Abstract
Scope
There are limited data on the metabolism of [6]-shogaol, a major bioactive component of ginger. This study demonstrates metabolism of [6]-shogaol in liver microsomes from mouse, rat, dog, monkey, and human.
Methods and results
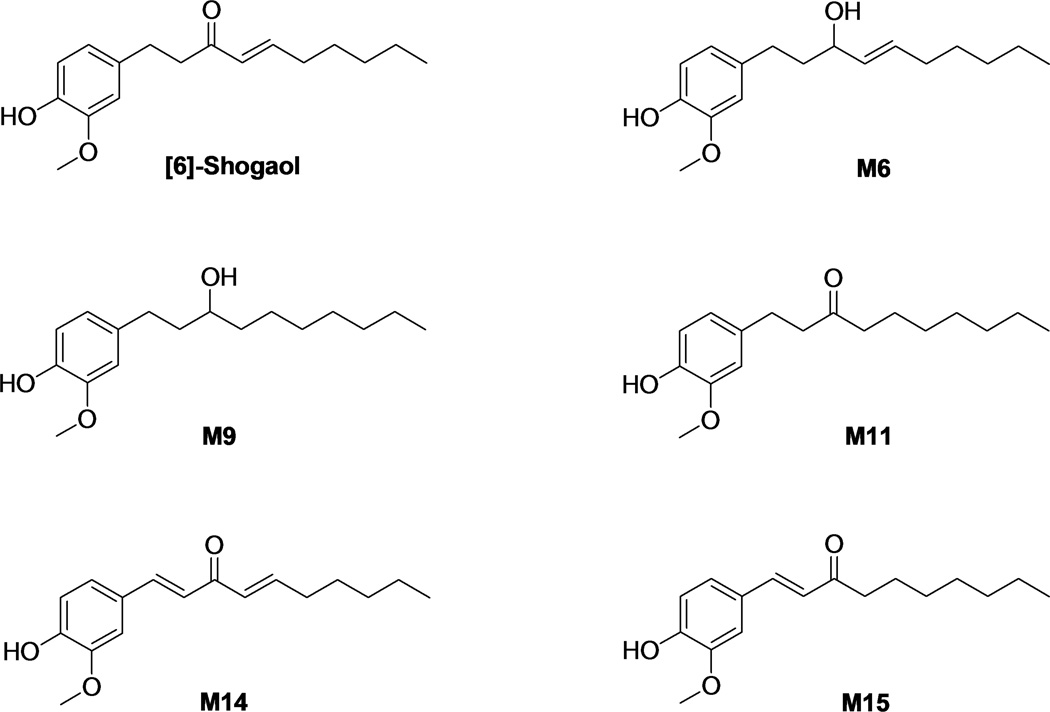
The in vitro metabolism of [6]-shogaol was compared among five species using liver microsomes from mouse, rat, dog, monkey, and human. Following incubations with [6]-shogaol, three major reductive metabolites 1-(4'-hydroxy-3'-methoxyphenyl)-4-decen-3-ol (M6), 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M9), and 1-(4'-hydroxy-3'-methoxyphenyl)-decan-3-one (M11), as well as two new oxidative metabolites (1E, 4E)-1-(4'-hydroxy-3'-methoxyphenyl)-deca-1,4-dien-3-one (M14) and (E)-1-(4'-hydroxy-3'-methoxyphenyl)-dec-1-en-3-one (M15) were found in all species. The kinetic parameters of M6 in liver microsomes from each respective species were quantified using Michaelis-Menten theory. A broad CYP-450 inhibitor, 1-aminobenzotriazole, precluded the formation of oxidative metabolites M14 and M15, and 18β-glycyrrhetinic acid, an aldo-keto reductase inhibitor, eradicated the formation of the reductive metabolites M6, M9, and M11 in all species. Metabolites M14 and M15 were tested for cancer cell growth inhibition and induction of apoptosis and both showed substantial activity, with M14 displaying greater potency than [6]-shogaol.
Conclusion
We conclude that [6]-shogaol is metabolized extensively in mammalian species mouse, rat, dog, monkey, and human, and that there are significant interspecies differences to consider when planning pre-clinical trials towards [6]-shogaol chemoprevention.
Keywords: [6]-Shogaol, Ginger, Metabolism, Liver microsomes, Cancer
1 Introduction
Ginger, the rhizome of Zingiber officinale is a commonly used spice and crude drug, a flavoring and curative utilized for many years throughout the world. Ginger is touted for its relief of nausea [1], anti-inflammatory properties [2] and putative anti-cancer activity [3–5]. The major pharmacologically active components of ginger are gingerols and shogaols [6–9]. Thermal processing of [6]-gingerol gives [6]-shogaol, the latter of which has been shown to exert higher growth-inhibitory effects against cancer cells than the former [10]. There have been several studies promoting the bioactivity of [6]-shogaol. For instance, Hu et al. demonstrated efficacy of [6]-shogaol against tumor growth in mouse SMMC-7721 xenografts by induction of apoptosis, activation of caspase-3, and inactivation of endoplasmic reticulum stress-related protein eIF2α [11]. Additionally, an investigation by Weng et al. demonstrated that [6]-shogaol exerts its antiinvasive activity against metastic hepatoma cells via inhibition of the transcription and translation pathways of matrix metalloproteinases (MMPs) and urokinase-type plasminogen activators (uPA) [12]. A study of structure-function activity identified [6]-shogaol as a potent inhibitor of PGE2 release, an indication of anti-inflammation, compared to other ginger components and hexahydrocurcumin [13]. Our group has also shown [6]-shogoal causes induction of programmed cell death in HCT-116 human colon cancer cells and H-1299 human lung cancer cells [14].
Over the past few decades, the pharmacokinetics of [6]-shogaol have been investigated in mouse, rat, and in human [15–19]. However, little attention has been paid to its metabolism, which may influence its efficacy and toxicity in vivo. Our group’s previous study reported the metabolism of [6]-shogaol in mice and cancer cells, in which 13 metabolites were identified [14]. By examining the biotransformation of [6]-shogaol, we found that the mercapturic acid pathway and a redox pathway are its major metabolic routes. Our results also indicated that [6]-shogaol’s redox metabolites (M9 and M11) triggered apoptosis in human colon and lung cancer cells, giving evidence that [6]-shogaol continues to be somewhat pharmacologically effective after being metabolized in vitro. However, significant differences in metabolic pathways among different species may exist, and the various metabolites and their concentrations can potentially affect in vivo bioactivities and toxicities. For example, coumarin is metabolized in human and in mouse to 7-hydroxy coumarin. However, coumarin is transformed to a carcinogenic epoxide in rat, a drastic consequence of differential metabolism in seemingly similar mammals [20]. This proven rationale of interspecies variation in terms of metabolic behavior was the impetus to investigate the biotransformation of [6]-shogaol among different species.
The current study therefore provides a comparison of [6]-shogaol metabolism in humans and in different experimental animals including mouse, rat, dog, and monkey, and compares the pharmacokinetic parameters in liver microsomes from these species.
2 Materials and Methods
2.1 Reagents
Microsomes and NADPH-regenerating systems were procured from BD Biosciences (Bedford, MA). [6]-Shogaol was prepared from ginger extract in our laboratory [10]. 1-(4′-Hydroxy-3′-methoxyphenyl)-4-decen-3-ol (M6), 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M9), and 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M11) were purified from mouse feces as described before [14]. 1-Aminobenzotriazole (ABT), 18β-glyccerhetinic acid (18β-GA), and 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) were purchased from Sigma Aldrich (St. Louis, MO). Analytical (250 µm thickness, 2–25 µm particle size) and preparative TLC plates (2000 µm thickness, 2–25 µm particle size) were purchased from Sigma Aldrich (St. Louis, MO) and Sorbent Technologies (Atlanta, GA), respectively. HPLC and LC/MS grade solvents were obtained from VWR Scientific (South Plainfield, NJ). HCT-116 human colon cancer cells and H-1299 human lung cancer cells were obtained from American Type Tissue Culture (Manassas, VA). McCoy’s 5A medium was purchased from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) and penicillin/streptomycin were purchased from Gemini Bio-Products (West Sacramento, CA). MTT (3-(4,5-dimethylthiaxol-2-yl)-2,5-diphenyltetrazolium bromide) was procured from Calbiochem-Novabiochem (San Diego, CA).
