Abstract
We set out to determine whether expansions in the C9ORF72 repeat found in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) families are associated with Parkinson disease (PD). We determined the repeat size in a total of 889 clinically ascertained patients (including PD and essential tremor plus Parkinsonism (ETP)) and 1144 controls using a repeat-primed PCR assay. We found that large C9ORF72 repeat expansions (>30 repeats) were not contributing to PD risk. However, PD and ETP cases had a significant increase in intermediate (>20 to 30+) repeat copies compared to controls. Overall, 14 cases (13 PD, 1 ETP) and three controls had >20 repeat copies (Fisher's exact test p = 0.002). Further, seven cases and no controls had >23 repeat copies (p = 0.003). Our results suggest that intermediate copy numbers of the C9ORF72 repeat contribute to risk for PD and ETP. This also suggests that PD, ALS and FTD share some pathophysiological mechanisms of disease. Further studies are needed to elucidate the contribution of the C9ORF72 repeat in the overall PD population and to determine whether other common genetic risk factors exist between these neurodegenerative disorders.
Keywords: Parkinson disease, C9ORF72 repeat, association, risk factor
Introduction
Parkinson disease (PD) is a neurodegenerative movement disorder characterized by bradykinesia, rigidity, postural instability and tremor. PD prevalence increases with age, with estimates of 4–5% of the population being affected at 85 years and older (Fahn, 2003; Tansey et al., 2007). While the diagnosis of “idiopathic” PD is well established, there are many disorders that contain the essential elements of the PD phenotype, often termed Parkinsonism, such as progressive supranuclear palsy (PSP) and multiple system atrophy (MSA). Wider overlap also occurs, including frontotemporal dementia with Parkinsonism (FTDP-17) and Lewy Body Dementia (Wszolek et al., 2006). However, minimal clinical overlap has been reported between PD and amyotrophic lateral sclerosis (ALS), a neurodegenerative disorder characterized by progressive muscular paralysis reflecting degeneration of the motor neuron in the primary motor cortex, brainstem and spinal cord (Wijesekera and Leigh, 2009). Previous observations have linked Parkinsonism with motor neuron disease (Brait–Fahn disease) or the Guam PD-ALS-dementia complex. This raised the hypothesis that ALS and PD might be related, as members of a spectrum of phenotypes with shared pathophysiological elements (Brait et al., 1973; Uitti et al., 1995; Williams et al., 1995; Qureshi et al., 1996; Pinkhardt et al., 2009). However, consumption of the unusual amino acid BAA has subsequently been thought to be the cause of Guam PD-ALS-dementia, and the incidence of the disorder in Guam has dropped since dietary modifications (Khabazian et al., 2002; Pablo et al., 2009). Nonetheless, recently, a study has shown that rare genetic variants in the Angiogenin gene (ANG) contribute to the occurrence of both PD and ALS (van Es et al., 2011). Mutations in the Valosin Containing Protein gene (VCP) are linked with ALS and also have been postulated to be a risk factor for PD, though data reported to date are inconsistent (Johnson et al., 2010; Chan et al., 2012; Majounie et al., 2012c). This raised the hypothesis that these two disorders share other common genes or pathways that contribute to the development of neurodegeneration. Therefore, when several reports recently demonstrated that large expansions of a hexanucleotide repeat in C9ORF72 were correlated with the development of ALS (Renton et al., 2011; DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012), we set out to examine the involvement of this complex repeat in PD. Interestingly, several subsequent publications (Boeve et al., 2012; Cooper-Knock et al., 2012; Mahoney et al., 2012; Simon-Sanchez et al., 2012) report an unexpected high presence of Parkinsonism and PD in ALS and FTD families of C9ORF72 repeat expansion carriers. Here, we demonstrate that intermediate C9ORF72 repeat copy numbers (>20–30+ repeat copies) are observed at significantly higher levels in PD patients than controls.
Materials and Methods
Samples
Initially, we evaluated 396 unrelated cases with PD (non-Hispanic/Latino Caucasians, range of age-at-onset (AAO): 10–85 year, average AAO: 53.6 year) and 12 cases of essential tremor with Parkinsonism (ETP) (Rocca et al., 1998). Patients were collected by 1 of 13 ascertainment centres in the PD Genetics Collaboration (Scott et al., 2001) or by the Morris K. Udall Parkinson Disease Center of Excellence (PI: Vance JM) ascertainment core. These participants were recruited primarily by participating movement disorder and neurology clinics. All individuals with PD and ET were examined by a board-certified neurologist. The diagnostic criteria used in evaluating ET patients are based on the Movement Disorders Society Consensus Criteria (Deuschl et al., 1998). Individuals are considered possibly affected with ET (types 1 and 2) when meeting criteria defined by the Tremor Investigation Group (Deuschl et al., 1995). Families with positive family history for FTD were excluded from the analysis. All patients were screened at ascertainment for cognitive status using the 3MS or short blessed test (Teng & Chui 1987; Katzman et al., 1983). Any cases with an onset of dementia within one year of the diagnosis of PD were excluded from the analysis. As positive controls for the hexanucleotide repeat, we included three ALS patients (M. Benatar, personal communication) with >50 C9ORF72 repeats (Renton et al., 2011). For the replication dataset, we used 481 patient DNA samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). Clinical data for these patients are available online. The submitters that contributed samples are acknowledged in detailed descriptions of each panel: NDPT083-085-087-088-089-102-104-105. Of the 481 samples reported here, 169 samples were reported before by Majounie et al. (2012a). We selected samples for whom clinical data were provided by a neurologist (Caucasians, range of AAO: 7–87 year, average AAO: 55.3 year) to minimize the difference in inclusion criteria between the two patient groups in the initial and replication datasets. Both patient groups have similar percentages of early onset age (<50 year; 39.5% versus 35%) and positive family history (36% versus 38%).
We evaluated 427 unaffected controls in the initial dataset (non-Hispanic/Latino Caucasians, range of age-at-exam (AAE): 60–101 year, average AAE: 77.2 year). In the replication analyses, an additional 717 controls (Caucasians, range of AAE: 60–100 year, average AAE: 71.7 year) were analysed. To lessen the chance of presymptomatic PD patients, we utilized controls older than 60 year that were collected for an ongoing study in Alzheimer disease (MPV PI). All controls scored within normal limits on the Modified Mini Mental State test and had normal neurological exams at age-of-exam. PD symptomology is an exclusion criterion for the controls in the Alzheimer disease study. The patients and controls have been included in GWAS before (Edwards et al., 2010) with no significant population stratification differences.
Genotyping: Repeat-Primed PCR and Haplotype SNP Genotyping
The repeat-primed PCR assay used to screen for the (GGGGCC)n repeat expansion limits the maximum number of repeats that can be detected to 60. The PCR cycling program of Renton et al. ( 2011) was modified to achieve more robust results on a Veriti 96-well Fast Thermal Cycler (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). A custom PCR cycling program was used (4 min at 94°C; 50 cycles of 1 min at 94°C, 1 min at 64°C and 2 min at 72°C; 10 min at 72°C). Genotypes for the C9ORF72 risk haplotype SNPs (Mok et al., 2012a) were available for this analysis from a previous GWAS study (Edwards et al., 2010) for 10 out of 14 intermediate repeat copy carriers. For the remaining four, the haplotype tag rs3849942 was genotyped by Sanger sequencing. Primer sequences flanking rs3849942 are F; 5′-GCAATTTCCTTGATTTGCAG and R; 5′-GGGGGAAAAGAGAAATGGAT. Fragment length analysis and Sanger sequencing were performed on an ABI 3730×l genetic analyser (Applied Biosystems, Life Technologies), and fragment analysis data were analysed using GeneMapper software (version 4.0, Applied Biosystems, Life Technologies).
