Abstract
Researchers working with rodent models of neurological disease often require an accurate map of the anatomical organization of the white matter of the rodent brain. With the increasing popularity of small animal MRI techniques, including diffusion tensor imaging (DTI), there is considerable interest in rapid segmentation methods of neurological structures for quantitative comparisons. DTI-derived tractography allows simple and rapid segmentation of major white matter tracts, but the anatomic accuracy of these computer-generated fibers is open to question and has not been rigorously evaluated in the rat brain. In this study, we examine the anatomic accuracy of tractography-based segmentation in the adult rat brain. We analysed 12 major white matter pathways using semi-automated tractography-based segmentation alongside manual segmentation of Gallyas silver-stained histology sections. We applied four fiber-tracking algorithms to the DTI data - two integration methods and two deflection methods. In many cases, tractography-based segmentation closely matched histology-based segmentation; however different tractography algorithms produced dramatically different results. Results suggest that certain white matter pathways are more amenable to tractography-based segmentation than others. We believe that these data will help researchers decide whether it is appropriate to use tractography-based segmentation of white matter structures for quantitative DTI-based analysis of neurologic disease models.
Keywords: MRI, DTI, automated segmentation, Gallyas silver myelin staining
Introduction
Three-dimensional Magnetic Resonance Imaging (MRI) and Diffusion Tensor Imaging (DTI) have become important research tools for studying the structure and function of the brain both in normal and diseased states. In recent years DTI has garnered significant attention as a non-invasive method for studying and diagnosing white matter pathologies in both patients and preclinical models. The use of high-resolution MRI to study fixed tissue is sometimes referred to as Magnetic Resonance Histology because it can be used to characterize tissue microstructure and pathology in a manner similar to conventional histology. Unlike conventional histology, MRH can be combined with DTI (referred to as DT-MRH) to create brain-wide quantitative maps of white matter integrity. With the increasing use of DT-MRH to study preclinical models of neurological disease, there is considerable interest in rapid automated white matter segmentation techniques to facilitate quantitative comparisons. Automated segmentation based on DTI tractography is an enticing possibility, but researchers debate the anatomic accuracy of computer-generated fiber tracts, particularly when derived from a simple, single-tensor model. Semi-automated tractography-based segmentation is relatively common in both clinical and preclinical MRI studies, as it provides rapid and qualitatively accurate delineations of major white matter pathways in 3D. There is considerable debate in the diffusion tractography literature on the best sampling strategy for generating reliable tracts. Much of this debate stems from the issue of intra-voxel fiber crossings, which cannot be resolved with a simple single tensor model. Several elegant solutions have been developed to resolve intra-voxel fiber crossings including High Angular Resolution Diffusion Imaging (HARDI) (Tuch et al. 2002), Q-ball imaging (Tuch 2004), and Diffusion Spectrum Imaging (DSI) (Wedeen et al. 2008). Each of these methods uses some combination of increased angular sampling or increased b-value sampling to reveal multiple fiber orientations within a single voxel. Another simple, although often impractical, method to resolve intra-voxel fiber crossings is to use smaller voxels and thereby attempt to avoid the problem altogether. In many ways, higher spatial resolution is preferable to higher angular resolution because it does not rely on complex computational methods or post-processing steps. Unfortunately achieving sufficiently high spatial resolution is often impossible due to limited acquisition time and/or limited MRI signal from the subject being imaged. For in vivo and human imaging increased angular resolution is usually preferable because signal is limited and prolonged scan times are impossible or unethical in living beings. However, ex vivo small animal imaging is not limited by signal or acquisition time, and high-spatial resolution can be a viable solution for resolving complex fiber structures. Several groups have attempted to use tractography to create automated segmentation algorithms (Savadjiev et al. 2008; Van Hecke et al. 2008; Hasan et al. 2007; O’Donnell and Westin 2007). Despite the widespread use of tractography-based white-matter segmentation in both clinical and small animal MR research, surprisingly little work has been done to validate its anatomic accuracy (Yendiki et al. 2011; Niogi et al. 2007). In particular, no rigorous comparison exists of tractography-based segmentation with the current anatomic “gold standard” for white matter segmentation—the manual delineation of myelin-stained histological sections.
In this study, we compare manual segmentation of silver-stained histology with semiautomated tractography based segmentation using four of the most common tractography algorithms. Using these segmentation methods, we mapped and analyzed 12 major fiber tracts in a single rat brain that was imaged with ex-vivo DTI and later processed for conventional histology with Gallyas silver stain. For the first time, we provide a quantitative and qualitative anatomical comparison of tractography-based segmentation versus conventional manual segmentation of 12 major fiber tracts in the rat brain. All analyses were done in the stereotaxic coordinate system of the rat brain created by Paxinos and Watson (2007), allowing us to determine the exact localizations of white matter tracts and surrounding structures in stereotaxic space.
Materials and methods
MR Specimen Preparation
All experiments and procedures were done with the approval of the Duke University Institutional Animal Care and Use Committee. A single adult male Wistar rat (Charles River Laboratories, Wilmington, MA, USA) weighing approximately 250 g was perfusion fixed using the active staining technique (Johnson et al. 2012b). Perfusion fixation was achieved using a 10% solution of Neutral Buffered Formalin (NBF) containing 10% (50 mM) Gadoteridol. After perfusion fixation, the head was removed and immersed in 10% NBF for 24 hours. Finally, the fixed rat head was transferred to a 0.1 M solution of Phosphate Buffered Saline containing 1% (5 mM) Gadoteridol at 4° C for 5–7 days to ensure equilibration of contrast agent. Prior to imaging, the specimen was placed in a custom-made, MRI-compatible tube and immersed in a 1H-MRI signal-free, liquid fluorocarbon (Fomblin perfluoropolyether, Ausimont, Thorofare, NJ, USA) for susceptibility matching and to prevent specimen dehydration. All imaging experiments were performed with the brain in-situ in the cranium to preserve its native shape.