2.2. Incubation with liver microsomes
2.2.1 Experiment 1
Human liver microsomes (HLM) (either 0.1 mg/mL or 0.5 mg/mL final concentration) were incubated with [6]-shogaol (50 µM) for several time points. The experimental incubation mixture consisted of 100 mM potassium phosphate buffer, a prepared NADPH-regenerating system, and human liver microsomes. In all experiments, [6]-shogaol was dissolved in dimethylsulfoxide (DMSO) with a final concentration not exceeding 1% (v/v). After 5 min preincubation in a 37°C water bath, the reaction was initiated by the addition of [6]-shogaol and was further incubated at 37°C. The reaction was terminated at 0, 30, 45, and 60 minutes by the addition of ice-cold acetonitrile (equal volume) containing 2% acetic acid. The mixture was vortexed and underwent centrifugation at 13,000g for 10 minutes. Aliquots of supernatant were stored at −20°C until analysis. Control incubations without NADPH-regenerating system, without substrate, or without microsomes were performed to ensure that metabolite formation was microsome- and NADPH- dependent.
2.2.2 Experiment 2
Based upon the optimized conditions in experiment 1, mouse liver microsomes (MLM), rat liver microsomes (RLM), dog liver microsomes (DLM), monkey liver microsomes (CyLM) and human liver microsomes (HLM) (each 0.5 mg/mL, respectively), were mixed with aforementioned incubation mixture and were held in 37°C water bath for 5 minutes before the reactions were initiated by the addition of [6]-shogaol (50 µM). Reactions were terminated after 30 minutes incubation in a 37°C water bath by the addition of ice-cold acetonitrile (equal volume) containing 2% acetic acid. The mixture was vortexed and underwent centrifugation at 13,000g for 10 minutes. Aliquots of supernatant were stored at −20°C until analysis.
2.3 Chemical inhibition studies
Inhibitors ABT (500 µM) and 18β-GA (500 µM) or DMSO controls were pre-incubated with microsomes in experimental incubation mixture at 37°C for 20 minutes before initiating the reaction by the addition of [6]-shogaol (50 µM). Reactions were terminated 30 minutes after substrate addition and incubation in a 37°C water bath by equal volume of ice-cold acetonitrile with 2% acetic acid. The mixture was vortexed and underwent centrifugation at 13,000g for 10 minutes. Aliquots of supernatant were stored at −20°C until analysis.
2.4 Sample preparation for LC/MS analysis
To elucidate the structures of the two previously uncharacterized metabolites of [6]-shogaol, samples from CyLM incubations were enriched by partitioning with EtOAc. Briefly, a large-scale reaction (1.0 mL) of CyLM incubated with [6]-shogaol was performed in which enzyme concentration was 0.5 mg/mL and [6]-shogaol concentration was 50 µM. All other reagent relative concentrations were kept constant. After 30 minutes incubation in a 37°C water bath, samples were treated in an identical fashion as described above to stabilize metabolites and precipitate proteins from solution. The supernatant was then extracted three times with ice-cold EtOAc (5x volumes each time). The pooled supernatant was dried under reduced pressure at 30 °C, and the residue was resuspended in MeOH with 0.2% acetic acid for LC/MS analysis.
2.5 Synthesis of (1E, 4E)-1-(4′-hydroxy-3′-methoxyphenyl)-deca-1,4-dien-3-one (M14)
[6]-Shogaol (276 mg, 1.0 mmol) was dissolved in tetrahydrofuran (THF) and cooled down to 0 °C. To this mixture, a solution of DDQ (181 mg, 0.8 mmol) in THF was added. The mixture was stirred at 0 °C for 30 minutes and then warmed to room temperature (RT) for 3 hours. Then water was added and the resulting mixture was extracted with EtOAc (× 3). The organic phase was washed with water (× 1) and brine (× 1), dried over anhydrous NaSO4, and filtered. The filtration was evaporated and the residue was purified by chromatography column (CC) (hexane/ethyl acetate = 8:1 and 6:1) to give the desired compound as a yellow oil (170 mg, yield: 62%); 1H NMR (600 MHz, CDCl3) δ 7.59 (1H, d, J = 15.9 Hz, H-1), 6.82 (1H, d, J = 15.9 Hz, H-2), 6.44 (1H, d, J = 15.6 Hz, H-4), 6.99 (1H, dt, J = 15.6, 7.0 Hz, H-5), 2.27 (2H, q, J = 7.0 Hz, H-6), 1.51 (2H, m, H-7), 1.34-1.29 (4H, m, H-8 and H-9), 0.90 (3H, t, J = 6.8 Hz, H-10), 7.07 (1H, d, J = 1.4 Hz, H-2'), 6.93 (1H, d, J = 8.2 Hz, H-5'), 7.14 (1H, dd, J = 8.2, 1.4 Hz, H-6'), and 3.93 (3H, s, OMe-3'); 13C NMR (150 MHz, CDCl3) δ 143.3 (d, C-1), 123.3 (d, C-2), 189.3 (s, C-3, C=O), 129.1 (d, C-4), 148.0 (d, C-5), 32.7 (t, C-6), 27.9 (t, C-7), 31.4 (t, C-8), 22.4 (t, C-9), 14.0 (q, C-10), 127.4 (s, C-1'), 109.7 (d, C-2'), 148.1 (s, C-3'), 146.8 (s, C-4'), 114.8 (d, C-5'), 122.9 (d, C-6'), and 56.0 (q, OMe-3'); positive ESI-MS, m/z 275 [M + H]+.
2.6 Synthesis of (E)-1-(4′-hydroxy-3′-methoxyphenyl)-dec-1-en-3-one (M15)
General procedure above was followed by using M11 (45 mg, 0.16 mmol), prepared previously in our lab, and DDQ (29 mg, 0.13 mmol) in THF (6 mL). The mixture was stirred at RT for 2 hours. The resulting residue was purified by preparative TLC (hexane/ethyl acetate = 5:1) to give the desired compound as a yellow oil (34 mg, yield: 77%); 1H NMR (600 MHz, CDCl3) δ 7.47 (1H, d, J = 16.1 Hz, H-1), 6.59 (1H, d, J = 16.1 Hz, H-2), 2.63 (2H, t, J = 7.5 Hz, H-4), 1.66 (2H, m, H-5), 1.33-1.25 (8H, m, ranged from H-6 to H-9), 0.87 (3H, t, J = 6.7 Hz, H-10), 7.08 (1H, d, J = 1.6 Hz, H-2'), 6.91 (1H, d, J = 8.2 Hz, H-5'), 7.09 (1H, dd, J = 8.2, 1.6 Hz, H-6'), and 3.92 (3H, s, OMe-3'); 13C NMR (150 MHz, CDCl3) δ 142.6 (d, C-1), 124.2 (d, C-2), 200.8 (s, C-3, C=O), 40.7 (t, C-4), 24.6 (t, C-5), 29.3 (t, C-6), 29.1 (t, C-7), 31.7 (t, C-8), 22.6 (t, C-9), 14.1 (q, C-10), 127.1 (s, C-1'), 109.4 (d, C-2'), 148.1 (s, C-3'), 146.8 (s, C-4'), 114.8 (d, C-5'), 123.4 (d, C-6'), and 56.0 (q, OMe-3'); positive ESI-MS, m/z 277 [M + H]+.
2.7 HPLC analysis
An HPLC-ECD (ESA, Chelmsford, MA) consisting of an ESA model 584 HPLC pump, an ESA model 542 autosampler, an ESA organizer, and an ESA electrochemical detector (ECD) coupled with two ESA model 6210 four sensor cells was used. A Gemini C18 column (150 mm × 4.6 mm, 5 µm; Phenomenex, Torrance, CA) was used for chromatographic analysis at a flow rate of 1.0 mL/min. The mobile phases consisted of solvent A (30 mM sodium phosphate buffer containing 1.75% acetonitrile and 0.125% tetrahydrofuran, pH 3.35) and solvent B (15 mM sodium phosphate buffer containing 58.5% acetonitrile and 12.5% tetrahydrofuran, pH 3.45). The gradient elution had the following profile: 20–62% B from 0 to 13 min; 62% B from 13 to 39 min; 62–100% B from 39 to 48 min; 100% B from 48 to 53 min; and 20%B from 53.1 to 58 min. The cells were then cleaned at a potential of 1000 mV for 1 minute. The injection volume of the sample was 10 µL. The eluent was monitored by the Coulochem electrode array system (CEAS) with potential settings at 0, 200, 250, 300, 350, 400 and 500 mV. Data for Figures 2, 3, and 5 were from the channel set at 300 mV of the CEAS.
