Abstract
Structural and functional neuroimaging have substantively informed the pathophysiology of numerous adult neurological and psychiatric disorders. While structural neuroimaging is readily acquired in sedated young children, pediatric application of functional neuroimaging has been limited by the behavioral demands of in-scanner task performance. Here, we investigated whether functional magnetic resonance imaging (fMRI) acquired during natural sleep and without experimental stimulation offers a viable strategy for studying young children. We targeted the lengthy epoch of non-Rapid Eye Movement, stage 3 (NREM3) sleep typically observed at sleep onset in sleep-deprived children. Seven healthy, preschool-aged children (24-58 months) were studied, acquiring fMRI measurements of cerebral blood flow (CBF) and of intrinsic connectivity networks (ICNs), with concurrent sleep-stage monitoring. ICN data (T2* fMRI) were reliably obtained during NREM3 sleep; CBF data (arterial spin labeled fMRI) were not reliably obtained, as scanner noises disrupted sleep. Applying independent components analysis (ICA) to T2* data, distinct ICNs were observed which corresponded closely with those reported in the adult literature. Notably, a network associated with orthography in adults was not observed, suggesting that ICNs exhibit a developmental trajectory. We conclude that resting-state fMRI obtained in sleep is a promising paradigm for neurophysiological investigations of young children.
Keywords: Magnetic Resonance Imaging, Intrinsic Connectivity Network, Sleep, pediatric, biomarker
1. Introduction
Pediatric-onset neurological disorders (PODs) increase medical and educational costs, worsen educational outcomes, and diminish life-time earning potential, relative to typically developing children (SEEP, 2004). Although PODs are steadily increasing in diagnostic frequency, the pathophysiology of many PODs is poorly understood (CDC, 2008; Kahn & Farone, 2006). Consequently, biomarkers enabling early diagnosis and intervention are few. Structural and functional neuroimaging investigations have increased our understanding of the pathophysiology of numerous adult neurological and psychiatric disorders, suggesting that the same could prove true for childhood-onset disorders. While structural neuroimaging is readily applied in young children (typically with sedation), pediatric application of functional neuroimaging has been limited by the behavioral demands of in-scanner task performance (Jones et al., 2009; Sava & Yurgelun-Todd, 2008; Mercadillo et al., 2012) The task-minus-control experimental paradigm of functional magnetic resonance imaging (fMRI), as typically performed, requires active participation by the subject and nearly complete between-scan head stability. Obtaining adequate task-control fMRI data in awake children under the age of 5 years, particularly those suspected of having a POD, is effectively impossible.
Natural sleep offers a promising alternative for fMRI investigations of young children. Natural sleep consists of a repeating progression of stages characterized by varying degrees of neuronal activity and physical immobility (Iber et al., 2007). The deepest sleep stage, Non-Rapid Eye Movement 3 (NREM3), is a plausible target for sleep imaging both because the arousal threshold is highest and because spontaneous motions are minimized. NREM3 typically occurs early in the night (1st third), with the longest epochs coming shortly after sleep onset. NREM3 is prominent in children and decreases in duration with advancing age (Gaudreau et al., 2001). In children, sleep deprivation significantly reduces the latency from sleep onset to NREM3 (Sheldon et al., 1992). Sleep has successfully been used to obtain immobility for anatomical MRI in pediatric populations (Giedd et al., 1999, 2000). Task-activation fMRI during sleep (using auditory stimuli) has been reported in infants and toddlers, but without sleep stage monitoring (Anderson et al., 2001; Eyler et al., 2012; Dehaene-Lambertz et al., 2002; Redcay et al., 2008; Redcay & Courchesne, 2008). Deeper sleep stages would be expected to exhibit weaker stimulus-driven neural responses, but also to be less confounded by motion artifact and arousal. As an alternative to task-activation fMRI, many neurophysiological variables can be meaningfully measured during eyes-closed rest, detecting developmental and disease-related regional alterations, as follows (Smith et al., 2009; Connor et al., 2012).
Resting-state alterations in cerebral blood flow (CBF), cerebral blood volume, blood transit time, cerebral metabolic rates of glucose and oxygen, oxygen extraction fraction, and other physiological variables have been employed for decades to detect regional abnormalities in neurological and psychiatric disorders (Drevets et al., 2002). For many years, these physiological variables could only be measured using radiotracers, i.e., with exposure to ionizing radiation. More recently, advances in fMRI methods allow several hemodynamic and metabolic variables to be quantified without radiation exposure, thereby making them acceptable for routine use in children (Silva & Paiva, 2009). Of these fMRI techniques, CBF as measured by arterial spin labeling (ASL) is the most well validated and widely available. Developmental changes in whole-brain and regional CBF can be detected using ASL fMRI, as well as with H215O-positron-emission tomography (Chugani, 1998).
