Abstract
Spider dragline silk is considered to be the toughest biopolymer on Earth due to an extraordinary combination of strength and elasticity. Moreover, silks are biocompatible and biodegradable protein-based materials. Recent advances in genetic engineering make it possible to produce recombinant silks in heterologous hosts, opening up opportunities for large-scale production of recombinant silks for various biomedical and material science applications. We review the current strategies to produce recombinant spider silks.
Introduction
Spider silks have been a focus of research for almost two decades due to their outstanding mechanical and biophysical properties. Spider silks are remarkable natural polymers that consist of three domains: a repetitive middle core domain that dominates the protein chain, and non-repetitive N-terminal and C-terminal domains. The large core domain is organized in a block copolymer-like arrangement, in which two basic sequences, crystalline [poly(A) or poly(GA)] and less crystalline (GGX or GPGXX) polypeptides alternate. At least seven different types of silk proteins are known for one orb-weaver species of spider (Lewis, 2006a). Silks differ in primary sequence, physical properties and functions (Hu et al., 2006). For example, dragline silks used to build frames, radii and lifelines are known for outstanding mechanical properties including strength, toughness and elasticity (Gosline et al., 1984). On an equal weight basis, spider silk has a higher toughness than steel and Kevlar (Vepari and Kaplan, 2007; Heim et al., 2009). Flageliform silk found in capture spirals has extensibility of up to 500%. Minor ampullate silk, which is found in auxiliary spirals of the orb-web and in prey wrapping, possesses high toughness and strength almost similar to major ampullate silks, but does not supercontract in water. Figure 1 depicts the location and structural elements of MaSp, MiSp and Flag silks.
Figure 1.
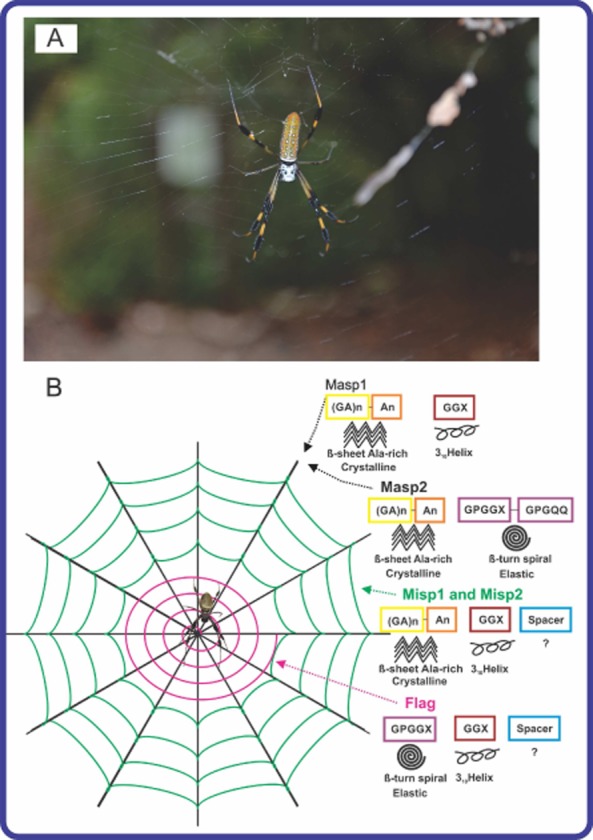
A. An adult female orb weaver spider Nephila clavipes and her web.
B. Schematic overview of N. clavipes web composed of three different spider silk proteins and their structures. The coloured boxes indicate the structural motifs in silk proteins. An empty box marked ‘?’ indicates that the secondary structure of the ‘spacer’ region is unknown. Note: MaSp1 or MaSp2: major ampullate spidroin 1 or 2; MiSp1 and 2: minor ampullate spidroin1 and 2; Flag: flagelliform protein. The photo was taken by Olena and Artem Tokarev in the Florida Keys.
Finally, there are other silk types such as aciniform, pyriform, aggregate and tubuliform (egg case) with unusual primary structure, composition and properties. Diverse and unique biomechanical properties together with biocompatibility and a slow rate of degradation make spider silks excellent candidates as biomaterials for tissue engineering, guided tissue repair and drug delivery, for cosmetic products (e.g. nail and hair strengthener, skin care products), and industrial materials (e.g. nanowires, nanofibres, surface coatings).
Recent advances in genetic engineering have provided a route to produce various types of recombinant spider silks (Prince et al., 1995; Fahnestock and Bedzyk, 1997; Rabotyagova et al., 2009; Xu et al., 2007). However, production of spider silk proteins at a larger scale remains challenging. Moreover, recombinant silk threads do not recapitulate the full potential of native fibres in terms of mechanical properties. Different heterologous host systems have been investigated to develop suitable production systems. In this review, we discuss recent advances in the production of recombinant spider silks in heterologous host systems with the main focus on microbial production. In particular, we focus on dragline silks. Current cloning strategies, expression systems and purification strategies will be discussed to help researchers to engineer customized synthetic spider silk-like proteins for various needs, including biomaterials and material science applications.
Structure of silk proteins
Spider silks are fascinating polymers, as is the spinning process that members of Araneidae family use to make these exceptional materials. Spiders use complex spinning to rapidly transform water soluble, high molecular weight, silk proteins into solid fibres at ambient temperature and pressure, giving rise to an environmentally safe, biodegradable and high performance material (Asakura et al., 2007; Lewicka et al., 2012; Teulé et al., 2012a). The details on anatomy and physiology of the spider spinning apparatus (N. clavipes) can be found elsewhere (Knight and Vollrath, 2001; 2002; Eisoldt et al., 2011; Rising et al., 2011).