Southern Blot
The 5 μg of DNA was digested with EcoRI and SacI. Digested samples were separated on a discontinuous 0.9% (4 cm)/1.5% (10 cm) agarose TAE gel, followed by partial depurination with HCl (0.4N) for 15 min and denaturation in NaOH (0.5N) for 30 min. DNA was transferred in 20× standard saline citrate (SSC) to a charged nylon membrane (Roche Applied Science, Indianapolis, USA) by capillarity. The 1 kb and 100 bp DNA ladders were used as size standards. The membranes were cross-linked and hybridized overnight at 47°C in roller bottles in Dig Easy Hybridization Buffer (Roche Applied Science) with a 241 bp probe synthesized by PCR from genomic DNA, using primers flanking exon 1 of C9ORF72 (5′-AGAACAGGACAAGTTGCC-3′ and 5′-AACACACACCTCCTAAACC-3′) labelled with Dig-11-dUTP by PCR (PCR Dig Synthesis Kit; Roche Applied Science). Filters were washed twice for 5 min in 2× SSC/1% SDS at room temperature and twice for 15 min in 1× SSC/0.1% SDS at 65°C. Filter blocking and signal detection were performed by incubating in blocking solution (DIG Nucleic Acid Detection Kit; Roche Applied Science) for 30 min, with the anti-DIG-AP antibody (1:5000 in blocking solution) for 30 min, and ultimately in the solution containing the NBT/BCIP substrate until the colour could be visualized. Images were captured with a Sony DSC-W350 digital camera and processed by Adobe Photoshop.
Statistical Methods
To test whether higher repeat copy numbers increase one's risk for PD, we conducted Fisher's exact tests using the a priori thresholds of greater than 20 or 23 repeat copies (RCs). The common maximum RCs reported in controls in this study and others is 23 repeats, while by far the majority (∼99.4%) of all reported controls have RCs lower than 20 (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Byrne et al., 2012; Cooper-Knock et al., 2012; Chio et al., 2012; Daoud et al., 2012; Dejesus-Hernandez et al., 2012; Dobson-Stone et al., 2012; Ferrari et al., 2012; Garcia-Redondo et al., 2012; Gijselinck et al., 2012; Konno et al., 2012; Majounie et al., 2012b; Millecamps et al., 2012; Mok et al., 2012b; Ogaki et al., 2012; Ratti et al., 2012; Rollinson et al., 2012; Rutherford et al., 2012; Sabatelli et al., 2012; Simon-Sanchez et al., 2012; van Rheenen et al., 2012; Xi et al., 2012; Yeh et al., 2012). Odds ratios represent the effect of intermediate repeat copies relative to lower repeat copies (≤20 or ≤23 RCs depending on analysis) and are calculated using a 2×2 contingency table. To test for overall distributional and mean differences, we used the Kolmogorov–Smirnov test and a standard t-test, respectively. Linear regression was used to test correlation between number of RCs and age-at-onset, age-at-exam or family history. To determine whether confounding influenced our pooling analysis, we applied the Cochran–Mantel–Haenszel test which performs pooled association analyses while controlling for datasets. All analyses were conducted using R software version 2.13.0. p-Values of 0.05 or below were considered statistically significant evidence of association.
Results
Scoring of the PCR electropherogram found inconsistencies between different researchers when attempting to call one or two peaks. Thus, all samples with one or two peaks were merged into a single group. To determine the correlation of the number of peaks in the repeat-primed PCR assay with actual repeat copy number (RC), we sequenced the repeat region in 34 samples with variable peak numbers, using the unlabeled genotyping primers reported in DeJesus–Hernandez et al. (2011). This demonstrated that the number of the actual RCs in our study equals the number of peaks plus 2.
The positive controls (ALS patients) had a maximum number of repeats greater than 50. The average maximum number of RCs in the initial clinical dataset was 6.9 in the controls and 8.2 in the cases (ranging ≤4 to ≥30) (Fig. 1). There was no significant contribution of repeats >30 in the PD cases. However, the PD cases had more intermediate size repeats (>20 repeats to 30+) versus controls. As the majority of controls in the literature are reported to have less than 20 repeats, we used this value as a first a priori threshold for normal versus intermediate RCs (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Millecamps et al., 2012; Sabatelli et al., 2012). We found eight PD patients, one ETP patient and one control with intermediate RCs (Table 1) in our initial dataset. The one-tailed Fisher's exact test in the initial dataset showed significant association of the intermediate RCs with an increased risk for PD (p = 0.008; two-tailed p = 0.010). The odds ratio for the intermediate RCs in this original dataset is 9.6 [95% CI: 1.32 – 421] (two-tailed). The confidence interval is very wide, likely due to the rarity of the intermediate repeat. Even when using the second, more stringent a priori threshold of 23 RCs—the common maximum number of repeats reported in controls—marginal significance was obtained (ratio patients:controls 4:0; one/two-tailed p = 0.05). The intermediate RC results in the clinical NINDS replication dataset are shown in Figure 2 and Table 2. Patient ND09224 was reported before by Majounie et al. (2012a); differences in interpretation of the peak pattern might account for the variance in reported repeat copies. Again, the number of PD cases with intermediate repeats was twice that of controls, although the overall number of individuals with intermediate repeats was not large enough to reach significance (>20 copies; ratio patients:controls 5:2, one-tailed p = 0.09, two-tailed p = 0.12; >23 copies; ratio patients:controls 3:0, one/two-tailed p = 0.06). However, importantly, the pooled analysis of both clinical datasets (patients N = 889/controls N = 1144) (Fig. 3) had more power and obtained a lower p-value than seen in either dataset: >20RCs; patients:controls 14:3 (risk OR 6.1 [95% CI: 1.68–33.1] (two-tailed); one-tailed p = 0.001, two-tailed p = 0.002), >23RCs; patients 7: controls 0 (one/two-tailed p = 0.003), indicating that the replication dataset strengthens the signal from the original dataset.
Figure 1.
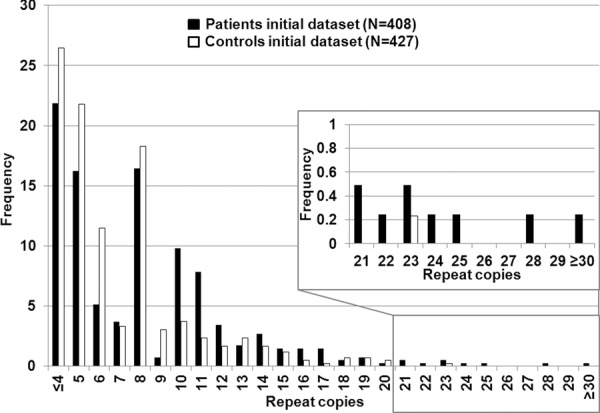
Histogram of original dataset.Histogram of the maximum number of C9ORF72 repeat copies (X axis) for 407 PD and ET-with Parkinsonism patients (black) and 427 controls (white).
