Abstract
Background
Genetic variants at the CLU, CR1 and PICALM loci associate with risk for late-onset Alzheimer’s disease (LOAD) in genome-wide association studies (GWAS). In this study, our aim was to determine whether the LOAD risk variants at these three loci influence memory endophenotypes in African-American and Caucasian subjects.
Methods
We pursued an association study between single nucleotide polymorphism (SNP) genotypes at the CLU, CR1 and PICALM loci and memory endophenotypes. We assessed African-American subjects (AA: 44 with LOAD, 224 controls) recruited at Mayo Clinic Florida and Caucasians recruited at Mayo Clinic Minnesota (RS: 372 with LOAD, 1,690 controls) and Florida (JS: 60 with LOAD, 529 controls). SNPs at the LOAD risk loci CLU (rs11136000), CR1 (rs6656401, rs3818361) and PICALM (rs3851179) were genotyped and tested for association with Logical Memory immediate recall (LMIR), delayed recall (LMDR) and percent retention (LMPR) and Visual Reproduction (VRIR, VRDR, VRPR) scores from Wechsler Memory Scale-Revised, using multivariable linear regression analysis, adjusting for age-at-exam, sex, education and APOE ε4 dosage.
Results
We identified nominally significant or suggestive associations between the LOAD risky CR1 variants and worse LMIR scores in the African-Americans (p=0.068 - 0.046, β= −2.7 to −1.2). The LOAD protective CLU variant is associated with better logical memory endophenotypes in the Caucasian subjects (p=0.099-0.027, β= 0.31 to 0.93). The CR1 associations persisted when the control subjects from the African-American series were assessed separately. The CLU associations appeared to be driven by one of the Caucasian series (RS) and were also observed when the control subset from RS was analyzed.
Conclusion
These results suggest for the first time that LOAD risk variants at CR1 may influence memory endophenotypes in African-Americans. Additionally, CLU LOAD protective variant may confer enhanced memory in Caucasians. Although these results would not remain significant after stringent corrections for multiple testing, they need to be considered in the context of the LOAD associations, with which they have biological consistency. They also provide estimates for effect sizes on memory endophenotypes that could guide future studies. The detection of memory effects for these variants in clinically normal subjects, implies that these LOAD risk loci might modify memory prior to clinical diagnosis of AD.
BACKGROUND
Recent genome wide association studies of late-onset Alzheimer’s disease (LOAD) case-control series identified single nucleotide polymorphisms (SNPs) at nine loci which associate with disease risk1–5. Understanding the mechanism of action of these variants or functional variants tagged by these SNPs and determining their contribution to LOAD is an important next step in translating these findings to potential drug targets and predictive biomarkers for this disease. Using endophenotypes as outcomes may be a powerful approach in testing the effects of disease risk variants on quantitative, biologically relevant outcomes6–9, which may provide valuable information regarding downstream biological consequence of such genetic variation. Further, genetic variants may display stronger association with the intermediate endophenotypes than the disease phenotype, and may be useful in detecting novel disease risk loci. Finally, if disease risk variants influence endophenotypes prior to onset of clinical disease, then these quantitative variables may be used as biomarkers in predicting disease course and onset.
Cognitive measures are proposed as highly relevant endophenotypes for neuropsychiatric conditions6, 8, including AD10. A conceptual model of AD pathogenesis posits that dynamic changes in cognition occur prior to clinical diagnosis of AD11. If correct, then risk factors, including genetic variants that influence AD risk should also associate with cognitive endophenotypes and these associations should be detected prior to the diagnosis of dementia. Likewise, genetic factors that influence cognitive endophenotypes should harbor variants that confer AD risk. This model, if accurate, will enable confirmation of candidate AD risk variants for their role in cognition, characterization of their mechanism of action for specific cognitive abilities and may lead to identification of novel genetic risk factors. Indeed, there is ample proof of principle for this model from studies investigating the influence of APOE, the strongest known LOAD genetic risk factor, on cognitive endophenotypes, which identified a stronger effect on cognition than disease risk10 and one that could be detected prior to development of clinical AD12, 13.
With the identification of additional risk variants from the large LOAD GWAS1–5, similar studies are beginning to emerge for these novel genetic factors. Chibnik et al. investigated SNPs at the CLU, CR1 and PICALM loci and identified significant associations between the LOAD risk allele of CR1 rs6656401 SNP and faster global cognitive decline, as well as increased amyloid pathology, in a study of two longitudinal, Caucasian cohorts14. This group subsequently identified a coding CR1 variant, rs4844609, in tight linkage disequilibrium (LD) with the LOAD GWAS SNP (rs6656401) and which associated with faster episodic memory decline, increased AD neuropathology and risk15. Barral et al. assessed 2-SNP genotypes at BIN1, CLU, CR1 and PICALM loci with or without APOE for their influence on episodic memory in a Caucasian, family-based sample and identified several genotype patterns with significant association, some of which were also significant in their unaffected subset of subjects16. In that study, the combination of the LOAD risky PICALM genotypes with either the LOAD protective CLU, CR1 or BIN1 genotypes associated with worse episodic memory estimates, rendering biologically-consistent interpretations difficult. Hamilton et al. assessed two Scottish cohorts without dementia for 158 SNPs from 11 genes, including BIN1, CLU, CR1 and PICALM, and detected an interaction between a BIN1 and an APP SNP which influenced Logical Memory scores in the APOE ε4 positive subset of one of their cohorts17. Finally, Verhaaren et al. constructed a risk score using genotypes of the nine LOAD GWAS variants and assessed their effects on baseline cognitive endophenotypes in a non-demented population-based cohort from the Netherlands18. They found only a marginal influence of the joint effect of the LOAD GWAS variants on memory, above APOE.