In order to understand the challenges and needs associated with biotechnological production of recombinant spider silks, primary protein motifs, composition and secondary structural elements must be discussed. As mentioned earlier, one spider is capable of producing up to seven different types of silks with varying mechanical properties. In spite of different mechanical and physiological properties, the majority of spider silks share a common primary structural pattern comprised of a large central core of repetitive protein domains flanked by non-repetitive N- and C-terminal domains. The most investigated silk is dragline silk, which shows a remarkable combination of strength and elasticity. The golden orb-weaver spider, N. clavipes, produces dragline silk in the major ampullate gland (Knight and Vollrath, 2001). Dragline silk is the protein complex composed of major ampullate dragline silk protein 1 (MaSp1) and major ampullate dragline silk protein 2 (MaSp2). Both silks are approximately 3500 amino acid long. MaSp1 can be found in the fibre core and the periphery, whereas MaSp2 forms clusters in certain core areas. The large central domains of MaSp1 and MaSp2 are organized in block copolymer-like arrangements, in which two basic sequences, crystalline [poly(A) or poly(GA)] and less crystalline (GGX or GPGXX) polypeptides alternate in core domain. The main difference between MaSp1 and MaSp2 is the presence of proline (P) residues accounting for 15% of the total amino acid content in MaSp2 (Hu et al., 2006), whereas MaSp1 is proline-free. By calculating the number of proline residues in N. clavipes dragline silk, it is possible to estimate the presence of the two proteins in fibres; 81% MaSp1 and 19% MaSp2 (Brooks et al., 2005). Different spiders have different ratios of MaSp1 and MaSp2. For example, a dragline silk fibre from the orb weaver Argiope aurantia contains 41% MaSp1 and 59% MaSp2 (Huemmerich et al., 2004). Such changes in the ratios of major ampullate silks can dictate the performance of the silk fibre (Vollrath and Knight, 1999). Specific secondary structures have been assigned to poly(A)/(GA), GGX and GPGXX motifs including β-sheet, 310-helix and β-spiral respectively (Humenik et al., 2011). The primary sequence, composition and secondary structural elements of the repetitive core domain are responsible for mechanical properties of spider silks; whereas, non-repetitive N- and C-terminal domains are essential for the storage of liquid silk dope in a lumen and fibre formation in a spinning duct (Ittah et al., 2006). The primary amino acid sequence, composition and secondary structural elements of other silk types are reviewed elsewhere (Lewis, 2006b; Humenik et al., 2011).
Production of recombinant silk proteins
Spiders cannot be farmed, in contrast to silkworms, due to their aggressive behaviour and territorial nature (Kluge et al., 2008). Collecting silk from webs is a time-consuming task. It took 8 years to make a golden spider silk cape from 1.2 million golden orb webs (Chung et al., 2012). Therefore, biotechnological production of recombinant spider silks is the only practicable solution to harvest silks on a larger scale and to meet growing needs of medicine and biotechnology. A variety of heterologous host systems have been explored to produce different types of recombinant silks (Table 1 and Table 2). Recombinant partial spidroins as well as engineered silks have been cloned and expressed in bacteria (Escherichia coli), yeast (Pichia pastoris), insects (silkworm larvae), plants (tobacco, soybean, potato, Arabidopsis), mammalian cell lines (BHT/hamster) and transgenic animals (mice, goats).
Table 1.
Summary of recombinantly expressed spider silks in bacteria and yeasts. Spider silk origin, number of monomers, molecular weight, cloning and expression plasmids as well as restriction enzymes and purification strategies used to produce recombinant silks are shown
| Type | Host | Origin | Protein | Number of monomers | MW (KDa) | Cloning plasmid | RE | Expression plasmid | Purification Strategy | Yield (mg L−1) | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | E. coli | Nephila clavipes | 16, 32, 64, 96 | 55, 100, 193, 285 | pET30a(+) | Nhe I/ Spe I | pET30a(+) | Ammonium sulphate | Xia et al., 2010 | ||
| 6; 15 | 16, 39.5 | pETNX | PA | 96.8 mg L−1; 200 mg L−1 | Dams-Kozlowska et al., 2012 | ||||||
| MaSp 1 | 8; 16 | 65–163 | pBR322 derived | Pst I | pFP202 (pET9a + pET11a) | 300 mg L−1 | Fahnestock and Irwin, 1997 | ||||
| 8; 16 | 65–163 | pFP202, pFP204, or pFP207 | IMAC | Fahnestock and Irwin, 1997 | |||||||
| 16, 24 | 46, 70 | pBSSKII+ | AvrII, Nhe1 | pET19k | N A−1 | An et al., 2011 | |||||
| poly(A) and GGX | 1, 2, 3, 6 | 10, 18 | pET30a(+) | Spe I/Nde I | pET30a(+) | 25 mg ml−1 | Rabotyagova et al., 2009 | ||||
| E. coli | Nephila clavipes | 8; 16 | 65–163 | pBR322 derived | Pst I | pFP202, pFP204, or pFP207 | 300 mg L−1 | Fahnestock and Irwin, 1997 | |||
| 8, 16, 32 | 31, 58, 112 | pBBSK | Sca/Xma/BspEI | pET19b | 10 mg g−1 | Lewis et al., 1996 | |||||
| Argiope aurantia | MaSp2 | 16 | 63 | IMAC | Brooks et al., 2008 | ||||||
| 12 | 71 | pET30a(+) | N A−1 | pET30a(+) | N A−1 | Brooks et al., 2008 | |||||
| 8 | 67 | Brooks et al., 2008 | |||||||||
| E. coli | Nephila clavipes | Masp1/Masp2 | 24/16 | 62/47 | pBSSKII+ | Xma1/Sca1/BspE1 | pET19K | IMAC | 120 mg L−1 | An et al., 2011 | |
| 1x-18x | 15, 23, 32, 41 | pUC18 | Spe I/Nde I | pQE-9 | 15, 7, 3, 2 mg L−1 | Prince et al., 1995 | |||||
| E. coli | Argiope trifasciata | AcSp1 | 2, 3, 4 | 19, 38, 51.7, 76.1 | pET32 | BamHI/ BsgI/ BseRI | pET32 | IMAC | 80 mg L−1; 22 mg L−1 | ||
| Nephila antipodiana | TuSp1 | 11 | 190 | Xma1/Pvu1/BspE1 | 40 mg L−1 | Xu et al., 2007 | |||||
| Salmonella | Araneus diadematus | ADF1 | 1x-3x | 30–56 | pJ2 | HindIII/XbaI | pTRC99a_Cm | SEC | N A−1 | Widmaier et al., 2009 | |
| ADF2 | 1x-3x | Widmaier et al., 2009 | |||||||||
| ADF3 | 1x-3x | Widmaier et al., 2009 | |||||||||
| Yeast | Pichia Pastori | Nephila clavipes | Masp 1 | 8, 16 | 65 | pBR322 derived | Pst I | pFP684 | Ammonium sulphate | 663 mg L−1 | Fahnestock and Bedzyk, 1997 |
Table 2.
