Abstract
The genetic etiology of late onset Alzheimer disease (LOAD) has proven complex, involving clinical and genetic heterogeneity and gene-gene interactions. Recent genome wide association studies (GWAS) in LOAD have led to the discovery of novel genetic risk factors; however, the investigation of gene-gene interactions has been limited. Conventional genetic studies often use binary disease status as the primary phenotype, but for complex brain-based diseases, neuroimaging data can serve as quantitative endophenotypes that correlate with disease status and closely reflect pathological changes. In the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort, we tested for association of genetic interactions with longitudinal MRI measurements of the inferior lateral ventricles (ILVs), which have repeatedly shown a relationship to LOAD status and progression. We performed linear regression to evaluate the ability of pathway-derived SNP-SNP pairs to predict the slope of change in volume of the ILVs. After Bonferroni correction, we identified four significant interactions in the right ILV (RILV) corresponding to gene-gene pairs SYNJ2-PI4KA, PARD3-MYH2, PDE3A-ABHD12B and OR2L13-PRKG1 and one significant interaction in the left ILV (LILV) corresponding to SYNJ2-PI4KA. The SNP-SNP interaction corresponding to SYNJ2-PI4KA was identical in the RILV and LILV and was the most significant interaction in each (RILV: p=9.10×10−12; LILV: p=8.20×10−13). Both genes belong to the inositol phosphate signaling pathway which has been previously associated with neurodegeneration in AD and we discuss the possibility that perturbation of this pathway results in a down-regulation of the Akt cell survival pathway and, thereby, decreased neuronal survival, as reflected by increased volume of the ventricles.
Keywords: Alzheimer Disease, neuroimaging, atrophy, epistasis, Magnetic Resonance Imaging, ADNI, inositol
Introduction
Late onset Alzheimer’s disease (LOAD) is a devastating, degenerative neurological disease that affects over 5 million people in the United States alone, an already substantial statistic that is expected to triple by 2050 (http://www.alz.org) [1]. The complex genetics of LOAD have proven difficult to unravel due to the disease’s clinical and genetic heterogeneity. To date, ten genes have been confirmed by replication and meta-analysis to be associated with LOAD [2]. Of these genes, only APOE has a large effect with an odds ratio of about 3.7 for one copy of the high risk ε4 allele; the remaining nine genes (CR1, CLU, PICALM, BIN1, EPHA1, MS4A, CD33, CD2AP and ABCA7) exhibit small effect sizes, with odds ratios closer to 1.2 [2]. In order to increase our power and biological interpretability of this complex disease, we expanded our analysis beyond the traditional approach of testing for single marker effects using binary disease status as the primary outcome. In this study, we include rich phenotypic information derived from magnetic resonance imaging (MRI) quantitative traits (QTs) as our outcomes, which addresses the problem of clinical heterogeneity, and we explicitly test for gene-gene interactions, which confronts the issue of genetic heterogeneity [3].
Interactions have shown significant associations in many other complex diseases such as schizophrenia [4], autism [5] and type 2 diabetes [6]. In LOAD, previous gene-gene interaction studies have implicated interactions between CR1 and APOE [7], cholesterol trafficking genes [8,9], tau phosphorylation genes [10], and calcium signaling and axon guidance genes [11]. These studies demonstrate that important mechanistic insight can be garnered from investigating higher order genetic relationships in complex diseases like LOAD.
For LOAD and other brain-based diseases, brain structure derived from imaging modalities can be the source of relevant QTs or endophenotypes. Endophenotypes are biological measurements that are more proximal to genetic function and pathology than disease status [12] and can provide increased statistical power (and therefore decreased sample size requirements) over dichotomous outcome variables [13]. Many measurements of brain structures have been shown to correlate with LOAD status and to have greater sensitivity in detecting early pathological changes [14]. QTs from structural MRI have been used as endophenotypes in LOAD GWAS previously [15], and in this study, we extend that work by investigating associations of an endophenotype of LOAD with gene-gene interactions.
The lateral ventricles have repeatedly shown a relationship to Alzheimer’s disease (AD) status and progression [16–19]. The lateral ventricles normally dilate over time with age, as brain tissue volume decreases, but in patients with mild cognitive impairment (MCI) or AD, the rate of ventricular dilation is much greater than in the normal aging population [20]. MRI measurements of lateral ventricle expansion correlate with disease status, with ventricular volumes and rates of dilation increasing from healthy controls (HC) to MCI and from MCI to AD [20]. The inferior horns of the lateral ventricles are surrounded by gray and white matter structures (corpus callosum, hippocampus, amygdala, caudate nucleus, deep white matter, and thalamus). These structures, particularly the hippocampus and amygdala, often deteriorate in AD, and patients with AD and MCI have significantly higher rates of tissue atrophy in these structures than normal aging adults [20], and ventricular dilation is inversely reflective of atrophy of these surrounding structures [21]. Ventricular dilation is evident 10 years before clinical symptoms, and dilation rate rapidly accelerates two years prior to initial MCI diagnosis, making longitudinal MRI measurement of ventricular dilation a plausible clinical trial biomarker for disease inclusion or progression criteria [20]. Because of the evidence demonstrating atrophy of brain structures surrounding the ILVs in LOAD and because changes in these structures are reflected and magnified in the ILVs, we chose to investigate genetic associations with longitudinal change in volume of these structures.