In our study we aimed to assess the influence of the LOAD GWAS variants at the CLU, CR1 and PICALM loci on episodic memory in one African-American and two Caucasian series from Mayo Clinic. These variants were the first to be identified from the LOAD GWAS1, 2 and were therefore the focus of our study. Given that to date all of the studies of cognitive endophenotypes and LOAD GWAS variants are focused on Caucasian subjects, one of our goals was to investigate these effects in a non-Caucasian series. Second, we sought to evaluate six related cognitive scores from two types of episodic memory domains as separate endophenotypes rather than a single combined score. Third, we aimed to replicate any of the previously reported cognitive endophenotype associations or identify new ones in two Caucasian series, for these three LOAD GWAS loci. Our findings support a role for CR1 and CLU loci variants in influencing episodic memory in African-Americans and Caucasians, respectively.
METHODS
Subjects
Study participants were selected from three established Mayo Clinic Alzheimer’s disease case-control series. Subjects included Caucasian LOAD patients with an age of diagnosis greater than 60 years and elderly controls older than 60 at the time of testing, as well as African-American LOAD patients who were slightly younger at age of onset, compared to Caucasians (mean=78.9, range=52.2–91.2) and elderly controls (mean=78.7, range = 60.5–96.4) (Table 1). All subjects were diagnosed by a Mayo Clinic neurologist and underwent neuropsychological testing. The African-American case-control series (AA) was collected at Mayo Clinic Florida in Jacksonville, where the cases were participants in the Mayo Clinic Alzheimer’s Disease Research Center and the controls were cognitively normal volunteers recruited at local churches and community centers. Two case control series recruited at Mayo Clinic Minnesota in Rochester (RS) and Mayo Clinic Florida in Jacksonville (JS) were composed of North American Caucasian adults. The RS cases and controls were participants in either the Mayo Clinic Alzheimer’s Disease Research Center or the population-based Mayo Clinic Study of Aging series19. The JS cases were recruited either as part of the Mayo Clinic Alzheimer’s Disease Research Center or were clinically diagnosed AD subjects from Mayo Clinic Florida, Department of Neurology. The JS controls were volunteers recruited from retirement communities around Jacksonville, Florida. All controls had a clinical dementia rating (CDR) score of 0 at the most recent time of testing and all LOAD cases had a diagnosis of probable or possible AD according to NINCDS-ADRDA criteria20. Additional series details are provided in Table 1. This study was approved by the Mayo Clinic institutional review board and informed consent was obtained from all participants.
Table 1.
Demographic characteristics of the series. Number of subjects (N); number and percentage of male participants; number and percentage of subjects with 0 (ApoE X/X), 1 (ApoE X/4) or 2 (ApoE 4/4) copies of the APOE ε4 allele, mean age at last Logical Memory test (standard deviation =SD) and mean number of years of education are shown. More subjects were tested for Logical Memory than Visual Reproduction, therefore the mean age at former test was used. Two-sided unpaired t-test assuming unequal variances was used to test significance of difference between at test or education years for each group against the largest RS control group. Differences in gender and APOE 4 dosage between the RS control and other groups was determined by chi-squared test.