Summary of recombinantly expressed spider silks in insects, plants and mammalians. Spider silk origin, number of monomers, molecular weight, cloning and expression plasmids as well as restriction enzymes and purification strategies used to produce recombinant silks are shown
| Type | Host | Origin | Protein | Number of monomers | MW (KDa) | Cloning plasmid | RE | Expression plasmid | Purification strategy | Yield (mg L−1) | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Insects | B. mori | Nephila clavipes | Masp1 | 2 | 83 | pSLfa1180fa | Spe1/Nde1 | pBac[3xP3-DsRedaf] | IMAC | N A−1 | Wen et al., 2010 |
| 4 | 70 | pSL1180 | pFastBacHT-C | 6 mg/larva | Zhang et al., 2008 | ||||||
| Masp1+Flag | multiple | 75–130 | pBSSKII+ and pSLfa1180fa | Sca1/Xma1/BspE1 | pBAC[3xP3-DsRedaf] | N A−1 | Teulé et al., 2012b | ||||
| Plants | Nicotiana tobaccum Solaum tubercum | Masp1 | multiple | 12.9–99.8 | pUC19 | NgoMIV/HindIII | pRTRA7/3 | (NH4)2SO4 10–50% saturation | 0.5-2 % total protein | Scheller et al., 2001 | |
| Nicotiana benthamiana | Nephila clavipes | Flag (intein) | 4; 10 | 47, 72, 100, 250 | pRTRA15 | splicing events | pCB301-Kan | IMAC | 1.8 mg/50 g leaft material 0.34%; 0.03% in leaves, 1.2%; 0.78% in seeds | Hauptmann et al., 2013 | |
| Arabdopsis thaliana Glycine max | Masp1 | 8, 16 | 64, 127 | pBSSK+ | BglII/BamH1 | Cong' + Pha3' | ammonium sulphate | 1% in somatic embryos | Barr et al., 2004 | ||
| Mammalians | Trangenic mice | Nephila clavipes | MaSP1 | 6 | 31–66 | pGEM-5zf | BamHI/NcoI | pBC1 | centrifugation | 11.7 mg L−1 | Xu et al., 2007 |
| COS-1 cells | Euprosthenops sp. | 25, 22 | pER1-14 | BamH1/EcoRV | pSecTag2/Hygro A | N A−1 | N A−1 | Grip et al., 2006 | |||
| Baby hamster kidney | Nephila clavipes | Masp1/ Masp2 | N A−1 | 59, 106/ 59 | pBSSK+ | ApaI/SapI | CMV promoter | ammonium sulfate | |||
| Baby hamster kidney | Araneus diadematus | ADF3 | 63, 60, 110, 140 | pSecTag-C | MscI/PvuII | 25–50 mg L−1 | Lazaris et al., 2002 |
Unicellular organisms as heterologous host systems
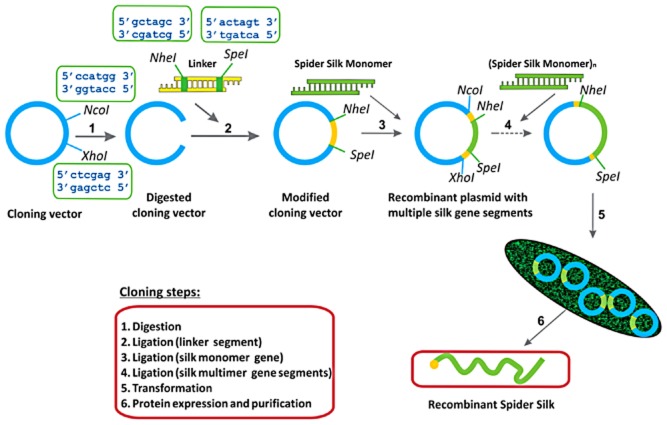
Unicellular organisms, such as bacteria and yeast, have been investigated as host systems for recombinant silks. A gram-negative, rod-shaped bacterium E. coli is a well-established host for industrial scale production of proteins. Therefore, the majority of recombinant spider silks have been produced in E. coli (Lewis et al., 2011; Fahnestock and Irwin, 1997; Wang et al., 2006; Rabotyagova et al., 2009; Rabotyagova et al., 2010; An et al., 2011; An et al., 2012; Teulé et al., 2012a). E. coli is easy to manipulate, has a short generation time, is relatively low cost and can be scaled up for larger amounts protein production. The recombinant DNA approach enables the production of recombinant spider silks with programmed sequences, secondary structures, architectures and precise molecular weight (Rabotyagova et al., 2011). There are four main steps in the process: (i) design and assembly of synthetic silk-like genes into genetic ‘cassettes’, (ii) insertion of this segment into a DNA vector, (iii) transformation of this recombinant DNA molecule into a host cell and (iv) expression and purification of the selected clones. Figure 2 summarizes the recombinant DNA approach used to prepare silk-like proteins.
Figure 2.
Recombinant DNA approach used to prepare silk-like proteins.
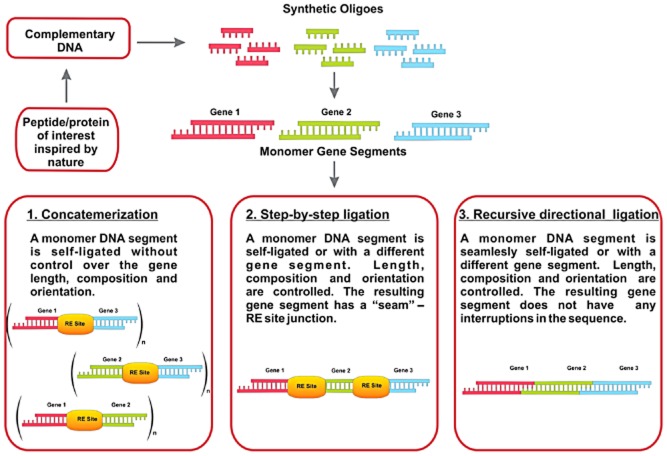
The monomeric silk-like gene sequences can be synthesized as short single-stranded oligonucleotides (up to 100 bp) by commercial oligonucleotide synthesis or used directly as polymerase chain reaction products from cDNA libraries. Large repetitive sequences can be constructed by using concatemerization, step-by-step directional approach and recursive ligation (Fig. 3). Concatemerization is a useful method when a library of genes of different sizes is desired but has limitations in the preparation of genes with specific sizes (Meyer and Chilkoti, 2002). To overcome limitations of concatemerization, recursive directional ligation or a step-by-step ligation is employed (Meyer and Chilkoti, 2002; Wright and Conticello, 2002). Recursive directional ligation allows for facile modularity, where control over the size of the genetic cassettes is achieved. Moreover, recursive directional ligation eliminates the restriction sites at the junctions between monomeric genetic cassettes without interrupting key gene sequences with additional base pairs that makes it different from the step-by-step ligation approach (Higashiya et al., 2007).