While correction for multiple testing in single-marker genome-wide association analysis is highly burdensome, due to combinatorics, in genome-wide interaction analyses it is essentially prohibitive except perhaps for very large datasets. However, interaction analyses limited to known candidate genes are unduly constrained by information from previously published studies. Alternative strategies may instead conduct intelligent variable selection based on prior biological knowledge assembled from a wide variety of scientific disciplines. In this study, we selected genes participating in common biological pathways for investigation of gene-gene interactions associated with the endophenotypes of ILV atrophy rate. By doing so, we aimed to increase the biological plausibility of interactions that are novel to AD, while decreasing computational burden. We hypothesized that novel gene-gene interactions would be significantly associated with the dilation of the ILVs and that the novel interactions will generate new or altered hypotheses regarding the etiology of this disease.
Materials and Methods
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see ww.adni-info.org. Further information on ADNI can be found here [22].
Subjects
Participants were enrolled based on the criteria outlined in the ADNI-1 protocols (http://www.adni-info.org/Scientists/AboutADNI.aspx). Information on ADNI subject protocols can be found here [23]. To minimize population stratification, only Caucasian subjects who had both genotype data and T1-weighted MRI data were included. Demographic data are presented in Table 1.
Table 1.
Demographic Information
| Clinical Diagnosis# | |||
|---|---|---|---|
| Normal Control | Mild Cognitive Impairment |
Alzheimer’s Disease | |
| Number of Patients | 187 | 191 | 352 |
| Number of APOE-ε4 Carriers | 45 | 81 | 231 |
| Number of Females | 85 | 61 | 146 |
| Mean Baseline Age (SD^) | 75.98 (5.61) | 75.79 (7.09) | 75.13 (7.33) |
| Mean Years of Education (SD^) | 16.11 (2.79) | 15.73 (3.01) | 15.32 (2.97) |
| Mean RILV slope* (SD^) | 70.55 (77.37) | 119.85 (123.15) | 237.67 (185.37) |
| Mean LILV slope* (SD^) | 68.52 (78.63) | 121.65 (110.92) | 237.55 (179.6) |
Clinical Diagnosis#
Normal Control subjects had a Mini-Mental Status Examination (MMSE) score between 24 and 30, a Clinical Dementia Rating (CDR) score of 0, and were not depressed (Geriatric Depression Scale score < 6).
Mild Cognitive Impairment subjects had a MMSE score between 24 and 30; objective memory impairment, subjective memory impairment, and a CDR score of 0.5
Alzheimer’s Disease subjects met clinical criteria for dementia, had an MMSE of between 20 and 26, and had CDR score of .5 or 1.
standard deviation.
Left/Right inferior lateral ventricles.
Genotyping
Genotyping was performed by the ADNI Genetics Core using the Illumina Infinium Human-610-Quad BeadChip. Further information about the ADNI Genetics Core efforts can be found here [24]. ADNI quality control (QC) steps included removing copy number variant probes, strand checking, base pair position checking, and allele specificity checking [25]. Further QC was performed using PLINK software (version 1.07; [26]), excluding SNPs with a genotyping efficiency < 95%, out of Hardy Weinberg Equilibrium (p<1×10−6), or with a minor allele frequency (MAF) of < 5%. Subjects were excluded if they had a call rate < 95%, if there was a reported versus genetic sex inconsistency, or if relatedness was established (PI_HAT > 0.5). After QC, 515,839 SNPs and 730 subjects remained available for discovery analyses.
Analysis of Imaging Data
Structural T1-weighted MRI scans were acquired on subjects at baseline, 12, 24, and 36 month follow-up appointments as per the ADNI protocol [27]. Further information on ADNI’s MRI protocols can be found here [27,28]. Cortical reconstruction and volumetric segmentation of these images were performed with the FreeSurfer [29] image analysis suite version 4.3 by the ADNI consortium which has been described in detail elsewhere [30]. An early version of the longitudinal image processing framework was used to process the sequential scans [31]. Volumes of the RILV and LILV were calculated in FreeSurfer (in mm3) for every scan available for each individual in the dataset and slopes of change in RILV and LILV volume over time were calculated in SAS 9.3 (SAS Institute Inc., Cary, NC) using mixed model regression (PROC MIXED) to leverage the longitudinal data available in ADNI-1. We used the slopes of change of the LILV and RILV as our primary outcome measurement (mm3/year) and included a measurement of intracranial volume (ICV in mm3) as a covariate in all volume analyses, which was also defined with FreeSurfer.
SNP-SNP Interaction Analysis
Genotype data that passed QC were analyzed in a pathway-based interaction analysis using the publicly available InterSNP program [32]. We took advantage of InterSNP’s pathway based option that tests SNP-SNP pairs between genes that belong to a common biological pathway derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). This option decreases the number of tests performed and also increases the interpretability of an interaction based on a priori information. SNP-SNP interaction effects were explored using a dominant model and a linear regression framework for QTs [32]. A total of 130,512,955 SNP-SNP interaction pairs were tested for each region of interest (ROI), and SNP-SNP pairs were considered significant after Bonferroni correction accounting for the number of pairs tested (p < 3.83×10−10). All SNPs were entered into the model as binary variables (minor allele absent or present) to attenuate the problem of data sparsity commonly confronted in interaction analyses.
Model covariates included: baseline age (in years), sex, education (in years), APOE status (number of ε4 risk alleles) and last diagnosis recorded as of January 2013 (1= Normal, 2=MCI, 3=AD). Each of these covariates was chosen to avoid confounding, with the goal of identifying interactions which explain variance beyond these known risk factors. The brain is known to atrophy over time even in normal aging [33], and the rate of change differs by sex [34]. Education level is correlated with age of disease onset [35]. APOE is a very strong genetic risk factor predisposing patients to LOAD, with even normal subjects who are carriers of the risk allele showing greater neurodegeneration before any symptom onset [36]. And finally, atrophy rates are known to trend with disease status, wherein AD>MCI>HC [16–18,37].
