1-mTORC2 effector’s functions and significantly reduces tumor proliferation.
Antitumor activity of CC214-2 in U87EGFRvIII flank xenograft models, following 6 days of oral dosing CC214-2 (50 mg/kg once a day) treatment resulted in greater than 50% tumor volume reduction (A). Representative xenografts are shown underneath the graph. Immunohistochemistry on U87EGFRvIII xenograft’s sections shows down-regulation of the main mTORC1, mTORC2 biomarkers (B). TUNEL staining did not change between controls and treated samples, whereas ki67, a marker for cell proliferation, was decreased upon CC214-2 treatment (B). P-S6 (Ser 235-236), P-4E-BP1 (Thr 37-46) denote mTORC1 effectors, P-Akt (Ser 473) represents mTORC2 activity. * indicates P<0.05; ** indicates P<0.01, Student’s t-Test; n=2 and n= 4 respectively for vehicle and CC214-2 treated flank xenografts; scale bar corresponds to 100µm.
CC214-1 and CC214-2 promote autophagy in vitro and in vivo
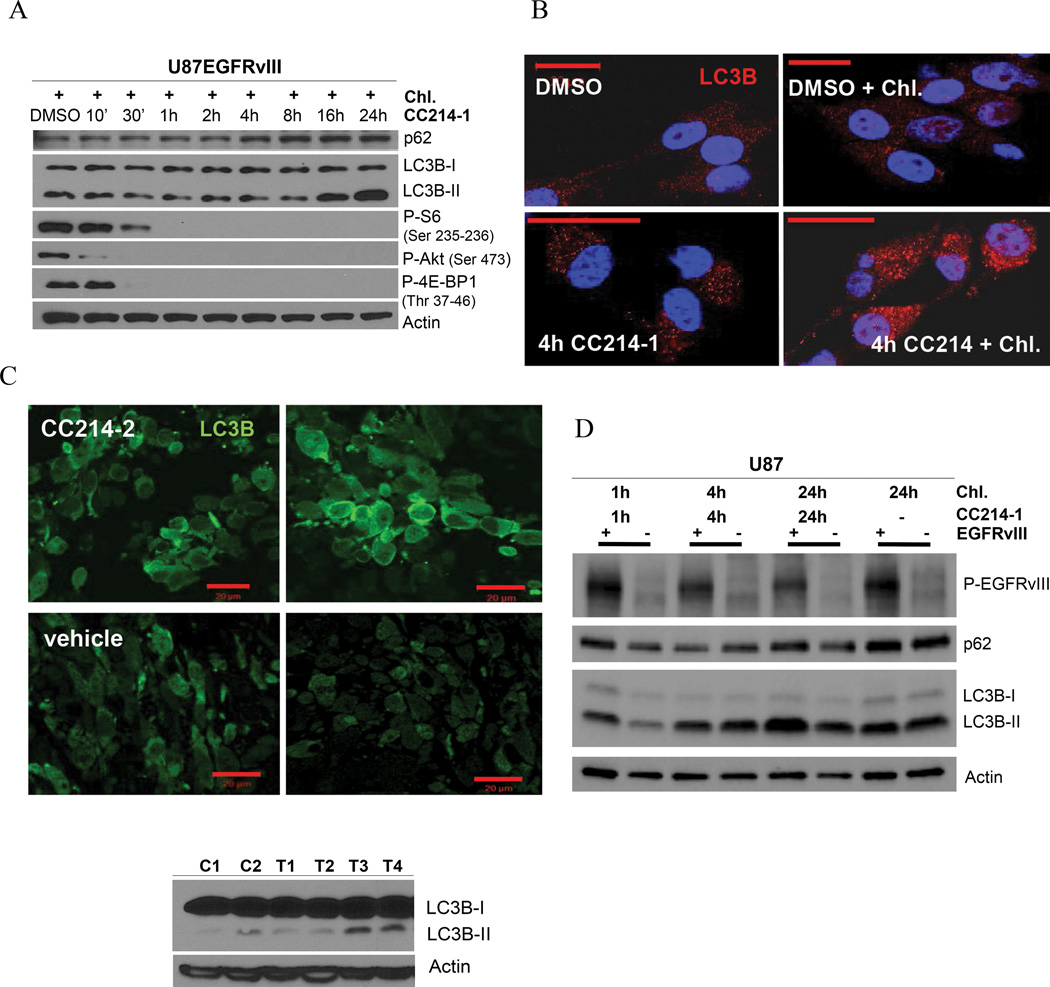
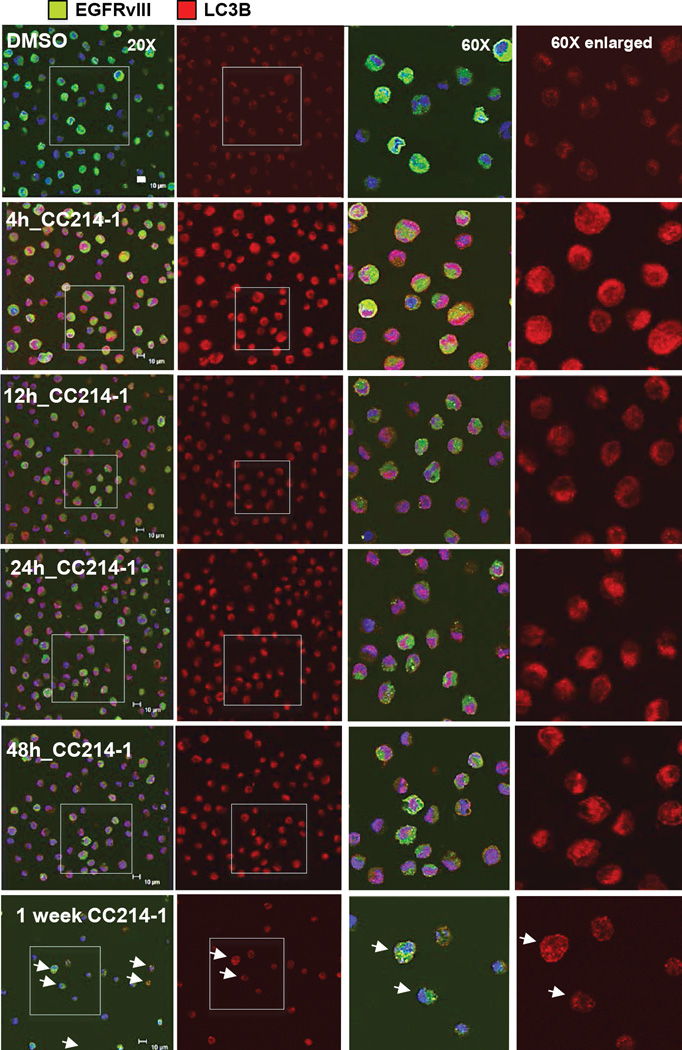
Inhibition of mTORC1 signaling has been linked to induction of autophagy, a state in which cells rely on intracellular degradative machineries to supply biomolecular building blocks to subvert stressful conditions (21). We hypothesized that induction of autophagy enabled U87EGFRvIII GBM cells to resist CC214-1 and CC214-2 mediated cell death. To test this hypothesis, we examined the effect of CC214-1 on autophagy induction, and then determined its functional consequences using genetic and pharmacologic approaches. We first examined the level of LC3B-II and Atg5-12, which are recognized as autophagic biomarkers (Supplementary Fig. S4B). CC214-1 induced a transient expression of LC3B-II isoform and the conjugation of Atg12 to Atg5 indicative of the stimulation of the autophagy flux in U87EGFRvIII cell line (Supplementary Fig. S4B). Further, in the presence of chloroquine, which inhibits the late stages of autophagy by preventing degradation of autophagosome contents, CC214-1 treatment led to markedly elevated levels of LC3B-II and p62, consistent with CC214-1-mediated autophagy (Fig. 4A, Supplementary Fig. S4C). Immunofluorescence analysis highlighted enrichment of LC3B with localization into autophagosomes depicted as puncta (Fig. 4B). In vivo, CC214-2 similarly activated autophagy in U87EGFRvIII xenografts (Fig. 4C).
Figure 4. U87EGFRvIII GBM cell line shows higher level of autophagy induction following CC214-1 exposure.
