Abstract
Centrosome duplication is critical for cell division, and genome instability can result if duplication is not restricted to a single round per cell cycle. Centrosome duplication is controlled in part by CP110, a centriolar protein that positively regulates centriole duplication while restricting centriole elongation and ciliogenesis. Maintenance of normal CP110 levels is essential, since excessive CP110 drives centrosome over-duplication and suppresses ciliogenesis, whereas its depletion inhibits centriole amplification and leads to highly elongated centrioles and aberrant assembly of cilia in growing cells1,2. CP110 levels are tightly controlled in part through SCFcyclin F-mediated ubiquitylation during G2 and M phase of the cell cycle3. Here we report a new mechanism for regulation of centrosome duplication that requires USP33, a de-ubiquitylating enzyme (DUB) able to regulate CP110 levels. USP33 interacts with CP110 and localizes to centrioles primarily during S and G2/M phase, the period during which centrioles duplicate and elongate. USP33 potently and specifically de-ubiquitylates CP110, but not other cyclin F substrates. USP33 activity antagonizes SCFcyclin F-mediated ubiquitylation and promotes generation of supernumerary centriolar foci, whereas ablation of USP33 destabilizes CP110 and thereby inhibits centrosome amplification and mitotic defects. To our knowledge, these studies have identified the first centriolar de-ubiquitinating enzyme whose expression regulates centrosome homeostasis by countering cyclin F-mediated destruction of a key substrate and suggest potential therapeutic strategies for inhibiting tumorigenesis associated with centrosome amplification.
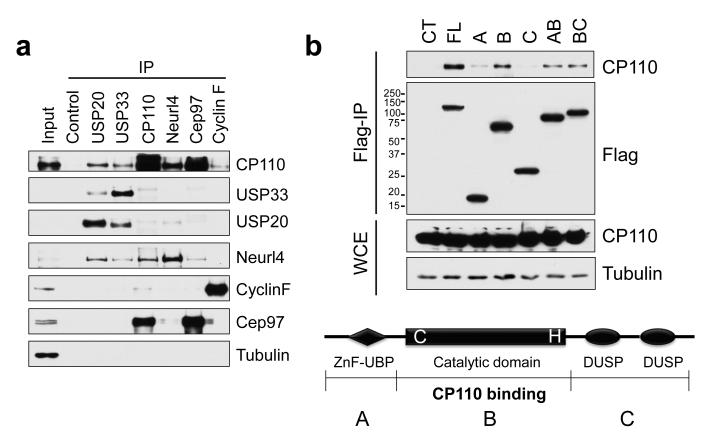
We have recently characterized a novel CP110-interacting protein, Neurl4, a daughter centriole-specific protein that inhibits formation of ectopic microtubule organizing centers4. During the course of these studies, Neurl4 was independently found to interact with USP20/VDU2 (VHL protein-interacting deubiquitinating enzyme 2) in a proteome-wide screen for DUB-associated proteins 5. USP20 is highly related to a second DUB, USP33/VDU1, which shares ~59% amino acid identity, explaining their overlapping functions 6,7. Thus, we sought to examine whether USP20 and USP33 interact with the CP110-Neurl4 complex to regulate centrosome function. We immunoprecipitated each protein from cell lysates and found that endogenous USP33 and USP20 specifically interact with CP110 and Neurl4, but not with two other CP110-interacting proteins, Cep97 and cyclin F (Fig. 1a). Moreover, these interactions were also observed in cells ectopically expressing Flag-CP110 or Flag-USP33 (Figs. 1b, S1a-e). The interactions with USP20/33 are most likely direct because (1) bacterially expressed GST-USP33 and USP20 bind to in vitro translated CP110 in binding assays (Fig. S1f) and (2) Neurl4 depletion does not affect the binding between USP33 and CP110 (Fig. S1g). We found that endogenous USP33 and CP110 co-fractionate in at least three distinct protein complexes ranging from 0.4-1.3 MDa. This suggests the existence of larger CP110-USP33 deubiquitylase complexes that include proteins other than Neurl4 (Fig. S2). Both USP33 and USP20 share three functional domains, a zinc-finger ubiquitin binding motif (ZnF-UBP), a catalytic domain, and two domains present in ubiquitin-specific proteases (DUSP). We investigated the interaction between USP33 and CP110 and found that CP110 binds to a region of USP33 within its catalytic domain (Fig. 1b).