Intrinsic connectivity networks (ICNs), also termed resting-state networks (RSNs), are a neurophysiological phenomenon first reported using (and still most readily detected by) blood oxygen level dependent (BOLD) fMRI (Biswal et al., 1995). ICNs are extracted from BOLD images by virtue of the temporal coherence of spontaneous fluctuations in T2* signal (a mixture of hemodynamic and metabolic variables) in structurally and functionally connected brain regions (Beckmann et al., 2005). ICN’s were originally detected by region seeding, i.e., selecting a brain region and computing its covariance with all other brain voxels. Alternatively, ICNs can be extracted by independent components analysis (ICA), a fully data-driven procedure that ranks networks by temporal coherence. Both approaches can distinguish between patient and control populations (Greicius et al., 2007; Hoffman et al., 2011). Furthermore, at least some ICNs are present during natural sleep in infants, suggesting that both approaches should work with pre-school aged children (Gao et al., 2009).
The purpose of the present investigation was to test the feasibility of obtaining useful resting-state fMRI data during natural sleep in preschool-aged children. In particular, we targeted the first epoch of NREM3 after sleep onset, using in-scanner electrophysiological monitoring to confirm sleep stage. Anatomical (T1-weighted) and two types of physiological (fMRI) MRI data were obtained: BOLD (T2*) image volumes for ICN analysis; and, ASL image volumes for CBF analysis. To our knowledge, this is the first report of in-scanner sleep dynamics in a pediatric population. Furthermore, we believe this is the first paper to investigate ICNs in preschool-aged children.
2. METHODS
2.1 Participants and Participant Preparations
Participants were typically developing children between the ages of 24 and 58 months serving as controls for a larger fMRI study on autism. Children were administered the Autism Diagnostic Observation Schedule- Revised. All scores were below diagnostic thresholds. To qualify, children had to score below diagnostic thresholds on both the language and social subscales. Additionally, language samples were obtained during ADOS-R testing and seven children used phrase speech. These seven all underwent sleep-onset fMRI with electrophysiological sleep-stage monitoring. Table 1 shows the ages and genders of the seven participants. This study was approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio. Parents provided permission for their children’s participation.
Table 1. Participants with In-scanner Sleep-stage Monitoring.
Seven subjects participated in this study, ranging in age from 24 to 58 months.
| Child | Age | Gender | Acquisition time (S1; S2) | Sessions | Sequences | Arousals (B; F) | Arousal Cause : |
|---|---|---|---|---|---|---|---|
| 1 | 24 | F | 49.1 (49.1; 0.0) | 2 | 5 | 0; 3 | P, ASL, P |
| 2 | 29 | M | 62.833 (0.0; 62.833) | 2 | 9 | 0; 1 | ASL |
| 3 | 35 | M | 52.983 (52.983; n/a) | 1 | 5 | 1; 3 | P, ASL, ASL |
| 4 | 39 | M | 39.183 (34.017; 5.167) | 2 | 7 | 4; 6 | P, T2*, P, P, ASL, ASL |
| †5 | †53 | F | 33.933 (33.933; 0.0) | 2 | 5 | 2; 6 | P, T2*, P, P, ASL, P |
| 6 | 56 | F | 82.033 (53.133; 28.9) | 2 | 9 | 2; 3 | T2*, ASL, ASL |
| 7 | 58 | M | 54.6 (41.683; 12.917) | 2 | 9 | 0; 2 | T2*, ASL |
Age = post-natal months. All seven subjects contributed to sleep-stage analysis. All seven participants underwent in-scanner sleep-stage monitoring with EEG, EOG and video monitoring. Acquisition time = minutes of fMRI data acquired in NREM3 sleep for Session 1 (S1) and Session 2 (S2). Sessions = number of scanning session per child. Sequences = number of image-acquisition epochs initiated per child. Arousals = number of confirmed arousals, either brief (B) or full (F). Arousal Cause = cause of full arousals, where P = subject preparation; ASL = ASL pulse-sequence acquisition; T2* = T2* pulse-sequence acquisition.
= subject excluded from ICN analysis due to insufficient T2* data.