| Series (Diagnosis) | N | Male (%) | p (Male) | ApoE X/X (%) | ApoE X/4 (%) | ApoE 4/4 (%) | p (ApoE) | Mean Age at Test (SD, Range) | p (Mean Age) | Mean Yrs Education (SD, Range) | p (Education yrs) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| African- Americans (AD) | 44 | 11 (25%) | 0.0043 | 11 (25%) | 21 (48%) | 12 (27%) | <0.0001 | 78.9 (7.8, 52.2–91.2) | 0.0046 | 12.8 (2.6, 4–20) | 0.074 |
| African-Americans (CON) | 224 | 51 (23%) | <0.0001 | 153 (68%) | 65 (29%) | 6 (3%) | 0.014 | 78.7 (7.0, 60.5–96.4) | <0.0001 | 12.6 (3.4, 2–20) | <0.0001 |
| RS (AD) | 372 | 145 (39%) | 0.0021 | 183 (49%) | 163 (44%) | 26 (7%) | <0.0001 | 83.2 (6.9, 61.6–101.1) | 0.0439 | 13.3 (3.0, 7–20) | 0.0019 |
| RS (Controls) | 1690 | 810 (48%) | NA | 1295 (77%) | 373 (22%) | 22 (1%) | NA | 82.4 (5.5, 65.9–100.2) | NA | 13.8 (2.9, 5–20) | NA |
| JS (AD) | 60 | 27 (45%) | 0.7529 | 15 (25%) | 38 (63%) | 7 (12%) | <0.0001 | 79.7 (6.1, 62.2–94.9) | 0.0011 | 14.5 (3.0, 8–20) | 0.0861 |
| JS (Control) | 529 | 178 (34%) | <0.0001 | 377 (71%) | 138 (26%) | 14 (3%) | 0.012 | 78.9 (6.5, 61.6–95.9) | <0.0001 | 15.1 (2.6, 4–20) | <0.0001 |
Memory Endophenotypes
Verbal and nonverbal episodic memory were evaluated using phenotypes from the Logical Memory (LM) and Visual Reproduction (VR) subtests from the Wechsler Memory Scale-Revised21. Specifically, raw scores from the immediate recall (LMIR, VRIR) and 30-minute delayed recall trials (LMDR, VRDR) were evaluated for association with each single nucleotide polymorphism (SNP). We also evaluated the percent of verbal or nonverbal material retained over the 30-minute interval (LMPR, VRPR). Logical Memory and Visual Reproduction were administered using standardized instructions. All SNPs were assessed for association with these memory endophenotypes measured at each subject’s most recent (last or proximal) visit. The descriptive statistics for these memory endophenotypes are depicted in Supplementary Table 1.
Genotyping
Genotypes for the four SNPs, rs11136000 (CLU), rs3818361 (CR1), rs6656401 (CR1) and rs3851179 (PICALM) were obtained by one of two approaches. Subjects from all series were genotyped for rs6656401 using Taqman® technology. A subset of the subjects from the RS (198 LOADs and 667 controls) and JS (33 LOADs and 230 controls) series were participants in the Mayo Clinic LOAD GWAS22. These subjects had genotypes for the rs11136000, rs3818361 and rs3851179 SNPs that were extracted from the GWAS genotypes. The remaining subjects from the RS (352 LOADs and 1,797 controls) and JS (33 LOADs and 427 controls), as well as all African-American subjects were genotyped for these three SNPs using Taqman® assays. The descriptive statistics for the genotyped SNPs are shown in Supplementary Table 2.
Statistical analyses
Each of the four SNPs were tested for association with the six memory endophenotypes obtained at the most recent visit for each subject, using multivariate linear regression analysis implemented in PLINK23. An additive model was employed for each SNP, where the dosage effect of each minor allele was evaluated, while controlling for the covariates, which were sex, age-at-examination, years of education, and number of APOE ε4 alleles for all analyses. When both LOADs and controls (All) were analyzed together an additional term for diagnosis was also included (LOAD = 1, control = 0). When both Caucasian series were analyzed together a further term was included in the model for series (JS=1, RS=0). In all analyses with the African-American series, we also included a reading score, the Reading subtest from the Wide-Range Achievement Test-324, as a proxy for quality of education25, 26.
Two sided, unpaired t-tests assuming unequal variances were conducted to compare age at test, years of education (Table 1) and cognitive scores (Supplementary Table 1) of the largest RS control series with each of the other series, using StatsDirect v2.7.8. Chi-squared test was used to compare APOE4 dose and gender between the RS controls and each of the other series (Table 1).
Power calculations were done for the Caucasian and African-American series separately, for sample sizes of 2,500 and 250, respectively, as approximations of the largest sample sizes for each ethnic group, and for 2,000 and 500, reflecting sample sizes for the Caucasian RS and JS series, respectively. Minor allele frequencies were chosen to reflect those of the four tested SNPs for each ethnic group and standard effect sizes for a range of 0.05–1 were utilized. Standardized effect sizes refer to the average increase (or decrease) in memory score with an increase in one copy of the minor allele divided by the standard deviation of the memory score. Power estimates were based on simulations with linear regression and additive effect for minor allele using α<0.05. Given the six memory endophenotypes tested with four SNPs, an uncorrected p value of 0.002 is required to achieve significance (24 tests, p required = 0.05/24 = 0.002), assuming completely independent tests. Since the memory endophenotypes are expected to correlate with each other; the two CR1 SNPs are in LD and given our study needs to be considered in the context of the prior LOAD GWAS findings, we focused on an α<0.05 for our power calculations and results.
Box plots for the cognitive scores vs. SNP genotypes, which were generated using the functionalities in R. Box plots, represent the residuals after accounting for the effects of covariates on the cognitive scores.
RESULTS
We had 268 African-American (44 LOAD, 224 controls) and a total of 2,651 Caucasian (432 LOAD, 2,219 controls) subjects with episodic memory endophenotypes (Table 1, Supplementary Table 1). Based on our power estimates, assuming a sample size of 250 African-Americans, minor allele frequencies (MAF) of 0.01, 0.1 or 0.4 (Supplementary Table 2), we expect to have 10%, 51% and 90% power, respectively, to detect a standardized effect size of 0.3 at α=0.05. For 2,500 or 2,000 Caucasians and MAF of 0.2 or 0.4, we can expect >99% power to detect a standardized effect size >0.2 (Supplementary Table 3). Under the same assumptions, a sample size of 500 will yield 72% or 87% power respectively for MAF of 0.2 or 0.4.