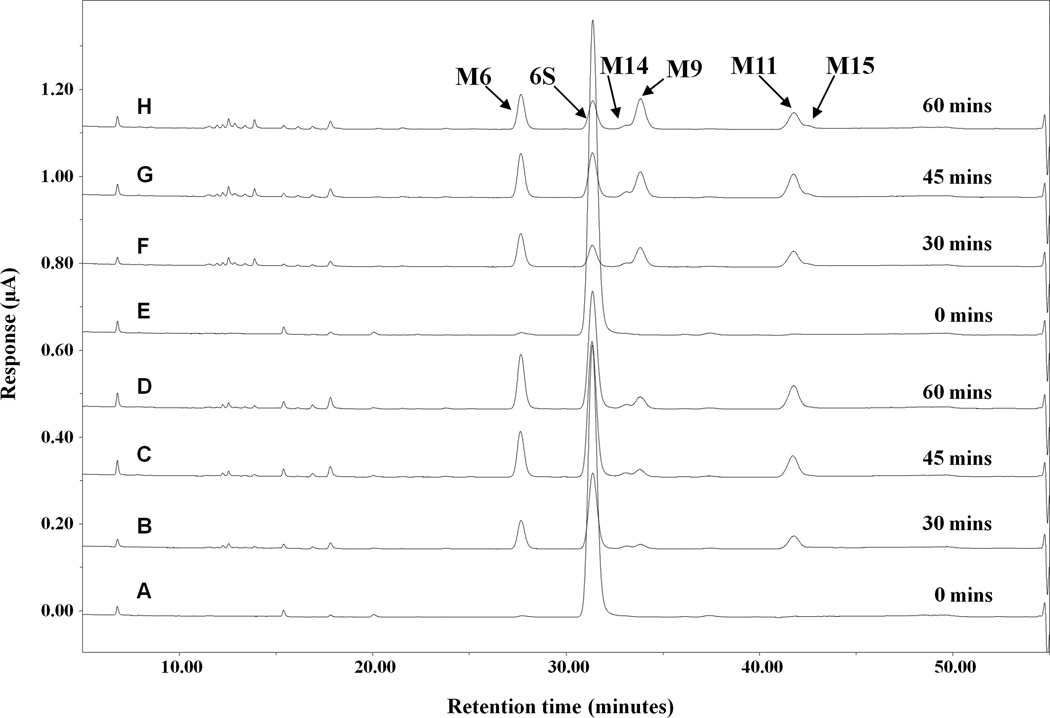
Figure 2.
Representative HPLC-ECD chromatograms of [6]-shogaol and its metabolites in liver microsomes from human. [6]-Shogaol (50 µM) was incubated with NADPH-regenerating system and liver microsomes at 0.1 mg/mL (A–D) or 0.5 mg/mL (E–H) at 37°C for 0, 30, 45, and 60 min.
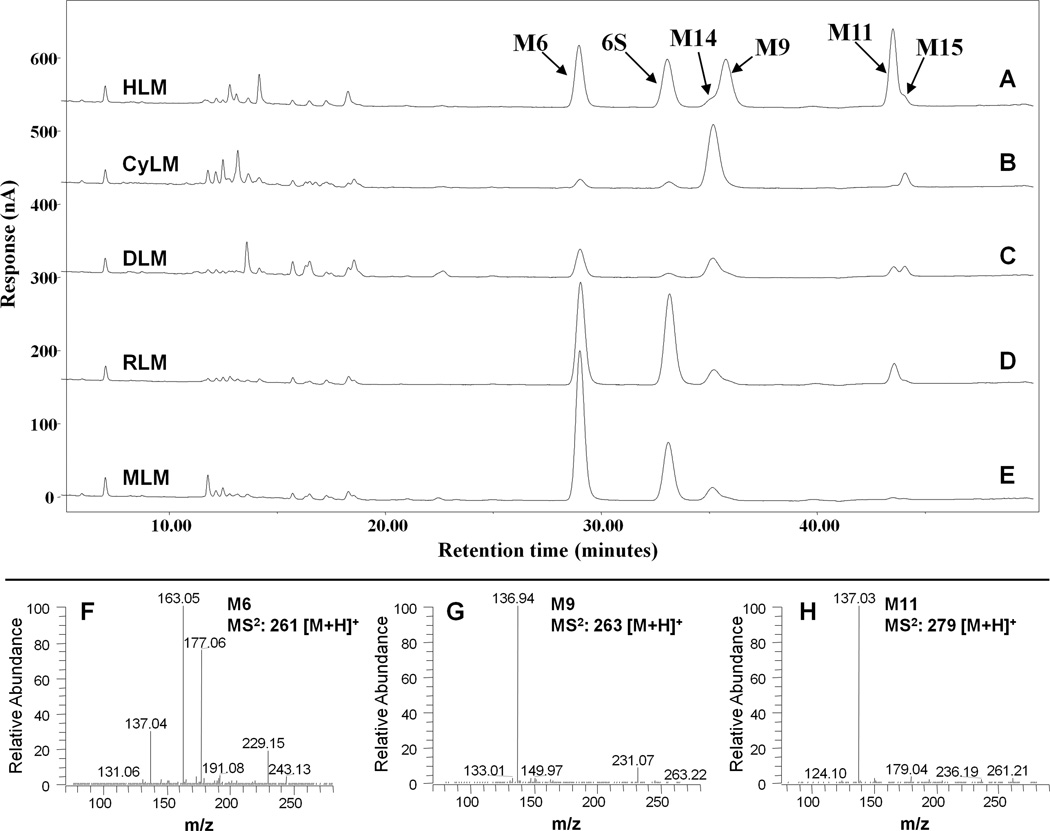
Figure 3.
(A–E) Representative HPLC-ECD chromatograms of [6]-shogaol and its metabolites in liver microsomes from mouse, rat, dog, monkey, and human; and (F–H) MS/MS (positive ion) spectra of authentic M6, M9, and M11. [6]-shogaol (50 µM) was incubated with liver microsomes (0.5 mg/ml) from different species and an NADPH-regenerating system at 37°C for 30 min.
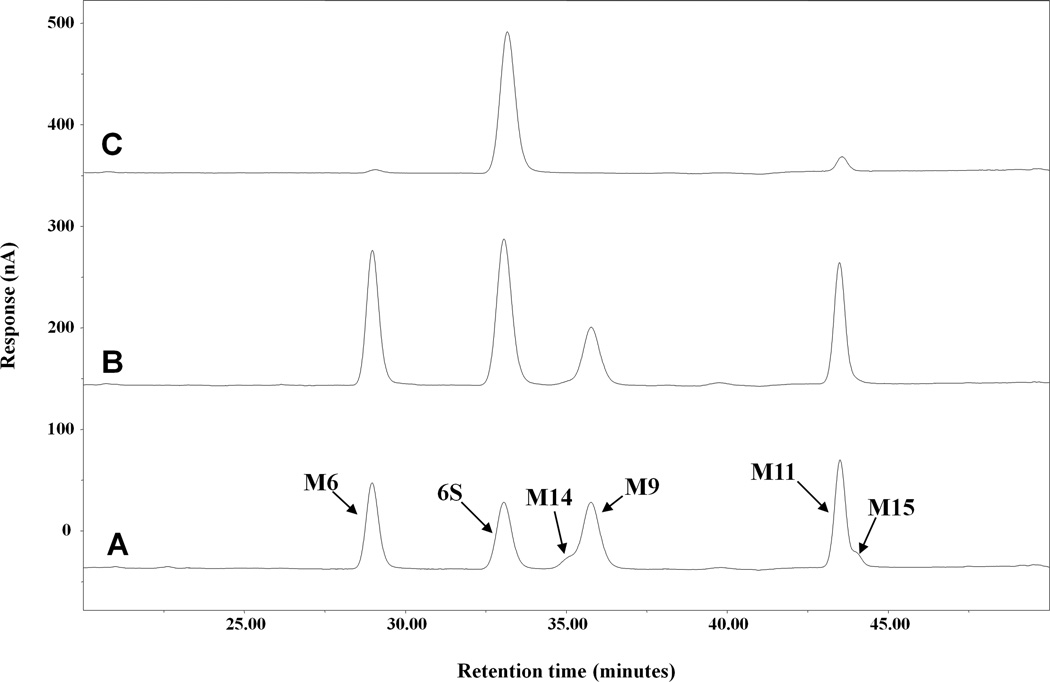
Figure 5.