Figure 1. Identification of USP33 as a CP110-associated protein.
(a) HEK293T cell extracts were immunoprecipitated with the indicated antibodies (top), and western blots were probed as shown (right). Non-specific rabbit IgG was used as control. α-tubulin is shown as a negative control. (b) (Bottom) Domains of USP33 are indicated. The catalytic domain of USP33 stably binds to CP110. Double cysteine and histidine point mutations in the catalytic domain (DM mutant) render the DUB inactive. (Top) 293T cells were transfected with HA-tagged CP110 and Flag-tagged full-length or truncated fragments of USP33, extracts were immunoprecipitated, and western blots of the resulting immunoprecipitates were probed with anti-Flag and CP110 antibodies. WCE, whole cell extract.
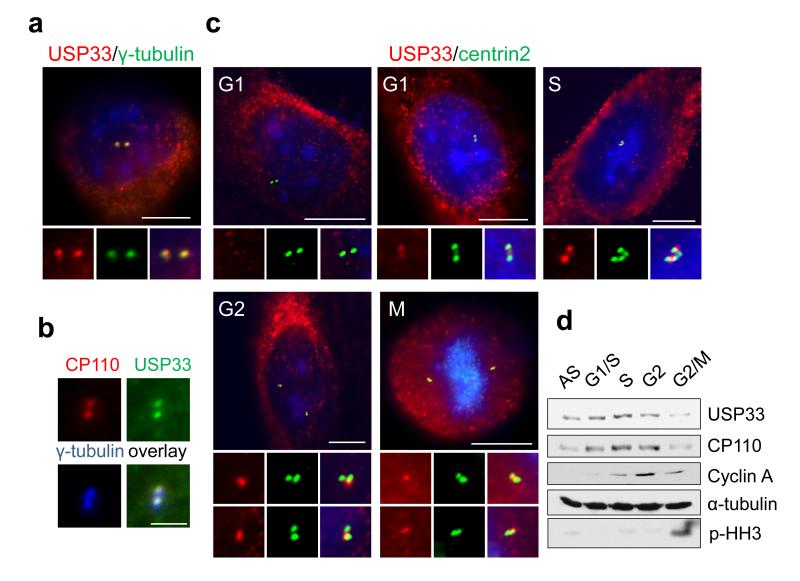
USP33 and USP20 have been shown to partition to the cytoplasm, including the endoplasmic reticulum8, and a short isoform of USP33 localizes to the Golgi apparatus9. Since CP110 is a centrosomal protein, we visualized endogenous USP33, USP20, CP110, and other centrosomal markers in human osteosarcoma (U2OS) cells by indirect immunofluorescence. We observed USP33 localization to the cytoplasm, as expected, and in addition, we found that USP33 co-localized with centrin-2 and γ-tubulin, centrosomal markers (Fig. 2a,b and S3a,b). Notably, USP33 partially co-localized with CP110 at centrioles. USP33 localization appeared more proximal than CP110, suggesting that the former protein interacts with a sub-population of CP110. A similar localization pattern was observed for USP20 (not shown). The centrosomal localization of USP33 is specific because it disappeared after USP33 was silenced with siRNAs (Fig. S3c). Since CP110 levels are high during S phase and low during G2/M due to cyclin F-mediated ubiquitylation, we sought to determine the levels of USP33 in synchronized U2OS cells. USP33 associated with centrosomes predominantly in S and G2 phases of the cell cycle but less prominently in G1 phase, mirroring the abundance of CP110 protein during the cell cycle (Fig. 2c and S3d)1,3. Strikingly, USP33 protein levels also oscillated in parallel during cell cycle progression, exhibiting elevated expression in S and early G2 phase and lower levels in late G2 and M phases (Fig. 2d, 3b and S3e). We found that centrosomal targeting of USP33 is partially dependent on CP110 because depletion of CP110 reduced USP33 localization to centrosomes (see below) (Fig. S3f).