Participants were brought to the imaging suite by their parent(s) at or after their typical bedtime, typically 8:00-9:00 p.m. Children were mildly sleep deprived by limiting daytime naps. Children were put to bed by their parent(s) in a child-friendly bedroom adjacent to the MRI suite. When judged by the parent(s) to be deeply asleep, the child was carried to the MRI suite and prepared for imaging. Preparations included placing noise-reduction ear plugs, adhesive electrodes and foam head restraints.
2.2 Sleep-Stage monitoring
MRI-compatible, carbon-fiber electrodes were placed frontally to monitor the electroencephalogram (EEG) and peri-orbitally to monitor the electro-oculogram (EOG). Two frontal channels were placed, one monitored and one used to quickly change monitored lead when needed. Data were acquired using a commercially available, digital recording system (Biopac™, Goleta, CA). Sleep stage was assessed prior to and immediately following each image-acquisition epoch, as recording channels saturated with scanner noise during image acquisition. Sleep-stage was determined in an ongoing manner by an investigator trained in pediatric sleep-stage scoring (JM). Research staff remained in the scanning suite and in tactile contact (hand on leg) with the children throughout image-data acquisition, to detect arousal. An in-scanner, infra-red video camera (with a monitor adjacent to the MRI console) was also used to detect arousal.
2.3 Image Acquisition
MRI data were obtained on a 3T Siemens TIM-Trio (Siemens Medical Solutions, Erlangen, Germany), using a standard 12-channel head coil as a RF receiver and the integrated circularly polarized body coil as the RF transmitter. T1-weighted images were acquired using the MPRAGE pulse sequence with TR/TE = 2200/ 2.72 ms, flip angle = 13°, TI = 766 ms, volumes = 208, 1mm3 voxel size. BOLD (T2*-weighted) images were acquired with TR/TE= 3000/30, flip angle = 90°, volumes = 100, 2×2×4 mm3 spatial resolution. CBF ASL images were obtained with TR/TE= 4780/23 ms, flip angle = 90°, 4 mm slice thickness, volumes = 32 with each time point taking 4.8 seconds.
Scans were obtained in prioritized order (Figure 1), with the localizer and T1-weighted anatomical scans being obtained first. Functional scans were obtained thereafter, in order of in-scanner loudness. The T2* protocol, with an in-scanner loudness of 106 dB(A), was obtained second. The ASL sequence, with an in-scanner loudness of 114 dB(A), was performed third. While all MRI sequences produce loud “banging” sounds as the gradients are energized, the T1 and T2* sequences are repetitive and rhythmic, making them less likely to cause arousal. The ASL sequence, by contrast, is more dysrhythmic, with abrupt loudness transitions at shifts between the labeling and recording phases.
Figure 1. Image-Data Acquisition Time line.

Prep = subject preparation, including placement of sound-attenuating ear plugs, adhesive electrodes, and foam head restraints. Localizer = anatomical localization scan to position the imaging volume. T1 = T1-weighted anatomical scan. T2* = 15 min BOLD image-volume acquisition for ICN analysis. ASL = 2.5 min arterial spin labeled acquisition. The number of epochs of T2* (1-3) and ASL data (3-5) varied across subjects, depending on sleep duration. SS = sleep-stage determination, which was performed between all imaging epochs.
2.4 Data Analysis
ICNs were computed using the FMRIB MELODIC software package, as follows. T1-weighted anatomical images were preprocessed using the Brain Extraction Tool (BET) to remove skull and non-brain tissues (Jenkinson et al., 2005). FMRIB’s MELODIC was then run on the T2* weighted BOLD images using a high pass filter of 100 seconds. MCFLIRT motion correction, brain extraction, and spatial smoothing of 5mm FWHM were conducted (Jenkinson et al., 2002). Multi-sesson temporal concatenation with a fixed dimensionality of 20 components was conducted on group-wise data (Beckmann et al., 2005; Smith et al., 2011). Registration of T2* BOLD images to anatomical images was done with a normal search and 7 degrees of freedom. Registered brains were then converted into standard space using a resampling resolution of 4mm. ICNs were behaviorally interpreted by reference to independent components analysis of the BrainMap database, as described by Smith et al. (2009).