MR Data Acquisition
All imaging experiments were performed on a 7-Tesla small animal MRI system (Magnex Scientific, Yarnton, Oxford, England) equipped with 670 mT/m Resonance Research gradient coils (Resonance Research, Inc., Billerica, MA, USA), and controlled with a General Electric Signa console (GE Medical Systems, Milwaukee, WI, USA). RF transmission and reception was achieved using a custom-built 25 mm diameter by 50 mm long solenoid RF coil.
Diffusion weighted images were acquired using a custom-designed spin-echo pulse sequence (TR = 100 ms, TE = 16.2 ms, NEX = 1). Diffusion preparation was accomplished using a modified Tanner-Stejskal diffusion-encoding scheme with a pair of unipolar, half-sine diffusion gradient waveforms (width [δ]= 3 ms, separation [Δ] = 8.5 ms, gradient amplitude = 600 mT/m). One b0 image and 6 high b-value images (b=1500 s/mm2) were acquired with diffusion sensitization along each of 6 non-colinear diffusion gradient vectors: [1, 1, 0], [1, 0, 1], [0, 1, 1], [−1, 1, 0], [1, 0, −1], and [0, −1, 1]. The acquisition matrix was 800 × 400 × 400 over a 40 × 20 × 20 mm field of view (FOV) for a Nyquist limited isotropic voxel size of (50 μm)3. Total acquisition time was approximately 38 hours per specimen.
Subsequently and without moving the specimen, high-resolution T2*-weighted structural images were acquired using a custom-designed 3D gradient recalled echo (GRE) sequence (TR = 50 ms, TE = 8.3 ms, NEX = 2) with a dynamic receiver gain strategy designed to amplify high-frequency signal content and prevent digitizer saturation at the center of k-space, described more completely in (Johnson et al. 2012b). The acquisition matrix was 1600 × 800 × 800 over a 40 × 20 × 20 mm field of view (FOV) for a Nyquist-limited isotropic voxel size of (25 μm)3. Approximate scan time was 20 hours per specimen.
Image Registration and Tensor Calculation
Prior to other manipulations, diffusion-weighted images were registered to the b0 image using a 6-parameter rigid-affine registration to correct for the linear component of eddy current distortions. Tensor estimation and calculation of derived images (i.e. Fractional Anisotropy [FA]) was accomplished using the Diffusion Toolkit software package (http://www.trackvis.org).
To take advantage of the wealth of anatomic information that exists in current histology atlases of the rat brain, we aligned the anatomic GRE image volume to the Paxinos and Watson stereotaxic space (Paxinos and Watson, 2007). The data was iteratively manually rotated in 3D until the coronal image plane matched the Paxinos and Watson coronal plane as closely as possible. The resulting rigid transformation was calculated and applied to DTI data using a log-Euclidean transformation to reorient the tensor with as little error as possible. We checked that the anatomic GRE image volume was inherently co-registered to the b0 volume, and confirmed that further rigid-affine registration did not improve alignment between the two. To assess alignment accuracy, we measured the average stereotaxic discrepancy between MR images and the Paxinos and Watson atlas for 10 coronal slices evenly spaced throughout the brain.
Tractography, Semi-Automated Segmentation, and Analysis
Whole-brain fiber tractography was performed using the Diffusion Toolkit implementations of two integration algorithms, FACT and 2nd-order Runge-Kutta, and two deflection algorithms, Interplated Streamline and Tensorline, with fractional anisotropy threshold = 0.25 and angle threshold = 45°. Semi-automated tractography-based segmentation was achieved by placing a single region-of-interest (ROI)-disk at the midpoint of the white matter pathway being segmented. Tract locations and midpoints were determined using two histology atlases—that of Paxinos and Watson (2007) and that of Konig and Klippel (1963) (Paxinos and Watson 2007; Konig and Klippel 1963). For fiber tracts with a major midline crossing (cc/ec, fi/f, opt/och, ac) a single ROI-disk was drawn on the mid-sagittal image. For all other tracts, a single ROI disc was placed at the midpoint of the tract on either side of the brain. All tracks less than 1 mm in length were excluded to reduce short track clustering around the ROI disc, but no other track modification or editing was performed. The same ROI’s were used for all four tractography methods, for their respective white matter tracts. Track length statistics were gathered for each white matter structure using each of the four-tractography algorithms. The mean tract length for all analyzed structures is provided in the supplement. Vector-based streamlines were converted to voxel-based ROIs using MATLAB. In each case, the ROI was generated from all voxels containing one or more tracts. Finally, ROIs were smoothed using a 3D Gaussian convolution kernel with σ = 3.
Immunohistochemistry
After MR imaging, the rat brain was removed from the cranium and immersed in 30% sucrose in 0.1M PB for cryoprotection for 12 hours. 40 μm coronal sections were cut of the entire brain by using a cryostat (Leica, Germany) in four series. Histological sections were taken in the same coronal plane as the Paxinos and Watson atlas by following the sectioning guidelines presented in the 6th edition (Paxinos and Watson 2007). Myelin silver staining, first described by Gallyas (Gallyas 1971, 1979), was carried out on the first two series. In short, after pre-treatment with a lipid solvent (2:1 pyridine: acetic anhydride) for 30 minutes, sections were rehydrated, then immersed into colloidal silver nitrate solution for impregnation. After the developing of the silver particles, sections were immersed into the bleaching solution for de-staining when necessary (Pistorio et al. 2006).
Modified Nissl staining was performed on the third series of sections. The slides were dehydrated with increasing concentration of ethanol up to 95%, with 5% Acetic Acid and warmed up to 65° C for about one hour. Subsequently, the slides were merged into 0.1% Cresyl Violet for 5 minutes at 65° C. Next, the sections were differentiated using 70% and 95% ethyl alcohol containing 0.5% Acetic Acid for 13 minutes. Then the slides were further dehydrated in 95% and 100% ethyl alcohol and xylene for 3 × 10 minutes.