Figure 2. USP33 localizes to centrosomes primarily during S and G2/M phase of the cell cycle.
(a-b) Co-localization of USP33 with CP110 and other centrosome and centriolar markers was visualized by immunofluorescence, as indicated. (c) USP33 (red) and CP110 (green) were visualized throughout the cell cycle as in panels a-b. DNA was stained with DAPI (blue). U2OS cells were treated with mimosine for G1 phase cells or were subject to double thymidine block and release to enrich for S and G2 phase cells. Cells with 2 centrin-2 dots were counted as G1 phase cells; those with 3-4 clustered centrin-2 dots as S phase cells; and those with 3-4 centrin-2 dots, in which two pairs of dots were at least 3μm apart, were counted as G2 phase cells. Localization of USP33 to centrosomes was most prominent in S and G2/M phases. (d) USP33 protein levels parallel those of CP110 during the cell cycle. Cells were synchronized by double-thymidine block and release and synchronization confirmed by FACS analysis (see Fig.S3e). Scale bar in a, c : 10μm. Scale bar in b: 2 μm.
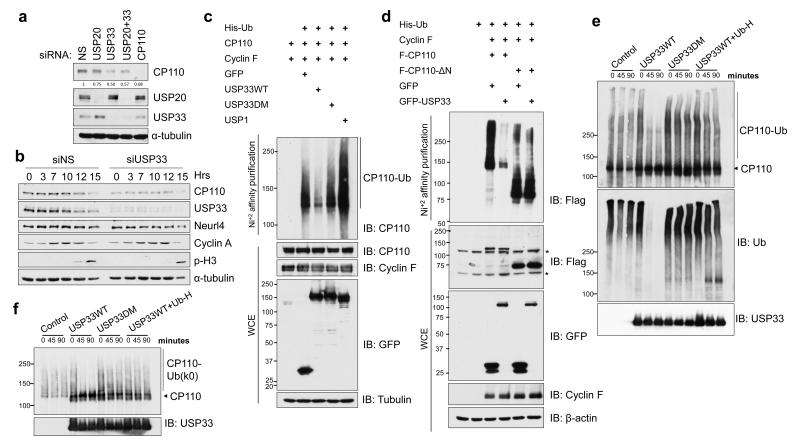
Figure 3. Recruitment of USP33 leads to de-ubiquitylation and stabilization of CP110.
(a) CP110 levels were monitored after silencing of USP20 and USP33 by transfection of siRNAs into HeLa cells. NS, non-specific siRNA control. (b) Cells were transfected with the indicated siRNAs and synchronized by double-thymidine block and release. Lysates of cells from indicated time points (hours) after release were probed with antibodies against CP110 and an interacting protein (Neurl4), as well as cell cycle markers (cyclin A, phospho-histone H3 (S10). (c) Enforced expression of GFP-tagged, wild-type USP33 (WT), but not a catalytically inactive version (DM), drives deubiquitylation of CP110. The indicated plasmids (top) were transfected into cells, the resulting tagged ubiquitylated proteins were enriched by nickel-NTA chelation chromatography, and western blots were probed with antibodies against CP110. An unrelated DUB (USP1) expressed as a control had no effect on CP110 ubiquitylation. (d) De-ubiquitylation by USP33 requires selective binding to an amino-terminal portion of CP110. 293T cells were transfected with indicated GFP- and Flag-tagged (F) plasmids and processed as in (c). * Denotes non-specific bands. (e, f) In vitro de-ubiquitylation reactions were performed using (e) wild-type Ubiquitin (Ub) or (f) a mutant form of ubiquitin (Ub-k0) in which all lysines have been mutated, and analyzed by western blotting with the indicated antibodies. The ubiquitin mutant (Ub-K0) in (g) lacks the ability to form poly-ubiquitin chains and can only be conjugated to substrate as monoubiquitin form. In vitro de-ubiquitylation of CP110 was blocked by simultaneous incubation with Ubiquitin-aldehyde (UB-H) as indicated.
