Abstract
Cyclic constraints have proven to be very effective for preorganizing β-amino acid residues and thereby stabilizing β- and α/β-peptide helices, but little is known about possible preorganization effects among γ residues. Here we assess and compare the impact of cyclic preorganization of β and γ residues in the context of a specific α/β/γ-peptide helix. The results show that β residue preorganization is critical for helix stability but that γ residue preorganization is less important.
The surfaces of folded proteins often bear identifying information that is encoded in specific side chain arrangements and recognized by other proteins.1 Oligomers that can mimic these molecular messages while displaying superiority to conventional peptides in other aspects (e.g., diminished susceptibility to enzymatic degradation, enhanced conformational stability) can be useful as antagonists of specific protein-protein associations or agonists of polypeptide-activated receptors.2 We have previously shown that α/β-peptides generated from a prototype sequence by replacing 25–33% of the original α-amino acid residues with β-amino acid residues, at sites that are dispersed along the backbone, can adopt a conformation very similar to the α-helix and bind tightly to recognition surfaces that evolved to bind natural α-helices.3 In some cases, affinity can be enhanced by use of cyclic β residues, presumably because the extra backbone bond introduced with each α→β replacement raises the entropic cost of folding unless that bond is appropriately constrained.3,4 The effective α-helix mimicry manifested by these α/β-peptides is somewhat surprising because they contain at least one additional backbone atom per helical turn relative to a true α-helix.