3. RESULTS
3.1 In-scanner Sleep Stage
MRI data were successfully acquired only in the NREM3 sleep stage. Our young, sleep-deprived participants tended to fall rapidly into NREM3 and to remain in that state for extended periods of time. Despite there being only a one EEG and one EOG, sleep stage was clearly detectable when the leads were not saturated with MRI radio-frequency energy. NREM3 image-acquisition epochs of 30-50 minutes were common (Table 1). When image acquisition was attempted in lighter sleep stages (NREM2, REM), children immediately aroused fully.
3.2 Arousals
Arousals of two types were observed: brief and full. Brief arousals were incomplete awakenings lasting 2-5 seconds and not requiring removal from the scanner. These were observed chiefly by EEG/EOG and were relatively uncommon (9 episodes; Table 1). Additional brief arousals may have occurred during image-acquisition epochs, but would not have been detected due to channel saturation.
Full arousals were complete awakenings, being observed behaviorally as well as by EEG/EOG. Full arousals were more common than brief arousals, with 24 full-arousals being recorded. The most common cause of full arousal was subject preparation, most notably placement of earplugs and adhesive electrodes. The second most common cause of full arousal was the initiation of an ASL sequence. Arousals during the T2* sequence were uncommon, despite their duration (multiple 15-minute epochs) being much longer than the ASL sequences (2.5-min epochs).
3.3 Sleep-state Image Acquisition
All seven children were successfully imaged during natural sleep, for at least part of the imaging protocol. In all seven children, T1-weighted anatomical scans suitable for spatial normalization of the functional images were obtained. Motion in all completed scans was minimal, with mean relative displacement ranging from 0.05 to 0.1 mm (mean= 0.072, sd= 0.012). In six of the seven children, at least two 15-minute epochs of T2* images suitable for ICN analysis were obtained. In the seventh child, 15 min of T2* data were obtained. ASL CBF image acquisition proved the most problematic to obtain, as all seven children were fully awakened by sequence onset in at least one imaging session. Useful ASL data were obtained only in three subjects.
3.4 Intrinsic Connectivity Networks
Each included subject provided 600 brain volumes for analysis. At a dimensionality of 20, ICA applied to twelve, 15-minute T2* image volumes from 6 subjects (2 per subject) extracted 18 non-artifactual ICNs. Of these, the ten accounting for the most variance are illustrated in Figure 2. The ICNs observed here in sleeping children corresponded closely both to those reported in 35 awake, resting-state in health adults and to those observed by applying ICA analysis the BrainMap database, a large (~ 33,000 subjects) task-state data set (Fox & Lancaster, 2002; Smith et al., 2009). Table 2 lists the observed pediatric ICNs ranked by variance accounted for and the closest analog (by spatial pattern) from adult networks. The rank order of the ICNs (based on % variance accounted for) differed between the groups. For example, ICN1 in the present study most closely matches the adult ICN9, as reported by Smith (2009). One notable difference between the pediatric ICNs reported here and the adult ICNs reported by Smith (2009) is the absence of Smith’s ICN 3, associated with orthographic processing.
Figure 2. Intrinsic Connectivity Networks (ICNs).
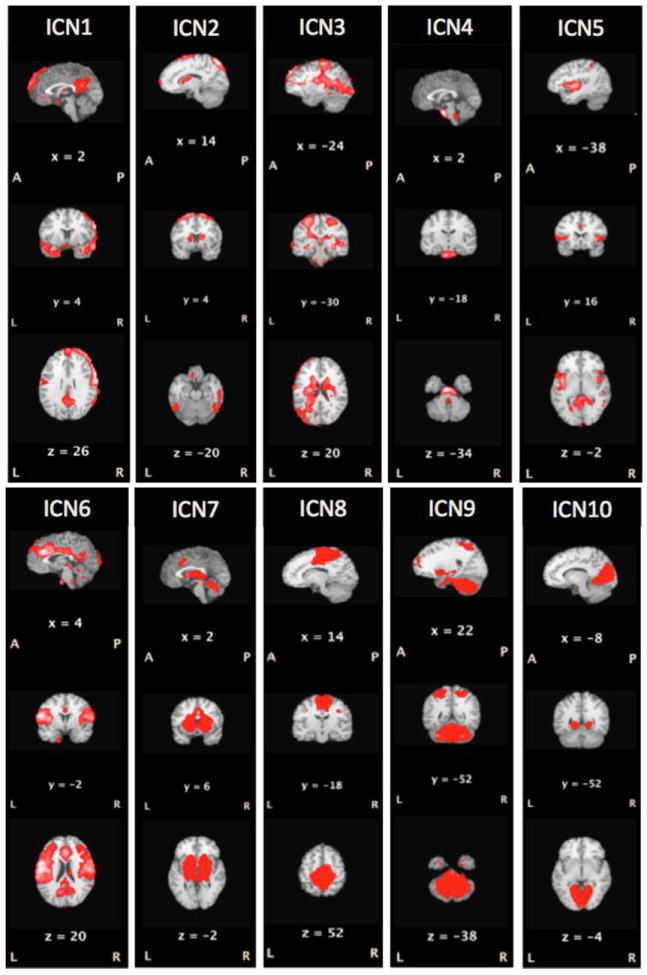
The ten ICNs accounting for the greatest variance in seven, soundly sleeping (NREM3), preschool-aged children (24-58 months) are illustrated (p≤ 0.05). The ICNs observed here in sleeping children corresponded closely both to those reported in 35 awake, resting-state in health adults and to those observed by applying ICA analysis the BrainMap database (Fox and Lancaster, 2002), a large (~ 33,000 subjects) task-state data set (Smith, Fox et al., 2009). Table 2 lists the observed pediatric ICNs ranked by variance accounted for and the closest analog (by spatial pattern) from adult networks.