Table 1.
Clinical information of the patients in the initial dataset with 20 or more repeat copies
| Patient | # of repeat copies | Gender | AAO | AAE | Family history | Phenotype | Tremor | Bradykinesia | Rigidity | Gait disturbance | Dementia |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 30 | F | 20 | 97 | Yes | Essential tremor with parkinsonism | Kinetic | Yes-(Mild) | Yes-(Mild) | No | No |
| P2 | 28 | M | 31 | 34 | No | Parkinson disease | Resting | Yes | Yes | No | Not assessed |
| P3 | 25 | F | 33 | 34 | Yes | Parkinsonism with predominant tremor | Resting | No | No | Yes | No |
| P4 | 24 | F | 56 | 71 | No | Parkinson disease | Resting | Yes | Yes | Yes | No |
| P5 | 23 | M | 46 | 51 | Yes | Parkinson disease | Resting | Yes | Yes | Yes | Yes |
| P6 | 22 | M | 61 | 68 | Yes | Parkinson disease | Resting | Yes | No | Yes | No |
| P7 | 21 | F | 44 | 56 | Yes | Parkinson disease | Resting | No | Yes | Yes | No |
| P8 | 21 | M | 64 | 66 | Yes | Parkinson disease | Resting | Yes | No | Yes | Not assessed |
| P9 | 21 | M | 53 | 53 | No | Parkinson disease | Resting | Yes | Yes | Yes | No |
AAO; age at onset, AAE; age-at-exam, Dementia; dementia at time of ascertainment.
Figure 2.
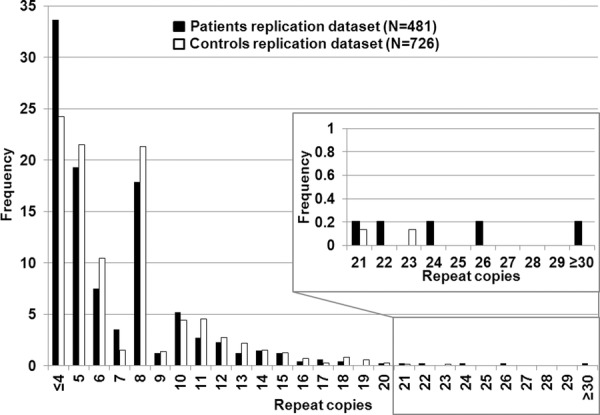
Histogram of replication dataset.Histogram of the maximum number of C9ORF72 repeat copies (X-axis) for 481 PD cases (black) and 726 controls (white).
Table 2.
Clinical information of the NINDS patients with 20 or more repeat copies
| Patient | # of repeat copies | Gender | AAO | AAE | Family history | Phenotype | Tremor | Bradykinesia | Rigidity | Gait disturbance | Dementia |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ND00236 | >30 | M | 40 | 60 | yes | Parkinson disease | No | Yes | Yes | Yes | NA |
| ND09224 | 26 | F | 68 | 77 | no | Parkinson disease | Resting | Yes | Yes | Yes | No |
| ND02556 | 24 | M | 65 | 77 | no | Parkinson disease | Resting | Yes | Yes | Yes | NA |
| ND03101 | 22 | F | 61 | 74 | no | Parkinson disease | Resting | Yes | Yes | Yes | NA |
| ND00277 | 21 | M | 40 | 65 | no | Parkinson disease | Resting | Yes | Yes | Yes | NA |
AAO; age at onset, AAE; age-at-exam, Dementia; dementia at time of ascertainment, Patient ND09224 was reported before by Majounie et al (2012a).
Figure 3.
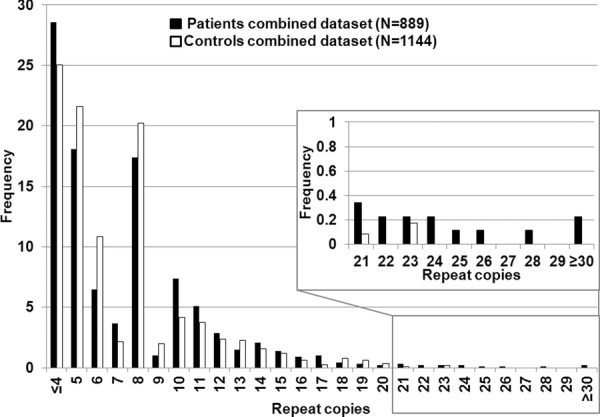
Histogram of combined dataset.Histogram of the maximum number of C9ORF72 repeat copies (X-axis) for 889 PD cases (black) and 1144 controls (white).
Overall 2% of the 889 clinical PD cases had RCs > 20. As the frequency of >20RCs carriers in our control groups (∼0.3%) is lower than the overall frequency in the literature (∼0.6%), we performed the same association analysis in the total dataset adjusting for a higher frequency of >20RCs as reported in controls. We still obtained evidence for significant association (>20 copies; ratio patients:controls 14:‘6’, p = 0.015).
The 20 SNPs reported by Mok et al. (2012a) were genotyped in a previous GWAS for 10 out of 14 clinical intermediate repeat copy carriers (Edwards et al., 2010). All 10 intermediate copy carriers had the alleles of the risk haplotype (N = 20), with the exception of the most extreme ends of the haplotype in three individuals (Table S1). All intermediate copy carriers carried the T allele of SNP rs3849942, which “tags” the haplotype as the least frequent allele on the haplotype. Next, we examined this tagSNP in our full PD dataset. We found that 95% of the clinical cases and controls with greater than 8 repeats carried the tagging T allele versus 10% of those individuals with <8 repeats (Fig. 4). This led us to test whether there was an overall significant shift in the distribution of repeat size between cases and controls. In the initial clinical dataset, the distribution of the RCs was significantly different between cases and controls (KS test p-value = 5.28e-07, t-test for means of the distribution p-value = 6.68e-07). However, the NINDS dataset did not have a significant distribution shift of RCs in PD versus controls or a mean difference (KS test p-value = 0.97, t-test p-value = 0.98). A comparison of the two control groups showed that there were no differences in either the overall distribution or the repeat count mean that would account for this difference. While the NINDS set by itself was not significant, a combined analysis of both datasets showed a significant difference between overall distribution and distribution means of RCs between cases and controls (KS test p-value = 0.01/t-test p-value = 0.01), p-values being notably less significant than in the original analysis. As the histograms of both case groups showed differences in distribution (Figs 1 and 2), we tested whether the results of the pooled analysis were due to confounding by the datasets. To control for having two different datasets, we performed the Cochran–Mantel–Haenszel test (CMH). The CMH test is analogous to a χ2 test, but can be used to control for differing datasets. The CMH results still showed significant results (p = 0.0064/p = 0.0074 for the two thresholds), indicating that results were not due to confounding.
Figure 4.