The APOE ε4 positive genotype frequencies were higher in the LOAD subjects in all three series (AA, RS, JS), as expected (Table 1). The genotype frequencies for APOE fall within the expected range for the different series27. The mean education was lowest in the AA series, followed by RS and JS series. Comparison of education years for each group, compared to the largest group, RS control subjects, revealed marginal to significant differences for lower education in the AA series and RS LOAD subjects and higher education for JS series. The years of education was also slightly lower in the LOAD subjects vs. controls from within the two Caucasian series, but not the African-American series. There were fewer male than female participants across all series and diagnoses, with lowest male frequencies in the AA controls (23%) and LOADs (25%) and highest in the RS controls (48%). Compared to the RS controls, RS LOAD subjects were slightly older and all other groups were somewhat younger.
Compared to the controls, the LOAD subjects had lower (worse) scores for all memory endophenotypes as expected (Supplementary Table 1). When the largest RS controls were used as the comparison group, the African-American controls had lower and JS controls had higher cognitive scores, in the same order as years of education.
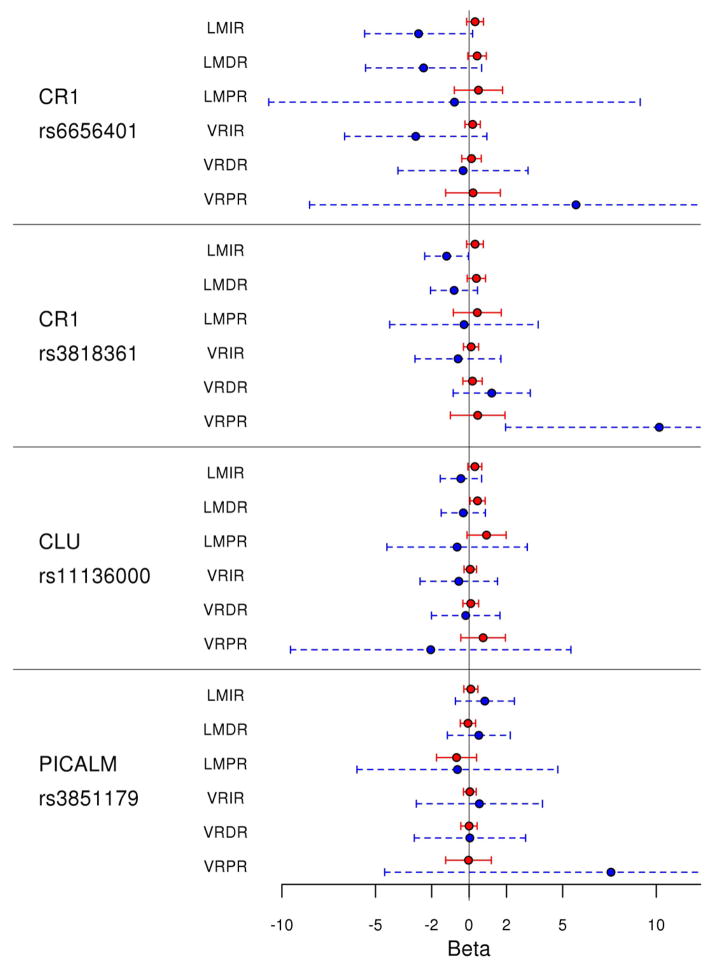
Our main analyses were confined to the largest possible African-American (i.e. LOAD cases + controls) and Caucasian (i.e. RS and JS combined with LOAD cases + controls) series. Table 1 depicts the results of these main analyses and Figure 1 contains the forest plots for the effect sizes of all SNPs tested for each memory endophenotype. In the analysis of the AA series, minor alleles of the CR1 locus SNPs rs6656401 and rs3818361 were both associated with worse LMIR scores (β= −2.7, p=0.068 and β= −1.2, p=0.046), respectively (Table 2, Supplementary Figure 1). The associations with the LMDR scores also showed similar trends for these two SNPs in the AA series (β= −0.80 to −2.43), however these results did not achieve significance (p=0.125–0.211). Similarly, although the LMPR scores for both SNPs had lower score estimates, these findings were not significant. As a secondary analysis, we also assessed the subset of AA subjects who were clinically non-demented, separately and determined that LMIR scores were also lower in the carriers of the CR1 risk allele who are controls (Supplementary Table 4, Supplementary Figure 2).
Figure 1. Forest plot of effect size for CLU, CR1 and PICALM loci variants and memory endophenotypes.
The circles represent the β coefficient of variation from the multivariable linear regression analysis results shown in Table 2. The lines represent the 95% confidence intervals (CI). The blue figures denote the effects for the African-Americans and the red figures are for the Caucasians. Some of the confidence intervals in the African-American group are truncated due to large CI.
Table 2. Association between CLU, CR1 and PICALM loci variants and memory endophenotypes in an African-American and Caucasian series.