This design strategy built upon pioneering studies of the folding of γ residue-containing oligomers by other groups.6–8 The αγααβα arrangement we developed allows the α residues to define one face of the helix. We showed via 2D NMR data that α/β/γ-peptide 12-mer 1 in aqueous solution adopts a conformation resembling an α-helix, and that this conformation is considerably more stable than the true α-helix formed by analogous α-peptide 14-mer 2.5 We originally postulated that this high conformational stability in water arises from the backbone preorganization of both the β and γ residues provided by the five- and six-membered rings, respectively. Here we test this hypothesis and show it to be only partially correct.
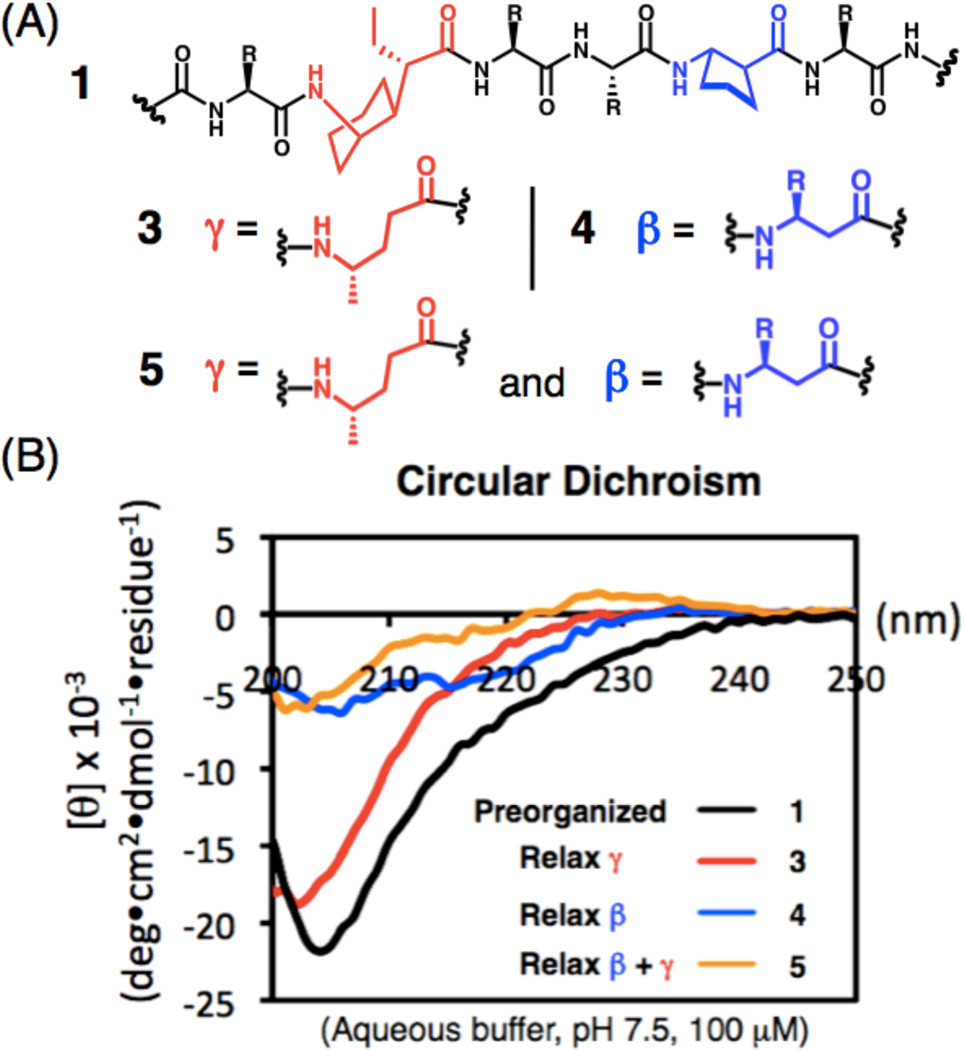
To evaluate the contribution of β residue and γ residue preorganization independently, we prepared three α/β/γ-peptide analogues of 1 (Figure 1c and 2a). In 3, the γ residue constraints have been relaxed, because γ4-hAla occurs in place of the two cyclohexane-based residues of 1. In 4, the β residue constraints have been relaxed, because β3-hGln and β3-hPhe replace the (S, S)-trans-2-aminocyclopentanecarboxylic acid (ACPC) residues. In 5, the β and γ residues constraints are relaxed simultaneously. Circular dichroism was used for an initial comparison of new α/β/γ-peptides 3–5 with 1 in aqueous buffer (Figure 2). The strong minimum at 204 nm seen for 1 was previously assigned to the β-helix-like conformation because 2D NMR data reveal a network of medium-range NOEs consistent with this conformation under the same conditions.5 For α/β/γ-peptide 5, the very limited CD intensity suggests little or no helix formation, as we expected for a backbone lacking β or γ residue constraints. Similar behavior is observed for 4, which indicates that the β residue constraint provided by the ring in ACPC is important for helical folding; this observation is consistent with previous demonstrations that incorporation of ACPC or similar cyclic β residues is crucial for helical folding of α/β-peptides in aqueous media.3a,b, 4,9 The CD data for 3, however, were surprising: the strong minimum near 202 nm implies a significant population of the helical conformation, despite the absence of γ residue constraints.
Figure 1.
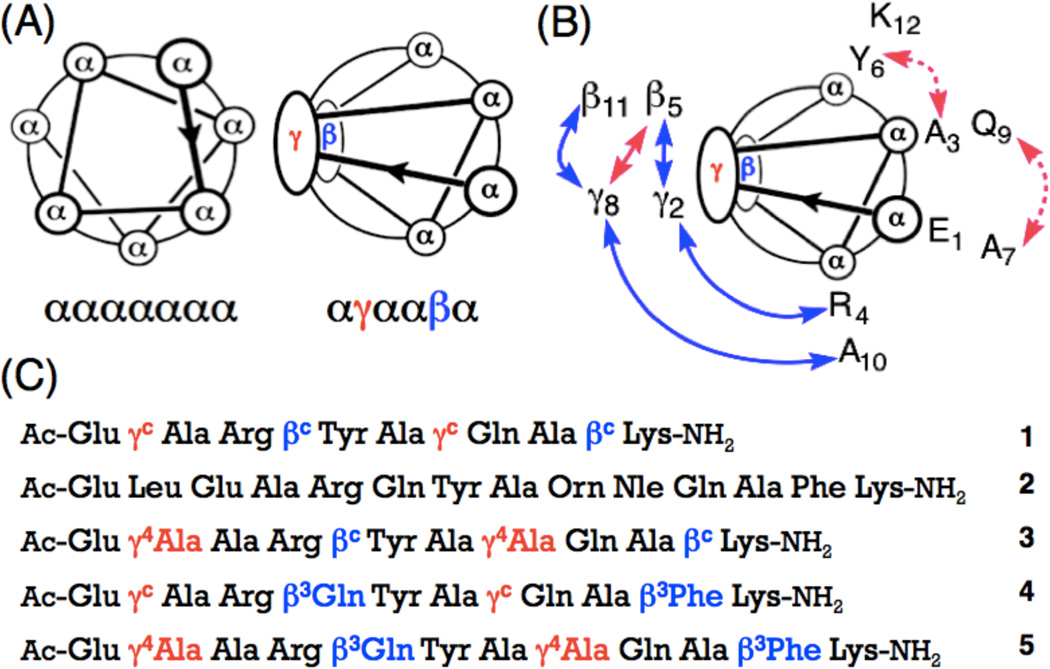