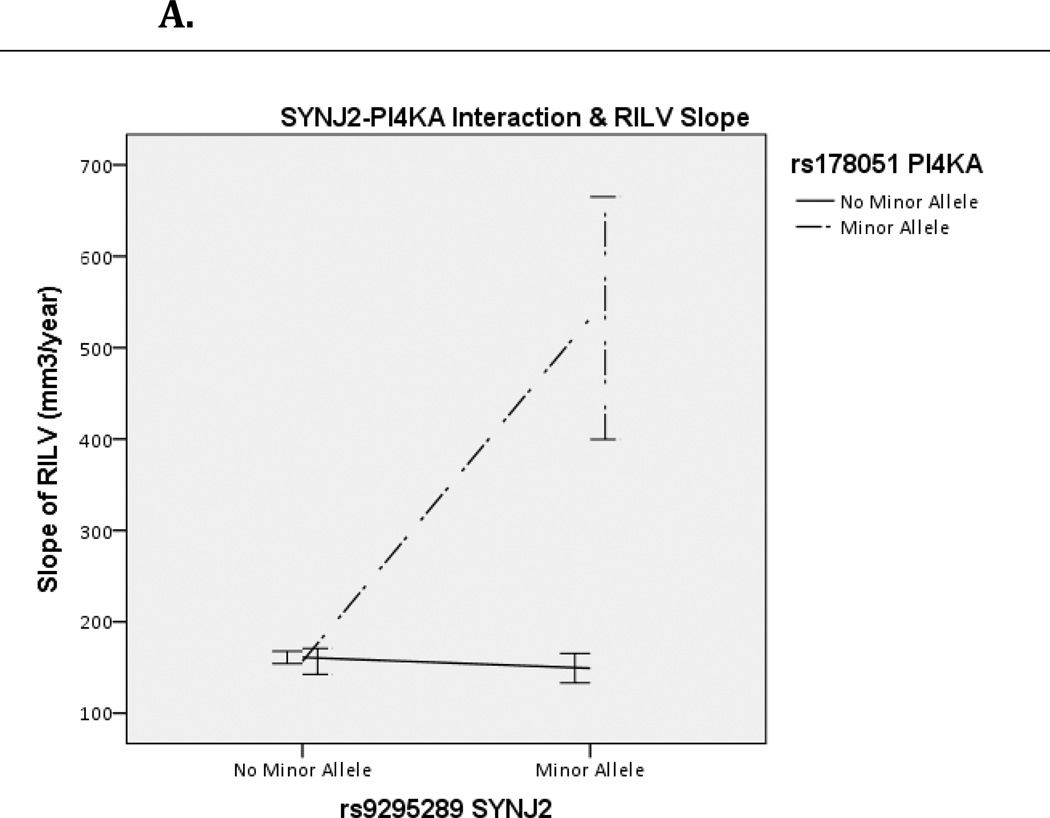
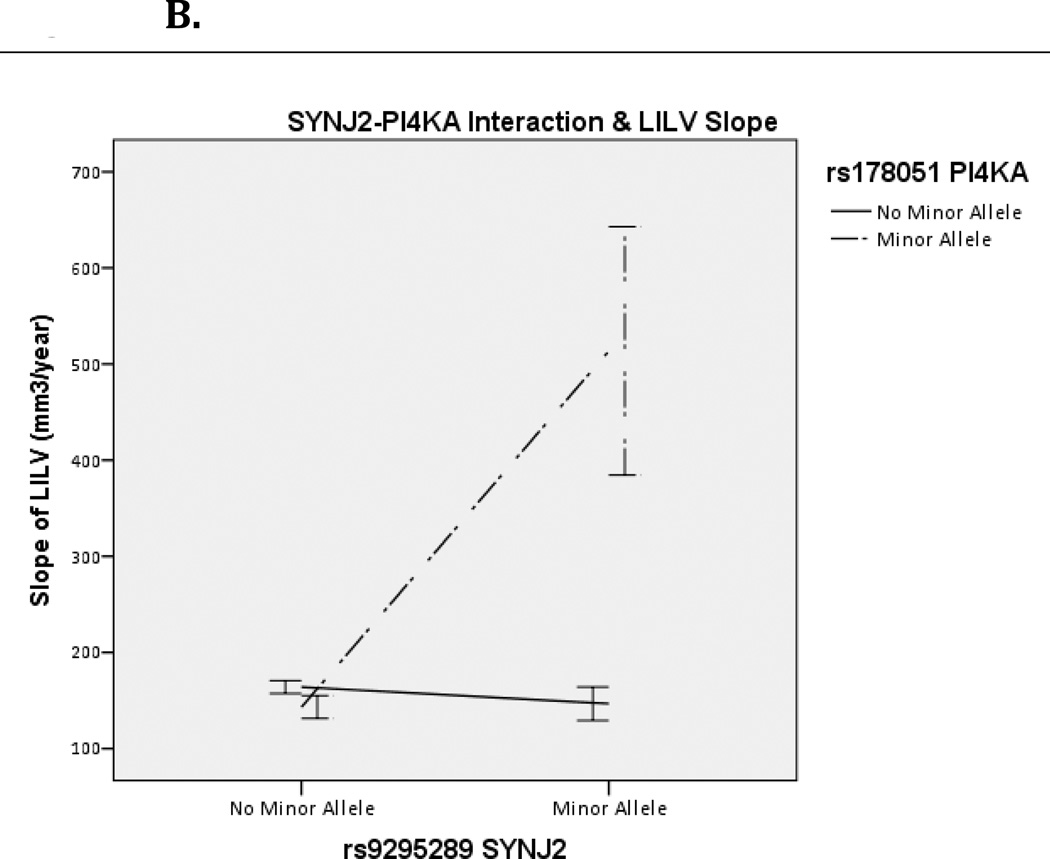
Significant SNP-SNP interactions were annotated to their gene-gene pairs using dbSNP (Homo sapiens, updated in build 137) and their common pathways using KEGG. InterSNP calculates contingency tables which can be seen in Supplemental Table 1. The difference in R2 for these models was calculated in SPSS as R2= R2(full model with interaction term)– R2(reduced model without interaction term). Visualization of interaction effects was created in SPSS as well, showing rate of change by allelic combination (Figure 1A/B, Figure 2).