Six episodic memory endophenotypes [Logical Memory immediate recall (LMIR), delayed recall (LMDR), and percent retention (LMPR), and Visual Reproduction immediate recall (VRIR), delayed recall (VRDR) and percent retention (VRPR) from the Wechsler Memory Scale-Revised] were used to assess association with four SNPs at three LOAD GWAS loci, using multivariable linear regression analysis assuming an additive model, and correcting for age-at-testing, sex, APOE ε4 dosage, education years and reading score. The number of subjects for each test (N), β coefficients of variation for each copy of the minor allele, standard error (SE) and 95% confidence interval of this effect (95%CI) and p values of association are depicted. Tests were done for the combined non-demented control and LOAD subjects, and include a covariate for LOAD. Caucasian series from Mayo Clinic Minnesota in Rochester (RS) and Mayo Clinic Florida in Jacksonville (JS) were analyzed. P values <0.1 are shown in bold and those <0.05 are shown in bold, red. The minor alleles tested for cognitive score associations and their known effects on LOAD risk are also shown.
| Trait | African American All Subjects | RS-JS-ALL | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Beta (SE) | 95% CI | P-value | N | Beta (SE) | 95% CI | P-value | ||
| CR1 rs6656401 Tested Allele=A (risky) | LMIR | 261 | −2.7 (1.47) | −5.58 to 0.19 | 0.068 | 2,587 | 0.32 (0.23) | −0.13 – 0.77 | 0.168 |
| LMDR | 260 | −2.43 (1.58) | −5.53 to 0.67 | 0.125 | 2,583 | 0.43 (0.25) | −0.06 – 0.92 | 0.088 | |
| LMPR | 260 | −0.78 (5.06) | −10.7 to 9.14 | 0.878 | 2,583 | 0.5 (0.66) | −0.79 – 1.79 | 0.448 | |
| VRIR | 92 | −2.85 (1.94) | −6.65 to 0.95 | 0.145 | 2,518 | 0.19 (0.21) | −0.22 – 0.6 | 0.358 | |
| VRDR | 92 | −0.32 (1.77) | −3.80 to 3.15 | 0.855 | 2,516 | 0.13 (0.27) | −0.39 – 0.65 | 0.628 | |
| VRPR | 92 | 5.71 (7.26) | −8.52 to 19.93 | 0.434 | 2,516 | 0.21 (0.75) | −1.25 – 1.67 | 0.777 | |
| CR1 rs3818361 Tested Allele=A (risky) | LMIR | 261 | −1.2 (0.60) | −2.37 to −0.03 | 0.046 | 2,568 | 0.32 (0.23) | −0.13 – 0.76 | 0.167 |
| LMDR | 260 | −0.80 (0.64) | −2.06 to 0.45 | 0.211 | 2,564 | 0.39 (0.25) | −0.1 – 0.88 | 0.119 | |
| LMPR | 260 | −0.27 (2.03) | −4.24 to 3.70 | 0.892 | 2,564 | 0.44 (0.65) | −0.84 – 1.72 | 0.502 | |
| VRIR | 88 | −0.59 (1.17) | −2.89 to 1.70 | 0.613 | 2,502 | 0.11 (0.21) | −0.3 – 0.51 | 0.601 | |
| VRDR | 88 | 1.21 (1.05) | −0.85 to 3.28 | 0.252 | 2,500 | 0.18 (0.26) | −0.33 – 0.7 | 0.487 | |
| VRPR | 88 | 10.16 (4.19) | 1.95 to 18.36 | 0.018 | 2,500 | 0.46 (0.74) | −1 – 1.92 | 0.534 | |
| CLU rs11136000 Tested Allele=C (risky) | LMIR | 267 | −0.44 (0.56) | −1.54 to 0.67 | 0.439 | 2,556 | 0.31 (0.19) | −0.06 – 0.68 | 0.099 |
| LMDR | 266 | −0.31 (0.60) | −1.49 to 0.87 | 0.608 | 2,552 | 0.45 (0.21) | 0.05 – 0.86 | 0.027 | |
| LMPR | 266 | −0.64 (1.92) | −4.39 to 3.11 | 0.738 | 2,552 | 0.93 (0.53) | −0.11 – 1.98 | 0.081 | |
| VRIR | 94 | −0.55 (1.06) | −2.62 to 1.52 | 0.603 | 2,490 | 0.06 (0.17) | −0.27 – 0.4 | 0.701 | |
| VRDR | 94 | −0.18 (0.93) | −2.01 to 1.65 | 0.847 | 2,488 | 0.09 (0.22) | −0.33 – 0.51 | 0.681 | |
| VRPR | 94 | −2.05 (3.82) | −9.54 to 5.44 | 0.593 | 2,488 | 0.75 (0.61) | −0.44 – 1.94 | 0.215 | |
| PICALM rs3851179 Tested Allele=A (protective) | LMIR | 267 | 0.84 (0.80) | −0.73 to 2.42 | 0.297 | 2,551 | 0.09 (0.19) | −0.28 – 0.47 | 0.626 |
| LMDR | 266 | 0.52 (0.86) | −1.16 to 2.20 | 0.547 | 2,547 | −0.06 (0.21) | −0.47 – 0.35 | 0.767 | |
| LMPR | 266 | −0.62 (2.74) | −5.99 to 4.74 | 0.820 | 2,547 | −0.67 (0.55) | −1.74 – 0.4 | 0.223 | |
| VRIR | 94 | 0.55 (1.72) | −2.82 to 3.92 | 0.749 | 2,485 | 0.04 (0.17) | −0.3 – 0.38 | 0.824 | |
| VRDR | 94 | 0.04 (1.52) | −2.93 to 3.02 | 0.978 | 2,483 | 0 (0.22) | −0.44 – 0.43 | 0.989 | |
| VRPR | 94 | 7.59 (6.17) | −4.51 to 19.68 | 0.222 | 2,483 | −0.03 (0.62) | −1.25 – 1.19 | 0.966 | |
Only about a third of the AA subjects who obtained the Logical Memory test also completed Visual Reproduction. Therefore, although the results from the latter are also shown, it is not possible to do a direct comparison for the two groups of memory endophenotypes in this series (Table 2). For the same reason, although there is a significant association with better VRPR scores for the CR1 rs3818361 SNP in all AA subjects (Table 2) and a marginal trend in their control subset (Supplementary Table 4), given the small sample sizes for these tests (n=57–88) and the absence of consistent results from other memory endophenotypes or the CR1 rs6656401 SNP, the validity of these VRPR endophenotype findings are questionable.