Representative HPLC-ECD chromatograms of [6]-shogaol and its metabolites in liver microsomes from human (0.5mg/mL) incubated with no inhibitor (A), 500µM ABT (B), or 500µM 18β-Glycyyrhetinic Acid (C).
2.8 LC/MS analysis
LC/MS analysis was carried out with a Thermo-Finnigan Spectra System, which consisted of an Accela high-speed MS pump, an Accela refrigerated autosampler, and an LTQ Velos ion trap mass detector (Thermo Electron, San Jose, CA, USA) incorporated with heated electrospray ionization (H-ESI) interfaces. A Gemini C18 column (50 × 2.0 mm i.d., 3 µm; Phenomenex, Torrance, CA, USA) was used for separation at a flow rate of 0.2 mL/min. The column was eluted with 100% solvent A (5% aqueous methanol with 0.2% acetic acid) for 3 minutes, followed by linear increases in B (95% aqueous methanol with 0.2% acetic acid) to 40% from 3 to 15 minutes, to 85% from 15 to 45 minutes, to 100% from 45 to 50 minutes, and then with 100% B from 50 to 55 minutes. The column was then re-equilibrated with 100% A for 5 minutes. The LC eluent was introduced into the H-ESI interface. The positive ion polarity mode was set for the H-ESI source with the voltage on the H-ESI interface maintained at approximately 4.5 kV. Nitrogen gas was used as the sheath gas and auxiliary gas. Optimized source parameters, including ESI capillary temperature (300 °C), capillary voltage (50 V), ion spray voltage (3.6 kV), sheath gas flow rate (30 units), auxiliary gas flow rate (5 units), and tube lens (120 V), were tuned using authentic [6]-shogaol. The collision-induced dissociation (CID) was conducted with an isolation width of 2 Da and normalized collision energy of 35 for MS2 and MS3. Default automated gain control target ion values were used for MS — MS3 analyses. The mass range was measured from 50 to 1000 m/z. Data acquisition was performed with Xcalibur 2.0 version (Thermo Electron, San Jose, CA, USA).
2.9 Nuclear magnetic resonance (NMR)
1H and 13C NMR spectra were recorded on a Bruker AVANCE 600 MHz spectrometer (Brucker, Inc., Silberstreifen, Rheinstetten, Germany). Compounds were analyzed in CDCl3. Multiplicities are indicated by s (singlet), d (doublet), t (triplet), q (quartet), and br (broad). The 13C NMR spectra are proton decoupled.
2.10 Kinetic study
To estimate kinetic parameters of metabolism of [6]-shogoal to major product M6 in liver microsomes from human and other species, the incubation conditions were optimized to ensure that formation rate of M6 was in the linear range in relation to incubation time and protein concentration. [6]-Shogaol (7.81, 15.63, 31.25, 62.5, 125, 250, 500, 1000, and 2000 µM) was incubated with MLM, RLM, DLM, CyLM, and HLM (0.1 mg/mL each, respectively) for 30 minutes. All incubations were performed in triplicate. The apparent Km and Vmax values were calculated from analysis of experimental data according to the Michaelis-Menten equation, and the results were graphically represented (for HLM) by an Eadie-Hofstee plot. Kinetic constants were reported as the value +/− S.E. of the parameter estimate.
2.11 Growth inhibitory effects of M14 and M15 against human colon and lung cancer cells
Cell growth inhibition was determined by a MTT colorimetric assay [21]. Human colon cancer cells HCT-116 and human lung cancer cells H-1299 were plated in 96-well microtiter plates with 3000 cells/well and allowed to attach for 24 hours at 37 °C. The test compounds (in DMSO) were added to cell culture medium to the desired final concentrations (final DMSO concentrations for control and treatments were 0.1 %). After the cells were cultured for 24 hours, the medium was aspirated and cells were treated with 200 µL fresh medium containing 2.41 mmol/L MTT. After incubation for 3 hours at 37 °C, the medium containing MTT was aspirated, 100 µL of DMSO was added to solubilize the formazan precipitate, and plates were shaken gently for an hour at room temperature. Absorbance values were derived from the plate reading at 550 nm on a Biotek microtiter plate reader (Winooski, VT). The reading reflected the number of viable cells and was expressed as a percentage of viable cells in the control. Both HCT-116 and H-1299 cells were cultured in McCoy’s 5A medium. All of the above media were supplemented with 10 % fetal bovine serum, 1 % penicillin/streptomycin, and 1 % glutamine, and the cells were kept in a 37 °C incubator with 95% humidity and 5% CO2.
2.12 Measurement of induction of apoptosis in human cancer cells by M14, M15, and [6]-shogaol
Human colon cancer cells HCT-116 and human lung cancer cells H-1299 were plated in 96-well plates at a density of 5000 cells/well and allowed to attach overnight at 37°C. M14, M15, or [6]-shogaol in DMSO, or DMSO control, diluted in media, were added to cells and incubated for an additional 24 hours at 37°C. After 24 hours, media containing compound was removed and cells were lysed in their respective wells with reagents from a Cell Death Detection ELISAPLUS kit from Roche Applied Science (Mannheim, Germany). Samples were harvested after cell lysates were spun down at 300 g for 10 minutes. To streptavidin coated microplates, 20 µL samples were added, and mixed with 80 µL Immunoreagent, which consisted of anti-histone-biotin, and anti-DNA-POD, and incubated with gentle shaking for 2 hours at room temperature. After incubation, Immunoreagent was removed and samples were washed 3 times with 250 µL incubation buffer. Substrate solution, ABTS, was added to each well and color was developed for 15 minutes before stopping the reaction with ABTS stop solution and reading absorbance on a microplate reader at 430 nm. Experiments were performed in triplicate and the average is given in comparison to DMSO control with standard deviation.
3 Results
3.1 Incubation conditions of [6]-shgoaol with human liver microsomes
As there are no reports of the metabolism of [6]-shogaol in liver microsomes and subsequent HPLC-ECD analysis, we found it was necessary to optimize the parameters of our experimental conditions by varying both the enzyme concentration relative to the reaction mixture and the incubation times (Figure 2). As expected, transformation of [6]-shogaol occurred more rapidly in the condition containing 0.5 mg/mL human liver microsomes than in the condition with 0.1 mg/mL microsomes. The profiles were comparable, with no noticeable difference besides the aforementioned metabolic velocity. After examining the profiles from different conditions, we fixed the incubation parameters optimized for metabolite identification and interspecies comparison with hepatic microsomes at 0.5 mg/mL along with NADPH-regenerating system for 30 minutes.
3.2 Structure elucidation of [6]-shogaol metabolites
Figures 3A–3E show the LC chromatograms of [6]-shogaol and its metabolites after 30 minutes incubation with mouse, rat, dog, monkey, and human liver microsomes. We observed five major product peaks when [6]-shogaol (50 µM) was incubated with the hepatic microsomes from these five species (Figure 3). All of the peaks were identified by comparing their MS2 spectra with those of authentic standards.
3.2.1 Metabolites M6, M9 and M11
Metabolites at the retention time of 29.3, 33.9, and 41.8 min were identified as 1-(4'-hydroxy-3'-methoxyphenyl)-4-decen-3-ol (M6), 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M9), and 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M11), respectively, by comparing their mass spectra with those of authentic standards (Figures 4F–4H) that were purified from mouse feces as described before [14].