The last series of sections was processed for acetylcholine esterase (AChE) staining. Sections were incubated with 0.1 M maleate buffer (pH 6.0) containing acethyltiocholine iodine, K3Fe(CN)6, CuSO4 and sodium citrate for 60 minutes at room temperature. Staining was developed by placing the sections in 50mM Tris-HCl Buffer containing diaminobenzidine, nickel chloride hexahydrate, and 0.3 % H2O2. Cells and fibers became visible in 10–15 minutes. Sections were then washed in 0.05 M Tris-HCl (pH7.6) (Tago et al. 1986).
After staining procedures, all sections were dehydrated and cover-slipped with DePex (Electron Microscopy Sciences, USA). Slides were digitally scanned with ScanScope XT (Aperio, USA) bright field scanner at 20x magnification.
Manual Segmentation and 3D reconstruction of the histological data
The first series of the Gallyas stained sections were used in our analysis. This series contained every fourth section of the entire brain (meaning that the distance between the mapped sections was 160 μm) and were manually segmented based on microscopic observation (Zeiss AxioImager.M2 upright research microscope equipped with a true-color digital CCD microscopy camera, using variable magnification of 2.5–10x). MBF Bioscience StereoInvestigator (MBF Bioscience, Williston, VT 05495 USA) was used to reconstruct the major white matter fiber tracts of the rat brain. Coronal serial sections of the brain were mapped by using NeuroLucida software. Rigid registration (rotation and translation) was used to align and match each consecutive segmented histological section with the previous one. Each section was registered to the previous section starting from the second. Since the histological sections were cut parallel to each other (without changing the angle of the blade) and the thickness of the sections was small (40 μm), this procedure aligns the tissue boundaries in consecutive sections successfully achieving a global alignment of all sections. After the alignment of histological sections, they were stacked to form a 3D histological image. The 3D reconstruction was analyzed by Neurolucida Explorer.
Results
Accuracy of data alignment to stereotaxic atlas space
After alignment of MRI data to the Paxinos and Watson histology atlas space, we assessed alignment accuracy by quantifying the differences in anatomic coordinates as measured by MRI and stereology. We analyzed 10 coronal MR images and their corresponding histology plates. These data are presented in Table 1. The average discrepancy between MR and histologic stereotaxic space was 0.393 mm, which is comparable with other such attempts (Johnson et al. 2012a).
Table 1.
Stereotaxic coordinates of the analyzed anatomical sections and MR images and their discrepancy
| Atlas Coordinate (mm from Bregma) | MR Coordinate (mm from Bregma) | Discrepancy (mm) |
|---|---|---|
| 7.5 | 7.15 | 0.35 |
| 5.6 | 5.3 | 0.3 |
| 3.72 | 3.3 | 0.42 |
| 2.3 | 1.8 | 0.5 |
| 0.36 | 0.27 | 0.09 |
| −1.3 | −1.43 | 0.13 |
| −2.64 | −3.13 | 0.49 |
| −5.4 | −5.28 | 0.12 |
| −7.44 | −6.93 | 0.51 |
| −8.9 | −8.66 | 0.24 |
| −10.1 | −10.285 | 0.185 |
| −11.4 | −12.785 | 1.385 |
| Mean Discrepancy (mm) | 0.393 |
Comparing MRH to conventional histology
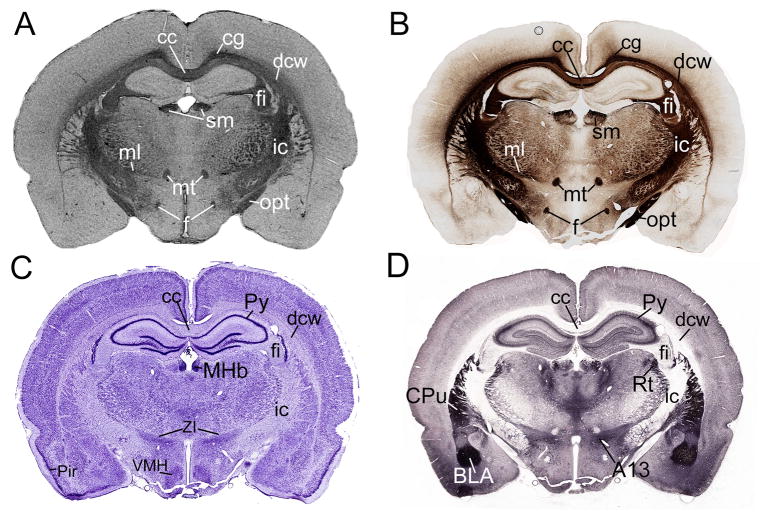
Figure 1 shows a direct comparison of MRH and conventional histology. A Gallyas silver stained section (Fig. 1A) is shown along side a T2*-weighted MRH coronal slice (Fig. 1B) and two cellular histology stains, Nissl and acetyl cholinesterase (Fig. 1C and D, respectively). Several prominent structures are identified on each modality. We found that several white matter structures including the fornix (f), the medial lemniscus (ml), the mammillothalamic tract (mt), the optic tract (opt), the internal capsule (ic), the deep cerebral white matter (dcw), the corpus callosum (cc), and the cingulum (cg) have a strikingly similar appearance in MRH and the Gallyas myelin staining of the same level (Fig. 1A, B), while cellular stains tended to have significantly different tissue contrast (Fig. 1C, D).
Fig. 1.
Comparison of myelinated structures with several well-defined nuclei on the same coronal plane of 4 different stains. A) On the Gallyas myelin-stained section, it is easy to recognize the cc, cg, ml, mt, sm, f, fi, opt, dcw. B) These same structures can be labeled on the MR image, which is has nearly the same contrast as the myelin stained section. C) On the Nissl-stained section, cellular structures are more evident, such as the pyramidal layer of the hippocampus (Py) and the ventromedial hypothalamus (VMH). D) The acetylcholine esterase stained section reveals different structures again, including caudate putamen (CPu) or the basolateral amygdala (BLA). Taken together, it is evident that a comprehensive, updated myelin atlas of the rat brain is needed that focuses on the white matter structures recognizable on MR images.