When the four SNPs for the three LOAD GWAS loci were assessed for association with memory endophenotypes in the Caucasian subjects, the LOAD-protective CLU rs11136000 minor allele showed association with higher (better) scores for the LMDR test (β=0.45, p=0.027) and similar trends for the immediate recall (β=0.31, p=0.099) and percent retention (β=0.93, p=0.081) endophenotypes (Table 2, Supplementary Figure 3). As a secondary analysis we assessed each Caucasian series (Supplementary Table 5) and control subjects (Supplementary Table 6), separately. In the RS series (LOAD + control), the LMIR and LMDR scores were significantly higher in the CLU rs11136000 LOAD-protective minor allele carriers, with suggestive trends for LMPR (Supplementary Table 5, Supplementary Figure 3), though no suggestive or significant results were observed for the JS series. The same trends were also observed for the control subset from the RS series with significant LMDR and suggestive LMIR and LMPR associations with CLU rs11136000 (Supplementary Table 6, Supplementary Figure 3). Again, no memory endophenotype associations were observed for the JS series.
The forest plots of the effect sizes for each SNP and memory endophenotype is depicted for the Caucasians (red) and African-Americans (blue) in Figure 1. The suggestive or significant CR1 SNPs in the African-American series, demonstrate trends in the opposite direction for the Caucasians. Similarly, CLU rs11136000 SNP with memory-enhancing associations in the Caucasians have non-significant trends in the opposite direction for the African-Americans. These results could be due to heterogeneity between these cohorts due to different functional variants, environmental effects, gene-gene, gene-environment interactions, inaccurate effect sizes due to low power or a combination of these factors. These results highlight the importance of assessing different ethnic groups, as there may be common as well as different disease variants that influence such populations.
DISCUSSION
In this study we assessed associations between four SNPs from three loci previously identified by the first large LOAD GWAS1, 2 and episodic memory endophenotypes. The underlying hypothesis is that genetic variants that confer risk of LOAD, which is typically characterized by memory dysfunction, will also influence cognitive endophenotypes. We tested six memory endophenotypes and four SNPs. If these tests were completely independent, our results would not hold up to multiple testing (24 tests, p required = 0.05/24 = 0.002). The memory endophenotypes, however are expected to correlate with each other. Furthermore, CR1 6656401 and rs3818361 are in LD, but were tested individually to allow for comparison with the available LOAD association studies which use one or the other. Given these aspects and the fact that our study needs to be considered in the context of the prior evidence for these three loci in LOAD risk, we focused on those associations that have nominal significance at p≤0.05. Although we did not find significant associations besides CR1 in the AA and CLU in the Caucasian series, this could be due to low power, testing of markers and not the functional variant, series-specific differences (locus or allelic heterogeneity) and/or uncaptured environmental or gene-gene interaction effects. The AA series composed of at most 260 subjects have 20–60% power to detect the CR1 cognition association (Supplementary Table 3). Our Caucasian series of ~2,500 subjects have 98% power to detect the CLU cognition association,. Nonetheless, given the “winner’s curse” phenomenon28, larger sample sizes are likely required to detect these effects in future studies. The results from our studies can be utilized to guide future, larger studies to test the memory effects for CR1 and CLU suggested by our paper.