Figure 3.
Gene multimerization approaches. Note: RE Site stands for a restriction enzyme site.
For example, we have employed step-by-step directional ligation to produce various partial recombinant spider silks as well as engineered silk-like proteins based on the sequences of dragline silk originated from N. clavipes (Prince et al., 1995; Wang et al., 2006; Huang et al., 2007; Rabotyagova et al., 2009; 2010; Mieszawska et al., 2010; Gomes et al., 2011; Numata et al., 2012). As one example, spider silk block copolymers were generated in E. coli (Rabotyagova et al., 2009; 2010). In the first cloning step, a commercially available pET30a(+) vector (Novagen, San Diego, CA, USA) was modified with an adaptor sequence, carrying NheI and SpeI restriction sites. The adaptor was inserted into XhoI and NcoI sites of a pET30a(+) to generate pET30L. The coding sequences of two spider silk-like monomers A (hydrophobic block) and B (hydrophilic) were designed to carry SpeI and NheI restriction sites at the ends of the sequences. This allowed ligation of the domains into a pET30L vector. By using a step-by-step directional ligation approach, direct control over the assembly of monomeric genes into complex sequences was achieved. Six different constructs were cloned and transformed into the bacterial host for expression. An N-terminal His-tag was used for protein purification by immobilized metal affinity chromatography (Rabotyagova et al., 2009).
Another genetic engineered strategy has been proposed by Lewis Laboratory to assemble long repetitive spider silk genes (Teule et al., 2009). This cloning strategy employs a one-step head-to-tail ligation that can produce large inserts in precise manner (Lewis et al., 1996; Brooks et al., 2008; Teule et al., 2009; Teulé et al., 2012a). The spider silk synthetic genes were optimized for codon usage in E. coli and were cloned into a plasmid vector pBluescriptII SK(+) (Stratagene). Each silk module was carrying compatible XmaI and BspEI restriction sites at the ends on the coding sequences. The vector also contained a unique restriction site (ScaI) in the ampicillin resistance gene. By simultaneously performing two double digestion reactions ScaI – XmaI and ScaI – BspEI two fragments each containing a copy of a silk monomer gene were obtained. The fragments were ligated together using T4 ligase resulting in the doubling of the size of silk genes and restoring the ampicillin resistance of the plasmid (Fig. 4). Several round of cloning were performed to obtain repetitive sequences of a desired size. Next, the multimeric synthetic genes were subcloned into an expression pET19b vector using NdeI and BamHI restriction sites. Since the expression vector was carrying NdeI and BamHI sites, the liberated inserts were cloned in-frame with pET19b. Similar to pET30L, silk genes in pET19b are under control of the T7 promoter and require the addition of isopropyl-β-D-1-thiogalactopyranoside to initiate protein expression. The expressed proteins can be purified by immobilized metal affinity chromatography (IMAC) due to the presence of an N-terminal His-tag. Several recombinant spider silk proteins from different species were produced using this genetic engineering strategy including silks from N. clavipes (Teule et al., 2009) Argiope aurantia (Brooks et al., 2008). Recombinant spider silk proteins from Nephylengys cruentata, Parawixia bistriata and Avicularia juruensis were produced employing this cloning strategy (Leopoldo et al., 2007) (US patent 20 100 311 645). Figure 4 summarizes the strategy.
Figure 4.
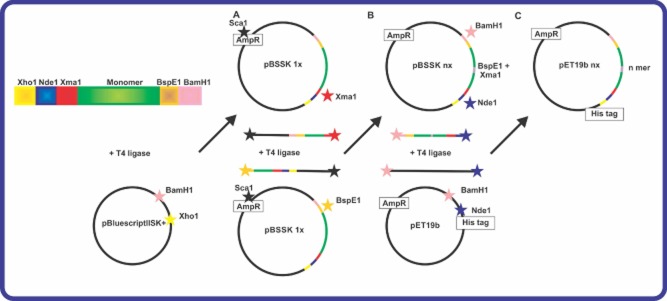
Cloning strategy used by the Lewis group to engineer long repetitive spider silk sequences (in green).
A. Cloning of a silk monomer into the vector pBluescript II SK+.
B. The resulting plasmid is double digested and fragments containing silk monomers are ligated again to produce longer sequences.
C. The synthetic spider silk multimer is ligated into pET19b expression vector. Note: Restriction digestion sites are indicated by star. Adapted from reference (Teule et al., 2009).
A three module cloning strategy based on the sequences of ADF-3 and ADF-4 was developed by Scheibel research group (Huemmerich et al., 2004), designed so that multiple modules can be combined. Moreover, additional coding sequences such as N- or C-terminal domains can be added if needed. The purification protocol is based on heat resistance of silk proteins followed by an ammonium sulphate precipitation that is different from Ni-NTA IMAC.
Different purification strategies have been employed recently to optimize small and large-scale production of recombinant silks. Most of the spider silk proteins are produced with an N- or C-terminal His-tags to make purification simple and produce enough amounts of the protein. However, the presence of this tag can affect protein secondary structure and interfere with the process of spider silk fibre formation. Dams-Kozlowska et al. (2012) proposed two strategies to purify spider silks from lysates without the use of a His-tag. These protocols are based on thermal treatment and organic acid resistance of silk proteins and do not require the presence of the His-tag. After purification, silk proteins based on MaSp1 gene sequence were formed into films that subsequently were used to grow murine fibroblast cell culture. The results demonstrated that silk films were non-toxic to the cells (Dams-Kozlowska et al., 2012).