Volumetric comparisons of tractography-based and histology-based segmentation
To assess the accuracy of tractography-based segmentation, we compared white matter structures segmented using semiautomatic tractography-based segmentation and manual histology-based segmentation. Our investigation focused on the 12 major white matter structures listed in Table 2. To quantify our results, we calculated the volumes of each structure as segmented using semiautomated tractography-based segmentation and manual histology-based segmentation. Tractography-based segmentation volume was calculated for all four tractography algorithms. These results are shown in Table 3. In general, tract volume was greatest for Tensorline, followed by interpolated streamline, 2nd-order Runge-Kutta. FACT-based segmentation generally yielded the smallest volume. The relationship between tractography-based and histology-based segmentation was not as straightforward. In some cases, histology-based segmentation yielded large volumes, and in other cases tractography-based segmentation was larger. Inter-modality agreement varied from tract to tract, and was good in some cases, and poor in others. For example, histology-based segmentation provided a larger volume estimate of the cingulum (cg) but a smaller volume for the corpus callosum/external capsule/forceps minor of the corpus callosum/deep cerebral white matter/alveus (cc/ec/fmi/dcw/alv), when compared to tractography-based segmentation. The medial longitudinal fasciculus (mlf) showed the best agreement between segmentation modalities. A poor inter-modality agreement was observed in the optic tract/optic chiasm (opt/och) where the closest tractography volume estimate was 45% larger than the histology-based segmentation.
Table 2.
List of myelin fiber tracts analyzed by both semi-automated DTI segmentation and manual 3D reconstruction of the mapped structures on serial sections using NeuroLucida and NeuroLucida Explorer by MBF Bioscience. Colors are used to indicate their corresponding structures hereafter.
| Abbreviation | Description | Color |
|---|---|---|
| ac/aci/acp | anterior commissure/intrabulbar part/posterior part |
|
| cc/ec/fmi/dcw/alv | corpus callosum/external capsule/forceps minor of the corpus callosum/deep cerebral white matter/alveus |
|
| cg | cingulum |
|
| fi/f/vhc | fimbria of the hippocampus/fornix/ventral hippocampal commissure |
|
| fr | fasciculus retroflexus |
|
| ic/cp/py/lfp | internal capsule/cerebral peduncle/pyramid/longitudinal fasciculus of the pons |
|
| lo | lateral optic tract |
|
| ml | medial lemniscus |
|
| mlf | medial longitudinal fasciculus |
|
| mt | mammillothalamic tract |
|
| opt/och | optic tract/optic chiasm |
|
| sm/st | stria medullaris of the thalamus/stria terminalis |
|
Table 3.
Volumes of segmented and mapped white matter structures and total brain volume (left and right hemisphere values combined) calculated by using FACT, Runge-Kutta, Interpolated Streamline and Tensorline fiber tract segmentation methods compared with anatomical, histological measurements.
| Name of structures | FACT (mm3) | Histology (mm3) | Difference (mm3) | Difference % |
|---|---|---|---|---|
| ac/aci/acp | 6.72 | 5.99 | 0.73 | 11.49 |
| cc/ec/fmi/dcw/alv | 69.75 | 67.59 | 2.16 | 3.15 |
| cg | 2.56 | 4.27 | −1.71 | −50.07 |
| fi/f/vhc | 20.62 | 17.53 | 3.09 | 16.20 |
| fr | 1.03 | 0.59 | 0.44 | 54.32 |
| ic/cp/py/lfp | 23.59 | 24.17 | −0.58 | −2.43 |
| lo | 5.23 | 7.44 | −2.21 | −34.89 |
| ml | 4.58 | 5.29 | −0.71 | −14.39 |
| mlf | 2.18 | 2.4 | −0.22 | −9.61 |
| mt | 0.52 | 0.64 | −0.12 | −20.69 |
| opt/och | 12.71 | 7.99 | 4.72 | 45.60 |
| sm/st | 3.67 | 3.44 | 0.23 | 6.47 |
| total brain | 2290.12 | 1645.55 | 644.57 | 32.76 |
| Name of structures | Runge Kutta (mm3) | Histology (mm3) | Difference (mm3) | Difference % |
| ac/aci/acp | 7.5 | 5.99 | 1.51 | 22.39 |