An important novel aspect of our study is the simultaneous investigation of this hypothesis in both Caucasian and African-American subjects. To our knowledge, this is the first study that shows associations between LOAD risk variants at the CR1 locus and memory endophenotypes in African-American subjects. The risky allele of both CR1 rs6656401 and rs3818361 variants were associated with lower (worse) memory scores, especially for immediate recall phenotypes. Although verbal delayed recall and percent retention phenotypes did not attain statistical significance, their direction of association with the CR1 variants is consistent with the immediate recall results. The association was stronger in the combined subjects, but was also significant or suggestive even when non-demented subjects were assessed separately. These findings are consistent with the biological expectation from a risk variant as well as a prior report, which identified faster global cognitive decline with the risky allele of the CR1 rs6656401 SNP in Caucasian cohorts14. Our results suggest that the CR1 locus variants may influence memory endophenotypes in a way that can be detected prior to clinical diagnosis of dementia, consistent with the prior report on CR114 as well as studies on the influence of APOE on cognition in non-demented series12, 13. Given the small size of our African-American cohort, these findings require replication. Nevertheless, they suggest that the cognitive effect of the CR1 locus previously obtained in Caucasian subjects may generalize to other ethnic groups. Though we did not identify significant associations with CLU or PICALM variants, this could be secondary to our small sample size in the African-American series and should be re-investigated in larger cohorts.
We also evaluated two Caucasian series, collectively composed of ~2,500 subjects. We identified higher (better) episodic memory scores for the CLU rs11136000 LOAD protective variant in the RS series. This association is biologically congruent and adds to the various lines of evidence for a protective role of CLU in AD. Functional evidence suggests multiple potentially protective roles in LOAD for clusterin, encoded by CLU, including Aβ clearance, mitigation of excess inflammation and apoptosis, and clearing of neuronal debris29. In an expression GWAS of brain transcript levels, our group recently determined that the CLU rs11136000 protective allele associates with higher brain CLU levels30, a finding also corroborated by others31. Thus, an emerging hypothesis is that this SNP, or more likely a functional variant(s) that is in linkage disequilibrium (LD) with it increases brain CLU levels, which confers a protective effect on cognition that can be detected prior to onset of clinical dementia, and ultimately decreases LOAD risk. It will be important to replicate the effect of this SNP on cognition. We observed this effect in the RS, but not the JS series. A potential reason for this discrepancy may be that the JS series is ~0.3 times the size of the RS series. Other differences are that the RS series is older, has lower education and worse memory scores compared to the JS series. If the cognitive effects of this variant become more pronounced with aging and in the context of unfavorable environmental effects, such as lower education, then it could have been easier to capture in the RS than JS series. We also note the differences between the recruitment strategies for the subjects in the Caucasian RS and JS series and the African-Americans, which could have contributed to the heterogeneity in the findings from these series.
For both the CR1 results in the AA and CLU results in the Caucasian series, the episodic memory associations were stronger in the LOAD cases and controls combined group than controls-only analyses. This improvement cannot be solely attributed to increased sample sizes, as LOAD subjects constitute less than a quarter of the controls. Likewise, this is unlikely to be an effect of the LOAD diagnosis itself, as we have controlled for this in our model. If we postulate that memory endophenotypes represent a continuum, with LOAD subjects representing the lower end of the spectrum, and that genetic variants influence cognition throughout this spectrum, then inclusion of LOAD subjects will increase the variance of these quantitative endophenotypes and the relative contribution of the genetic variants to this variance, thereby increasing the power to detect the association in the combined group of LOAD and control subjects. Indeed, studies by others on APOE10 and CR114 showed stronger association with combined LOAD and control series compared to controls alone. The fact that an association can be detected for controls only in our study and others, albeit weaker, suggests that these genetic variants indeed influence cognition and that the memory associations are not a mere reflection of LOAD risk association only.
A shortcoming of our study is that we focused on memory endophenotypes at a single time point, the last available exam, rather than assessing longitudinal outcomes. We opted for a cross-sectional analysis to maximize our sample size, as not all subjects had longitudinal testing and many subjects, especially from the AA cohort, had a single cognitive assessment. We focused on the last available examination as the most accurate assessment of each individual’s latest cognitive state corresponding to their diagnosis at the time and also to increase our ability to capture memory endophenotype changes secondary to genetic factors, based on the premise that such changes become more pronounced with aging. Indeed, in the longitudinal Chibnik et al. study, the cognitive score differences between the different CR1 genotypes are much smaller at baseline, compared to last examination14. This may be the reason that assessment of baseline cognitive endophenotype associations in a large population-based cohort from the Netherlands revealed only marginal associations with LOAD GWAS variants18. It may be that in the absence of longitudinal data on many subjects within cohorts, using the last available examination, while carefully controlling for age and education, is the next, most powerful approach to capture associations with cognitive phenotypes.
Contrasting the asssociations obtained from the Logical Memory (LM) versus Visual Reproduction (VR) tests in our Caucasian series, we found a significant association between LM phenotypes and the CLU SNP, but no significant findings for any phenotype from VR. This may suggest that episodic memory as captured by LM is either more susceptible to genetic variation than VR or that it is a more sensitive test in detecting more subtle cognitive changes. Alternatively, CLU variants may have a role in verbal but not nonverbal memory.