Figure 1.
Effect of SYNJ2 and PI4KA interaction on Right (A) and Left (B) Inferior Lateral Ventricle (RILV, LILV respectively). Bars represent one standard error.
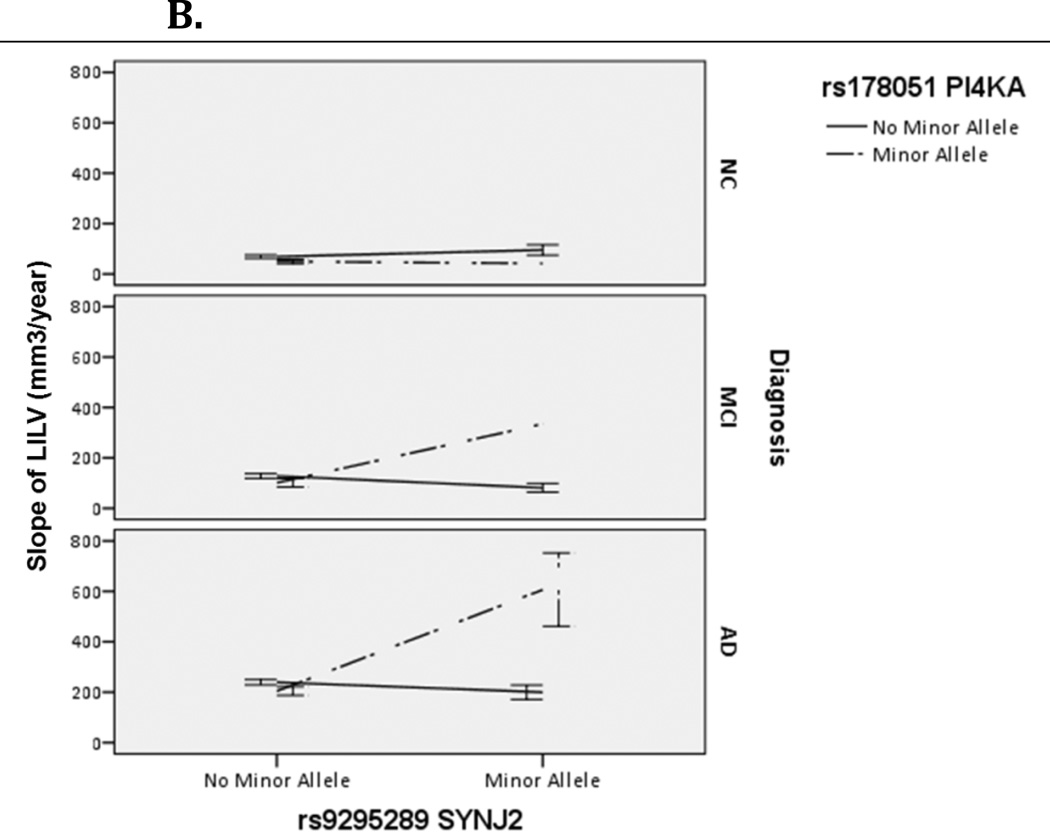
Figure 2.
Effect of SYNJ2 and PI4KA interaction on Right (A) and Left (B) Inferior Lateral Ventricle (RILV, LILV respectively) across diagnoses. Bars represent one standard error.
Results
Pathway Based Interaction Analysis
Demographic information is presented in Table 1. The study sample included 187 NC, 191 MCI, and 352 AD subjects. The number of APOE-ε4 carriers is enriched in the AD and MCI subjects. The slopes of the RILV and LILV increased from NC to MCI to AD subjects. We identified four Bonferroni-corrected significant SNP-SNP interactions in the RILV corresponding to the gene-gene pairs: SYNJ2-PI4KA, PARD3-MYH2, PDE3A-ABHD12B, and OR2L13-PRKG1, in order of significance. None of the SNPs involved had significant main effects (uncorrected p≥0.01).