Because of the highly repetitive core sequence of spider silk genes, frequent homologous recombination, deletions, transcription errors, translation pauses, accumulation in inclusion bodies and low yields were observed during the production of recombinant silks in E. coli. Moreover, when the protein size was increased from 43 kDa to higher (the size of native spidroins is between 300 and 350 kDa), protein yields decreased dramatically. Codon optimization for the specific host expression system helped maximize the translation of the foreign gene transcripts and thus, improved protein yields (Fahnestock and Bedzyk, 1997, Lewis, 2006b). It was also suggested that depletion of tRNA pools upon protein expression resulted in transcription and translation errors (Rosenberg et al., 1993). Recently, Xia et al. (2010) employed a metabolic engineered strategy to enhance the production of recombinant spider silks. The authors reported production of full length (284.9 kDa) recombinant N. clavipes dragline silk proteins that were rich in glycine (43–45%). Production of these silk proteins was enhanced by the use of the metabolically engineered expression host within which the glycyl-tRNA pool was elevated. The fibres spun with the native-sized recombinant spider silk protein showed tenacity, elongation and Young's modulus of 508 MPa, 15% and 21 GPa, respectively, comparable to those of native spider dragline silk (Xia et al., 2010). Through extensive proteomic analysis, serine hydroxymethyltransferase (GlyA) and β-subunit of glycly-tRNA synthetase (GlyS) were found to be upregulated to meet the high cellular demand for glycly-tRNA when expressing glycine-rich silk proteins. Increased glycine biosynthetic flux by overexpressing glycyl-tRNA synthetase elevated the total tRNAGly pool and resulted in enhanced production of high molecular weight recombinant spider silks.
Recently, large spider recombinant egg case silk protein from Nephila antipodiana, 378 kDa, was engineered using E. coli, where gene multimers were chemically linked by cysteine disulfide bonds. The recombinant silk sequence consisted of two silk proteins: tubuliform spidroin 1 (TuSp1) and C-terminal domain of MisP1. Non-repetitive C-terminal domain of MiSp1 was chosen due to its higher water solubility and stability compared with the C-terminal domain of TuSp1. A disulfide linkage between two C-terminal domains was formed by introducing a point mutation (S76 to S76C). This link allowed the formation of a hybrid DNA construct that was expressed in E. coli (DE3). The recombinant protein was expressed in E. coli. Moreover, the artificial fibres spun from this protein showed higher tensile strength and Young' modulus than natural egg case protein (Lin et al., 2013).
The highly repetitive silk gene arrangement and the unusual mRNA secondary structure result in inefficient translation that limits the size of the silks produced in E. coli. To minimize the presence of truncated silk proteins and allow the extracellular secretion of silks, the mythylotropic yeast P. pastoris has been used. Fahnestock and Bedzyk (1997) produced N. clavipes spider dragline silks in yeast P. pastoris. Synthetic genes were expressed at high levels under control of the methanol-inducible AOX1 promoter. Transformants containing multiple gene copies produced elevated levels of silk protein. Results demonstrated that P. pastoris can be used to successfully produced produce long repetitive proteins (Fahnestock and Bedzyk, 1997).
Spider silks from Araneus diadematus (ADF-1, 2 and 3) have also been expressed using the type III secretion system of a gram-negative, non-spore-forming, enterobacterium Salmonella. The authors reported yield values range from 90 to 410 nmol L−1h−1 that is similar to 10 mg L−1 h−1 for a protein the size of ADF-2. The results demonstrated the feasibility to use Salmonella for the large-scale spider silk production (Widmaier et al., 2009). Mammalian cell lines, such as bovine mammary epithelial alveolar and baby hamster kidney cells, were used to express MaSp1 and MaSp2 (Lazaris et al., 2002). The cells expressed recombinant proteins; however, as size of silk gene increased, the yield decreased dramatically due to inability of mammalian cells to cope with large repetitive sequences. Several factors have attributed to the decreased yields including, but not limited to, inefficient transcription, insufficient secretion, low copy numbers and translational limitations. The produced silk proteins were spun into fibres, and their mechanical properties were tested. It was noted that those recombinant silks that were produced without a His-tag demonstrated better mechanical properties compared with fibres made of silk proteins with a His-tag (i.e. fibres were brittle). Similar problems (i.e. transcription and translation limitations) have been reported when green monkey kidney fibroblast-like cell lines (COS-1) were used to express a 636-base pair gene fragment of MaSp1 from the African spider Euprosthenops sp. (Grip et al., 2006). Table 1 summarizes genetic engineering approaches, cloning strategies, and production yields of recombinant silk proteins produced in unicellular heterologous host systems.
Multicellular organisms as heterologous host systems
Due to the low production rate and instability (i.e. frequent homologous recombination, deletions, transcription errors, translation pauses) of spider silk repetitive genes in unicellular organisms, multicellular organisms such as insects, plants and mammals have been studied for production of recombinant spider silk proteins.
Silkworms (B. mori) can be farmed and produce cocoons containing large quantities of silkworm silk known as fibroin (Vepari and Kaplan, 2007; Hu and Kaplan, 2011). Moreover, to produce a solid thread, silkworms employ a spinning process that is similar to that used by spiders to make dragline silk. The presence of a natural silk production system in silkworms makes them excellent candidates to investigate as heterologous hosts for spider silk production. There have been several reports of the transfer of silk genes from spiders to silkworms (Motohashi et al., 2005; Zhang et al., 2011; Teulé et al., 2012b).
Baculovirus-based expression systems have been used to introduce silk genes into a heterologous host. Baculovirus infects silkworms and allows for production of large quantities of heterologous proteins in a short period of time (Motohashi et al., 2005). Using this expression system, MaSp1 from N. clavipies linked with an enhanced green fluorescent protein (EGFP) fusion protein was cloned and expressed in the B. mori cell line (BmN) and larvae (Zhang et al., 2008). The authors reported successful production of a recombinant EGFP-MaSp1 fusion protein in both systems. In the silkworm larvae, a total of 6 mg of fusion protein was expressed, whereas in the BmN cells, 5% of the cell total protein was occupied by this recombinant silk. The major limitations of this expression system were low solubility of silk proteins and inability to assemble spider silk fibres. It was shown that more than 60% of the fusion proteins formed aggregates via self-assembly. To overcome solubility issues, MaSp1 C-terminal domain is to be incorporated due to its role to prevent aggregate formation. To produce fibres, germline-transgenic silkworms (B. mori) were produced by injecting silkworm eggs with a piggyBac transformation vector carrying MaSp1 sequence (Wen et al., 2010). The insects were capable of spinning fibres and forming cocoons containing recombinant spider silk. However, the mechanical properties of the fibres were lower than dragline MaSp1 silk due to the low ratio of MaSp1 in the total silk protein.
In a recent effort to develop tough fibres, transgenic silkworms encoding chimeric silkworm/spider silk proteins were produced using piggyBac vectors (Teulé et al., 2012b). The vector, used previously by the Tamada group (Kojima et al., 2007) included the B. mori fibroin heavy chain promoter and enhancer, a genetic sequencing encoding a 78 kDa synthetic spider silk protein, and an EGFP tag. Strong EGFP signals were observed by fluorescence (Fig. 5). The composite fibres were tougher than the parental silkworm silk fibres and as tough as native dragline spider silk fibres.