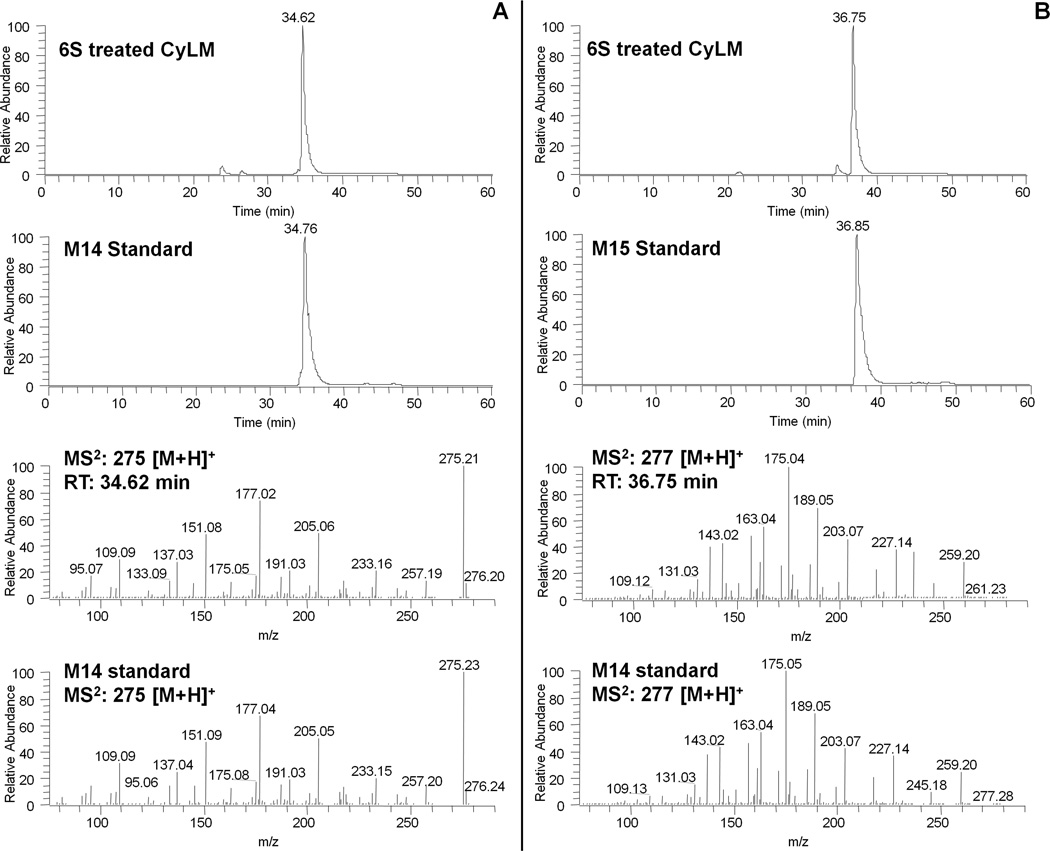
Figure 4.
(A) LC chromatograms of 6-shogaol (6S) treated CyLM as well as M14 (GA) authentic standard obtained by positive ESI/MS interface extracted with m/z 275 [M+H]+ and MS/MS (positive ion) spectra of the major metabolite (M14) corresponding to m/z 275 [M+H]+ in 6S treated CyLM and M14 authentic standard; (B) LC chromatograms of 6S treated CyLM as well as M15 (GA) authentic standard obtained by positive ESI/MS interface extracted with m/z 277 [M+H]+ and MS/MS (positive ion) spectra of the major metabolite (M15) corresponding to m/z 277 [M+H]+ in 6S treated CyLM and M15 authentic standard.
3.2.2 Metabolite M14
In order to identify the other two unknown peaks (M14 and M15), we ran the sample obtained from CyLM on LC/MS. Our results indicated that M14 had a molecular weight of m/z 274 as determined by the mass ions at m/z 275 [M+H]+, 297 [M+Na]+, and 571 [2M+Na]+, which was 2 mass units less than that of [6]-shogaol. This metabolite showed m/z 177 but not 137 (product ion of [6]-shogaol) as a major product ion, which led us to suspect that this unknown peak was a 1,2-dehydrogenated product of [6]-shogaol. In order to confirm this deduction, we synthesized the compound as described in the experimental section. This synthetic compound had a molecular formula C17H22O3 on the basis of positive ESI-MS at m/z 275 [M + H]+ and its 1H and 13C NMR data. The molecular weight of this compound (m/z: 274) was 2 mass units less than that of [6]-shogaol (m/z: 276) indicating it was dehydrogenated [6]-shogaol. This was also supported by the observation of the absence of two methenes and the presence of two more olefinic methines (δH 7.59 and δH 6.82; and δC 143.1 and δC 123.3) in its 1H and 13C NMR spectra. Additionally, geometry of the double bond between C-1 and C-2 was determined as E-configuration by the coupling constant J1,2 = 15.9 Hz. Therefore, this compound was confirmed as (1E,4E)-1-(4-hydroxy-3-methoxyphenyl)deca-1,4-dien-3-one. M14 had almost the same retention time as well as the same mass fragment as those of the synthetic (1E, 4E)-1-(4′-hydroxy-3′-methoxyphenyl)-deca-1,4-dien-3-one (Figure 4A). Therefore, M14 was identified as (1E, 4E)-1-(4′-hydroxy-3′-methoxyphenyl)-deca-1,4-dien-3-one (Figure 1).
Figure 1.
Structures of [6]-shogaol and its major metabolites.
3.2.3 Metabolite M15
In the same way, we found another peak showed a molecular weight of m/z 276 as determined by the mass ions at m/z 277 [M+H]+, 299 [M+Na]+, and 575 [2M+Na]+. M15 had the same molecular weight as that of [6]-shogaol, which let us suspect that it was a double bond transferred metabolite of [6]-shogaol. In order to confirm this deduction, we also synthesized this compound using M11 as described in the experimental section. This synthetic compound had a molecular formula C17H24O3 on the basis of positive ESI-MS and its 1H and 13C NMR data. The molecular weight of this compound was 2 mass units less than that of M11, coinciding with the fact that it was a dehydrogenated product of M11, and also supported by the appearance of two olefinic methines (δH 7.47 and δH 6.59; and δC 142.6 and δC 124.2) in its 1H and 13C NMR spectra. Additionally, the E-configuration between C-1 and C-2 was determined by the coupling constant J1,2 = 16.1 Hz. Therefore, this compound was confirmed to be (E)-1-(4′-hydroxy-3′-methoxyphenyl)-dec-1-en-3-one. M15 had almost the same retention time as well as the same mass fragment as those of the synthetic (E)-1-(4′-hydroxy-3′-methoxyphenyl)-dec-1-en-3-one (Figure 4B). Therefore, M15 was identified as (E)-1-(4′-hydroxy-3′-methoxyphenyl)-dec-1-en-3-one (Figure 1).
3.3 Metabolism of [6]-shogaol by liver microsomes from different species
The metabolic profiles of [6]-shogaol in MLM, RLM, DLM, and CyLM were similar to those in HLM as the five major metabolites, M6, M9, M11, M14, and M15 were detected in all five species (Figure 3). Monkey liver microsomes gave the highest abundance of oxidized metabolites after 30 minutes incubation, in stark contrast to human liver microsomes, which gave the least. These results suggest stronger interspecies differentiation in the enzymes responsible for [6]-shogaol oxidative metabolism than its reductive metabolism, which is seemingly conserved.
3.4 Chemical inhibition studies
To determine the inclusion of CYP-450 enzymes in [6]-shogaol metabolism, ABT, a broad-specificity P450 inactivator [22], was applied to incubations of liver microsomes with [6]-shogaol. In human (Figures 5A and 5B), oxidative metabolism was abrogated yet reductive metabolism still occurred. This result was similar in mouse, rat, dog, and monkey (data not shown). Hence, CYP-450 enzymes were implicated in oxidative metabolism of [6]-shogaol. As the broad-specificity P450 suicide substrate ABT did not inhibit reductive metabolism in all species, it was clear that non-P450 enzymes were implicated. Thus, we applied 18β-glycyrrhetinic acid, a known aldo-keto reductase inhibitor [23] to our reaction mixture of HLM and found that reductive metabolism of [6]-shogaol was decimated (Figure 5C).