One Bonferroni-corrected significant SNP-SNP interaction was discovered in the LILV corresponding to SYNJ2-PI4KA. The specific SNP-SNP interaction corresponding to SYNJ2-PI4KA was the same in both the RILV and LILV and was the top hit in both hemispheres (RILV: p=9.10×10−12; LILV: p=8.20×10−13). This gene-gene pair belongs to the metabolic, the inositol phosphate metabolism and the phosphatidylinositol signaling system pathways in KEGG (hsa001100, hsa00562 and hsa04070, respectively), and neither gene is involved in additional KEGG pathways. The effect of this interaction was in the same direction for both hemispheres (βLILV=352.44 and βRILV=351.88) and explained >4% of the variance in both the RILV and LILV (R2RILV= .043, R2LILV=.046). As seen in Figure 1A and 1B, having the minor allele for both genes corresponded to an increased rate of change in both ILVs. This interaction was consistent across subjects with MCI and AD diagnoses (Figure 2). Contingency tables showing sample sizes by genotype combination are presented in Supplemental Table 1.
The PDE3A-ABHD12B interaction that was significantly associated at the Bonferroni-corrected level with change in the RILV did not pass Bonferroni correction in the LILV (p=1.59×10−8). The other SNP-SNP interactions associated with the RILV did not show a strong trend in the LILV. As a post-hoc analysis, we looked at the effect of the significant interactions between SYNJ2-PI4KA, PARD3-MYH2, PDE3A-ABHD12B, and OR2L13-PRKG1 on the average volume of RILV and LILV combined (p=1.52×10−13, 3.49×10−8, 1.34×10−10, 1.85×10−7, respectively).
Discussion
In this study, we focused on the ILVs, which are used frequently as a source of quantitative endophenotypes for LOAD [16–18,37], and we limited hypothesis testing to SNP-SNP pairs within KEGG pathways. Our quantitative trait and pathway-based interaction analysis yielded several interesting candidate gene-gene interactions, one of which was significantly associated with change in both the right and left ILV. Using existing biological knowledge, we were able to deduce a plausible biological context for these significant interactions.
The SNP-SNP interaction rs9295289-rs178051 corresponds to the gene-gene pair SYNJ2-PI4KA and was significantly associated with change in the RILV and LILV, and with the average rate of change in the combined ILV. Below we present evidence suggesting that the biological mechanism for this statistical interaction may involve the perturbation of the phosphatidylinositol (PI) signaling system that results in a down-regulation of the Akt cell survival pathway, resulting in decreased neuronal survival as reflected by increased volume of the ventricles.
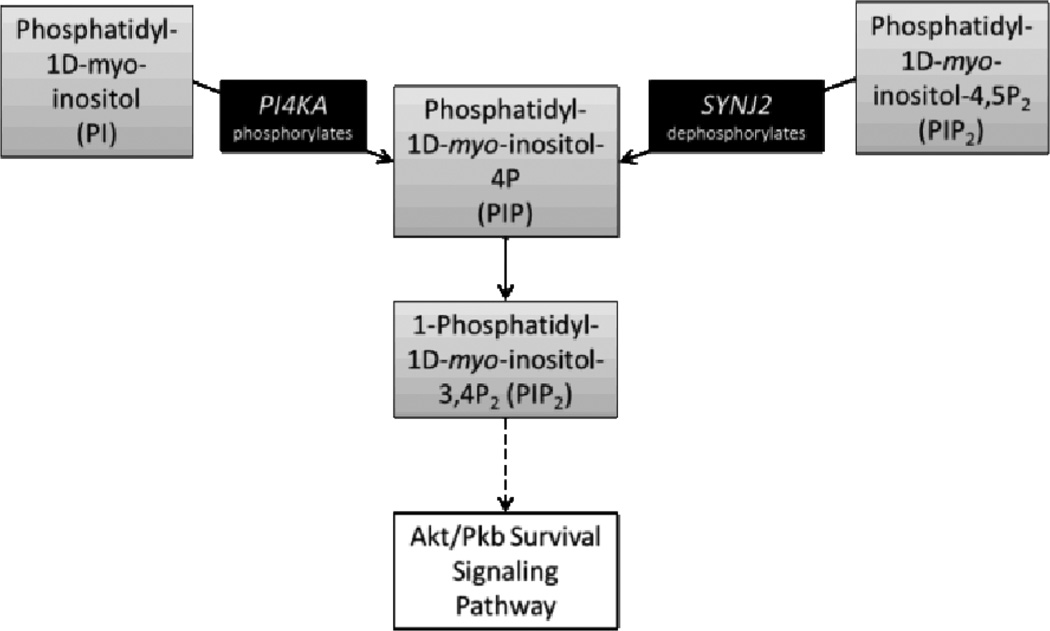
Both SYNJ2 and PI4KA are involved in the synthesis of 1-Phosphatidyl-1D-myo-inositol-4P (PIP) (Figure 3). SYNJ2 (MIM: 609410) encodes synaptojanin-2, which is a ubiquitously expressed inositol polyphosphate 5-phosphatase that dephosphorylates 1-Phosphatidyl-1D-myo-inositol-4,5P2 (PI-4,5P2) to PIP and PI4KA (MIM: 600286) encodes PI 4-kinase, which phosphorylates phosphatidylinositol (PI) to PIP (Figure 3).
Figure 3.