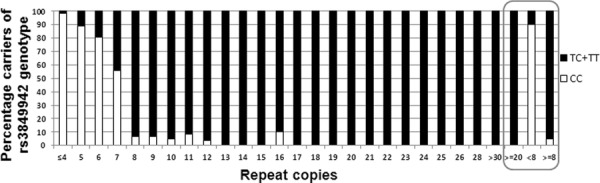
Percentages of rs3849942 genotypes corresponding to C9ORF72 repeat copy carriers.
No evidence was seen for correlation of RCs with AAO (p = 0.792) for the clinical cases, or with AAE for the two clinical case and two control groups (p = 0.8246/p = 0.1624/p = 0.2513/p = 0.8284, respectively). No correlation of repeat length with dementia was observed in the patients of the initial dataset, for which data were available (N = 230) (p = 0.267). RC distributions of PD patients with early (<40 year) versus late AAO and positive versus negative family history were not significantly different.
Genotype of >20 Repeat Individuals Using Southern Blot
We performed Southern blot analysis to determine whether the clinically affected individuals with >20 RCs are heterozygous or homozygous carriers of two intermediate RC alleles, as the repeat-primed PCR for Patient 1 (P1) indicated two possible alleles with intermediate repeat copies (Fig. 5B). All but two of the intermediate RC carriers of the original dataset were shown to be heterozygous for the intermediate allele and a low copy number allele (data not shown). The two other samples (P1; P9, Fig. 6) were further analysed with additional control samples (see below). For Southern blot analysis of the NINDS intermediate samples, we obtained additional DNA from cultured cells. However, the DNA quality was insufficient to obtain a distinct band on Southern blot after multiple attempts.
Figure 5.
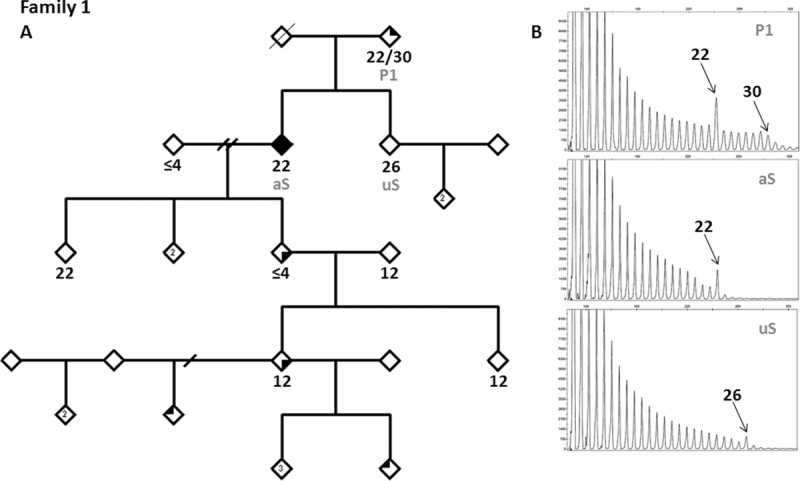
Segregation analysis in Family 1. (A) Pedigree. Symbols: Top right: ET type I; bottom right: possible ET type II; top left: ET by history; fully blackened: definite ET. (B) Electropherogram results of repeat-primed PCR of P1, aS and uS.
Figure 6.
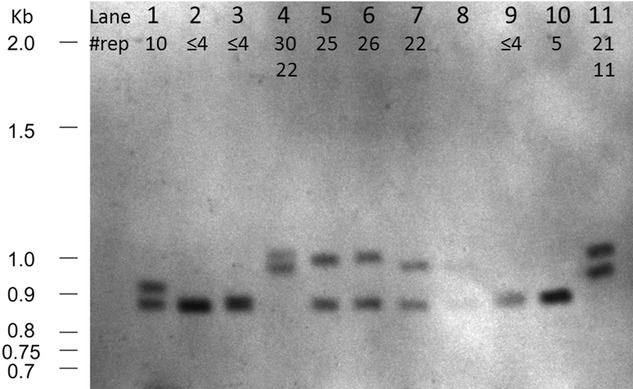
Southern blot analysis. (1) Control, 10 repeat copies (RCs). (2) Control, ≤4RCs. (3) Control ≤4RCs. (4) Patient 1, 30RCs. (5) Patient 3, 25RCs. (6) Unaffected sib (uS) from Fam1, 26RCs. (7) Affected sib (aS) from Fam1, 22RCs. (8) Overflow from lane (7). (9) Control, ≤4RCs. (10) Control, 5RCs. (11) Patient 9, 21RCs.
One sample (P1; lane 4, Fig. 6) was homozygous for intermediate repeats on Southern blot. Segregation analysis showed three different maximum intermediate RC alleles in her two offspring and herself (Fig. 5). Southern blot analysis confirmed the repeat-primed PCR results and excluded the possibility of the other parent (no sample available) also being a carrier of an intermediate RC allele, as both sibs carry low copy alleles not originating from P1. This suggests that the allele with 26RC in the unaffected sib is derived from one of the alleles in patient P1, implying instability in the repeat. By her own history, P1 developed ET at the age of 20. For this study, the patient underwent a neurological exam when she was 97 year old. She presented with mild bradykinesia and rigidity, moderate postural and intention tremor in both upper extremities.
Thirteen of the intermediate patients with >20 RCs had a phenotypic presentation that was compatible with classical PD (Tables 1 and 2). Eleven out of 14 patients have data available on levodopa response; all are responsive to L-dopa. A family history for ALS was not ascertained in these intermediate families. The intermediate RCs segregated genetically in a stable manner in several additional families. However, the intermediate alleles did not always segregate with PD (Figs S1–S8).
Discussion
Renton et al. suggested pathogenic alleles for the C9ORF72 repeat in ALS to be more than 30 RCs and benign alleles fewer than 20 RCs (Renton et al., 2011). Subsequent reports have identified rare controls in the 20 to 30RC range and more (DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012; Simon-Sanchez et al., 2012), with the most commonly reported maximum RC number in controls as 23. We demonstrate here that intermediate expansions of the C9ORF72 gene (>20RCs) are associated with increased risk for clinically diagnosed PD. Moreover, combining two different clinical datasets, we found seven PD but no controls having over 23RCs. Six more reports have identified clinical PD patients with intermediate/expanded RCs (DeJesus-Hernandez et al., 2012; Majounie et al., 2012a; Xi et al., 2012; Daoud et al., 2013; Harms et al., 2013; Lesage et al., 2013), with frequencies up to 2% amongst cases, similar to the work presented here. For five out of six reports, however, no risk assessment for the C9ORF72 gene and PD relative to controls was performed. In the sixth study (DeJesus-Hernandez et al., 2012), a lack of association with increased PD risk was reported. However, in contrast to the other reports, no intermediate repeat copies above 23 were identified in the PD patients. For single allele analysis, they used cutoffs based on sample quintiles and percentiles, so no meaningful comparison can be evaluated with this report. However, it is interesting to note that the 95th percentile (over 13 RCs) was the group with the highest increased odds ratio, though it was not significant.