Table 2. Intrinsic Connectivity Networks (ICNs).
| ICN Rank | Explained Variance | Total Variance | Smith ICN | Behavioral domain |
|---|---|---|---|---|
| 1 | 16.1299 | 7.51616 | 9 | Pain Perception |
| 2 | 7.40104 | 3.4487 | 8 | Executive Control |
| 3 | 7.14941 | 3.33145 | 6 | Sensorimotor |
| 4 | 6.77173 | 3.15546 | X | Pons & Medulla Oblongata |
| 5 | 6.32618 | 2.94785 | 10 | Cognitive & Language |
| 6 | 5.42852 | 2.52956 | 7 | Auditory |
| 7 | 5.32772 | 2.48259 | 14 | Basal Ganglia and Cerebellum |
| 8 | 5.30576 | 2.47236 | 4 | Default Mode Network |
| 9 | 5.26252 | 2.4522 | 5 | Cerebellum |
| 10 | 5.04266 | 2.34976 | 1 | Vision: Motion & Space |
ICN Rank = ICN rank order by explained variance (p≤0.05). Smith ICN = rank order of ICN’s reported by Smith (et al, 2009) based on concurrent ICA analysis of 35 healthy controls and the BrainMap database (Fox and Lancaster, 2009). Behavioral Domain = functional properties of each ICN, as determined by behavioral meta-data in the BrainMap database associated with each ICN.
4. DISCUSSION
The most fundamental observation of the present study is that interpretable resting-state functional MRI images can be obtained during natural, unsedated sleep and without experimental interventions (i.e. without stimulation or task performance) in preschool-aged children (25-58 months). Structural MRI was also successfully obtained during sleep NREM3 sleep, allowing for successful registration to the BOLD images. For approximately 1 hour following sleep onset in mildly sleep-deprived toddlers, sleep consisted almost entirely of stage NREM3. In this sleep stage, T2* BOLD fMRI images were readily acquired. T2* BOLD data during NREM3 provided ICNs quite similar to those observed in adults, with exceptions which suggest a development trajectory. Acquisition of the louder, less rhythmic ASL CBF fMRI sequence was problematic, typically interrupting sleep. This suggests that fMRI acquired during natural sleep, and particularly ICN analysis of T2* BOLD images, offer a promising paradigm for investigating normal brain development and the neuropathophysiology of PODs.
4.1 Sleep-stage Monitoring
To our knowledge, this is the first report of sleep-stage monitoring during MRI in children. Sleep-stage monitoring during anatomical MRI is unnecessary, as level of sleep would not alter anatomical studies, other than by affecting subject mobility. Functional studies, on the other hand, will almost certainly be affected by sleep stage. For task-activation fMRI (e.g., auditory stimulation during sleep), depth of sleep will likely determine the amplitude of neural responses and, thereby, the magnitude of the observed fMRI activations. For fMRI ICN studies, the spatial pattern of the observed networks will likely be unaltered by depth of sleep, as networks observed at rest mimic those used during task performance. Resting-state and task-state ICA analyses provide similar network decompositions. On the other hand, the temporal coherence of individual networks may well differ between wakefulness and deep sleep, potentially explaining the differences in ICN rank order observed here (Table 2).