Phosphatidylinositol and Akt/PKB Survival Signalling Pathway, adapted from the KEGG Phosphatidylinositol signaling system (hsa04070, found at http://www.genome.jp/dbget-bin/www_bget?hsa04070)
In multiple cohorts, PIP levels were observed to be reduced in the temporal cortices of patients with LOAD [39,40]. Further, PI kinase activity was decreased while PIP kinase activity remained stable, suggesting that PI kinases, like that encoded by PI4KA, have a specific functional relevance to LOAD [39,41]. PIP is important because it can be phosphorylated to form 1-Phosphatidyl-1D-myo-inositol-3,4P2 (PI-3,4P2), which activates the protein Akt, also known as Protein Kinase B (PKB) [42]. Akt/PKB has proliferative and anti-apoptotic cell response function [42] and has been shown to regulate neuronal survival [43], protect from the neurotoxic effects of AD associated amyloid beta protein [44], and mediate neuronal cell death when its activation is inhibited [45]. Thus, a decrease in PIP synthesis could disrupt a vital mechanism of neuroprotection. Variation in either SYNJ2 or PI4KA might modulate the efficiency of PIP synthesis, but perhaps disruption of both routes of PIP synthesis is required to have detrimental effects seen in our study as increased ventricle dilation.
Evidence that PI4KA is involved in LOAD derives primarily from its role in the synthesis of PIP. However, there is some additional literature linking SYNJ2 to cognitive function, which would lend further support for a role in Alzheimer disease. SYNJ2 is localized at nerve terminals in the brain [46]. It is differentially expressed in hippocampal sub-regions of the marmoset primate [47] and shows decreased expression in the human temporal cortex in persons with major depressive disorder [48]. SYNJ2 has been associated with cognitive abilities in two independent elderly cohorts [49]. Finally, haploinsufficiency of SYNJ2 due to a microdeletion on 6q is associated with a syndrome that presents with microencephaly, developmental delay and agenesis of the corpus callosum [50].
Thus, the genetic interactions associated with ILV atrophy rate in this study may be mapping variants in SYNJ2 and PI4KA that interact to decrease synthesis of PIP and its phosphorylated form (PI-3,4P2), which is required for activation of the neuroprotective, Akt-mediated, cell survival signaling pathway.
In conclusion, by using a pathway based approach, we identified four SNP-SNP interactions significantly associated with AD related quantitative endophenotypes, one of which was significantly associated with bilateral volume change of the inferior lateral ventricles. Focusing on this interaction, we used existing biological knowledge of within-pathway interactions and proposed a plausible biological context for this statistical interaction, which suggests that volume change in the LOAD brain might be mediated by alterations of the inositol signaling pathway, leading to deficits in neuroprotective mechanisms.
The most common variable selection strategy for interaction studies only selects SNPs with main effects to test for interactions. It is important to emphasize that such an approach would not have discovered any of the Bonferroni-significant interactions presented here, highlighting the strength of the pathway-based approach we took.
In the future, the interactions found in this study should be replicated in independent datasets to confirm the SNP-SNP associations. Furthermore, functional analyses could help clarify the basis of these statistical genetic interactions and provide greater specificity for identification of targets for clinical intervention or diagnosis. Further molecular studies on the relationship between SYNJ2 and PI4KA and the PI / PI-3,4P2 / Akt balance are warranted to draw definite conclusions about the relationship between risk variants SYNJ2 and PI4KA causing downstream decreases in PI-3,4P2, Akt mediated cell survival signaling and, ultimately, increased neurodegeneration.
The present results must be interpreted within the framework of our statistical models. In all cases, we included covariates related to disease status and progression including age, sex, education, diagnosis and APOE status. Thus, all significant interactions are explaining variance beyond known predictors of risk, and while the contributions of these interactions appear to be meaningful, the implications should not be extended without considering the variance accounted for by the other factors in our model. The interactions in this study represent dominant effects (carriers versus non-carriers), and the results were interpreted accordingly.
We limited our studies to interactions within pathways, but interactions between genes across pathways may be related to disease risk as well and warrant further exploration. By using KEGG pathways, we also biased our results toward mechanisms that are well-studied. As a result, there may be other novel interactions in other realms of biology that this strategy did not discover. While the sample size utilized in this study is considered large for imaging studies, it is still modest compared with most case-control genetic association studies being conducted at this time, and this is a limitation of the current study as well. We advocate for similar analyses in other complex neurological or neuropsychiatric disorders to improve our understanding of the mechanisms underlying genetic risk for disease.
Supplementary Material
Table 2.
Full results with Bonferroni corrected Significant SNP-SNP interactions
| Gene 1 | Gene 2 | Interaction Term | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ROI | SNP | Gene | Main Effect p- value |
SNP | Gene | Main Effect p- value |
pvalue | R2 | t |
| RILV | rs9295289 | SYNJ2 | 0.10 | rs178051 | PI4KA | 0.04 | 9.10E-12 | 0.04 | 7 |
| rs11596284 | PARD3 | 0.30 | rs3744566 | MYH2 | 0.89 | 3.20E-10 | 0.04 | 6 | |
| rs11614805 | PDE3A | 0.78 | rs7154732 | ABHD12B | 0.91 | 3.60E-10 | 0.04 | 6 | |
| rs11590865 | OR2L13 | 0.01 | rs1922127 | PRKG1 | 0.19 | 3.60E-10 | 0.04 | 6 | |
| LILV | rs9295289 | SYNJ2 | 0.14 | rs178051 | PI4KA | 0.34 | 8.20E-13 | 0.05 | 7 |
ROI: region of interest. RILV: Right inferior lateral ventricle. LILV: Left inferior lateral ventricle. Chr: Chromosome. SNP: reference SNP number of first SNP in SNP-SNP pair. Gene: gene corresponding to SNP. Main: Main effect of SNP on ROI. p: nominal p value. α: Bonferroni corrected p-value. β: beta for SNP-SNP interaction term. R2= R2 for full model −R2 for reduced model (without interaction term). t: t-statistic for interaction term. Bolded interaction was significant at Bonferroni corrected levels in both the RILV and LILV.