(a) Helical wheel diagrams for the ααααααα heptad and the αγααβα hexad. (b) NOEs observed between nonadjacent residues for α/β/γ-peptide 3 in CD3OH (blue and pink, peptide concentration 6 mM), or in aqueous condition (blue, peptide concentration 2 mM) at 10 °C. Medium (2.6–3.5 Å) and weak (3.6–5.5 Å) NOEs are shown as curved arrows on the helical wheel diagram, (c) Sequences of α/β/γ-peptides 1, 3–5 and α-peptide 2. Superscript ‘c’ indicates a ring-constrained residue; the structures of βc and γc are shown in Figure 2 (All stereocenters are S).
Figure 2.
(a) Backbones of αγααβα hexads from α/β/γ-peptides 1 and 3–5, β- and γ-residues are shown in blue and red, respectively, (b) Circular dichroism data for α/β/γ-peptides 1 and 3–5 measured in PBS buffer, pH 7.5 at 20 °C. Peptide concentration is 0.1 mM for all spectra.
NMR analysis was undertaken to probe the behavior of α/β/γ-peptide 3 in more detail (Figure 1b, SI), because CD can provide only low-resolution insight on molecular structure. Previously we observed that fully preorganized α/β/γ-peptide 1 displays numerous NOEs between protons from residues with i,i+2 and i,i+3 sequence relationships in both aqueous buffer and methanol; these medium-range NOEs were all consistent with an α-helix-like conformation.5 For 3, in which the cyclic γ residues have been replaced by unconstrained γ4-hAla residues, a smaller set of i,i+2 and i,i+3 NOEs was observed in methanol relative to 1 in methanol, and an even smaller set of medium-range NOEs was detected for 3 in aqueous solution. However, the NOEs observed for 3 in water included both of the possible γ residue CγH(i) -- β residue NH(i+3) NOEs and both γ residue CγH(i) -- α residue NH(i+2) NOEs, which provides strong evidence for a significant population of the helical state. Thus, the 2D NMR data indicate that removing the γ residue constraints, to generate 3, causes some diminution in helix population relative to fully preorganized 1, but that the unconstrained γ4-hAla residues nevertheless manifest a significant helical propensity, more so than unconstrained β3 residues.