Time course experiment with CC214-1 (2 µM) plus the autophagy inhibitor chloroquine (20 µM), in U87EGFRvIII cells, reveals high degree of autophagic flux, registered as accumulation of LC3B-II, p62 proteins (A). Immunofluorescent staining for LC3B protein in U87EGFRvIII cell line (B). CC214-1 treatment (2 µM) shows typical autophagosome’s puncta intra-cellular pattern (B). Autophagy induction was registered also in CC214-2 treated (50 mg/kg qd for 6 days) U87EGFRvIII flank xenografts as indicated by the higher level of LC3B marker, in IF staining and immunoblotting (C), 24 hours following last dose as compared in the vehicle control samples. Time course experiment with CC214-1 (2 µM) in U87EGFRvIII cells compared to U87 cell line shows that EGFRvIII sensitizes cells to CC214-1 mediated induction of autophagy, as shown by the major increase of LC3B protein and the higher p62 turnover (D). Chl.: Chloroquine. Error bar: 20 µm.
To determine whether EGFRvIII expression sensitized GBM cells to CC214-mediated autophagy, we compared the level of autophagic flux in three different isogenic GBM cell lines, with or without EGFRvIII expression. In U87 GBM cells, or in U373 or LN229 GBM cells in which EGFRvIII expression was under the control of a tetracycline-regulatable promoter, EGFRvIII enhanced autophagic flux in response to CC214-1 treatment (Fig. 4D, Supplementary Fig. S5).
To further understand the role of endogenous EGFRvIII expression in promoting autophagic flux in response to mTOR kinase inhibition in GBM, we used DEAL technology (19), to capture and analyze individual EGFRvIII expressing tumor cells from the heterogeneous-patient-derived GBM model, GBM39 (20). GBM39 cells were treated with CC214-1 and the EGFR, EGFRvIII positive tumor cells were captured by DEAL as previously described (Fig. 5). The captured GBM cells were stained using double immunofluorescence for EGFRvIII and LC3B proteins (Fig. 5). The DEAL captured EGFRvIII positive cell were highly positive for punctate LC3B immunostaining upon CC214-1 treatment (Fig. 5). Western blot analysis confirmed the induction of autophagy in GBM39 cells, characterized by massive lipidation of LC3B-I to LC3B-II isoform (Supplementary Fig. S6A).
Figure 5. EGFRvIII positive cells from the heterogeneous GBM39 primary neurospheres rely on autophagy as mechanism to resist to CC214-1 treatment.
Double immunofluorescent staining for EGFRvIII (green) and LC3B (red) on DEAL captured EGFR wt, EGFRvIII positive GBM39 cells highlights the high level of staining of the autophagy marker LC3B in the EGFRvIII positive cells, upon CC214-1 (5 µM) time course treatment (scale bar corresponds to 10 µm).
These results raised the possibility that EGFRvIII, while sensitizing GBM cells to mTOR kinase growth inhibition, may also protect them from cell death by engaging autophagy. High levels of autophagy have been previously described in oncogene addicted tumor models, with activating mutations in Ras (22), and in GBM cells treated with PI3K/mTOR kinase inhibitors (23). In fact, induction of autophagy has been shown to protect GBM cells from PI3K/mTOR kinase inhibitor induced tumor cell death in a chloroquine reversible fashion (23). Therefore, we set out to use genetic and pharmacological approaches to determine whether inhibition of autophagy could sensitize GBM cells to CC214-1-dependent cell death. Chloroquine abrogated CC214-1-mediated autophagy, significantly sensitizing GBM cells to CC214-1-dependent cell death, particularly in EGFRvIII-expressing tumor cells (Supplementary Fig. S7A-B). Consistent with this model, genetic inhibition of autophagy achieved through ATG5 knockdown similarly sensitized GBM cells to massive tumor cell death in response to CC214-1 (Supplementary Fig. S6B-D).