In summary, we provide here, for the first time, evidence of an effect of the LOAD risky CR1 locus on memory endophenotypes in African-Americans. We also demonstrate better memory endophenotypes for the LOAD protective CLU variant rs11136000 in a Caucasian series. These findings provide further support for the role of these loci in LOAD through mechanisms that influence cognition prior to development of clinical dementia. These results have implications for the utility of genetic variants and cognitive endophenotypes in studies of the mechanism of action of these factors as well as their potential future application in disease prediction paradigms.
Supplementary Material
Systematic Review
In this study, our aim was to determine whether the LOAD risk variants at three genome-wide association study (GWAS) loci for late-onset Alzheimer’s disease (LOAD) influence memory endophenotypes in African-American and Caucasian subjects. We reviewed the literature for the novel LOAD GWAS loci and memory endophenotype associations, and detected several papers which identified associations in Caucasian cohorts for some of these loci.
Interpretations
Our study identifies for the first time associations between LOAD-risky CR1 variants and worse Logical Memory scores in African-Americans. We also describe associations between the LOAD-protective CLU variant and better Logical Memory scores in Caucasians. The memory effects for both loci could be detected in both the entire cohorts which include both LOAD cases and controls, and also in just the control subsets.
Future Directions
These findings need to be confirmed in well-powered additional studies and also tested with longitudinal memory endophenotypes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009 Sep 6; doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics. 2009 Oct;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 3.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010 May 12;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011 May;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature genetics. 2011 May;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003 Apr;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 7.Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006 Jun;22(6):306–313. doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Glahn DC, Almasy L, Blangero J, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007 Mar 5;144(2):242–249. doi: 10.1002/ajmg.b.30446. [DOI] [PubMed] [Google Scholar]
- 9.Ertekin-Taner N. Gene expression endophenotypes: a novel approach for gene discovery in Alzheimer’s disease. Mol Neurodegener. 2011 May 14;6(1):31. doi: 10.1186/1750-1326-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009 Apr 28;72(17):1495–1503. doi: 10.1212/WNL.0b013e3181a2e87d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010 Jan;9(1):119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caselli RJ, Reiman EM, Locke DE, et al. Cognitive domain decline in healthy apolipoprotein E epsilon4 homozygotes before the diagnosis of mild cognitive impairment. Archives of neurology. 2007 Sep;64(9):1306–1311. doi: 10.1001/archneur.64.9.1306. [DOI] [PubMed] [Google Scholar]
- 13.Caselli RJ, Dueck AC, Osborne D, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. The New England journal of medicine. 2009 Jul 16;361(3):255–263. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chibnik LB, Shulman JM, Leurgans SE, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011 Mar;69(3):560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keenan BT, Shulman JM, Chibnik LB, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Human molecular genetics. 2012 May 15;21(10):2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barral S, Bird T, Goate A, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012 May 8;78(19):1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton G, Harris SE, Davies G, et al. Alzheimer’s disease genes are associated with measures of cognitive ageing in the lothian birth cohorts of 1921 and 1936. International journal of Alzheimer’s disease. 2011;2011:505984. doi: 10.4061/2011/505984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhaaren BF, Vernooij MW, Koudstaal PJ, et al. Alzheimer’s Disease Genes and Cognition in the Nondemented General Population. Biological psychiatry. 2012 May 14; doi: 10.1016/j.biopsych.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Roberts RO, Geda YE, Knopman DS, et al. The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics. Neuroepidemiology. 2008;30(1):58–69. doi: 10.1159/000115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 21.Wechsler DA. Wechsler Memory Scale-Revised. New York: Psychological Corporation; 1987. [Google Scholar]
- 22.Carrasquillo MM, Zou F, Pankratz VS, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009 Feb;41(2):192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson GS. Wide-Range Achievement Test Revision-3. Jastak Association; 1993. [Google Scholar]
- 25.Manly JJ, Jacobs DM, Touradji P, Small SA, Stern Y. Reading level attenuates differences in neuropsychological test performance between African American and White elders. Journal of the International Neuropsychological Society: JINS. 2002 Mar;8(3):341–348. doi: 10.1017/s1355617702813157. [DOI] [PubMed] [Google Scholar]
- 26.Pedraza O, Mungas D. Measurement in cross-cultural neuropsychology. Neuropsychology review. 2008 Sep;18(3):184–193. doi: 10.1007/s11065-008-9067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward A, Crean S, Mercaldi CJ, et al. Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer’s disease: a systematic review and meta-analysis. Neuroepidemiology. 2012;38(1):1–17. doi: 10.1159/000334607. [DOI] [PubMed] [Google Scholar]
- 28.Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. American journal of human genetics. 2007 Apr;80(4):605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer’s disease. Brain research reviews. 2009 Oct;61(2):89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Allen M, Zou F, Chai HS, et al. Novel late-onset Alzheimer’s disease loci variants associate with brain gene expression. Neurology. 2012 doi: 10.1212/WNL.0b013e3182605801. in-press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling IF, Bhongsatiern J, Simpson JF, Fardo DW, Estus S. Genetics of clusterin isoform expression and Alzheimer’s disease risk. PLoS One. 2012;7(4):e33923. doi: 10.1371/journal.pone.0033923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.