Figure 5.
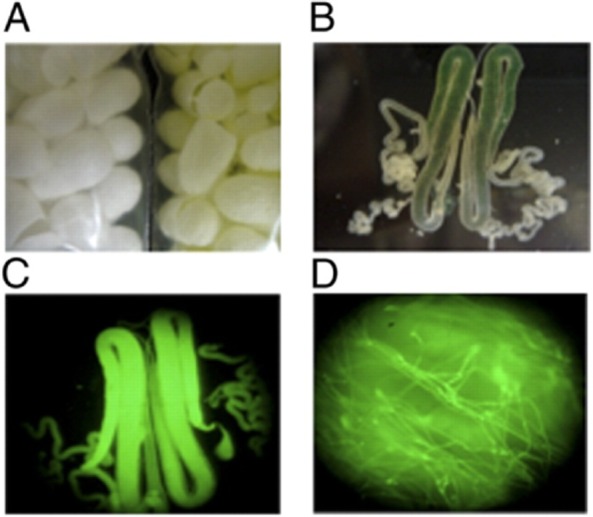
Expression of the chimeric silkworm/spider silk/EGFP protein in (A) cocoons, (B and C) silk glands and (D) silk fibres from spider 6-GFP silkworms. Reproduced with permission from (Teulé et al., 2012b).
These results demonstrate that silkworms can be engineered to generate composite silk fibres containing stably integrated spider silk protein sequences, which significantly improved overall mechanical properties.
Transgenic plants have also been investigated as heterologous host systems to produce recombinant spider silks. Advances in genetic engineering technology and transformation methods make it possible to produce non-plant proteins in plants (Yang et al., 2005; Rech et al., 2008). Moreover, one plant offers several different expression systems, such as seeds, leaves, tubers and roots with potential for organelle-specific accumulation of recombinant proteins (Scheller and Conrad, 2005).
Stable transgenic tobacco and potato lines were engineered to express MaSp1 genes from N. clavipes ranging from 420 to 3600 bp (Scheller et al., 2001). Recombinant spider silk proteins were found in the endoplasmic reticulum (ER) of tobacco and potato leaves at the accumulation of 2% of total soluble protein. Moreover, the production levels were independent of the size of silk genes. Purification was performed using high temperature treatment followed by acidification and ammonium sulphate precipitation. Additionally, recombinant MaSp1-like proteins were also produced in the leaves and seeds of Arabidopsis (small flowering plants related to cabbage) as well as in somatic soybean embryos (Barr et al., 2004). The expression of recombinant silks was driven by the 35S promoter in leaves and the β-conglycinin α' subunit promoter in seeds and somatic soybean embryos. The results demonstrated that recombinant spider silk proteins had higher accumulation levels in seeds than in the leaves. Recently, a native-sized FLAG protein from N. clavipes was cloned and expressed in the ER of tobacco plant (Nicotiana benthamiana) leaf cells using an intein-based posttranslational protein fusion technology (Hauptmann et al., 2013). This method avoids the need for highly repetitive transgenes resulting in a higher genetic and transcriptional stability. Additional details on production of fibrous proteins in plants can be found elsewhere (Scheller and Conrad, 2005).
Transgenic production of recombinant silk proteins in mammary glands and secretion of them into milk has been investigated in mice and goats (Williams, 2003; Xu et al., 2007). In case of transgenic mice production, MaSp1 and MaSp2 synthetic genes (40 and 55 kDa) were synthesized and cloned into the pBC1 expression vector (Invitrogen, Carlsbad, CA, USA) together with a goat β-casein signal sequence. The chimeric gene construct was microinjected into pronuclei of fertilized eggs of Kunming white mice (Xu et al., 2007). Southern blot analysis was used to identify mice containing transgene construct as well as a copy number of transgene. The expression of dragline silk in milk was confirmed by Northern blot followed by Western blot analysis. The results revealed that transgenic mice were capable of expressing recombinant silk proteins in their milk. Genetically engineered (transgenic) goats capable of expressing spider silk proteins based on the sequences of MaSp1 and MaSp 2 were produced by Nexia Biotechnologies, and later by the Lewis group (Lazaris et al., 2002; Service, 2002). Silk protein expression was controlled by the β-casein promoter and was expressed in the milk of transgenic goats. Silk proteins were observed only in mammary tissues as confirmed by Western blot (Steinkraus et al., 2012). Maximum yields observed for the recombinant silk production in transgenic animals were low (11.7 mg l−1) when compared with bacterial expression (Table 1 and Table 2). Today, the large-scale production of recombinant silk proteins from transgenic animals is relatively expensive and challenging in terms of animal breeding.
Future outlook
Over the last decade there has been considerable progress in understanding the genetic organization encoding spider silks. Cloning, expression and purification of spider silks has improved, and the self-assembly and processing of spider silk into many material formats is now better understood. Recently a native-sized (285 kDa) recombinant protein of the spider N. clavipes was produced and spun into a fibre displaying mechanical properties comparable to those of the native silk, indicating a breakthrough in standard recombinant production of spider silks. Moreover, a variety of heterologous host systems have been explored to produce different types of recombinant silks. For example, transgenic silkworm/spider silk production systems have been developed to produce tough fibres. It is possible to mix and match key modules via recombinant approaches, providing additional insights into the role of individual modules and effects of neighbouring elements on properties. This approach should lead to the development of custom structures built from specific silk elements. Future challenges will include development of tailor-made production systems for recombinant silks keeping in mind differences in chemical and physical properties of individual silk modules, scaling up silk production, prevention of the formation of aggregates and matches to the mechanical properties of silk fibres.
Acknowledgments
The authors thank the NIH (EB014976, EB002520; EY020856; DE017207; EB014283) and the BSF for support of the studies reviewed here. This study was supported in part by the National Council for Scientific and Technological Development (CNPq), Brazil, Embrapa Genetics Resources and Biotechnology, Brasilia, DF, Brazil and Foundation for Research Support (FAP-DF), Brazil.