As anticipated, our sleep-deprived, young (24-58 months) subject sample exhibited prolonged epochs of NREM3 sleep starting rapidly after sleep onset. In this sleep stage, anatomical MRI and T2* fMRI (for ICN analysis) were reliably obtained. ASL fMRI (for CBF analysis) was not reliably obtained, as the noise generated by this substantially louder and less rhythmic sequence (with distinct labeling and read-out phases) routinely aroused the children.
An unanticipated observation was that intermediate sleep stages were not observed. When children were roused by scanner noise, they roused fully and needed to be returned to their parents for comforting and (at the parents discretion) re-establishment of sleep. This observation suggests that sleep-stage monitoring may be unnecessary, as a sleep-deprived child sleeping during fMRI can be presumed to be in NREM3 sleep, at least based on our 7-subject sample. If this is a reliable generalization, eliminating sleep-stage monitoring in future studies could substantially decrease the subject burden of this study as well as increase success rate. The majority of full arousals were caused by subject preparation (Table 1), most notably the placement of adhesive electrodes for sleep-stage monitoring. It is possible, albeit unlikely, that a child exited NREM3 briefly and returned to NREM3 during an imaging sequence. This is most likely to have occurred during the longer T2* acquisition. Fortunately, when sleep stage transitions occur in a sleep center setting, they are frequently preceded by a brief arousal (Sheldon et al., 1992). We believe that this brief arousal, when it occurred during data acquisition in the MRI, resulted in full awakening (see Table 1.). However, if REM sleep did occur, undetected, during the resting state MRI, we would expect the order of ICNs to be altered with visual networks accounting for more variability and frontal, executive networks accounting for less variability. We did not see visual networks increasing in ICN rank.
4.2. ICNs
The spatial patterns of the ICNs observed in deeply sleeping young children corresponded closely to those observed in awake adults, with some exceptions. The rank order observed was markedly different (Table 2). Given that the temporal coherence of individual ICN’s varies with cognitive state (Fransson et al., 2009) it would be expected that it would vary with depth of sleep and, potentially, with fluctuations in the cognitive content (including dreams) of sleep. Additionally, there were some variations in ICN pattern. For example, ICN4 in our study including pons and medulla oblongata was not found in Smith et al. (2009). We believe that it is a representation of the reticular activating system’s role in sleep maintenance. Additionally, a visual-system ICN strongly implicated in orthography (ICN3 from Smith et al., 2009) was not observed. This suggests a developmental trajectory of ICNs – e.g., with orthographic-network connectivity being augmented by the process of learning to read – which could potentially be used to characterize normal development as well investigate developmental delays and therapeutic interventions.
The similarities found in the networks of our sleeping children to established networks in adults (Smith et al., 2009) are striking. In terms of the variance explained by each ICN, ICN1 corresponds well to pain perception in both fMRI activation studies and resting-state imaging in adults. ICN2 corresponds to executive control tasks, ICN3 to sensorimotor demands, ICN5 to cognitive and language tasks, ICN6 to Audition, ICN7 to basal ganglia and cerebellum, ICN8 to the Default Mode Network, ICN9 to cerebellum and ICN10 to visual processing of spatial and motion information. ICN4, the reticular activating system, was not represented in the published adult data, likely because is not needed to maintain rest or to perform in-scanner tasks.
Two additional possible sources of differences between the rank order of the ICNs observed here and those reported in adults deserve mention. First, our subject sample is admittedly small (n = 6). Mitigating this, the acquisition duration (30 min) is substantially longer than the 8-10 min acquisitions often reported in the literature. The longer acquisition duration adds data points, increasing statistical strength without increasing sample size. We have 600 brain volumes per child for a total of 3600 brain volumes. The ICN calculation is based upon total volumes, not sample size. Second, because of the smaller brain/head size of pediatric subject sample, we were able to obtain the entire brain in all subjects. Functional MRI studies in adults frequently fail to obtain the more caudal portions of the cerebellum and brainstem.
The current version of the ICN analysis package used here (FSL, MELODIC) does not accommodate between-subject variation in scan duration. In the present study, two 15-min epochs of T2* data were acquired in 6 of 7 subjects, with the 7th subject having only one epoch. Based on the observation that most arousals occur at transitions in pulse sequences, we would recommend a single, 30-minute T2* acquisition epoch. Thirty minutes should provide sufficient data for robust (potentially per-subject) analysis, fit comfortably within the typical NREM3 duration observed (Table 1), and minimize the risk of arousal.