3.5 Kinetics
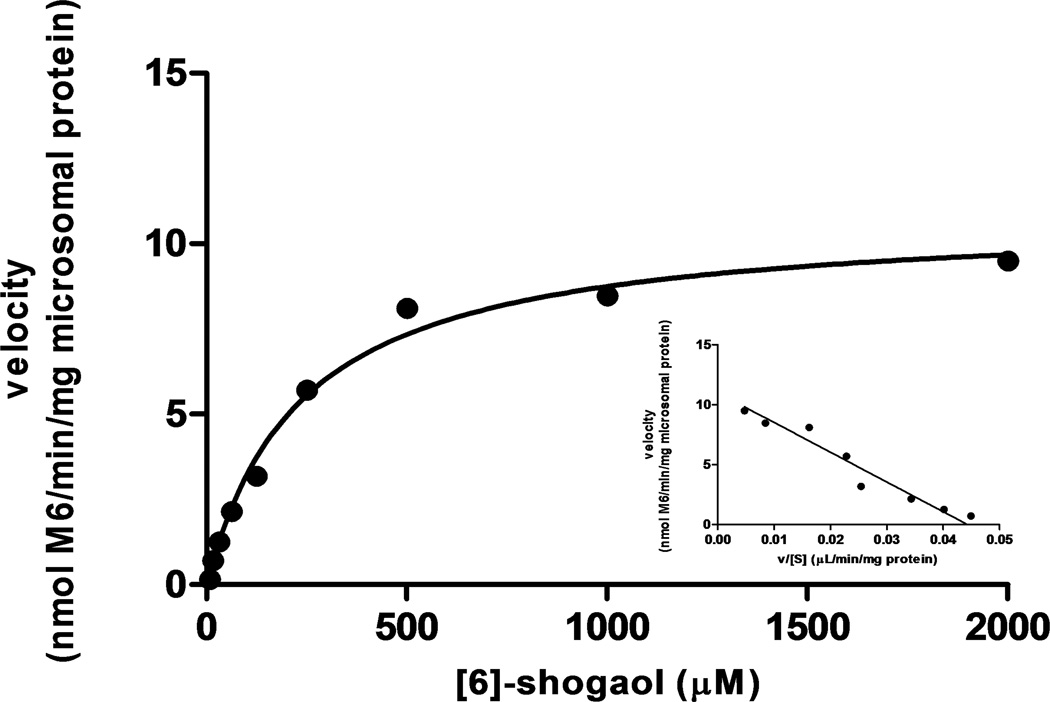
Over the concentration range tested, metabolism of [6]-shogaol to M6 in liver microsomes from humans and four animal species obeyed Michaelis-Menten kinetics, as evidenced by the Eadie-Hofstee plot (Figure 6). The kinetic parameters, including Km, Vmax, and the intrinsic clearance (Vmax/Km), were determined and are listed in Table 1. In human liver microsomes, the Km value for M6 formation was 45.6 µM and the Vmax was 10.82 +/− 0.47 nmoles per minute per milligram microsomal protein. In liver microsomes from four experimental animals, the Km values for M6 formation ranged from 75.7 µM (mouse) to 424.2 µM (monkey), displaying vast interspecies variation. The Vmax values for the remaining four species varied as well, with 2.99 +/− 0.28 (monkey) to 8.19 +/− 0.48 (rat) nmoles per minute per milligram enzyme, a kinetic trend that was consistent with the perceived relative formation rates of M6 across five species in the HPLC-ECD chromatogram of Figure 3. Intrinsic clearance values varied significantly across all five species, from 7.0 µL per minute per milligram protein (monkey) to 80.5 µL per minute per milligram protein (rat), indicating the relative affinities of M6 formation from lowest to highest in CyLM and RLM, respectively.
Figure 6.
Michaelis-Menten plot of [6]-shogaol metabolism to M6 in human liver microsomes. [6]-Shogaol was incubated with HLM at 37°C for 30 minutes with an NADPH-regenerating system. An Eadie-Hofstee plot is shown as an insert to illustrate monophase kinetics. Data points represent the mean of triplicate determinations.
Table 1.
Kinetic parameters of M6 metabolism in liver microsomes: Km values are in micromolar values. Vmax values are nanomoles per minute per milligram liver microsomes. The range of substrate concentrations was 7.8 to 2000 µM. Each value is the mean +/− S.D. of three independent experiments.
| Species | Vmax | Km | Vmax/Km |
|---|---|---|---|
| Human | 10.82 +/− 0.47 | 237.2 +/− 32.7 | 45.6 |
| Monkey | 2.99 +/− 0.28 | 424.2 +/− 108.3 | 7.0 |
| Dog | 7.33 +/− 0.43 | 251.1 +/− 42.99 | 29.2 |
| Rat | 8.19 +/− 0.48 | 101.7 +/− 22.62 | 80.5 |
| Mouse | 4.10 +/− 0.19 | 75.7 +/− 12.17 | 54.2 |
3.6 Growth inhibitory effects against human cancer and normal cells
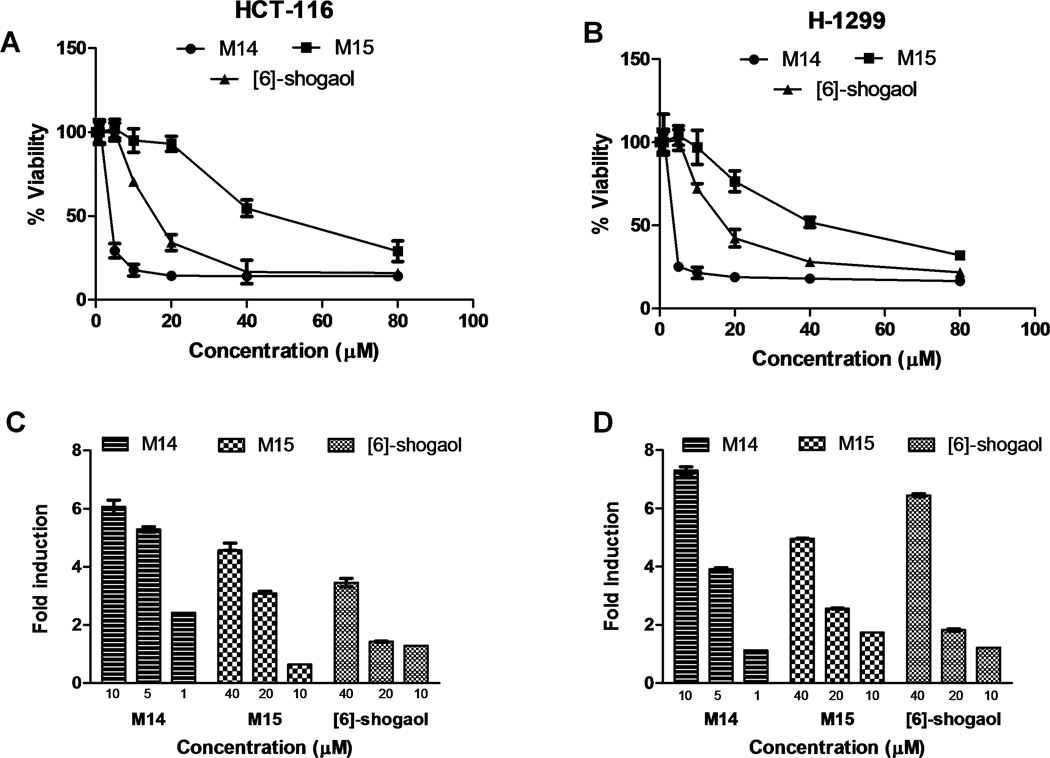
Two human cancer cell lines, HCT-116 and H-1299, were treated with [6]-shogaol or its synthetic metabolites M14 and M15, with concentrations ranging from 0 to 80 µM. Cell viability assays utilizing MTT resulted in high potency of M14 and moderate potency of M15, relative to [6]-shogaol, with IC50 values of 3.22, 43.02, and 19.94 µM, respectively, against colon cancer cells HCT-116 (Figure 7A). In lung cancer cells H-1299, a similar trend was observed with M14, M15, and [6]-shogaol, with IC50 values of 3.04, 41.59, and 17.32 µM, respectively (Figure 7B).
Figure 7.
Growth inhibition and induction of apoptosis by M14, M15 and [6]-shogaol against human cancer cells. Human colon cancer (HCT-116) (A) and human lung cancer (H-1299) (B) cells were incubated with respective metabolites or [6]-shogaol for 24 hours and tested for cell viability. Values are given as percent of positive control, where n = 6. Induction of apoptosis by of M14 and M15. Human colon cancer cells (HCT-116) (C) and human lung cancer cells (H-1299) (D) were incubated with respective metabolites or [6]-shogaol for 24 hours and tested for induction of apoptosis. Values are given as fold induction against DMSO control, where n = 3.
3.7 Induction of apoptosis in human cancer cells by M14, M15, and [6]-shogaol
Following the implications of cytotoxicity in cancer cells induced by M14, M15, and [6]-shogaol, the pro-apoptotic properties of these compounds were measured (Figure 7C and 7D). M14 still remained the most potent compound, with induction of apoptosis of about 6-fold in HCT-116 cells and about 7~8 fold in H-1299 cells, compared to DMSO control after administration of 10 µM compound and 24 hours incubation. Even treatment of 1 µM M14 gave a 2-fold induction of apoptosis in HCT-116 cells and 1-fold induction in H-1299 cells, compared to DMSO control, further verifying the strong bioactivity of this metabolite. Metabolite M15 gave similar, if somewhat greater apoptotic effects against both cancer cell lines after treatment for 24 hours, as parent compound [6]-shogaol. Administration of 10 µM M15 or [6]-shogaol was sufficient in both cell lines to induce apoptosis, by 1-fold in HCT-116 and almost 2-fold in H-1299. The similarities in pro-apoptotic potencies between [6]-shogaol and M15 was not entirely expected, as [6]-shogaol was more than twice as toxic in the previous MTT assays. However, as apoptosis is a specific form of cell death, the results are not unreasonable.