| cc/ec/fmi/dcw/alv | 83.47 | 67.59 | 15.88 | 21.02 |
| cg | 2.86 | 4.27 | −1.41 | −39.55 |
| fi/f/vhc | 26.26 | 17.53 | 8.73 | 39.87 |
| fr | 1.21 | 0.59 | 0.62 | 68.89 |
| ic/cp/py/lfp | 31.17 | 24.17 | 7 | 25.30 |
| lo | 5.72 | 7.44 | −1.72 | −26.14 |
| ml | 6.96 | 5.29 | 1.67 | 27.27 |
| mlf | 2.49 | 2.4 | 0.09 | 3.68 |
| mt | 0.76 | 0.64 | 0.12 | 17.14 |
| opt/och | 14.31 | 7.99 | 6.32 | 56.68 |
| sm/st | 5.08 | 3.44 | 1.64 | 38.50 |
| total brain | 2290.12 | 1645.55 | 644.57 | 32.76 |
| Name of structures | Interpolated Streamline (mm3) | Histology (mm3) | Difference (mm3) | Difference % |
| ac/aci/acp | 8.16 | 5.99 | 2.17 | 30.67 |
| cc/ec/fmi/dcw/alv | 91.27 | 67.59 | 23.68 | 29.81 |
| cg | 2.96 | 4.27 | −1.31 | −36.24 |
| fi/f/vhc | 27.49 | 17.53 | 9.96 | 44.25 |
| fr | 1.31 | 0.59 | 0.72 | 75.79 |
| ic/cp/py/lfp | 37.4 | 24.17 | 13.23 | 42.98 |
| lo | 6.06 | 7.44 | −1.38 | −20.44 |
| ml | 13.02 | 5.29 | 7.73 | 84.43 |
| mlf | 2.77 | 2.4 | 0.37 | 14.31 |
| mt | 0.93 | 0.64 | 0.29 | 36.94 |
| opt/och | 14.41 | 7.99 | 6.42 | 57.32 |
| sm/st | 5.3 | 3.44 | 1.86 | 42.56 |
| total brain | 2290.12 | 1645.55 | 644.57 | 32.76 |
| Name of structures | Tensorline (mm3) | Histology (mm3) | Difference (mm3) | Difference % |
| ac/aci/acp | 9.01 | 5.99 | 3.02 | 40.27 |
| cc/ec/fmi/dcw/alv | 111.92 | 67.59 | 44.33 | 49.39 |
| cg | 3.5 | 4.27 | −0.77 | −19.82 |
| fi/f/vhc | 39.29 | 17.53 | 21.76 | 76.59 |
| fr | 1.59 | 0.59 | 1 | 91.74 |
| ic/cp/py/lfp | 48.72 | 24.17 | 24.55 | 67.36 |
| lo | 6.88 | 7.44 | −0.56 | −7.82 |
| ml | 17.57 | 5.29 | 12.28 | 107.44 |
| mlf | 3.19 | 2.4 | 0.79 | 28.26 |
| mt | 1.33 | 0.64 | 0.69 | 70.05 |
| opt/och | 16.61 | 7.99 | 8.62 | 70.08 |
| sm/st | 7.71 | 3.44 | 4.27 | 76.59 |
| Total brain | 2290.12 | 1645.55 | 644.57 | 32.76 |
Assessment of 2D location in the coronal plane
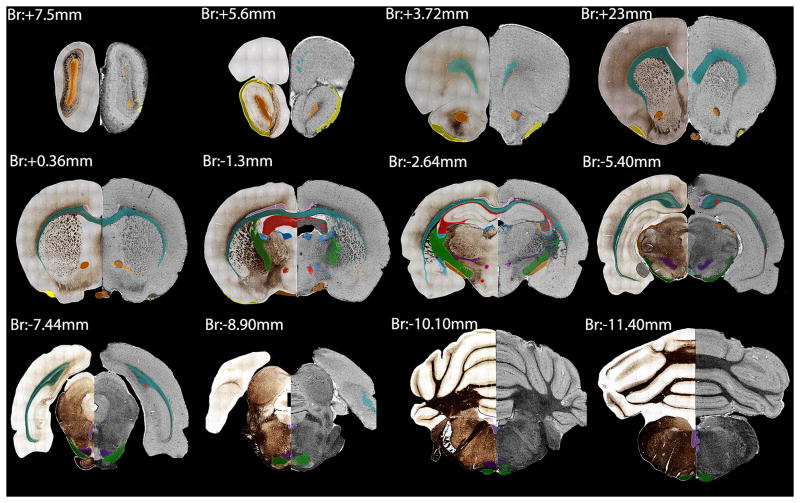
Segmentation results were qualitatively assessed in 2D as color overlays on coronal brain images. We analyzed slices throughout the entire brain starting from the olfactory bulb (anterior: +7.1mm from Bregma) ending at the caudal aspect of the cerebellum (posterior: −15.00mm from Bregma) covering a distance of about 21 mm, or 120 histology sections. Figure 2 displays every 10th section of those that were analyzed in 2D, such that displayed sections are 1600 μm apart from the olfactory bulbs to the cerebellum. The left half of each image provides a Gallyas myelin stain with manual segmentation displayed as a color overlay, while the right half shows MR histology with semi-automated FACT tractography segmentation displayed using the same color scheme. Semiautomated tractography-based segmentation yielded qualitatively similar segmentation results and was significantly faster and easier to perform than histology-based segmentation. In some cases, tractography-based segmentation generated spurious selections that were spatially distant from the expected structure location. In addition, tractography-based segmentation often resulted in incomplete structure selections, in which part or parts of the tract are not included in the final segmentation. Figure 2 (posterior: −1.3mm from Bregma) shows spurious tractography-based segmentation of the anterior commissure (ac) located lateral to the caudate/putamen at the most ventral extent of the corpus callosum along with an incomplete segmentation of the fimbria/fornix/ventral hippocampus (fi/f/vhc).
Fig. 2.
Representative coronal sections of the delineated and segmented structures through the brain. This figure shows 12 out of the 120 delineated sections, approximately 2 mm apart from each other. We matched the silver-stained sections (left hemisphere) with the MR images (right hemisphere) and labeled the segmented structures. The color-coded list of the structures that we labeled and analyzed is presented in Table 2.