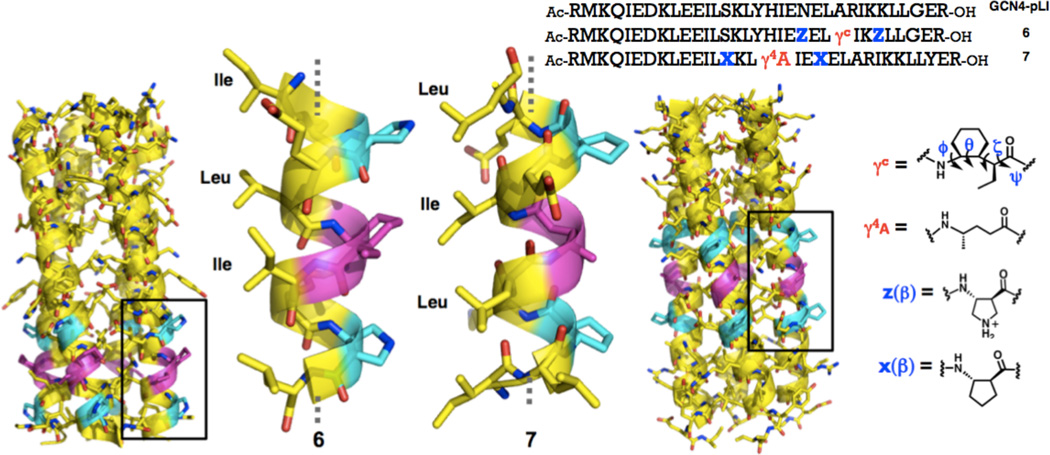
In order to gain higher-resolution insight regarding accommodation of the αγααβα motif in an α-helix-like conformation, we designed oligomers intended to provide crystallographic data. GCN4-pLI is a 33-residue α-peptide derived from the dimerization domain of the yeast transcriptional regulator GCN4.10 The pLI version forms a very stable tetrameric α-helix bundle, while the native sequence prefers lower oligomerization states.11 Previously we found that many α/β-peptides derived from GCN4-pLI crystallize in four-helix bundle assemblies that are remarkably similar to the quaternary structure of GCN4-pLI itself.6a, 12 We therefore prepared a series α/β/γ-peptides based on this sequence, containing varying numbers of αγααβα hexads in different locations; two of these oligomers provided high-quality crystals from aqueous solutions (Figure 3). The resulting structures show that an αβααγααβα unit can indeed adopt an α-helix-like conformation, whether the γ residue has the cyclic constraint we previously employed (6) or not (7).
Figure 3.
Four-helix bundles formed by 6 (left, PDB 4HJB) and 7 (right, PDB 4HJD), as determined via x-ray crystallography; β and γ residues are highlighted in purple. Sequences of GCN4-pLI and α + α/β/γ-chimeric peptides 6 and 7 and structures of β- and γ-residues.
The H-bonding pattern among backbone amide groups within the α/β/γ segments of 6 and 7 is somewhat more complex than the H-bonding pattern of the canonical α-helix, which features i,i+4 C=O--H-N H-bonds (13-atom rings)13. All backbone carbonyl groups in the α/β/γ segments of 6 and 7 form i,i+3 C=O--H-N H-bonds, which occur in 10-, 11- or 12-atom rings, depending upon the types of residues between the amide groups.13 Such H-bonds are typically identified based on O--N distances, with 3.3 Å as a standard upper limit;14 in the α/β/γ segments of 6 and 7, a few i,i+3 O--N distances exceed this limit (3.5–3.9 Å). Some backbone carbonyls in 6 and 7 participate in bifurcated H-bonds, involving i,i+4 C=O--H-N interactions (14-atom rings) in addition to the i,i+3 interactions.
The crystallographic data show that NOEs attributed to the α-helix-like conformation are indeed associated with pairs of backbone hydrogen atoms that are brought into proximity in this folded state, including the two medium range NOEs mentioned above for 3 in aqueous solution (Figure 1b, SI). In 6 the distance between the α residue CαH (position 24) and the β residue NH at position 27 is 3.0 Å, while in 7 the distance between the γ residue CγH (position 18) and the β residue NH at position 21 is 2.9 Å (H--H distance). In 6, the distance between the γ residue CγH (position 24) and the α residue NH at position 26 is 2.5 Å, and the same separation is observed in 7 between the γ residue CγH (position 18) and the α residue NH at position 20. We note, however, that some comparable backbone i,i+2 or i,i+3 H-H distances in the crystal structures do not give rise to NOEs for 3 in aqueous solution,13 perhaps because this conformation is only partially populated.