4 Discussion
The possible cancer-preventive activity of ginger is receiving more and more attention. Information on the metabolism of ginger components such as [6]-shogaol is important for understanding the biological effects of ginger. Previously, we reported the metabolism of [6]-shogaol in mice and in cancer cells. In general, animal models are used in the development of new drugs, as pre-clinical models to predict metabolic behavior in human. Thus far, the differences in the metabolic pathways and behaviors of [6]-shogaol between human and common experimental animals have not been revealed. Hence, in this study a comparison of metabolic profiles, enzymes involved, and catalytic efficiency for [6]-shogaol metabolism in liver microsomes from different species was performed.
Our study revealed a comparative metabolism of [6]-shogaol in liver microsomes from mouse, rat, dog, monkey, and human (Figures 3A–3E). The spectra were similar across all animal species, with a few distinct differences. For instance, the ratios and relative abundances of the major metabolites were not conserved, although the major metabolites were found in all spectra. Metabolite M6 was the dominant peak in spectra from the rodent species after 30 minutes incubation of [6]-shogaol with microsomes, was found in an intermediate relative amount in dog and in human, and was a minor metabolite in monkey. In addition, M11 was the major product of [6]-shogaol metabolism in HLM, but a very minor product of incubation from microsomes of other species, with minute amounts in MLM and CyLM. However, the greatest species variegation noted was with metabolites M14 and M15. In particular, M14 was the major metabolite in CyLM under our experimental parameters, while the peak produced from this compound was minor in all other species, with the smallest amount produced from HLM incubation. Metabolite M15, although present in all species after [6]-shogaol metabolism, was very minor in mouse, rat, and human, but was abundant in dog and in monkey.
For further exploration of the similarity of the enzymes responsible for the metabolism of [6]-shogaol from various species, chemical inhibition studies with use of selective general inhibitors were administrated. Using a general CYP-450 inhibitor ABT [22], it was shown that both M14 and M15 were catalyzed predominantly by CYP-450 enzymes (Figure 5B). This evidence suggests that the P450 enzymes involved in [6]-shogaol metabolism are not conserved as a function of evolutionary similarity, given the disparity between CyLM and HLM. Genetically, monkey and human have a low relative divergence [24]. However, the minute differences in the composition of the CYP enzymes in seemingly familiar species have very apparent metabolic consequences, as noted in this study. In planning preliminary in vivo experiments with animal models to scale for human dosing, this is an important consideration. Further work to elucidate the specific CYP-450 isoform(s) implicated in [6]-shogaol biotransformation would be necessary to uncover possible drug-interactions or other metabolic consequences of [6]-shogaol consumption.
While CYP-450 enzymes are implicated in the metabolism of many xenobiotics, non-P450 enzymes have also been characterized in this pursuit, and are an important consideration of [6]-shogaol biotransformation. Reduction of xenobiotic carbonyls is a significant metabolic route to produce more hydrophilic and often less toxic compounds, which can be substrates for phase II conjugation by UDP-glucuronosyl transferases or sulfotransferases, leading ultimately to excretion of the products [25]. An α,β-unsaturated ketone such as [6]-shogaol is likely to undergo selective reductive metabolism with initial chain saturation and subsequent reduction to the alcohol metabolite by an aldo-keto reductase [4, 26–28]. An easily accessible and well known dehydrogenase inhibitor, licorice root derivative 18β-glycyrrhetinic acid (18β-GA) [29], was employed in this study to investigate the role of aldo-keto reductase enzymes in [6]-shogaol metabolism in liver microsomes. Administration of 18β-GA inhibited [6]-shogaol metabolism in all species, thereby verifying our assumption that [6]-shogaol reductive metabolism was a function of aldo-keto reductase(s).
Once we confirmed the implication of an aldo-keto reductase in [6]-shogaol metabolism in the five species tested, it was necessary to address the observed discrepancies of catalytic efficiencies amongst the animal liver microsomes. In order to quantify a portion of [6]-shogaol metabolism, we investigated the pharmacokinetics of major metabolite M6 formation in MLM, RLM, DLM, CyLM, and HLM, using Michaelis-Menten parameters. Our calculated results were consistent with the observed spectra engendered from incubations of each respective species and [6]-shogoal. That is, mouse and rat showed significant preference for M6 formation, both qualitatively and quantitatively, with the largest peak areas for this metabolite (Figure 3) and the lowest Km values (Table 1) after 30 minute incubations with [6]-shogaol. Intrinsic clearance values for these species were also the highest, indicating greater catalytic efficiency. Conversely, monkey liver microsomes produced a small amount of M6, with a matching high Km value and a low intrinsic clearance value. Human liver microsomes gave intermediate values for Km and intrinsic clearance, compared to the four species. Hence, qualitative inspection of the entire metabolic profiles was necessary to find the animal model most appropriate for human scaling. In preclinical studies to evaluate pharmacokinetics and toxicity in humans, surrogate models should have both similar metabolic profiles and close kinetic parameters [30]. In our study of [6]-shogaol metabolism, it appears that rodent species are the most suitable models for human scaling, compared to dog and monkey. Due to our inability to develop a method to separate the other metabolites analytically, we were only able to investigate the kinetic parameters of metabolite M6. Future work with an improved HPLC detection method would be necessary to investigate the kinetic parameters of other metabolites.
Another advantage of observing metabolism of [6]-shogaol in different species in this study was the opportunity to discover two new oxidative metabolites, M14 and M15, and identify these compounds as (1E,4E)-1-(4-Hydroxy-3-methoxyphenyl)deca-1,4-dien-3-one(M14) and (E)-1-(4-Hydroxy-3-methoxyphenyl)dec-1-en-3-one (M15) (Figure 1). Bioactivity assays showed M14 had a significantly increased potency over [6]-shogaol, in both killing cancer cells and inducing apoptosis, while M15 displayed moderate activity. By observing the structures of the metabolites tested in this study (Figure 1), it appears that the drastically increased potency of M14 can be attributed to the additional double bond on the α,β-unsaturated ketone [6]-shogaol. The double bonds in this compound are thought to be specifically available to sulfhydryl groups via Michael addition [31] and may react by depleting antioxidants such as glutathione. Drastic glutathione depletion is an indication of cellular distress and may induce apoptosis [32]. Similarly, as M15 retains an α,β-unsaturated ketone composition similar to [6]-shogaol, it is likely the activity retained by this metabolite can be attributed to this structure. However, in examining the placement of the double bond, the reduced activity is putatively a result of steric hindrance of the benzyl ring against bonding of sulfhyldryl groups and subsequent glutathione depletion. Metabolites M14 and M15 induced apoptosis in these cancer cells lines (Figure 7), indicative of their means of effect against cancer cells. As these metabolites possess much greater activity than the reduced metabolites previously assessed in [6]-shogaol biotransformation in cancer cells and in mice [14], their discovery in this study was of great utility.
This report gave an in-depth exploration of [6]-shogaol metabolism in liver microsomes from mouse, rat, dog, monkey, and human, with respect to metabolic profiles, enzymes involved, and catalytic efficacy. This novel investigation can contribute significant information towards planning animal selection in pharmacokinetic and toxicological studies of [6]-shogaol in vivo. Additionally, previously unknown metabolites of [6]-shogaol were discovered, both displaying significant anti-cancer activity. These data are fundamental towards future work in [6]-shogaol chemoprevention.
Acknowledgments
This work was supported by NIH grants CA138277 and CA138277S1 to S. Sang.
Abbreviations
- 6S
[6]-shogaol
- 18β-GA
18β-glycyrrhetinic acid
- ABT
1-aminobenzotriazole
- AKR
aldo-keto reductase
- CyLM
cynomolgus monkey liver microsomes
- CYP-450
cytochrome P450 enzyme
- DLM
beagle dog liver microsomes
- HLM
human liver microsomes
- HPLC-ECD
high performance liquid chromatrography/ESA electrochemical detector
- LC/MS
liquid chromatrography/mass spectrometry
- MLM
CD-1 mouse liver microsomes
- RLM
Sprague-Dawley rat liver microsomes
Footnotes
Conflict of interest statement
The authors declare no competing financial interest.