DUBs are able to remove ubiquitin from proteins and regulate substrate abundance or function5. Since USP33 and USP20 interact with CP110 at centrosomes, and CP110 protein levels correlated with USP33 and USP20 during the cell cycle, we investigated whether USP33 or USP20 might stabilize CP110. Interestingly, depletion of USP33 with siRNAs reduced CP110 levels by ~50%, whereas depletion of USP20 had a very modest impact on CP110 levels (Fig. 3a), and co-depletion of USP20 with USP33 did not accentuate the impact of USP33 silencing. These results indicate that USP33 may have a more prominent role than USP20 in regulating CP110 stability, and in the remainder of this study, we focused on the functional role of USP33. Importantly, cells treated with two different siRNAs targeting USP33 exhibited reduced levels of CP110, but not other CP110-interacting proteins, including Neurl4 and cyclin F (Fig. S4a). Moreover, USP33 depletion led to decreased CP110 intensity at centrioles in both U2OS and hTERT-immortalized retinal pigment epithelial cells (Fig. S4b and c). USP33 also stabilized CP110 in cells enriched in S and G2 phases (Fig. 3b, S4e and f). Further, USP33 suppression significantly reduced CP110 protein half-life from ~4.5 hours to ~2 hours (Fig. S4g and h). Since CP110 is a substrate of SCFCyclin F ubiquitin ligase complexes3, we examined the impact of USP33 on another SCFCyclin F substrate, RRM210. We observed that USP33 depletion did not lead to a reduction in RRM2 levels (Fig. S4e and f), indicating that USP33 specifically stabilizes CP110.
Next, we examined whether stabilization of CP110 by USP33 is mediated through deubiquitylation, since USP33 acts as a DUB. Remarkably, USP33 over-expression diminished CP110 ubiquitylation provoked by ectopic production of cyclin F in cells (Fig. 3c). CP110 de-ubiquitylation was markedly attenuated when a catalytically inactive USP33 mutant (USP33-DM), in which cysteine 163 in the active site was replaced with a serine (C163S) and histidine 683 was modified to glutamine (H642Q), was transfected11. In addition, over-expression of two unrelated DUBs, USP1 and USP37, did not promote CP110 de-ubiquitylation (Fig. 3c and data not shown). To test whether de-ubiquitylation by USP33 requires CP110 binding, we first mapped the region of CP110 that binds USP33 and found that the binding domain was located within the first 200 amino-terminal residues (Fig. S1d). Strikingly, when we tested a truncation mutant unable to bind USP33 (CP110 ΔN), we found that USP33 was unable to de-ubiquitylate this substrate (Fig. 3d). In addition, we tested the activity of USP33 in an in vitro deubiquitylation assay. We purified Flag-tagged USP33 and CP110 and the SCFCyclin F complex from insect cells infected with baculoviruses10 (Fig. S5a and b). CP110 was ubiquitylated in vitro by SCFCyclinF complex specifically via a K48 ubiquitin linkage (not shown). Ubiquitylated CP110 was de-ubiquitylated by wild-type USP33 protein (USP33-WT), but not by a catalytically inactive mutant (USP33-DM) (Fig. 3e and f). Moreover, USP33 activity was inhibited by ubiquitin-aldehyde, a DUB inhibitor. Thus, USP33 specifically de-ubiquitylates CP110 in vitro and in vivo, explaining how USP33 stabilizes CP110.