Acknowledgments
We are very grateful to all the individuals and their families who participated in the Alzheimer's Disease Neuroimaging Initiative (ADNI). The current study was funded in part by the following: Recruitment for Genetic Aging Research (P30 AG036445), the Vanderbilt Medical Scientist Training Program (T32 GM07347) and the Vanderbilt NIMH Neurogenomics Training grant (T32 MH65215). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Data collection and sharing for this project was funded by the ADNI (U01 AG024904). ADNI is funded by the NIA, the NIBIB, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org/). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
References
- 1.Latest Facts & Figures Report. [Accessed 5/13/2013]; http://www.alz.org/alzheimers_disease_facts_and_figures.asp. [Google Scholar]
- 2.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, DeJager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, StGeorge-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, VanDeerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature Genetics. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo I. Epistasis: Gene Interaction and the Phenotypic Expression of Complex Diseases Like Alzheimer’s. Nature Education. 2008;1 [Google Scholar]
- 4.Qin S, Zhao X, Pan Y, Liu J, Feng G, Fu J, Bao J, Zhang Z, He L. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. European journal of human genetics : EJHG. 2005;13:807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- 5.Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, Mei H, Ritchie MD, DeLong GR, Abramson RK, Wright HH, Cuccaro ML, Hussman JP, Gilbert JR, Pericak-Vance MA. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. American Journal of Human Genetics. 2005;77:377–388. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YM, Ritchie MD, Moore JH, Park JY, Lee KU, Shin HD, Lee HK, Park KS. Multifactor-dimensionality reduction shows a two-locus interaction associated with Type 2 diabetes mellitus. Diabetologia. 2004;47:549–554. doi: 10.1007/s00125-003-1321-3. [DOI] [PubMed] [Google Scholar]
- 7.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, Singleton AB, Wong DF, Ferrucci L, Saykin AJ, Resnick SM. Effect of Complement CR1 on Brain Amyloid Burden During Aging and Its Modification by APOE Genotype. Biological psychiatry. 2012 doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Rodríguez E, Vázquez-Higuera J, Sánchez-Juan P, Mateo I, Pozueta A, Martínez-García A, Frank A, Valdivieso F, Berciano J, Bullido M, Combarros O. Epistasis between intracellular cholesterol trafficking-related genes (NPC1 and ABCA1) and Alzheimer’s disease risk. Journal of Alzheimer’s disease : JAD. 2010;21:619–625. doi: 10.3233/JAD-2010-100432. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Rodríguez E, Mateo I, Infante J, Llorca J, Garcí-Gorostiaga I, Vázquez-Higuera J, Sánchez-Juan P, Berciano J, Combarros O. Interaction between HMGCR and ABCA1 cholesterol-related genes modulates Alzheimer’s disease risk. Brain research. 2009;1280:166–171. doi: 10.1016/j.brainres.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Mateo I, Vázquez-Higuera JL, Sánchez-Juan P, Rodríguez-Rodríguez E, Infante J, García-Gorostiaga I, Berciano J, Combarros O. Epistasis between tau phosphorylation regulating genes (CDK5R1 and GSK-3beta) and Alzheimer’s disease risk. Acta neurologica Scandinavica. 2009;120:130–133. doi: 10.1111/j.1600-0404.2008.01128.x. [DOI] [PubMed] [Google Scholar]
- 11.Meda SA, Koran MEI, Pryweller JR, Vega JN, Thornton-Wells TA. Genetic interactions associated with 12-month atrophy in hippocampus and entorhinal cortex in Alzheimer’s Disease Neuroimaging Initiative. Neurobiology of aging. 2012 doi: 10.1016/j.neurobiolaging.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman II, Gould T. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 13.Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DBDB, Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cognitive Neuropsychiatry. 2009;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tartaglia MC, Rosen HJ, Miller BL. Neuroimaging in Dementia. Neurotherapeutics. 2011;8:82–92. doi: 10.1007/s13311-010-0012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson PM, Hayashi KM, DeZubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Jack CR, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O’Brien PC, Cha R, Knopman D, Petersen RC. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60:253–260. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridha BH, Anderson VM, Barnes J, Boyes RG, Price SL, Rossor MN, Whitwell JL, Jenkins L, Black RS, Grundman M, Fox NC. Volumetric MRI and cognitive measures in Alzheimer disease : comparison of markers of progression. Journal of Neurology. 2008;255:567–574. doi: 10.1007/s00415-008-0750-9. [DOI] [PubMed] [Google Scholar]
- 19.Jack CR, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiner MW. Expanding ventricles may detect preclinical Alzheimer disease. Neurology. 2008;70:824–825. doi: 10.1212/01.wnl.0000304743.72127.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou Y-Y, Leporé N, Chiang M-C, Avedissian C, Barysheva M, McMahon KL, De Zubicaray GI, Meredith M, Wright MJ, Toga AW, Thompson PM. Mapping genetic influences on ventricular structure in twins. NeuroImage. 2009;44:1312–1323. doi: 10.1016/j.neuroimage.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging clinics of North America. 2005;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter WZ, Weiner MW, Jack CR, Jagust W, Toga AW, Lee VM-Y, Shaw LM. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6:230–238. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saykin AJ, Shen L, Foroud TMTM, Potkin SGSG, Swaminathan S, Kim S, Risacher SLSL, Nho K, Huentelman MJMJ, Craig DWDW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR, Weiner MW. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s and Dementia. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Archives of neurology. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of magnetic resonance imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack CR, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schuff N, Krueger G, Killiany RJ, Decarli CS, Dale AM, Carmichael OW, Tosun D, Weiner MW. Update on the magnetic resonance imaging core of the Alzheimers' disease neuroimaging initiative. Alzheimer’s & dementia : the journal of the Alzheimer's Association. 2010;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischl B. FreeSurfer. NeuroImage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, Koeppe RA, Mathis CA, Weiner MW, Jagust WJ. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain : a journal of neurology. 2009;132:1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-Subject Template Estimation for Unbiased Longitudinal Image Analysis. NeuroImage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herold C, Steffens M, Brockschmidt FF, Baur MP, Becker T. INTERSNP: genomewide interaction analysis guided by a priori information. Bioinformatics. 2009;25:3275–3281. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- 33.Takeda S, Matsuzawa T. Age-Related Brain Atrophy: A Study With Computed Tomography. J Gerontol. 1985;40:159–163. doi: 10.1093/geronj/40.2.159. [DOI] [PubMed] [Google Scholar]
- 34.Gur RC, Mozley PD, Resnick SM, Gottlieb GL, Kohn M, Zimmerman R, Herman G, Atlas S, Grossman R, Berretta D. Gender differences in age effect on brain atrophy measured by magnetic resonance imaging. Proceedings of the National Academy of Sciences. 1991;88:2845–2849. doi: 10.1073/pnas.88.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brayne C, Ince PG, Keage HAD, McKeith IG, Matthews FE, Polvikoski T, Sulkava R. Education, the brain and dementia: neuroprotection or compensation? Brain : a journal of neurology. 2010;133:2210–2216. doi: 10.1093/brain/awq185. [DOI] [PubMed] [Google Scholar]
- 36.Wishart HA, Saykin AJ, McAllister TW, Rabin LA, McDonald BC, Flashman LA, Roth RM, Mamourian AC, Tsongalis GJ, Rhodes CH. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- 37.Jack CR, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panagiotou OA, Ioannidis JPA. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. International journal of epidemiology. 2012;41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 39.Jolles J, Bothmer J, Markerink M, Ravid R. Phosphatidylinositol kinase is reduced in Alzheimer’s disease. Journal of neurochemistry. 1992;58:2326–2329. doi: 10.1111/j.1471-4159.1992.tb10981.x. [DOI] [PubMed] [Google Scholar]
- 40.Stokes CE, Hawthorne JN. Reduced Phosphoinositide Concentrations in Anterior Temporal Cortex of Alzheimer-Diseased Brains. Journal of Neurochemistry. 1987;48:1018–1021. doi: 10.1111/j.1471-4159.1987.tb05619.x. [DOI] [PubMed] [Google Scholar]
- 41.Bothmer J, Markerink M, Jolles J. Phosphoinositide kinase activities in synaptosomes prepared from brains of patients with Alzheimer’s disease and controls. Neuroscience Letters. 1994;176:169–172. doi: 10.1016/0304-3940(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 42.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt protooncogene product by phosphatidylinositol-3,4-bisphosphate. Science (New York, N. Y.) 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 43.Dudek H. Regulation of Neuronal Survival by the Serine-Threonine Protein Kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 44.Martín D, Salinas M, López-Valdaliso R, Serrano E, Recuero M, Cuadrado A. Effect of the Alzheimer amyloid fragment Aβ(25–35) on Akt/PKB kinase and survival of PC12 cells. Journal of Neurochemistry. 2001;78:1000–1008. doi: 10.1046/j.1471-4159.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 45.Malagelada C, Jin ZH, Greene LA. RTP801 is induced in Parkinson’s disease and mediates neuron death by inhibiting Akt phosphorylation/activation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:14363–14371. doi: 10.1523/JNEUROSCI.3928-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemoto Y, Wenk MR, Watanabe M, Daniell L, Murakami T, Ringstad N, Yamada H, Takei K, De Camilli P. Identification and characterization of a synaptojanin 2 splice isoform predominantly expressed in nerve terminals. The Journal of biological chemistry. 2001;276:41133–41142. doi: 10.1074/jbc.M106404200. [DOI] [PubMed] [Google Scholar]
- 47.Datson NA, Morsink MC, Steenbergen PJ, Aubert Y, Schlumbohm C, Fuchs E, De Kloet ER. A molecular blueprint of gene expression in hippocampal subregions CA1, CA3, and DG is conserved in the brain of the common marmoset. Hippocampus. 2009;19:739–752. doi: 10.1002/hipo.20555. [DOI] [PubMed] [Google Scholar]
- 48.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Molecular psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 49.Lopez LM, Harris SE, Luciano M, Liewald D, Davies G, Gow AJ, Tenesa A, Payton A, Ke X, Whalley LJ, Fox H, Haggerty P, Ollier W, Pickles A, Porteous DJ, Horan MA, Pendleton N, Starr JM, Deary IJ. Evolutionary conserved longevity genes and human cognitive abilities in elderly cohorts. European journal of human genetics : EJHG. 2012;20:341–347. doi: 10.1038/ejhg.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagamani SCS, Erez A, Eng C, Ou Z, Chinault C, Workman L, Coldwell J, Stankiewicz P, Patel A, Lupski JR, Cheung SW. Interstitial deletion of 6q25.2-q25.3: a novel microdeletion syndrome associated with microcephaly, developmental delay, dysmorphic features and hearing loss. European journal of human genetics : EJHG. 2009;17:573–581. doi: 10.1038/ejhg.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.