The crystal structures allow a detailed comparison of α/β/γ-peptide hexads with the α heptads they are meant to replace. In addition, the crystallographic data for 6 and 7 enable comparisons with several recently reported crystal structures of peptidic oligomers that contain γ residues and display C=O(i)--H-N(i+3) H-bonded helical secondary structures. Table 1 compares these standard helix parameters determined for the αβααγα hexads in 6 and in 7 with analogous parameters for the α-helix (as represented by GCN4-pLI; PDB 1GCL), for a canonical 310-helix,14 for α-helix-mimetic α/β-peptides (PDB 3O43, 3F87 and 3C3H), and for various oligomers containing the cyclic γ residue we have employed15 or γ4 residues.16,17 Although all of these helices have generally similar geometries, the αβααγα unit does best at matching the α-helix radius. Table 2 compares the four backbone torsion angles of cyclic γ residues and γ4 residues in various helical contexts. As might be expected, the cyclohexane-based γ residue generally prefers gauche torsion angles (~60°) about the backbone Cα-Cβ and Cβ-Cγ bonds.15 The α-helix-like conformation observed in α/β/γ-peptide 6, however, seems to require a somewhat smaller Cα-Cγ torsion angle (45.9°) in the cyclohexyl ring. Crystal structures of helical α/γ-peptides containing α4 residues that have recently emerged from the Gopi17 and Balaram16 laboratories show that in these cases the Cα-Cγ torsion angles are frequently in the range of 50°, but an even smaller torsion angle is observed for the corresponding backbone bond in the γ4-hAla residue of 7 (38.9°).
Table 1.
Helical parameters for α/β/γ-, γ-, α/β- and α-peptides.
Table 2.
Backbone torsion angles (degrees) for γ-residues in various peptides.
The results reported here suggest that β3- and γ4-amino acid residues, which bear a single side chain and are therefore homologous to γ-amino acid residues, differ in their intrinsic tendencies to adopt helical secondary structure, with γ4 residues displaying the higher propensity. Previous work has shown that short peptidic oligomers containing β3 residues or γ4 residues, either exclusively or in combination with α residues, can adopt helical conformations in the crystalline state or in organic solvents.6–18,16–19 Aqueous solution represents a more challenging environment in terms of peptidic secondary structure formation,20 and we have used comparisons in water to show that cyclic β residues display substantially higher helical propensities than do β3 residues.4,9,21 Against this background, the relatively large helical propensity of γ4 residues revealed by our comparisons among α/β/γ-peptides 1 and 3–5 in aqueous solution is unexpected. The helical propensity of the cyclic γ residue in 1 appears to be somewhat higher than that of γ4-hAla, but the relatively small θ backbone torsion angles documented crystallographically for the γ residues in 6 and 7 raise the possibility that the cyclohexyl-constrained γ backbone may not be ideally suited to the α-helix mimetic conformation. We are currently exploring the hypothesis that alternative types of γ residue constraint could lead to improved stability of the α-helix-like conformation available to α/β/γ-peptides.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Science Foundation (CHE-0848847) and the Wisconsin Alumni Research Foundation. D.E.M. was supported in part by a Molecular Biophysics Training Grant (T32GM008293). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. NMR spectrometers were purchased with partial support from the NSF and NIH.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental details, including polypeptide characterization, CD, NMR and X-ray crystallography. This material is available free of charge via the Internet at http://pubs.acs.org. Model coordinates and structure factors have been deposited in the Protein Data Bank as entries 4HJB (peptide 6) and 4HJD (peptide 7).