We confirmed the presence of the tagging allele (T) of rs3849942 in 100% of all intermediate carriers and the full previously reported haplotype in all carriers where genotype data were available. In addition, the tagging allele was in 95% of all individuals with eight repeats or greater, versus 10% in individuals with less than 8 RC, which is consistent with a recent report by van der Zee et al. (2013). These data indicate that genotyping this SNP could be used as a screening test to identify samples with possible higher repeat copy numbers. The reasons for this correlation are unknown, but it could be due to a specific haplotype background that contributes to the expansion. Alternatively, the initial expansion may have occurred only once on this background, leading to further expansion. Our previously reported GWAS (Edwards et al., 2010) showed no evidence for association with PD for rs3849942 (p = 0.852), indicating that the effect we report here is specific to the repeat, not the underlying haplotype.
Interestingly, several studies have suggested an excess of clinical PD or Parkinsonism cases in C9ORF72 repeat expansion families (Boeve et al., 2012; Cooper-Knock et al., 2012; Mahoney et al., 2012; Simon-Sanchez et al., 2012). This finding could be explained if, indeed, the intermediate allele described here is a “premutation” allele for ALS. The clinical phenotype of the majority of the patients with greater than 20 RCs appears to be consistent with “typical” PD with no clinical evidence of motor neuron disease. Furthermore, we saw no evidence for ALS in our family histories. As ALS is much less frequent than PD, if such a relationship existed, it is more likely to be observed in the rarer ALS families than vice versa. The PD patients with ≥30 RCs did not have symptoms observed (Udall patient) or reported (NINDS patient) indicative of ALS or FTD. Two other reports of intermediate/expanded repeats in clinical PD cases, however, indicate that some of these carriers present with a family history of atypical Parkinsonism or other neurodegenerative disorders (AD, ALS) (Xi et al., 2012; Lesage et al., 2013).
The lack of segregation of the intermediate RC allele with clinical PD in the small number of families studied here (with asymptomatic carriers at an age that exceeds the AAO of their affected relative) and the occurrence of intermediate/expanded repeats in some controls suggests that it is most likely a susceptibility risk factor, rather than causal for PD/Parkinsonism.
Although the research to elucidate the disease mechanism of this intermediate/expanded repeat is still in its infancy, the first reports suggested binding of the repeat to other RNAs, sequestering both from normal processing (Renton et al., 2011), inclusion of the repeat in nuclear foci, also supporting RNA binding as a possible mechanism, and reduced protein expression and affected splicing (DeJesus-Hernandez et al., 2011). Subsequent functional studies report on the presence of di-peptides—translated from the repeat in the mRNA through repeat associated non-ATG (RAN) translation—in the pathology of large, expanded repeat carriers (Ash et al., 2013; Mori et al., 2013). This RAN translation is possible due to formation of stable secondary structures, hairpins or G-quadruplex structures, by the repeat in the mRNA (Fratta et al., 2012; Reddy et al., 2013). Additionally, analyses of the effect of the repeat in cell and Drosophila models indicated that repeat expression (30 RCs used in the report) is sufficient to cause neurodegeneration (Xu et al., 2013). The authors identified one RNA-binding protein (Pur α) binding to the repeat in a concentration-dependent manner. Overexpression of Pur α seemed to rescue the phenotype caused by the repeat, supporting the hypothesis of RNA sequestration as a possible disease mechanism. It has been shown that multiple other genes contributing to ALS risk (FUS, TDP43, ANG) are involved in RNA metabolism as well (Emara et al., 2010; Lagier-Tourenne et al., 2010).
The finding of an abnormally expanded number of repeats in PD, but fewer than that observed in ALS, is reminiscent of Fragile X-associated Tremor/Ataxia Syndrome (FXTAS)/Fragile X (Garcia-Arocena & Hagerman 2010) and ATXN2 (SCA2) (Kim et al., 2007). In these disorders, intermediate RCs (premutations) are associated with Parkinsonism. The mechanism proposed for FXTAS is that the repeat, also G-C rich, binds local RNA, creating local cell stress and damage. Interestingly, ATXN2 has also been reported to be involved in RNA metabolism (Nonhoff et al., 2007), so this is another area of common overlap.
Finally, initial evidence suggested that the repeat may become unstable with increasing numbers (Renton et al., 2011). We did observe instability of the repeat in blood between patient 1 and her offspring with a loss of repeat copies. Repeats of intermediate length are reported to be prone to both gaining and losing copies as opposed to longer repeats whose length mostly increases (Li et al., 2002). The instability of the intermediate repeats might allow them to reach a threshold where further elongation to pathogenic lengths is more likely.
In summary, these data suggest that not only do the C9ORF72 repeats contribute to significant risk of developing PD/Parkinsonism, but that abnormal RNA metabolism may also be an important pathogenesis factor in PD as well, and should be further investigated.
Acknowledgments
This work was supported by National Institute of Health grants NS39764, 5P50NS071674-03, R01AG019085, R01AG027944-02, R01AG028786-02, RC2AG036528, U01AG032984-02, the Alzheimer's Association and the American Health Assistance Foundation. Some of the samples used in this study were collected when authors were at Duke University. Dr. Benatar's sources relevant to this work are the MDA (Muscular Dystrophy Association), the ALS Association and the ALS Recovery Fund.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Disclaimer: Supplementary materials have been peer-reviewed but not copyedited.
Haplotype analysis in intermediate carriers and pedigrees displaying segregation analysis in families 2–9 can be found online.
Table S1 Analysis of 20 SNP risk haplotype in C9ORF72 intermediate repeat carriers.
Figure S1 Segregation analysis in Family 2.
Figure S2 Segregation analysis in Family 3.
Figure S3 Segregation analysis in Family 4.
Figure S4 Segregation analysis in Family 5.
Figure S5 Segregation analysis in Family 6.
Figure S6 Segregation analysis in Family 7.
Figure S7 Segregation analysis in Family 8.