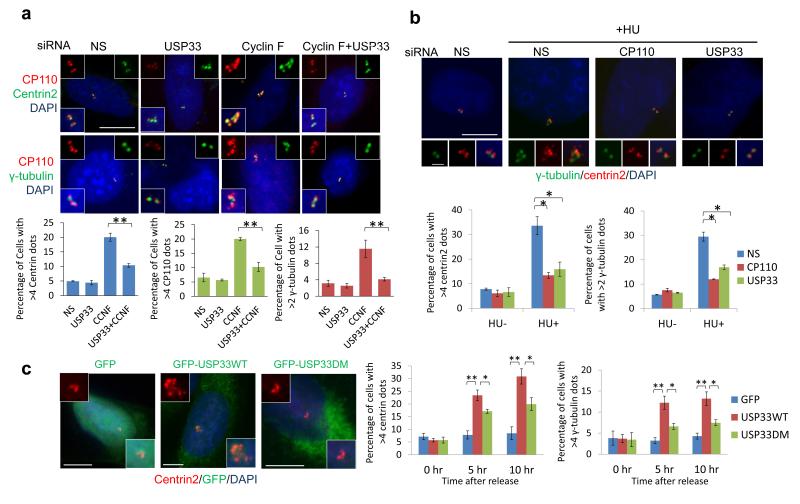
To study the biological significance of USP33-mediated stabilization of CP110, we investigated whether USP33 depletion affects centrosome duplication. It has been shown that cyclin F ablation leads to CP110 up-regulation, which markedly induces centrosome amplification3. Indeed, cyclin F knock-down induced centrosome amplification, visualized by the acquisition of >4 centrin-2 or CP110 foci and >2 γ-tubulin foci, whereas co-depletion of USP33 with cyclin F reversed this phenotype in U2OS cells (Fig. 4a and S6a). This phenotype was also observed in another cell line (HeLa) and was similarly reversed by treatment with distinct USP33 siRNAs (Fig. S6b). The centrosome phenotype provoked by USP33 silencing was not due to altered cell cycle progression (Fig. S6c). Further, we investigated whether USP33 depletion could affect centrosome reduplication in cells treated with hydroxyurea to induce prolonged S-phase. Similar to CP110 ablation2, depletion of USP33 significantly inhibited hydroxyurea (HU)-induced centrosome amplification in two cell lines (Fig. 4b and S6d). Thus, although USP33 depletion did not affect normal centriole duplication (Fig. 4a), silencing of USP33 specifically inhibited centrosome over-duplication, reminiscent of the CP110 depletion phenotype. Importantly, USP33 co-depletion rescued the mitotic defects observed in cyclin F-depleted cells: whereas amplified centrosomes resulting from cyclin F depletion led to multipolar and clustered bipolar (pseudo-bipolar) asymmetric spindles, co-silencing of USP33 and cyclin F reverted the frequency of mitotic aberrations to control levels (Fig. S6e). These results suggest that destabilization of CP110 through USP33 silencing can inhibit mitotic errors wrought by centrosomal defects.
Figure 4. Depletion of USP33 inhibits centrosome amplification and antagonizes cyclin F function.
(a) Following cyclin F knock-down, cells express small, but measurable, amounts of residual cyclin F, whose activity may be largely counteracted by USP33. (Left) Silencing of USP33 reverses the over-duplication of centrosomes observed upon cyclin F knock-down. Immunofluorescence detection of centrin (left, green), CP110 (red), and γ-tubulin (right, green) in U2OS cells depleted of USP33, cyclin F, or both USP33 and cyclin F. (Right) Quantitation of results in left panels. Scale bar, 10 μm. Error bar indicates SEM. ** p<0.01 (n=3). (b) (Left) Cells were depleted of USP33 and treated with HU. USP33 depletion suppresses HU-mediated centrosome amplification in U2OS cells. (Right) Quantitation of centrin-2 and γ-tubulin foci at left. Error bars indicate SEM. * p<0.05 (n=3). (c) Cells were transfected with the indicated plasmids and synchronized with double-thymidine block and release. GFP and centrin-2 fluorescence was visualized (left), and populations of cells with the indicated number of centrin-2 and γ-tubulin dots were counted (right). Scale bar, 10 μm. Error bars indicate SEM. * p<0.05; ** p<0.01 (n=3).