In an intracranial GBM model, CC214-2 inhibits mTORC1 and mTORC2 signaling and suppresses tumor growth, and addition of chloroquine promotes tumor cell death
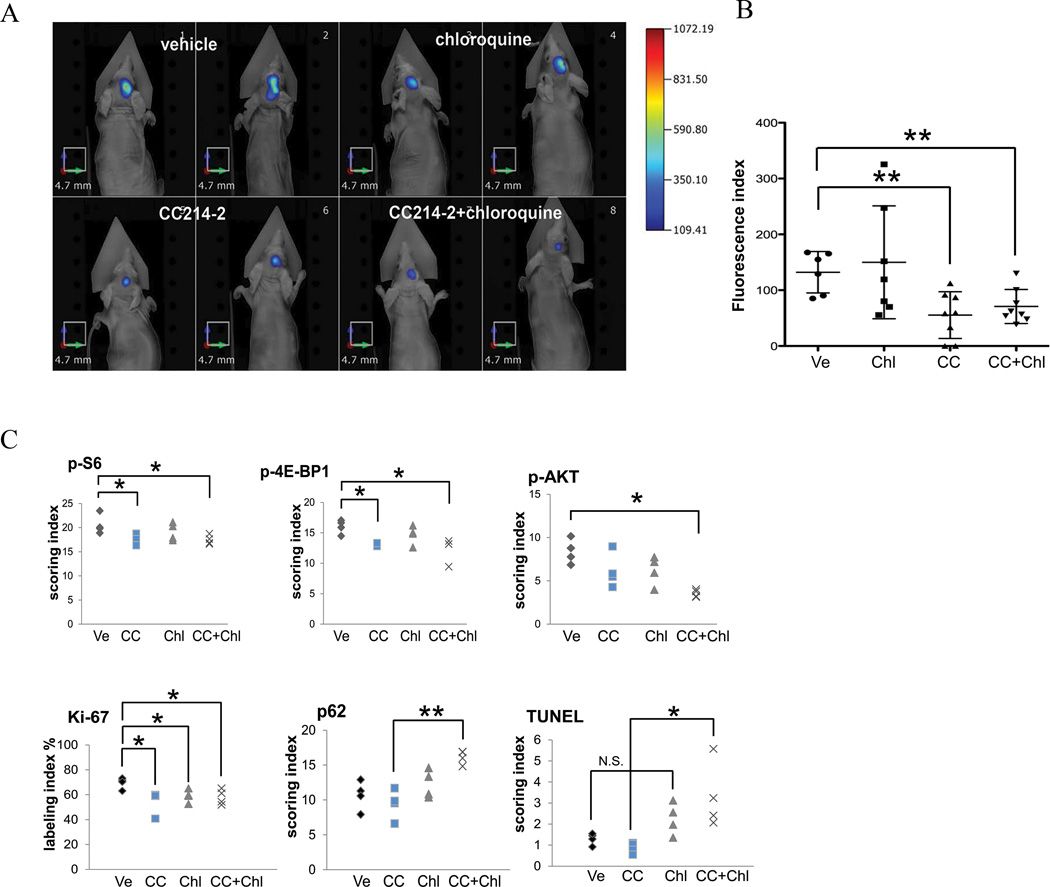
Finally, to assess the potential efficacy of mTOR kinase inhibition in an intracranial GBM model, and to assess the relative benefit of combining pharmacological inhibition of autophagy, we developed U87EGFRvIII intra-cranial xenografts in which the tumor cells were labeled with far-red protein TurboFP635 to enable non-invasive fluorescence molecular tomography (FMT) imaging (14). We then assessed the effect of mTOR kinase inhibition with CC214-2 (100 mg/kg once every two days by oral gavage for 6 days) on mTOR pathway inhibition and tumor growth, and we examined the effect of the autophagy inhibitor chloroquine (30 mg/kg once every two days by intraperitoneal injection), alone or in combination. CC214-2 treatment resulted in highly significant inhibition of intracranial GBM growth, concomitant with significant inhibition of mTORC1 and mTORC2 signaling (p<0.05), and significant inhibition of the Ki67 proliferation index (p<0.05) (Fig. 6A-C; Supplementary Fig. S7C). Chloroquine treatment had no effect on mTOR pathway inhibition or tumor growth as measured by FMT index (Fig. 6A-C), but did reduce the Ki67 index and elevated p62 staining (p<0.05). Importantly, combined treatment with CC214-2 and chloroquine did not alter tumor mTOR pathway inhibition or decrease tumor growth, but it did enhance p62 staining (p<0.01) and significantly increased TUNEL staining (p<0.05) (Fig. 6, Supplementary Fig. S7C). Taken together, these results demonstrate that CC214-2 inhibits mTORC1 and mTORC2 signaling in an intracranial GBM model, and suggests that inhibition of autophagy promotes cell death in response to CC214-2 treatment.
Figure 6. In vivo combinatory treatment in U87EGFRvIII orthotopic xenografts with CC214-2 and chloroquine significantly reduced tumor growth and induced tumor cell death.