Figure S8 Segregation analysis in Family 9.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW., 3rd Rademakers R, Boylan KB, Dickson DW. Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeve BF, Boylan KB, Graff-Radford NR, DeJesus-Hernandez M, Knopman DS, Pedraza O, Vemuri P, Jones D, Lowe V, Murray ME, Dickson DW, Josephs KA, Rush BK, Machulda MM, Fields JA, Ferman TJ, Baker M, Rutherford NJ, Adamson J, Wszolek ZK, Adeli A, Savica R, Boot B, Kuntz KM, Gavrilova R, Reeves A, Whitwell J, Kantarci K, Jack CR, Jr, Parisi JE, Lucas JA, Petersen RC. Rademakers R. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135:765–783. doi: 10.1093/brain/aws004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brait K, Fahn S. Schwarz GA. Sporadic and familial parkinsonism and motor neuron disease. Neurology. 1973;23:990–1002. doi: 10.1212/wnl.23.9.990. [DOI] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, McLaughlin RL, Iyer PM, O'Brien C, Phukan J, Wynne B, Bokde AL, Bradley DG, Pender N, Al-Chalabi A. Hardiman O. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N, Le C, Shieh P, Mozaffar T, Khare M, Bronstein J. Kimonis V. Valosin-containing protein mutation and Parkinson's disease. Parkinsonism Relat Disord. 2012;18:107–109. doi: 10.1016/j.parkreldis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Chio A, Borghero G, Restagno G, Mora G, Drepper C, Traynor BJ, Sendtner M, Brunetti M, Ossola I, Calvo A, Pugliatti M, Sotgiu MA, Murru MR, Marrosu MG, Marrosu F, Marinou K, Mandrioli J, Sola P, Caponnetto C, Mancardi G, Mandich P, La Bella V, Spataro R, Conte A, Monsurro MR, Tedeschi G, Pisano F, Bartolomei I, Salvi F, Lauria Pinter G, Simone I, Logroscino G, Gambardella A, Quattrone A, Lunetta C, Volanti P, Zollino M, Penco S, Battistini S ITALSGEN consortium. Renton AE, Majounie E, Abramzon Y, Conforti FL, Giannini F, Corbo M. Sabatelli M. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012;135:784–793. doi: 10.1093/brain/awr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, Martindale J, Hartley J, Walsh T, Gelsthorpe C, Baxter L, Forster G, Fox M, Bury J, Mok K, McDermott CJ, Traynor BJ, Kirby J, Wharton SB, Ince PG, Hardy J. Shaw PJ. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135:751–764. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud H, Noreau A, Rochefort D, Paquin-Lanthier G, Gauthier MT, Provencher P, Pourcher E, Dupre N, Chouinard S, Jodoin N, Soland V, Fon EA, Dion PA. Rouleau GA. Investigation of C9orf72 repeat expansions in Parkinson's disease. Neurobiol Aging. 2013;34:1710. doi: 10.1016/j.neurobiolaging.2012.11.025. e7–1710.e9. [DOI] [PubMed] [Google Scholar]
- Daoud H, Suhail H, Sabbagh M, Belzil V, Szuto A, Dionne-Laporte A, Khoris J, Camu W, Salachas F, Meininger V, Mathieu J, Strong M, Dion PA. Rouleau GA. C9orf72 hexanucleotide repeat expansions as the causative mutation for chromosome 9p21-linked amyotrophic lateral sclerosis and frontotemporal dementia. Arch Neurol. 2012;69:1159–1163. doi: 10.1001/archneurol.2012.377. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR. Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Rayaprolu S, Soto-Ortolaza AI, Rutherford NJ, Heckman MG, Traynor S, Strongosky A, Graff-Radford N, Van Gerpen J, Uitti RJ, Shih JJ, Lin SC, Wszolek ZK, Rademakers R. Ross OA. Analysis of the C9orf72 repeat in Parkinson's disease, essential tremor and restless legs syndrome. Parkinsonism Relat Disord. 2012;19:198–201. doi: 10.1016/j.parkreldis.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschl G, Bain P, Brin M Ad Hoc Scientific Committee. Consensus statement of the Movement Disorder Society on tremor. Mov Disord. 1998;13:2–23. doi: 10.1002/mds.870131303. [DOI] [PubMed] [Google Scholar]
- Deuschl G, Lauk M. Timmer J. Tremor classification and tremor time series analysis. Chaos. 1995;5:48–51. doi: 10.1063/1.166084. [DOI] [PubMed] [Google Scholar]
- Dobson-Stone C, Hallupp M, Bartley L, Shepherd CE, Halliday GM, Schofield PR, Hodges JR. Kwok JB. C9ORF72 repeat expansion in clinical and neuropathologic frontotemporal dementia cohorts. Neurology. 2012;79:995–1001. doi: 10.1212/WNL.0b013e3182684634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, Wang L, Zuchner S, Konidari I, Wang G, Singer C, Nahab F, Scott B, Stajich JM, Pericak-Vance M, Haines J, Vance JM. Martin ER. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M, Obaid L, Johnson S, Bigam DL. Cheung PY. Angiostatins decrease in the kidney of newborn piglets after hypoxia-reoxygenation. Eur J Pharmacol. 2010;644:203–208. doi: 10.1016/j.ejphar.2010.06.051. [DOI] [PubMed] [Google Scholar]
- Fahn S. Description of Parkinson's disease as a clinical syndrome. Ann N Y Acad Sci. 2003;991:1–14. doi: 10.1111/j.1749-6632.2003.tb07458.x. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Mok K, Moreno JH, Cosentino S, Goldman J, Pietrini P, Mayeux R, Tierney MC, Kapogiannis D, Jicha GA, Murrell JR, Ghetti B, Wassermann EM, Grafman J, Hardy J, Huey ED. Momeni P. Screening for C9ORF72 repeat expansion in FTLD. Neurobiol Aging. 2012;33:1850. doi: 10.1016/j.neurobiolaging.2012.02.017. e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G. Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arocena D. Hagerman PJ. Advances in understanding the molecular basis of FXTAS. Hum Mol Genet. 2010;19:R83–89. doi: 10.1093/hmg/ddq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Redondo A, Dols-Icardo O, Rojas-Garcia R, Esteban-Perez J, Cordero-Vazquez P, Munoz-Blanco JL, Catalina I, Gonzalez-Munoz M, Varona L, Sarasola E, Povedano M, Sevilla T, Guerrero A, Pardo J, de Munain AL, Marquez-Infante C, de Rivera FJ, Pastor P, Jerico I, de Arcaya AA, Mora JS, Clarimon J The C9ORF72 Spanish Study Group. Gonzalo-Martinez JF, Juarez-Rufian A, Atencia G, Jimenez-Bautista R, Moran Y, Mascias J, Hernandez-Barral M, Kapetanovic S, Garcia-Barcina M, Alcala C, Vela A, Ramirez-Ramos C, Galan L, Perez-Tur J, Quintans B, Sobrido MJ, Fernandez-Torron R, Poza JJ, Gorostidi A, Paradas C, Villoslada P, Larrode P, Capablo JL, Pascual-Calvet J, Goni M, Morgado Y, Guitart M, Moreno-Laguna S, Rueda A, Martin-Estefania C, Cemillan C, Blesa R. Lleo A. Analysis of the C9orf72 gene in patients with amyotrophic lateral sclerosis in Spain and different populations worldwide. Hum Mutat. 2012;34:79–82. doi: 10.1002/humu.22211. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M. Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Harms MB, Neumann D, Benitez BA, Cooper B, Carrell D, Racette BA, Perlmutter JS, Goate A. Cruchaga C. Parkinson disease is not associated with C9ORF72 repeat expansions. Neurobiol Aging. 2013;34:1519. doi: 10.1016/j.neurobiolaging.2012.10.001. e1–1519.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G ITALSGEN Consortium. Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A. Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman R, Brown T, Fuld P, Peck A, Schechter R. Schimmel H. Validation of a short orientation-memory-concentration test of cognitive impairment. Am J Psychiatry. 1983;140:734–739. doi: 10.1176/ajp.140.6.734. [DOI] [PubMed] [Google Scholar]