Assessment of 3D location and orientation
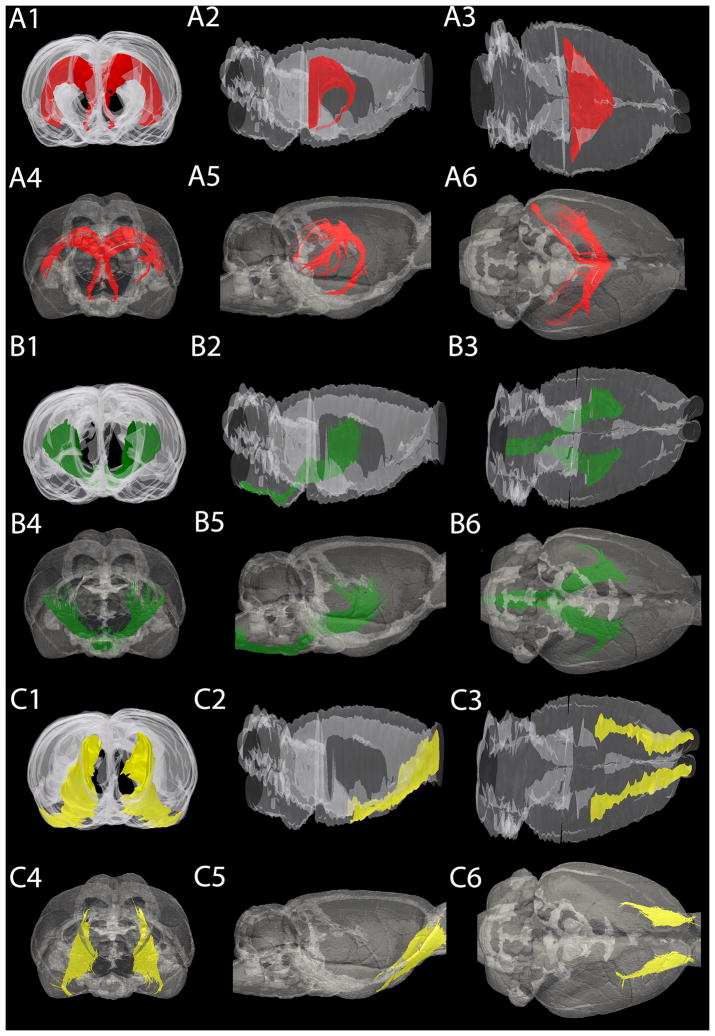
To assess the 3D location and orientation of tractography-based segmentation, we performed a qualitative comparison with 3D reconstructions of corresponding manually segmented tracts from histologic sections. We analyzed frontal, lateral and dorsal views of each tract. Figure 3 shows a three-view comparison between 3D reconstructions from histology sections and FACT tractography for three select tracts. Figure 3A is the opt/och, B is the internal capsule/cerebral peduncle/pyramid/longitudinal fasciculus of the pons (ic/cp/py/lfp) and C is the lateral olfactory tract (lo). Number 1–3 corresponds to anterior, lateral and dorsal views of 3D rendered histology-based segmentation, respectively. Number 4–6 corresponds to similar views from FACT tractography-based segmentations. Structures with good spatial agreement did not necessarily have similar volumes. Our examples show the optic tract (opt/och) (Fig. 3A), which was determined as a poor match in terms of volume between the two techniques. However, the similarity between the two reconstructed 3D structures on both the frontal, lateral and dorsal view is striking (Fig. 3A 1–6). The ic/cp/py/lfp (Fig. 3B) showed excellent correspondence between modalities, both in terms of volume and 3D location/orientation. For long, straight tracts, like the lateral olfactory tract, FACT tractography underestimated volume (by 34.89% in this case); however, visual comparison showed good agreement between modalities (Fig. 3C 1–6). Similar views for all of the white matter structures analyzed in this study are provided as supplementary material.
Fig. 3.
Three-dimensional rendering of the rat brain, showing three different structures. (A) Frontal (A1 A4), lateral (A2, A5) and dorsal (A3, A6) view of the fornix/fimbria of the hippocampus/ventral hippocampal commissure. These structures were found to be significantly different based on their volume calculations however; the location and the orientation of the fibers are in agreement. (B) Frontal (B1, B4), lateral (B2, B5) and dorsal (B3, B6) view of the internal capsule. The internal capsule volumes are quite similar between the reconstructed histology and the MR segmentation, and the position and the expansion of the structure shows sufficient overlap. (C) Frontal (C1, C4), lateral (C2, C5) and dorsal (C3, C6) view of the lateral olfactory tract. The lateral olfactory tract shows high inter-modality similarities in terms of volume and spatial extent. In all cases, the outline of the entire brain is shown in transparency for spatial reference.
Discussion
Alignment to stereotaxic atlas space
In this study we validated semi-automated tractography-based white matter segmentation by comparison with anatomical mapping of Gallyas silver-stained histology sections. MRI and histology data were aligned to a common reference space—the Paxinos and Watson histology atlas—to facilitate these comparisons and guide manual segmentation of histology sections. The Paxinos and Watson atlas does not include silver-stained sections; however it is currently the most complete anatomic reference available for the rat brain. After alignment of the MRI data to the stereotaxic space used in the Paxinos and Watson atlas, a slight deviation was noted. There was a 0.393 mm difference between MR and histologic stereotaxic space; however, this discrepancy is relatively small compared to similar studies, and is likely a result of tissue distortion and shrinkage during sectioning (Johnson et al. 2012b).
Comparisons between MR and histology
We used T2*-weighted images for all direct comparisons between MR and silver-stained histology because of their striking similarities. In general, white matter appears dark on T2*-weighted MRI due to its relatively short T2* relaxation time, which may in turn be a result of the diamagnetic properties of myelin (Renshaw et al. 1986; Koenig 1990). Because of this phenomenon, T2*-weighted MRI looks very similar to myelin silver-stained histology where myelin is heavily positively stained. In contrast, the Nissl and AChE-stained sections show white matter pathways largely based on their lack of staining, as is the case with the fornix, mammillothalamic tract, internal capsule, corpus callosum, fimbria of the hippocampus and optic tract (Fig. 1C, D). On the Nissl-stained section, the outstanding structures, such as the pyramidal layer of the hippocampus (Py) and the ventromedial hypothalamus (VMH), are not readily apparent on T2* MRI. Similarly, in the AChE-stained sections it is fairly easy to identify the caudate putamen (CPu) and the zona incerta (ZI), but these structures are almost invisible on T2* MRI. Areas that are relatively cell dense and myelin poor tend to stain darkly due to high concentrations of ribosomes (Nissl) and/or AChE, neither of which is present in high concentrations in white matter tracts. Figure 1 is intended to emphasize that existing histological atlases based on cellular staining provide only limited guidance and help for identifying white matter structures. For this reason, direct comparison of tractography-based segmentation to the Paxinos and Watson atlas is of limited utility.
Volumetric comparison between tractography-based and histology-based segmentation
White matter structures made up of large coherent fiber bundles, such as the corpus callosum and the internal capsule, showed almost identical volume measurements between modalities when using FACT tractography. These structures appear to be the most amenable to FACT tractography-based segmentation. There are several possible explanations for this observation. First, these white matter structures are relatively large compared to the other structures that we analyzed. Large volume structures are represented by a greater number of voxels on MRI and are less likely to be affected by partial volume effects. Second, these pathways consist largely of heavily myelinated, coherent axon bundles. Such structures result in tensors with high fractional anisotropy and relatively accurate primary eigenvectors, which in turn leads to FACT tractography results that more closely recapitulate the underlying white matter structure. Finally, these structures are centrally located in the brain, and therefore are less prone to susceptibility artifacts and edge effects, which can affect diffusion tensor accuracy (Mukherjee et al. 2008).