References
- An B, Hinman MB, Holland GP, Yarger JL, Lewis RV. Inducing β-sheets formation in synthetic spider silk fibers by aqueous post-spin stretching. Biomacromolecules. 2011;12:2375–2381. doi: 10.1021/bm200463e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Jenkins JE, Sampath S, Holland GP, Hinman M, Yarger JL, Lewis R. Reproducing natural spider silks' copolymer behavior in synthetic silk mimics. Biomacromolecules. 2012;13:3938–3948. doi: 10.1021/bm301110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T, Umemura K, Nakazawa Y, Hirose H, Higham J, Knight D. Some observations on the structure and function of the spinning apparatus in the silkworm Bombyx mori. Biomacromolecules. 2007;8:175–181. doi: 10.1021/bm060874z. [DOI] [PubMed] [Google Scholar]
- Barr LA, Fahnestock SR, Yang J. Production and purification of recombinant DP1B silk -like protein in plants. Mol Breed. 2004;13:345–356. [Google Scholar]
- Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. An investigation of the divergence of major ampullate silk fibers from Nephila clavipes and Argiope aurantia. Biomacromolecules. 2005;6:3095–3099. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- Brooks AE, Stricker SM, Joshi SB, Kamerzell TJ, Middaugh CR, Lewis RV. Properties of synthetic spider silk fibers based on Argiope aurantia MaSp2. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- Chung H, Kim TY, Lee SY. Recent advances in production of recombinant spider silk proteins. Curr Opin Biotechnol. 2012;23:957–964. doi: 10.1016/j.copbio.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Dams-Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. J Biomed Mater Res A. 2012;101A:456–464. doi: 10.1002/jbm.a.34353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisoldt L, Smith A, Scheibel T. Decoding the secrets of spider silk. Mater Today. 2011;14:80–86. [Google Scholar]
- Fahnestock SR, Bedzyk LA. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl Microbiol Biotechnol. 1997;47:33–39. doi: 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- Fahnestock SR, Irwin SL. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl Microbiol Biotechnol. 1997;47:23–32. doi: 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- Gomes SC, Leonor IB, Mano JF, Reis RL, Kaplan DL. Antimicrobial functionalized genetically engineered spider silk. Biomaterials. 2011;32:4255–4266. doi: 10.1016/j.biomaterials.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosline JM, Denny MW, DeMont ME. Spider silk as rubber. Nature. 1984;309:551–552. [Google Scholar]
- Grip S, Rising A, Nimmervoll H, Storckenfeldt E, McQueen-Mason S, Pouchkina-Stantcheva N, et al. Transient expression of a major ampullate spidroin 1 gene fragment from Euprosthenops sp. in mammalian cells. Cancer Genomics and Proteomics. 2006;3:83–87. [PubMed] [Google Scholar]
- Hauptmann V, Weichert N, Menzel M, Knoch D, Paege N, Scheller J, et al. Native-sized spider silk proteins synthesized in planta via intein-based multimerization. Transgenic Res. 2013;22:369–377. doi: 10.1007/s11248-012-9655-6. [DOI] [PubMed] [Google Scholar]
- Heim M, Keerl D, Scheibel T. Spider silk: from soluble protein to extraordinary eiber. Angew Chem Int Edit. 2009;48:3584–3596. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- Higashiya S, Topilina NI, Ngo SC, Zagorevskii D, Welch JT. Design and Preparation of b-Sheet Forming Repetitive and Block-Copolymerized Polypeptides. Biomacromolecules. 2007;8:1487–1497. doi: 10.1021/bm061098y. [DOI] [PubMed] [Google Scholar]
- Hu X, Kaplan DL. Silk Biomaterials. In: Ducheyne P, editor. Comprehensive Biomaterials. Oxford, UK: Elsevier; 2011. pp. 207–219. [Google Scholar]
- Hu X, Vasanthavada K, Kohler K, McNary S, Moore A, Vierra C. Molecular mechanisms of spider silk. Cell Mol Life Sci. 2006;63:1986–1999. doi: 10.1007/s00018-006-6090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wong C, George A, Kaplan DL. The effect of genetically engineered spider silk-dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation. Biomaterials. 2007;28:2358–2367. doi: 10.1016/j.biomaterials.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Huemmerich D, Helsen CW, Quedzuweit S, Oschmann J, Rudolph R, Scheibel T. Primary structure elements of spider dragline silks and their contribution to protein solubility. Biochemistry. 2004;43:13604–13612. doi: 10.1021/bi048983q. [DOI] [PubMed] [Google Scholar]
- Humenik M, Scheibel T, Smith A. Spider silk understanding the structure-function relationship of a natural fiber. Prog Mol Biol Transl Sci. 2011;103:131–185. doi: 10.1016/B978-0-12-415906-8.00007-8. [DOI] [PubMed] [Google Scholar]
- Ittah S, Cohen S, Garty S, Cohn D, Gat U. An essential role for the C-terminal domain of a dragline spider silk protein in directing fiber formation. Biomacromolecules. 2006;7:1790–1795. doi: 10.1021/bm060120k. [DOI] [PubMed] [Google Scholar]
- Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Spider silks and their applications. Trends Biotechnol. 2008;26:244–251. doi: 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Knight DP, Vollrath F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften. 2001;88:179–182. doi: 10.1007/s001140100220. [DOI] [PubMed] [Google Scholar]
- Knight DP, Vollrath F. Spinning an elastic ribbon of spider silk, Philosophical Transactions of the Royal Society B. Biol Sci. 2002;357:219–227. doi: 10.1098/rstb.2001.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Kuwana Y, Sezutsu H, Kobayashi I, Uchino K, Tamura T, Tamada Y. A new method for the modification of fibroin heavy chain protein in the transgenic silkworm. Biosci Biotechnol Biochem. 2007;71:2943–2951. doi: 10.1271/bbb.70353. [DOI] [PubMed] [Google Scholar]
- Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- Leopoldo RFE, Cristina VFN, Rodrigues VG, Rodrigues DSF, Jose LAF, Alberto CL, et al. 2007. US20100311645 – Proteins from the webs of Nephilengys cruentata, Avicularia juruensis and Parawixia bistriata spiders isolated from brazilian biodiversity. In: US20100311645 A1 20101209: Empresa Brasileira de Pesquisa Agropecuaria- EMBRAPA (Brasilia – DF, BR); Fundacao Universidade de Brasilia (Brasilia – DF, BR)
- Lewicka M, Hermanson O, Rising AU. Recombinant spider silk matrices for neural stem cell cultures. Biomaterials. 2012;33:7712–7717. doi: 10.1016/j.biomaterials.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Lewis RV. Spider silk: ancient ideas for new biomaterials. Chem Rev. 2006a;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- Lewis RV. Spider silk production. In: Renugopalakrishnan V, Lewis RV, editors. Bionanotechnology. Proteins to Nanodevices. Dordrecht, the Netherlands: Springer; 2006b. pp. 61–78. [Google Scholar]