- Khabazian I, Bains JS, Williams DE, Cheung J, Wilson JM, Pasqualotto BA, Pelech SL, Andersen RJ, Wang YT, Liu L, Nagai A, Kim SU, Craig UK. Shaw CA. Isolation of various forms of sterol beta-D-glucoside from the seed of Cycas circinalis: Neurotoxicity and implications for ALS-parkinsonism dementia complex. J Neurochem. 2002;82:516–528. doi: 10.1046/j.1471-4159.2002.00976.x. [DOI] [PubMed] [Google Scholar]
- Kim JM, Hong S, Kim GP, Choi YJ, Kim YK, Park SS, Kim SE. Jeon BS. Importance of low-range CAG expansion and CAA interruption in SCA2 Parkinsonism. Arch Neurol. 2007;64:1510–1518. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- Konno T, Shiga A, Tsujino A, Sugai A, Kato T, Kanai K, Yokoseki A, Eguchi H, Kuwabara S, Nishizawa M, Takahashi H. Onodera O. Japanese amyotrophic lateral sclerosis patients with GGGGCC hexanucleotide repeat expansion in C9ORF72. J Neurol Neurosurg Psychiatry. 2012;84:398–401. doi: 10.1136/jnnp-2012-302272. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M. Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Le Ber I, Condroyer C, Broussolle E, Gabelle A, Thobois S, Pasquier F, Mondon K, Dion PA, Rochefort D, Rouleau GA, Durr A, Brice A French Parkinson's Disease Genetics Study Group. C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain. 2013;136:385–391. doi: 10.1093/brain/aws357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A. Nevo E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC, Rossor MN, Hardy J, Collinge J, Revesz T, Mead S. Warren JD. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: Clinical, neuroanatomical and neuropathological features. Brain. 2012;135:736–750. doi: 10.1093/brain/awr361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Abramzon Y, Renton AE, Keller MF, Traynor BJ. Singleton AB. Large C9orf72 repeat expansions are not a common cause of Parkinson's disease. Neurobiol Aging. 2012a;33:2527.e1–2. doi: 10.1016/j.neurobiolaging.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O Chromosome 9-ALS; /FTD Consortium, French research network on FTLD/FTLD/ALS, ITALSGEN Consortium. Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S. Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012b;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Traynor BJ, Chio A, Restagno G, Mandrioli J, Benatar M, Taylor JP. Singleton AB. Mutational analysis of the VCP gene in Parkinson's disease. Neurobiol Aging. 2012c;33:209.e1–209.e2. doi: 10.1016/j.neurobiolaging.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Boillee S, Le Ber I, Seilhean D, Teyssou E, Giraudeau M, Moigneu C, Vandenberghe N, Danel-Brunaud V, Corcia P, Pradat PF, Le Forestier N, Lacomblez L, Bruneteau G, Camu W, Brice A, Cazeneuve C, Leguern E, Meininger V. Salachas F. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. J Med Genet. 2012;49:258–263. doi: 10.1136/jmedgenet-2011-100699. [DOI] [PubMed] [Google Scholar]
- Mok K, Traynor BJ, Schymick J, Tienari PJ, Laaksovirta H, Peuralinna T, Myllykangas L, Chio A, Shatunov A, Boeve BF, Boxer AL, DeJesus-Hernandez M, Mackenzie IR, Waite A, Williams N, Morris HR, Simon-Sanchez J, van Swieten JC, Heutink P, Restagno G, Mora G, Morrison KE, Shaw PJ, Rollinson PS, Al-Chalabi A, Rademakers R, Pickering-Brown S, Orrell RW, Nalls MA. Hardy J. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2012a;33:209. doi: 10.1016/j.neurobiolaging.2011.08.005. e3–209.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KY, Koutsis G, Schottlaender LV, Polke J, Panas M. Houlden H. High frequency of the expanded C9ORF72 hexanucleotide repeat in familial and sporadic Greek ALS patients. Neurobiol Aging. 2012b;33:1851. doi: 10.1016/j.neurobiolaging.2012.02.021. e1–1851.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C. Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H. Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki K, Li Y, Atsuta N, Tomiyama H, Funayama M, Watanabe H, Nakamura R, Yoshino H, Yato S, Tamura A, Naito Y, Taniguchi A, Fujita K, Izumi Y, Kaji R, Hattori N, Sobue G Japanese Consortium for Amyotrophic Lateral Sclerosis research (JaCALS) Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527. doi: 10.1016/j.neurobiolaging.2012.05.011. e11–2527.e16. [DOI] [PubMed] [Google Scholar]
- Pablo J, Banack SA, Cox PA, Johnson TE, Papapetropoulos S, Bradley WG, Buck A. Mash DC. Cyanobacterial neurotoxin BMAA in ALS and Alzheimer's disease. Acta Neurol Scand. 2009;120:216–225. doi: 10.1111/j.1600-0404.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- Pinkhardt EH, Sperfeld AD, Gdynia HJ, Ludolph AC. Kassubek J. The combination of dopa-responsive parkinsonian syndrome and motor neuron disease. Neurodegener Dis. 2009;6:95–101. doi: 10.1159/000207795. [DOI] [PubMed] [Google Scholar]
- Qureshi AI, Wilmot G, Dihenia B, Schneider JA. Krendel DA. Motor neuron disease with parkinsonism. Arch Neurol. 1996;53:987–991. doi: 10.1001/archneur.1996.00550100061015. [DOI] [PubMed] [Google Scholar]
- Ratti A, Corrado L, Castellotti B, Del Bo R, Fogh I, Cereda C, Tiloca C, D'Ascenzo C, Bagarotti A, Pensato V, Ranieri M, Gagliardi S, Calini D, Mazzini L, Taroni F, Corti S, Ceroni M, Oggioni GD, Lin K, Powell JF, Soraru G, Ticozzi N, Comi GP, D'Alfonso S, Gellera C, Silani V SLAGEN Consortium. C9ORF72 repeat expansion in a large Italian ALS cohort: Evidence of a founder effect. Neurobiol Aging. 2012;33:2528.e7–2528. doi: 10.1016/j.neurobiolaging.2012.06.008. 14. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB., Jr Pearson CE. The Disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ. Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca WA, Maraganore DM, McDonnell SK. Schaid DJ. Validation of a telephone questionnaire for Parkinson's disease. J Clin Epidemiol. 1998;51:517–523. doi: 10.1016/s0895-4356(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Rollinson S, Halliwell N, Young K, Callister JB, Toulson G, Gibbons L, Davidson YS, Robinson AC, Gerhard A, Richardson A, Neary D, Snowden J, Mann DM. Pickering-Brown SM. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiol Aging. 2012;33:1846.e5–6. doi: 10.1016/j.neurobiolaging.2012.01.109. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Knopman DS, Kretzschmar HA, Neumann M, Caselli RJ, White CL, 3rd, Mackenzie IR, Petersen RC, Strong MJ, Miller BL, Boeve BF, Uitti RJ, Boylan KB, Wszolek ZK, Graff-Radford NR, Dickson DW, Ross OA. Rademakers R. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol Aging. 2012;33:2950. doi: 10.1016/j.neurobiolaging.2012.07.005. e5–2950.