4.3 ASL
The success rate of acquiring ASL CBF data was fairly poor. This pulse sequence is significantly louder and less rhythmic than the T1-weighted and T2* pulse sequences, routinely arousing the subjects from sleep. While our experience might be taken to suggest that pursuing ASL CBF imaging in children is unproductive, it is known that brain hemodynamics and metabolism change dramatically throughout childhood (Chugani, 1998). We would argue that fMRI studies of brain blood flow in sleeping children should be pursued. Acquiring ASL CBF data immediately after sleep onset (rather that after 30 min or more of T2* imaging, as we did) may well improve the likelihood of success. Additionally, any pulse-sequence modifications that could lower the decibel level of the in-scanner noise would be highly beneficial.
4.4 Imaging Biomarker Development
A significant priority for many PODs is development of reliable diagnostic biomarkers. Structural and functional brain imaging offers considerable promise in this regard. To date, however, imaging studies in POD populations have chiefly enrolled adults. This is problematic on two counts. First, imaging in adults does not contribute to the development of imaging-based diagnostic tools useful at disease onset. Second, compensatory strategies employed over a lifetime almost certainly will modify brain structure and function, making studies in adults even less representative. Consequently, we encourage the continued development of fMRI-based measures of brain development in young children, with the expectation that some will yield diagnostic and therapeutic biomarkers. For very young children, imaging during sleep appears to be a viable way forward.
Supplementary Material
Acknowledgments
This work was supported by pilot grants funds from: The Research Imaging Institute Utilization Review Committee (MRI126) and The Palmaz Endowment (Pilot award 48004). JM was supported by Translational Science Training (TST) Across Disciplines program at the University of Texas Health Science Center at San Antonio, with funding provided by the University of Texas System’s Graduate Programs Initiative (128223), and the Kronkosky Foundation. PTF was supported by NIH grants MH074457, MH084812 and RR024387. EC was supported by Supported by NIMH Autism Center of Excellence grant P50-MH081755 and NIMH R01- MH036840.
We thank Elizabeth King, Robert Vargas, and Reagan Costello-White for assistance with participant preparation and data collection. We thank John Li, M.D. and Feng Gao, M.D. for training in scanner operation. We thank Crystal Franklin for training in data analysis. We thank Steven Plizka, M.D. for helpful comments on the manuscript. We thank Christus Santa Rosa Children’s Sleep Lab for training in pediatric sleep scoring.
Abbreviations
- ICA
Independent Component Analysis
- CBF
Cerebral Blood Flow
- fMRI
functional Magnetic Resonance Imaging
- POD
Pediatric-Onset Neurological Disorder
- ASL
Arterial Spin Labeling
- BOLD
Blood Oxygen Level Dependent
- EEG
electroencephalogram
- EOG
Electro-oculogram
- NREM
Non-Rapid Eye Movement
Footnotes
CONFLICTS OF INTEREST.
The authors have no conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson AW, Marois R, Colson ER, et al. Neonatal auditory activation detected by functional magnetic resonance imaging. MagnREson Imaging. 2001;19:1–5. doi: 10.1016/s0730-725x(00)00231-9. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, De Luca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Society. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Prevalence of Autism Spectrum Disorders — Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States, 2008. MMWR. 2012;61(SS03):1–19. [PubMed] [Google Scholar]
- Chugani HT. A critical period of brain development: studies of cerebral glucose utilization with PET. Prev Med. 1998;27:184–188. doi: 10.1006/pmed.1998.0274. [DOI] [PubMed] [Google Scholar]
- Connor CM, Crawford BC, Akbarian S. White matter neuron alterations in schizophrenia and related disorders. Int J Dev Neurosci. 2012;29:325–34. doi: 10.1016/j.ijdevneu.2010.07.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaene-Lambertz G, Dehaene S, Hertz-Pannier L. Functional neuroimaging of speech perception in infants. Science. 2002;6:2013–2015. doi: 10.1126/science.1077066. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Bogers W, Raichle ME. Functional anatomical correlates of antidepressant drug treatment assessed using PET measures of regional glucose metabolism. Eur Neuropsychopharmacol. 2002;12:527–44. doi: 10.1016/s0924-977x(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Eyler LT, Pierce K, Courchesne E. A failure of left temporal cortex to specialize for language is an early emerging and fundamental property of autism. Brain. 2012;135:949–60. doi: 10.1093/brain/awr364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL. Opinion: Mapping context and content: the BrainMap model. Nat Rev Neuroscience. 2002;3:319–21. doi: 10.1038/nrn789. [DOI] [PubMed] [Google Scholar]