Fluorescence Molecular Tomography (FMT) images demonstrating the antitumor activity of CC214-2 in single treatment or in combination with chloroquine in U87EGFRvIII intra-cranial xenograft models, following 6 days of dosing (A). CC214-2 treatment resulted in greater than 50% reduction of tumor volume as assessed by FMT imaging (B). CC214-2 treatment significantly inhibited mTORC1 and mTORC2 signaling, and reduced the ki67 proliferation index. Chloroquine treatment suppressed autophagy, as measured by enhanced p62 staining, promoting tumor cell death (TUNEL positive staining) of CC214-2 - treated mice (C). * indicates P<0.05; ** indicates P<0.01, Student’s t-Test. (n= 8 for the orthotopic xenografts; CC214-2, 100 mg/kg, once every two days by gavage; chloroquine, 30 mg/kg, once every two days intraperitoneally; Ve: vehicle; CC: CC214-2; Chl: Chloroquine).
Discussion
mTOR is a critical nexus in tumor cells, linking growth factor receptor signaling through the PI3K-Akt signaling pathway, with protein translation, cellular metabolism and nutrient and energy status (24, 25). However, the allosteric mTOR inhibitor rapamycin has failed in the clinic for GBM patients, potentially as a consequence of persistent mTORC2 signaling (6) and because of the inability of rapamycin to durably suppress 4E-BP1 phosphorylation (8, 26). The findings presented here, suggest that ATP-competitive mTOR kinase inhibitors such as CC214-1 and CC214-2 may be more effective than rapamycin because of their ability to suppress mTORC2 signaling and mTORC1-dependent 4E-BP1 phosphorylation.
Currently, the molecular determinants of mTOR kinase inhibitors are not well understood. Our data show that EGFRvIII, and to a lesser extent wild type signaling through EGFR, enhance the sensitivity to mTOR kinase inhibitors, whereas the presence of PTEN diminishes that requirement. This finding occurs in the absence of evidence that CC214 compounds are more effective at blocking mTORC1/2 signaling in the context of EGFR/PI3K pathway activation. Rather, these findings suggest that molecular context exerts significant effects on the dependence of tumor cells on mTOR signaling, such that EGFR and PTEN status could potentially be used to stratify patients for treatment. Of note, these data indicate that PI3K pathway activation may be the point of convergence, suggesting that other PI3K activating lesions in GBM may be critical determinants of response to ATP-competitive mTOR kinase inhibitors.
These results also highlight the importance of autophagy as a targeted therapy resistance mechanism in cancer. Our findings are consistent with the work of Weiss group, showing that PI3K/mTOR kinase inhibition induces autophagy in GBM cells (23). Similarly, our evidences, using genetic and pharmacologic autophagy inhibition, indicate that blocking autophagy could potentially transform mTOR kinase inhibitors into much more successful anti-cancer agents by converting a cytostatic to a cytotoxic response.
Lastly, the introduction of the DEAL technology to monitor autophagy in single cancer cells, captured from heterogeneous patient-derived tumors, clearly demonstrates that endogenous EGFRvIII is an active promoter of autophagy in mTOR kinase inhibited tumors.
Autophagy in cancer has a context dependent function, representing both, a tumor suppressor role in the early stage of tumor formation, and conversely, a tumor survival process in response to perturbations, such as chemotherapy treatment (27). As shown by Lock et al., oncogenic Ras addicted cells have higher level of basal autophagy (28), making the combination of Ras and autophagy inhibitors a potentially efficacious treatment for this cancer phenotype (22). Similarly, PTEN null GBM cells also exhibit higher basal levels of autophagy and combining PI3K/mTOR kinase inhibitors with chloroquine causes massive tumor cell death in that genetic context. Here we identify EGFRvIII as a key determinant of mTOR kinase inhibitor sensitivity in GBM, demonstrate that CC214-2 can inhibit mTORC1 and mTORC2 signaling in an intracranial tumor model and we show that chloroquine, or genetic autophagy inhibition, dramatically sensitizes GBMs to CC214-mediated tumor cell death, providing compelling rationale for combination therapy, coupled to a strategy for stratifying patients most likely to benefit.
Supplementary Material
Statement of translational relevance.