REFERENCES
- 1.(a) Arkin MR, Wells JA. Nat. Rev. Drug. Discov. 2004;3:301. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]; (b) Jochim AL, Arora PS. Mol. Biosyst. 2009;5:924. doi: 10.1039/b903202a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bullock BN, Jochim AL, Arora PS. J. Am. Chem. Soc. 2011;133:14220. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Azzarito V, Long K, Murphy NS, Wilson AJ. Nat. Chem. 2013;5:161. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]; (b) Guichard G, Huc I. Chem. Commun. 2011;47:5933. doi: 10.1039/c1cc11137j. [DOI] [PubMed] [Google Scholar]; (c) Goodman CM, Choi S, Shandler S, De-Grado WF. Nat. Chem. Biol. 2007;3:252. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Schafmeister CE, Brown ZZ, Gupta S. Ace. Chem. Res. 2008;41:1387. doi: 10.1021/ar700283y. [DOI] [PubMed] [Google Scholar]; (e) Yin H, Hamilton AD. Angew. Chem. Int. Ed. 2005;44:4130. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]; (f) Hecht S, Huc I, editors. Foldamers: Strucutre, Properties, and Applications. Weinheim, Germany: Wiley-VCH; 2007. [Google Scholar]; (g) Horne WS. Expert Opin. Drug Discov. 2011;6:1247. doi: 10.1517/17460441.2011.632002. [DOI] [PubMed] [Google Scholar]
- 3.(a) Johnson LM, Gellman SH. Methods Enzymol. 2013;523:407. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad Sci. USA. 2009;106:14751. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Johnson LM, Mortenson DE, Yun HG, Horne WS, Ketas TJ, Lu M, Moore JP, Gellman SH. J. Am. Chem. Soc. 2012;134:7317. doi: 10.1021/ja302428d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Boersma MD, Haase HS, Peterson-Kaufman KJ, Lee EF, Clarke OB, Colman PM, Smith BJ, Horne WS, Fairlie WD, Gellman SH. J. Am. Chem. Soc. 2012;134:315. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lee EF, Smith BJ, Horne WS, Mayer KN, Evangelista M, Colman PM, Gellman SH, Fairlie WD. ChemBioChem. 2011;12:2025. doi: 10.1002/cbic.201100314. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Horne WS, Boersma MD, Windsor MA, Gellman SH. Angew. Chem. Int. Ed. 2008;47:2853. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price JL, Hadley EB, Steinkruger JD, Gellman SH. Angew. Chem. Int. Ed. 2010;49:368. doi: 10.1002/anie.200904714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawada T, Gellman SH. J. Am. Chem. Soc. 2011;133 doi: 10.1021/ja202175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Hanessian S, Luo X, Schaum R, Michnick S. J. Am. Chem. Soc. 1998;120:8569. [Google Scholar]; (b) Hintermann T, Gademann K, Jaun B, Seebach D. Helv. Chim. Acta. 1998;81:983. [Google Scholar]; (c) Seebach D, Brenner M, Rueping M, Schweizer B, Jaun B. Chem. Commun. 2001;2:207. [Google Scholar]; (d) Baldauf C, Gunther R, Hofmann HJ. J. Org. Chem. 2006;71:1200. doi: 10.1021/jo052340e. [DOI] [PubMed] [Google Scholar]; (e) Claudon P, Violette A, Lamour K, Decossas M, Fournel S, Heurtault B, Godet J, Mely Y, Jamart-Gregoire B, Averlant-Petit M-C, Briand JP, Duportail G, Monteil H, Guichard G. Angew. Chem. Int. Ed. 2010;49:333. doi: 10.1002/anie.200905591. [DOI] [PubMed] [Google Scholar]
- 7.(a) Karle IL, Pramanik A, Banerjee A, Bhattacharjya S, Balaram P. J. Am. Chem. Soc. 1997;119:9087–9095. [Google Scholar]; (b) Araghi RR, Jäckel C, Cöfen H, Salwiczek M, Völkel A, Wagner SC, Wieczorek S, Baldauf C, Koksch B. ChemBio-Chem. 2010;11:335–339. doi: 10.1002/cbic.200900700. [DOI] [PubMed] [Google Scholar]