References
- 1.Ozgoli G, Goli M, Simbar M. Effects of ginger capsules on pregnancy, nausea, and vomiting. J Altern Complement Med. 2009;15:243–246. doi: 10.1089/acm.2008.0406. [DOI] [PubMed] [Google Scholar]
- 2.Rani MP, Padmakumari KP, Sankarikutty B, Cherian OL, et al. Inhibitory potential of ginger extracts against enzymes linked to type 2 diabetes, inflammation and induced oxidative stress. Int J Food Sci Nutr. 2011;62:106–110. doi: 10.3109/09637486.2010.515565. [DOI] [PubMed] [Google Scholar]
- 3.Shukla Y, aLS Cancer preventative properties of ginger: a brief review. Food Chem Toxicol. 1992;45:683–690. doi: 10.1016/j.fct.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Surh Y. Anti-tumor promoting potential of selected spice ingredients with antioxidative and anti-inflammatory activities: a short review. Food Chem Toxicol. 2002;40:1091–1097. doi: 10.1016/s0278-6915(02)00037-6. [DOI] [PubMed] [Google Scholar]
- 5.Manju V, Nalini N. Chemopreventive efficacy of ginger, a naturally occurring anticarcinogen during the initiation, post-initiation stages of 1,2 dimethylhydrazineinduced colon cancer. Clin Chim Acta. 2005;358:60–67. doi: 10.1016/j.cccn.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Masada Y, IT, Hashimoto K, Fujioka M, Uchino C. Studies on the constituents of ginger (Zingiber officinale Roscoe) by GC-MS (author's transl) Yakugaku Zasshi. 1974;94:735–738. doi: 10.1248/yakushi1947.94.6_735. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, HT, Yang B, Liu X, Duan G. Development of gas chromatography-mass spectrometry with microwave distillation and simultaneous solid-phase microextraction for rapid determination of volatile constituents in ginger. J Pharm Biomed. 2007;43:24–31. doi: 10.1016/j.jpba.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Solyom AM, Timmermann BN, Gang DR. Characterization of gingerolrelated compounds in ginger rhizome (Zingiber officinale Rosc.) by high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2957–2964. doi: 10.1002/rcm.2140. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Xie Z, Koo HJ, McLaughlin SP, et al. Metabolic profiling and phylogenetic analysis of medicinal Zingiber species: Tools for authentication of ginger (Zingiber officinale Rosc) Phytochemistry. 2006;67:1673–1685. doi: 10.1016/j.phytochem.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Sang S, HJ, Wu H, Liu J, Yang CS, Pan MH, Badmaev V, Ho CT. Increased growth inhibitory effects on human cancer cells and anti-inflammatory potency of shogaols from Zingiber officinale relative to gingerols. J Agric Food Chem. 2009;57:10645–10650. doi: 10.1021/jf9027443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu R, Zhou P, Peng YB, Xu X, et al. 6-Shogaol induces apoptosis in human hepatocellular carcinoma cells and exhibits anti-tumor activity in vivo through endoplasmic reticulum stress. PLoS One. 2012;7:e39664. doi: 10.1371/journal.pone.0039664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng CJ, Chou CP, Ho CT, Yen GC. Molecular mechanism inhibiting human hepatocarcinoma cell invasion by 6-shogaol and 6-gingerol. Mol Nutr Food Res. 2012;56:1304–1314. doi: 10.1002/mnfr.201200173. [DOI] [PubMed] [Google Scholar]
- 13.Bak MJ, Ok S, Jun M, Jeong WS. 6-shogaol-rich extract from ginger up-regulates the antioxidant defense systems in cells and mice. Molecules. 2012;17:8037–8055. doi: 10.3390/molecules17078037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Lv L, Soroka D, Warin RF, et al. Metabolism of [6]-shogaol in mice and in cancer cells. Drug Metab Dispos. 2012;40:742–753. doi: 10.1124/dmd.111.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, LC, Wen XD, Li P, Qi LW. Simultaneous determination of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol in rat plasma by liquid chromatography-mass spectrometry. Application to pharmacokinetics. J Chromatgr B ANalyt Technol Biomed Life Sci. 2009;877:671–679. doi: 10.1016/j.jchromb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 16.Asami A, Shimada T, Mizuhara Y, Asano T, et al. Pharmacokinetics of [6]-shogaol, a pungent ingredient of Zingiber officinale Roscoe (Part I) J Nat Med. 2010;64:281–287. doi: 10.1007/s11418-010-0404-y. [DOI] [PubMed] [Google Scholar]
- 17.Iwabu J, Watanabe J, Hirakura K, Ozaki Y, et al. Profiling of the compounds absorbed in human plasma and urine after oral administration of a traditional Japanese (kampo) medicine, daikenchuto. Drug Metab Dispos. 2010;38:2040–2048. doi: 10.1124/dmd.110.033589. [DOI] [PubMed] [Google Scholar]
- 18.Zick SM, Ruffin MT, Djuric Z, Normolle D, et al. Quantitation of 6-, 8- and 10- Gingerols and 6-Shogaol in Human Plasma by High-Performance Liquid Chromatography with Electrochemical Detection. Int J Biomed Sci. 2010;6:233–240. [PMC free article] [PubMed] [Google Scholar]
- 19.Zick SM, Djuric Z, Ruffin MT, Litzinger AJ, et al. Pharmacokinetics of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol and conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prev. 2008;17:1930–1936. doi: 10.1158/1055-9965.EPI-07-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis DFV, LB Molecular modelling of members of hte P4502A subfamily: application to studies of enzyme specificity. Xenobiotica. 1995;25:585–598. doi: 10.3109/00498259509061877. [DOI] [PubMed] [Google Scholar]
- 21.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 22.Su P, Kaushal KM, Kroetz DL. Inhibition of renal arachidonic acid omegahydroxylase activity with ABT reduces blood pressure in the SHR. Am J Physiol. 1998;275:R426–R438. doi: 10.1152/ajpregu.1998.275.2.R426. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Abdelrahman DR, Fokina VM, Hankins GD, et al. Metabolism of bupropion by baboon hepatic and placental microsomes. Biochem Pharmacol. 2011;82:295–303. doi: 10.1016/j.bcp.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opperman U. Carbonyl reductases: the complex relationships of mammalian carbonyland quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- 26.Surh YJ, Lee SS. Enzymatic reduction of shogaol: a novel biotransformation pathway for the alpha,beta-unsaturated ketone system. Biochem Int. 1992;27:179–187. [PubMed] [Google Scholar]
- 27.Surh YJ, Lee SS. Enzymatic reduction of xenobiotic alpha,beta-unsaturated ketones: formation of allyl alcohol metabolites from shogaol and dehydroparadol. Res Commun Chem Pathol Pharmacol. 1994;84:53–61. [PubMed] [Google Scholar]
- 28.Surh YJ, Lee SS. Enzymic reduction of [6]-gingerol, a major pungent principle of ginger, in the cell-free preparation of rat liver. Life Sci. 1994;54:PL321–PL326. doi: 10.1016/0024-3205(94)00602-4. [DOI] [PubMed] [Google Scholar]
- 29.Itoh K, YK, Adachi M, Kosaka T, Tanaka Y. Leukotriene B4 12-hydroxydehydrogenase/15-ketoprostaglandin Δ13-reductase (LTB4 12-HD/PDR) responsible for the reduction of a double-bond of the α,β-unsaturated ketone of an aryl propionic acid non-steroidal anti-inflammatory CS-670. Xenobiotica. 2008;38:249–263. doi: 10.1080/00498250701767667. [DOI] [PubMed] [Google Scholar]
- 30.Martignoni M, GG, de Kanter R. Species differences between mouse, rat, dog, monkey, and human CYP-mediated drug metabolism, inhibition, and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 31.Dick RA, Kwak MK, Sutter TR, Kensler TW. Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J Biol Chem. 2001;276:40803–40810. doi: 10.1074/jbc.M105487200. [DOI] [PubMed] [Google Scholar]
- 32.Merendino N, Loppi B, D'Aquino M, Molinari R, et al. Docosahexaenoic acid induces apoptosis in the human PaCa-44 pancreatic cancer cell line by active reduced glutathione extrusion and lipid peroxidation. Nutr Cancer. 2005;52:225–233. doi: 10.1207/s15327914nc5202_12. [DOI] [PubMed] [Google Scholar]