Despite the compelling nature of mTOR as a drug target in glioblastoma, the allosteric inhibitor rapamycin has failed to show benefit for glioblastoma patients, potentially because of a failure to fully suppress mTOR signaling. Here we show that the newly described ATP-competitive mTOR kinase inhibitors CC214-1 and CC214-2 suppress rapamycin-resistant mTORC1 signaling; block mTORC2 signaling and significantly inhibit the growth of glioblastomas in vitro and in vivo, and we identify EGFRvIII and PTEN loss as molecular determinants of response. We further demonstrate autophagy as resistance mechanism, which can be overcome with the addition of chloroquine. Taken together this study identifies potentially efficacious new mTOR kinase inhibitors; couples it to a strategy to direct it to patients most likely to benefit, and suggests a combinatorial pharmacological approach to suppress autophagy-dependent drug resistance.
Acknowledgements
Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA; confocal laser scanning microscopy was performed at the CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, supported with funding from NIH-NCRR shared resources grant (CJX1-443835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519); National Institute of Health (NS072838 to D.G.); The European Commission (PIOF-GA-2010-271819 to B.G.).
Funding
This work is supported by grants from National Institute for Neurological Diseases and Stroke (NS73831), the National Cancer Institute (CA119347), The Ben and Catherine Ivy Foundation, and generous donations from the Ziering Family Foundation in memory of Sigi Ziering. WKC is a Fellow of the National Foundation for Cancer Research.
Drs. Mischel and Cloughesy served as advisors to Celgene on the mTOR kinase inhibitor program and participated in a research contract to assess the pre-clinical and clinical efficacy of its compounds. Drs. Chopra, Mortensen, Raymon and Hege are Celgene employees.
Footnotes
All authors report no conflict of interest.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo DL, Gini B, et al. Oncogenic EGFR Signaling Activates an mTORC2-NF-kappa B Pathway That Promotes Chemotherapy Resistance. Cancer Discovery. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galanis E, Buckner JC, Maurer MJ, Kreisberg JI, Ballman K, Boni J, et al. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: a North Central Cancer Treatment Group Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:5294–5304. doi: 10.1200/JCO.2005.23.622. [DOI] [PubMed] [Google Scholar]
- 8.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortensen DS, Sapienza J, Lee BG, Perrin-Ninkovic SM, Harris R, Shevlin G, et al. Use of core modification in the discovery of CC214-2, an orally available, selective inhibitor of mTOR kinase. Bioorganic & medicinal chemistry letters. 2013;23:1588–1591. doi: 10.1016/j.bmcl.2013.01.110. [DOI] [PubMed] [Google Scholar]
- 11.Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, et al. An LXR Agonist Promotes Glioblastoma Cell Death through Inhibition of an EGFR/AKT/SREBP-1/LDLR-Dependent Pathway. Cancer Discov. 2011;1:442–456. doi: 10.1158/2159-8290.CD-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortensen DS, Perrin-Ninkovic SM, Harris R, Lee BG, Shevlin G, Hickman M, et al. Discovery and SAR exploration of a novel series of imidazo[4,5-b]pyrazin-2-ones as potent and selective mTOR kinase inhibitors. Bioorganic & medicinal chemistry letters. 2011;21:6793–6799. doi: 10.1016/j.bmcl.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 13.Shah K, Hingtgen S, Kasmieh R, Figueiredo JL, Garcia-Garcia E, Martinez-Serrano A, et al. Bimodal viral vectors and in vivo imaging reveal the fate of human neural stem cells in experimental glioma model. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:4406–4413. doi: 10.1523/JNEUROSCI.0296-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M, Muller F, Aquilanti E, Hu B, DePinho R. Stereotactic Orthotopic Xenograft Injections into the Mouse Brain. Protocol Exchange. 2012 [Google Scholar]
- 15.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 16.Gini B, Lovato L, Cianti R, Cecotti L, Marconi S, Anghileri E, et al. Novel autoantigens recognized by CSF IgG from Hashimoto's encephalitis revealed by a proteomic approach. J Neuroimmunol. 2008;196:153–158. doi: 10.1016/j.jneuroim.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins. Journal of the American Chemical Society. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Science translational medicine. 2012;4:139ra84. doi: 10.1126/scitranslmed.3003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong GA, Radu CG, Hwang K, Shu CJ, Ma C, Koya RC, et al. Modular nucleic acid assembled p/MHC microarrays for multiplexed sorting of antigen-specific T cells. Journal of the American Chemical Society. 2009;131:9695–9703. doi: 10.1021/ja9006707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Molecular cancer therapeutics. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 21.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Science signaling. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mTOR signaling. Cancer research. 2011;71:2815–2820. doi: 10.1158/0008-5472.CAN-10-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012 doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.