- Lewis RV, Hinman M, Kothakota S, Fournier MJ. Expression and purification of a spider silk protein: a new strategy for producing repetitive proteins. Protein Expr Purif. 1996;7:400–406. doi: 10.1006/prep.1996.0060. [DOI] [PubMed] [Google Scholar]
- Lin Z, Deng Q, Liu XY, Yang D. Engineered large spider eggcase silk protein for strong artificial fibers. Adv Mater. 2013;25:1216–1220. doi: 10.1002/adma.201204357. [DOI] [PubMed] [Google Scholar]
- Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- Mieszawska AJ, Fourligas N, Georgakoudi I, Ouhib NM, Belton DJ, Perry CC, Kaplan DL. Osteoinductive silk–silica composite biomaterials for bone regeneration. Biomaterials. 2010;31:8902–8910. doi: 10.1016/j.biomaterials.2010.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi T, Shimojima T, Fukagawa T, Maenaka K, Park EY. Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochem Biophys Res Commun. 2005;326:564–569. doi: 10.1016/j.bbrc.2004.11.060. [DOI] [PubMed] [Google Scholar]
- Numata K, Mieszawska-Czajkowska AJ, Kvenvold LA, Kaplan DL. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromol Biosci. 2012;12:75–82. doi: 10.1002/mabi.201100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry. 1995;34:10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10:229–236. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in β-sheet formation in spider silk block copolymers. Macromol Biosci. 2010;10:49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- Rabotyagova OS, Cebe P, Kaplan DL. Protein-based block copolymers. Biomacromolecules. 2011;12:269–289. doi: 10.1021/bm100928x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech EL, Vianna GR, Aragao FJL. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protocols. 2008;3:410–418. doi: 10.1038/nprot.2008.9. [DOI] [PubMed] [Google Scholar]
- Rising A, Widhe M, Johansson J, Hedhammar M. Spider silk proteins: recent advances in recombinant production, structure–function relationships and biomedical applications. Cell Mol Life Sci. 2011;68:169–184. doi: 10.1007/s00018-010-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J, Conrad U. Plant-based material, protein and biodegradable plastic. Curr Opin Plant Biol. 2005;8:188–196. doi: 10.1016/j.pbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Scheller J, Guhrs KH, Grosse F, Conrad U. Production of spider silk proteins in tobacco and potato. Nat Biotechnol. 2001;19:573–577. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- Service RF. Materials science. Mammalian cells spin a spidery new yarn. Science. 2002;295:419–421. doi: 10.1126/science.295.5554.419b. [DOI] [PubMed] [Google Scholar]
- Steinkraus HB, Rothfuss H, Jones JA, Dissen E, Shefferly E, Lewis RV. The absence of detectable fetal microchimerism in nontransgenic goats (Capra aegagrus hircus) bearing transgenic offspring. J Anim Sci. 2012;90:481–488. doi: 10.2527/jas.2011-4034. [DOI] [PubMed] [Google Scholar]
- Teule F, Cooper AR, Furin WA, Bittencourt D, Rech EL, Brooks A, Lewis RV. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat Protoc. 2009;4:341–355. doi: 10.1038/nprot.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teulé F, Addison B, Cooper AR, Ayon J, Henning RW, Benmore CJ, et al. Combining flagelliform and dragline spider silk motifs to produce tunable synthetic biopolymer fibers. Biopolymers. 2012a;97:418–431. doi: 10.1002/bip.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teulé F, Miao Y-G, Sohn B-H, Kim Y-S, Hull JJ, Fraser MJ, et al. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc Natl Acad Sci U S A. 2012b;109:923–928. doi: 10.1073/pnas.1109420109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vepari C, Kaplan DL. Silk as a biomaterial. Prog Polym Sci. 2007;32:991–1007. doi: 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath F, Knight DP. Structure and function of the silk production pathway in the spider Nephila edulis. Int J Biol Macromol. 1999;24:243–249. doi: 10.1016/s0141-8130(98)00095-6. [DOI] [PubMed] [Google Scholar]
- Wang C, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, et al. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc Natl Acad Sci U S A. 2006;103:9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Lan X, Zhang Y, Zhao T, Wang Y, Kajiura Z, Nakagaki M. Transgenic silkworms (Bombyx mori) produce recombinant spider dragline silk in cocoons. Mol Biol Rep. 2010;37:1815–1821. doi: 10.1007/s11033-009-9615-2. [DOI] [PubMed] [Google Scholar]
- Widmaier DM, Tullman-Ercek D, Mirsky EA, Hill R, Govindarajan S, Minshull J, Voigt CA. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol Syst Biol. 2009;5:1–9. doi: 10.1038/msb.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. Sows' ears, silk purses and goats' milk: new production methods and medical applications for silk. Med Device Technol. 2003;14:9–11. [PubMed] [Google Scholar]
- Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv Drug Delivery Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- Xia XX, Qian ZG, Ki CS, Park YH, Kaplan DL, Lee SY. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc Natl Acad Sci U S A. 2010;107:14059–14063. doi: 10.1073/pnas.1003366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HT, Fan BL, Yu SY, Huang YH, Zhao ZH, Lian ZX, et al. Construct synthetic gene encoding artificial spider dragline silk protein and its expression in milk of transgenic mice. Anim Biotechnol. 2007;18:1–12. doi: 10.1080/10495390601091024. [DOI] [PubMed] [Google Scholar]
- Xu L, Rainey JK, Meng Q, Liu X-Q. Recombinant minimalist spider wrapping silk proteins capable of native-like fiber formation. PLoS ONE. 2012;7:e50227. doi: 10.1371/journal.pone.0050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Barr LA, Fahnestock SR, Liu ZB. High yield recombinant silk-like protein production in transgenic plants through protein targeting. Transgenic Res. 2005;14:313–324. doi: 10.1007/s11248-005-0272-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hu J, Miao Y, Zhao A, Zhao T, Wu D, et al. Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. Mol Biol Rep. 2008;35:329–335. doi: 10.1007/s11033-007-9090-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao T, Zhao A, Nakagaki M. 2011. Stably express spider flagelliform silk protein in Bombyx mori cell line by piggyBac transposon-derived vector. Tianjin. 779–782.