- 8.(a) Hagihara M, Anthony NJ, Stout TJ, Clardy J, Schreiber SL. J. Am. Chem. Soc. 1992;114:6568. [Google Scholar]; (b) Baldauf C, Gunther R, Hofmann HJ. Helv. Chim. Acta. 2003;86:2573. [Google Scholar]; (c) Farrera-Sinfreu J, Zaccaro L, Vidal D, Salvatella X, Giralt E, Pons M, Albericio F, Royo MA. J. Am. Chem. Soc. 2004;126:6048. doi: 10.1021/ja0398621. [DOI] [PubMed] [Google Scholar]; (d) Sharma GVM, Jadhav VB, Ramakrishna KVS, Narsimulu K, Subash V, Kunwar AC. J. Am. Chem. Soc. 2006;128:14657. doi: 10.1021/ja064875a. [DOI] [PubMed] [Google Scholar]; (e) Qureshi MKN, Smith M. Chem. Commun. 2006:5006. doi: 10.1039/b611882h. [DOI] [PubMed] [Google Scholar]; (f) Baruah PK, Sreedevi NK, Gonnade R, Ravindranathan S, Damodaran K, Hofmann HJ, Sanjayan GJ. J. Org. Chem. 2007;72:636. doi: 10.1021/jo062032w. [DOI] [PubMed] [Google Scholar]; (g) Sharma GV, Chandramouli N, Choudhary M, Nagendar P, Ramakrishna KV, Kunwar AC, Schramm P, Hofmann HJ. J. Am. Chem. Soc. 2009;131:17335. doi: 10.1021/ja907074u. [DOI] [PubMed] [Google Scholar]
- 9.(a) Horne WS, Price JL, Gellman SH. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9151. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schmitt MA, Choi SH, Guzei IA, Gellman SH. Am. Chem. Soc. 2005;127:13130. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
- 10.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 11.(a) O'Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]; (b) Oshaben KM, Salari R, McCaslin DR, Chong LT, Horne WS. Biochemistry. 2012;51:9581. doi: 10.1021/bi301132k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Price JL, Horne WS, Gellman SH. J. Am. Chem. Soc. 2010;132:12378. doi: 10.1021/ja103543s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Horne WS, Price JL, Keck JL, Gellman SH. J. Am. Chem. Soc. 2007;129:4178. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]
- 13.Please see the Supporting Information.
- 14.Barlow DJ, Thornton JM. J. Mol. Biol. 1988;201:601. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 15.(a) Guo L, Zhang W, Reidenbach AG, Giuliano MW, Guzei IA, Spencer LC, Gellman SH. Angew. Chem. Int. Edit. 2011;50:5843. doi: 10.1002/anie.201101301. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Guo L, Almeida AM, Zhang W, Reidenbach AG, Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2010;132:7868. doi: 10.1021/ja103233a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Guo L, Chi Y, Almeida AM, Guzei IA, Parker BK, Gellman SH. J. Am. Chem. Soc. 2009;131:16018. doi: 10.1021/ja907233q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Guo L, Zhang W, Guzei IA, Spencer LC, Gellman SH. Org. Lett. 2012;14:2582. doi: 10.1021/ol3008815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Basuroy K, Dinesh B, Shamala N, Balaram P. Angew. Chem. Int. Ed. 2012;51:8736. doi: 10.1002/anie.201204436. [DOI] [PubMed] [Google Scholar]; (b) Basuroy K, Dinesh B, Shamala N, Balaram P. Angew. Chem. Int. Ed. 2013;52:3136. doi: 10.1002/anie.201209324. [DOI] [PubMed] [Google Scholar]; (c) Dinesh B, Basuroy K, Shamala N, Balaram P. Tetrahedron. 2012;68:4374. [Google Scholar]
- 17.(a) Jadhav SV, Bandyopadhyay A, Gopi HN. Org. Biomol. Chem. 2013;11:509. doi: 10.1039/c2ob26805a. [DOI] [PubMed] [Google Scholar]; (b) Bandyopadhyay A, Jadhav SV, Gopi HN. Chem. Commun. 2012;57:7170. doi: 10.1039/c2cc32911e. [DOI] [PubMed] [Google Scholar]
- 18.(a) Seebach D, Overhand M, Kuehnle FNM, Martinoni B. Helv. Chim. Acta. 1996;79:913. [Google Scholar]; (b) Seebach D, Ciceri PE, Overhand M, Jaun B, Rigo D, Oberer L, Hommel U, Amstutz R, Widmer H. Helv. Chim. Acta. 1996;79:2043. [Google Scholar]
- 19.Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J. Am. Chem. Soc. 2007;129:1532. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 21.(a) Appella DH, Barchi JJ, Jr, Durell SR, Gellman SH. J. Am. Chem. Soc. 1999;121:2309. [Google Scholar]; (b) LePlae PR, Fisk JD, Porter EA, Weisblum B, Gellman SH. J. Am. Chem. Soc. 2002;124:6820. doi: 10.1021/ja017869h. [DOI] [PubMed] [Google Scholar]; (c) Raguse TL, Lai JR, Gellman SH. Am. Chem. Soc. 2003:125–5592. doi: 10.1021/ja0341485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.