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Conforti FL, Zollino M, Mora G, Monsurro MR, Volanti P, Marinou K, Salvi F, Corbo M, Giannini F, Battistini S, Penco S, Lunetta C, Quattrone A, Gambardella A, Logroscino G, Simone I, Bartolomei I, Pisano F, Tedeschi G, Conte A, Spataro R, La Bella V, Caponnetto C, Mancardi G, Mandich P, Sola P, Mandrioli J, Renton AE, Majounie E, Abramzon Y, Marrosu F, Marrosu MG, Murru MR, Sotgiu MA, Pugliatti M, Rodolico C ITALSGEN Consortium. Moglia C, Calvo A, Ossola I, Brunetti M, Traynor BJ, Borghero G, Restagno G. Chio A. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol Aging. 2012;33:1848. doi: 10.1016/j.neurobiolaging.2012.02.011. e15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Slotterbeck B, Booze MW, Ribble RC, Rampersaud E, West SG, Gibson RA, Middleton LT, Roses AD, Haines JL, Scott BL, Vance JM. Pericak-Vance MA. Complete genomic screen in Parkinson disease: Evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, Smits M, Raaphorst J, van den Berg LH, Schelhaas HJ, De Die-Smulders CE, Majoor-Krakauer D, Rozemuller AJ, Willemsen R, Pijnenburg YA, Heutink P. van Swieten JC. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012;135:723–735. doi: 10.1093/brain/awr353. [DOI] [PubMed] [Google Scholar]
- Tansey MG, McCoy MK. Frank-Cannon TC. Neuroinflammatory mechanisms in Parkinson's disease: Potential environmental triggers, pathways, and targets for early therapeutic intervention. Exp Neurol. 2007;208:1–25. doi: 10.1016/j.expneurol.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng EL. Chui HC. The modified mini-mental state (3MS) examination. J Clin Psychiatry. 1987;48:314–318. [PubMed] [Google Scholar]
- Uitti RJ, Berry K, Yasuhara O, Eisen A, Feldman H, McGeer PL. Calne DB. Neurodegenerative ‘overlap’ syndrome: Clinical and pathological features of Parkinson's disease, motor neuron disease, and Alzheimer's disease. Parkinsonism Relat Disord. 1995;1:21–34. doi: 10.1016/1353-8020(95)00004-p. [DOI] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, Baumer V, Maes G, Corsmit E, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Pimentel J, Nacmias B, Bagnoli S, Sorbi S, Graff C, Chiang HH, Westerlund M, Sanchez-Valle R, Llado A, Gelpi E, Santana I, Almeida MR, Santiago B, Frisoni G, Zanetti O, Bonvicini C, Synofzik M, Maetzler W, Vom Hagen JM, Schols L, Heneka MT, Jessen F, Matej R, Parobkova E, Kovacs GG, Strobel T, Sarafov S, Tournev I, Jordanova A, Danek A, Arzberger T, Fabrizi GM, Testi S, Salmon E, Santens P, Martin JJ, Cras P, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Philtjens S, Sleegers K, Baumer V, Maes G, Corsmit E, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Philtjens S, Theuns J, Sleegers K, Baumer V, Maes G, Cruts M, Van Broeckhoven C, Engelborghs S, De Deyn PP, Cras P, Engelborghs S, De Deyn PP, Vandenbulcke M, Vandenbulcke M, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Firmo C, Pimentel J, Sanchez-Valle R, Llado A, Antonell A, Molinuevo J, Gelpi E, Graff C, Chiang HH, Westerlund M, Graff C, Kinhult Stahlbom A, Thonberg H, Nennesmo I, Borjesson-Hanson A, Nacmias B, Bagnoli S, Sorbi S, Bessi V, Piaceri I, Santana I, Santiago B, Santana I, Helena Ribeiro M, Rosario Almeida M, Oliveira C, Massano J, Garret C, Pires P, Frisoni G, Zanetti O, Bonvicini C, Sarafov S, Tournev I, Jordanova A, Tournev I, Kovacs GG, Strobel T, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, Jessen F, Matej R, Parobkova E, Danel A, Arzberger T, Fabrizi GM, Testi S, Ferrari S, Cavallaro T, Salmon E, Santens P, Cras P European Early-Onset Dementia Consortium. A pan-European study of the C9orf72 repeat associated with FTLD: Geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363–373. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, Schulte C, Blauw HM, Koppers M, Diekstra FP, Fumoto K, LeClerc AL, Keagle P, Bloem BR, Scheffer H, van Nuenen BF, van Blitterswijk M, van Rheenen W, Wills AM, Lowe PP, Hu GF, Yu W, Kishikawa H, Wu D, Folkerth RD, Mariani C, Goldwurm S, Pezzoli G, Van Damme P, Lemmens R, Dahlberg C, Birve A, Fernandez-Santiago R, Waibel S, Klein C, Weber M, van der Kooi AJ, de Visser M, Verbaan D, van Hilten JJ, Heutink P, Hennekam EA, Cuppen E, Berg D, Brown RH, Jr, Silani V, Gasser T, Ludolph AC, Robberecht W, Ophoff RA, Veldink JH, Pasterkamp RJ, de Bakker PI, Landers JE, van de Warrenburg BP. van den Berg LH. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol. 2011;70:964–973. doi: 10.1002/ana.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen W, van Blitterswijk M, Huisman MH, Vlam L, van Doormaal PT, Seelen M, Medic J, Dooijes D, de Visser M, van der Kooi AJ, Raaphorst J, Schelhaas HJ, van der Pol WL, Veldink JH. van den Berg LH. Hexanucleotide repeat expansions in C9ORF72 in the spectrum of motor neuron diseases. Neurology. 2012;79:878–882. doi: 10.1212/WNL.0b013e3182661d14. [DOI] [PubMed] [Google Scholar]
- Wijesekera LC. Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TL, Shaw PJ, Lowe J, Bates D. Ince PG. Parkinsonism in motor neuron disease: Case report and literature review. Acta Neuropathol. 1995;89:275–283. doi: 10.1007/BF00309344. [DOI] [PubMed] [Google Scholar]
- Wszolek ZK, Tsuboi Y, Ghetti B, Pickering-Brown S, Baba Y. Cheshire WP. Frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) Orphanet J Rare Dis. 2006;1:30. doi: 10.1186/1750-1172-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Grinberg Y, Moreno D, Sato C, Bilbao JM, Ghani M, Hernandez I, Ruiz A, Boada M, Moron FJ, Lang AE, Marras C, Bruni A, Colao R, Maletta RG, Puccio G, Rainero I, Pinessi L, Galimberti D, Morrison KE, Moorby C, Stockton JD, Masellis M, Black SE, Hazrati LN, Liang Y, van Haersma de With J, Fornazzari L, Villagra R, Rojas-Garcia R, Clarimon J, Mayeux R, Robertson J, St George-Hyslop P. Rogaeva E. Investigation of C9orf72 in 4 neurodegenerative disorders. Arch Neurol. 2012;69:1583–1590. doi: 10.1001/archneurol.2012.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS. Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1219643110. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TH, Lai SC, Weng YH, Kuo HC, Wu-Chou YH, Huang CL, Chen RS, Chang HC, Traynor B. Lu CS. Screening for C9orf72 repeat expansions in parkinsonian syndromes. Neurobiol Aging. 2012;34:1311.e3–4. doi: 10.1016/j.neurobiolaging.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Analysis of 20 SNP risk haplotype in C9ORF72 intermediate repeat carriers.
Figure S1 Segregation analysis in Family 2.
Figure S2 Segregation analysis in Family 3.
Figure S3 Segregation analysis in Family 4.
Figure S4 Segregation analysis in Family 5.
Figure S5 Segregation analysis in Family 6.
Figure S6 Segregation analysis in Family 7.
Figure S7 Segregation analysis in Family 8.
Figure S8 Segregation analysis in Family 9.