- Fransson P, Sklold B, Engstrom M, et al. Spontaneous brain activity in the newborn brain during natural sleep- An fMRI study in infants born at full term. Pediatr Res. 2009;66:301–5. doi: 10.1203/PDR.0b013e3181b1bd84. [DOI] [PubMed] [Google Scholar]
- Gao W, Zhu H, Giovanello K, Smith JK, et al. Evidence on the emergence of the brain’s default network from 2-week-old to 2-year-old healthy pediatric subjects. Proc Natl Acad Sci U S A. 2009;106:6790–5. doi: 10.1073/pnas.0811221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau H, Carrier J, Montplaisir J. Age-related modifications of NREM sleep EEG: From childhood to middle age. J Sleep Res. 2001;10:165–172. doi: 10.1046/j.1365-2869.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries C, et al. Development of the human corpus callosum during childhood and adolescence: a longitudinal MRI study. Prof Neuropsychopharmacol Biol Psychiatry. 1999;23:571–88. doi: 10.1016/s0278-5846(99)00017-2. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Rapoport JL, Garvey MA, et al. MRI assessment of children with obsessive-compulsive disorder or tics associated with streptococcal infection. AM J Psychiatry. 2000;157:281–3. doi: 10.1176/appi.ajp.157.2.281. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, et al. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry. 2007;62:429–37. doi: 10.1016/j.biopsych.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RE, Fernandez T, Pittman B, Hampson M. Elevated functional connectivity along a corticostriatal loop and the mechanism of auditory/verbal hallucinations in patients with schizophrenia. Biol Psychiatry. 2011;69:407–14. doi: 10.1016/j.biopsych.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber C, Ancoli-Israel S, Chesson A, Quan SF for the American Academy of Sleep Medicine. The AASM manual for the scoring of sleep and associated events: Rules, terminology and technical specifications. 1. Winchester, Il: American Academy of Sleep Medicine; 2007. [Google Scholar]
- Jenkinson M, Pechaud M, Smith S. BET2: MR-based estimation of brain, skull and scalp surfaces. Eleventh Annual Meeting of the Org for Hum Brain Mapp.2005. [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–41. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jones AP, Laurens KR, Herba CM, Barker GJ, Viding E. Amygdala hypoactivity to fearful faces in boys with conduct problems and callous-unemotional traits. Am J Psychiatry. 2009;166:95–102. doi: 10.1176/appi.ajp.2008.07071050. [DOI] [PubMed] [Google Scholar]
- Khan SA, Faraone SV. The genetics of attention-deficit/hyperactivity disorder: A literature review of 2005. Curr Psychiatry Rep. 2006;8:393–7. doi: 10.1007/s11920-006-0042-y. [DOI] [PubMed] [Google Scholar]
- Mercadillo ME, Trujillo C, Sanchez-Cortazar J, Barrios FA. In ADHD patients performing the counting stroop task: A social neuroscience approach. Psychol Rep. 2012;111:652–68. doi: 10.2466/15.10.19.PR0.111.5.652-668. [DOI] [PubMed] [Google Scholar]
- Redcay E, Courchesne E. Deviant functional magnetic resonance imaging patterns of brain activity to speech in 2-3-year-old children with autism spectrum disorder. Biol Psychiatry. 2008;64:589–98. doi: 10.1016/j.biopsych.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redcay E, Haist F, Courchesne E. Functional neuroimaging of speech perception during a pivotal period in language acquisition. Dev Sci. 2008;11:237–52. doi: 10.1111/j.1467-7687.2008.00674.x. [DOI] [PubMed] [Google Scholar]
- Sava S, Yurgelun-Todd DA. Functional magnetic resonance in psychiatry. Top Magn Reson Imaging. 2008;19:71–9. doi: 10.1097/RMR.0b013e318184187c. [DOI] [PubMed] [Google Scholar]
- Sheldon SH, Spire J-P, Levy HD. Pediatric Sleep Medicine. Philadelphia, PA: W. B. Saunders Company; 1992. [Google Scholar]
- Silva AC, Paiva FF. Dynamic magnetic resonance imaging of cerebral blood flow using arterial spin labeling. Methods Mol Biol. 2009;489:277–95. doi: 10.1007/978-1-59745-543-5_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–5. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi, et al. Network modelling methods for FMRI. NeuroImage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- What are we spending on special education services in the United States, 1999-2000. Special Education Expenditure Project. 2004:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