Although the observed quantitative and qualitative similarities for these structures is quite high, especially in the case of the corpus callosum where the observed volume difference was null, it is important to realize that this does not indicate that the two modalities resulted in the exact same segmentation. First of all, considerable distortion and brain shrinkage occurs during removal from the cranium and preparation for histology. In this study for example, total brain volume decreased by 33% between MRI and histologic measurements. Although it is certainly possible that the majority of volume loss occurs in cell body dense regions as opposed to cell body poor white matter regions, it is still reasonable to expect some degree of white matter volume loss. Additionally, both the corpus callosum and the anterior commissure yielded quite long tracks when reconstructed with FACT tractography. For example mean tract length of the cc was 11.06 mm and that of the ac was 13.40 mm. The FACT algorithm is known to accumulate a certain degree of error as it propagates, the result of which is that longer tracts are generally less anatomically accurate than shorter ones. While this effect cannot be discounted, it can be minimized by collecting high SNR data and minimizing potential MRI artifacts, as we have done in the study described here. Each of these important caveats likely affects the observed measurements to some degree, so perfect volume agreement between modalities should be regarded as somewhat of a coincidence. Nonetheless, it is clear that several structures, including the corpus callosum, internal capsule, and anterior commissure can be semi-automatically segmented using FACT tractography with a high degree of anatomical accuracy in the adult rat brain.
Some smaller tracts, such as the fasciculus retroflexus (fr) and the mammillothalamic tract (mt), showed a high percentage difference in their volume but a low absolute volume difference using all four types of tract analysis. Both the fr and mt are relatively thin and predominantly dorsoventral in orientation. The large percentage volume differences in these structures are likely a result of a small total tract volume and the difficulty reconstructing volumes of thin dorsoventrally oriented structures on serial coronal histology slices. It is likely that the histology-based segmentation of these structures could be substantially improved by segmentation in the horizontal plane; however, we chose the coronal plane because it is the most commonly used in neuropathology studies in the rat.
Non-FACT tractography
Based on the calculated tract volumes from histology and results from the four different tractography algorithms we have drawn conclusions about the accuracy of the algorithms, and on which structures they can be accurately used. In general, larger structures (>15 mm3) show greater volume variability between algorithms with Tensorline providing the largest estimate. With regards to the morphology of white matter fibers, a distinct pattern was noticed when comparing the volumes computed by tractography algorithms to histology; for white matter tracts with more curvature, FACT provided an estimate closest to histology while Tensorline yielded a volume significantly greater than all others. Integration methods like FACT better follow the angular changes in these tracts by directly following the primary eigenvector. In contrast, Tensorline does not faithfully follow the primary eigenvector but rather treats it as a deflection. As a result, the Tensorline algorithm is more likely to deflect into adjacent tracts outside the structure of interest, resulting in much larger segmentation volumes.
Long, straight tracts show the greatest similarity between deflection methods and histology, while integration methods underestimate these volumes. For these types of white matter structures, integration methods are more susceptible to early termination due to small eigenvector errors from noise or other artefacts. Deflection methods effectively smooth out small eigenvector errors by deflecting towards, rather than exactly following the primary eigenvector. For this reason, deflection methods like Tensorline are less prone to early termination errors in long straight tracts, and therefore yield more accurate volumetric segmentation results.
Based on these results, tractography-based segmentation accuracy depends on the complexity of the structure being segmented. Integration methods like FACT seem to provide more accurate volume estimates for structures with considerable curvature, including the corpus callosum and internal capsule, while deflection methods like Tensorline are more accurate for long straight tracts, such as the lateral olfactory tracts and the cingulum bundle.
3D location and orientation
While the volumes from FACT tractography vary from 2–50% compared to histology, the 3D representations of the white matter tracks are consistently similar. This discrepancy in volume may be due to a variety of factors including stray fibers generated in tractography, which are non-existent in histology reconstructions. Stray fibers are a result of incorrect propagation of tract along an erroneous primary eigenvector. This can result in track propagation into an adjacent white matter structure or other anatomic structure with sufficient fractional anisotropy (i.e. a blood vessel). Error in the primary eigenvector is multifactorial and can be a result of image noise, MR artefacts, adjacent anatomy and/or insufficient angular sampling of the diffusion tensor. In addition, manual placement of ROI-disks on the MRI data can cause accidental inclusion of unwanted voxels that provide means for inaccurate track propagation. In this study we used relatively high SNR data (SNR=60 for b0) and careful ROI-placement to minimize these effects. Qualitative assessment of the orientation and location of the white matter is necessary to ensure that similarities in volume between tractography-based segmentation and histology-based segmentation are not merely by coincidence. Despite the fact that the quantitative analysis showed significant differences in the case of a few structures (e.g. opt/och), qualitatively the two techniques yield very similar results.
Semi-automated tractography using MRH
The 12 structures we have labeled and analyzed in this study represent only a fraction of the entire white matter volume of the rat brain; however not all white matter pathways are readily visible on MRH images. We deliberately chose relatively large and discrete white matter structures that could be easily identified on MRH. Certain structures, such as the caudate putamen (CPu), are relatively easy to identify on MRH, but the multiple fiber bundles running throughout make it difficult to place a central ROI disk for semi-automated segmentation. Other structures, such as the median forebrain bundle (mbf), contain myelinated fibers that are not organized in a single well-defined bundle, but rather fan outward in many different directions making tractography difficult. It is also important to note that most of the analyzed structures are located in the forebrain and relatively few are located in the brainstem. The brainstem is strongly populated with heavily myelinated axons, ascending and descending from and to the spinal cord, which also makes discrete segmentation difficult.
It is often possible to identify white matter pathways in existing stereology atlases using surrounding cellular landmarks; however, the Gallyas silver staining clearly allows more accurate white matter delineation. To date there is only one major rat brain atlas which was based on myelin staining—that of Konig and Klippel (1963). This atlas has some major shortcomings (such as the use an unusual stereotaxic system, non-standard choice of animal size, and no coverage of the hindbrain), but it still provides a very detailed guide to manual white matter segmentation. MRH-based atlases are evolving fast and together with the conventional histology atlases they provide even more detail and information to the users. Semi-automated tractography-based segmentation will almost certainly play a role in future MRH-based brain atlases and preclinical studies, and we hope that the results of this study will help guide researchers when this technique is appropriate, and perhaps more importantly, when it is problematic. As these MRH and conventional histology continue to co-evolve, we hope and anticipate that researchers will continue to explore the strengths and weaknesses of each, just as we have attempted to do with the present study.
Supplementary Material
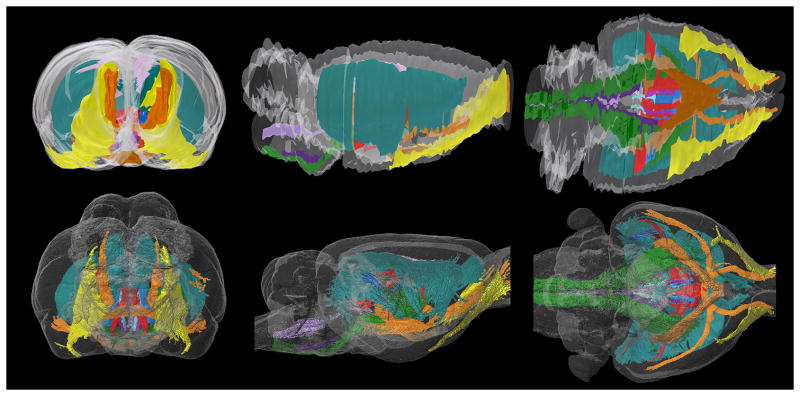
Fig. 4.
Three-dimensional rendering of all structures analyzed in this report. Although we only analyzed a small fraction of the entire white-matter volume of the brain, we captured many of the largest and most prominent tracts in the forebrain. This figure illustrates the complex intertwined structure of the major white matter tracts in the forebrain.
Acknowledgments
This work was supported by an Australia Fellowship awarded to Professor George Paxinos by the National Health and Medical Research Council (NHMRC) (466028) and the Duke Center for In Vivo Microscopy, an NIH/NCRR/NIBIB national Biomedical Technology Resource Center (P41 EB015897).
References
- Hasan KM, Halphen C, Sankar A, Eluvathingal TJ, Kramer L, Stuebing KK, Ewing-Cobbs L, Fletcher JM. Diffusion tensor imaging-based tissue segmentation: validation and application to the developing child and adolescent brain. Neuroimage. 2007;34(4):1497–1505. doi: 10.1016/j.neuroimage.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Calabrese E, Badea A, Paxinos G, Watson C. A multidimensional magnetic resonance histology atlas of the Wistar rat brain. Neuroimage. 2012a;62(3):1848–1856. doi: 10.1016/j.neuroimage.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Calabrese E, Badea A, Paxinos G, Watson C. A multidimensional magnetic resonance histology atlas of the Wistar rat brain. Neuroimage. 2012b doi: 10.1016/j.neuroimage.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig SH. Paramagnetic agents as tracers in magnetic resonance imaging. Extrapolations from Gd-DTPA to everything. Acta Radiol Suppl. 1990;374:17–23. [PubMed] [Google Scholar]
- Konig JFR, Klippel RA. The Rat Brain A Stereotaxic Atlas of the Forebrain and the Lower Parts of the Brain Stem. The Williams and Wilkins Company; Baltimore: 1963. [Google Scholar]
- Mukherjee P, Chung SW, Berman JI, Hess CP, Henry RG. Diffusion tensor MR imaging and fiber tractography: technical considerations. AJNR Am J Neuroradiol. 2008;29(5):843–852. doi: 10.3174/ajnr.A1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niogi SN, Mukherjee P, McCandliss BD. Diffusion tensor imaging segmentation of white matter structures using a Reproducible Objective Quantification Scheme (ROQS) Neuroimage. 2007;35(1):166–174. doi: 10.1016/j.neuroimage.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin CF. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Trans Med Imaging. 2007;26(11):1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 6. Elsevier Inc; 2007. [DOI] [PubMed] [Google Scholar]
- Pistorio AL, Hendry SH, Wang X. A modified technique for high-resolution staining of myelin. J Neurosci Methods. 2006;153(1):135–146. doi: 10.1016/j.jneumeth.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Renshaw PF, Owen CS, McLaughlin AC, Frey TG, Leigh JS., Jr Ferromagnetic contrast agents: a new approach. Magn Reson Med. 1986;3 (2):217–225. doi: 10.1002/mrm.1910030205. [DOI] [PubMed] [Google Scholar]
- Savadjiev P, Campbell JS, Pike GB, Siddiqi K. Streamline flows for white matter fibre pathway segmentation in diffusion MRI. Med Image Comput Comput Assist Interv. 2008;11 (Pt 1):135–143. doi: 10.1007/978-3-540-85988-8_17. [DOI] [PubMed] [Google Scholar]
- Tago H, Kimura H, Maeda T. Visualization of detailed acetylcholinesterase fiber and neuron staining in rat brain by a sensitive histochemical procedure. J Histochem Cytochem. 1986;34 (11):1431–1438. doi: 10.1177/34.11.2430009. [DOI] [PubMed] [Google Scholar]
- Tuch DS. Q-ball imaging. Magn Reson Med. 2004;52(6):1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48(4):577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- Van Hecke W, Leemans A, Sijbers J, Vandervliet E, Van Goethem J, Parizel PM. A tracking-based diffusion tensor imaging segmentation method for the detection of diffusion-related changes of the cervical spinal cord with aging. J Magn Reson Imaging. 2008;27(5):978–991. doi: 10.1002/jmri.21338. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D’Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41(4):1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Yendiki A, Panneck P, Srinivasan P, Stevens A, Zollei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, Jbabdi S, Gollub R, Fischl B. Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front Neuroinform. 2011;5:23. doi: 10.3389/fninf.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.