Next, to further investigate the role of USP33 in regulating centrosome duplication, we ectopically expressed USP33 in U2OS cells synchronized by double-thymidine block and release. We found that expression of GFP-tagged wild-type USP33 significantly increased the percentage of cells with amplified centrin-2, CP110, and γ-tubulin foci in comparison with the GFP control and USP33-DM mutant, and correlative light-electron microscopy confirmed the presence of bona fide, ectopic centrioles and electron-dense structures, likely to be centriolar precursors or intermediates, in a subset of cells expressing GFP-USP33 (Figs. 4c, S7a-f). Interestingly, we observed that the USP33-DM mutant also increased the percentage of cells with amplified centrioles. This may be due to either residual activity of the mutant in vivo, additional, non-catalytic roles of USP33, or a combination of both. Alternatively, excessive amounts of USP33-DM might bind CP110, blocking it from accessing SCFCyclin F. USP33 promotes the generation of excessive centriolar foci due to elevated levels of CP110 because (1) over-expression of human or murine USP33 led to CP110 up-regulation in multiple cell lines (Fig. S7c and d) and (2) depletion of CP110 overrode USP33-mediated generation of centriolar foci (Fig. S7 g-i). In sum, elevated levels of USP33 positively regulate centrosome duplication by augmenting CP110 levels, and loss of USP33 can prevent centrosome amplification and mitotic defects.
Centrosome amplification is frequently detected in human cancers, and it has been implicated as a contributing factor during cellular transformation12,13. Interestingly, a search of an integrated cancer microarray database (Oncomine) revealed that USP33 gene expression is significantly up-regulated in pancreatic cancer14,15 (Fig. S9a). This was of particular interest because centrosome amplification can be detected in primary pancreatic ductal carcinomas (PDAC)16,17 and because a mouse model suggested that supernumerary centrosomes can promote chromosomal aberrations and pancreatic cancer progression18. Therefore, we examined USP33 and CP110 levels in human pancreatic cell lines and tissue samples. Remarkably, USP33 and CP110 were both substantially up-regulated in cultured PDAC cell lines as compared to normal pancreatic epithelial cells, and USP33 and CP110 levels were strongly correlated (Fig. S8a). Further, depletion of USP33 in two PDAC cell lines (Panc1 and MiaPaCa2) led to a reduction in CP110 levels (Fig. S4d and S6d), indicating that USP33 stabilizes CP110 in PDAC cells. Notably, CP110 and USP33 levels also correlated with centrosome amplification in pancreatic cells (Fig. S8a and b), and we found that USP33 and CP110 depletion inhibited prolonged S-phase centrosome amplification in MiaPaCa2 cells (Fig. S6d).
Next, we examined USP33 and CP110 protein levels using human tissue arrays that included PDAC and histologically normal pancreatic samples. Two separate arrays were scored using a 4-tier grading system. Strikingly, we found that both USP33 and CP110 were highly expressed in PDAC but not in normal pancreas, and the levels of USP33 and CP110 were significantly correlated in the same tissue samples (Fig. S9b). In addition, we also visualized CP110, acetylated tubulin, γ-tubulin, and cytokeratin 19, a marker of pancreatic ducts and PDAC cells, using a tissue microarray containing 30 additional, independent cases of PDAC. In histologically normal ducts, a normal pattern of centriolar CP110 staining and ciliary acetylated tubulin staining was most frequently encountered (Fig. S9c). In contrast, CP110 over-expression was observed in PDAC cells in 29 of 30 cases. Our results suggest that the abundance of centrosome regulators, such as USP33 and CP110, may be mis-regulated in human tumors that are associated with centrosomal defects, although additional studies will be needed to elucidate the temporal connections between de-regulated expression of these proteins, centrosome aberrations, and PDAC pathogenesis.
We have for the first time identified USP33 as a centrosomal DUB that stabilizes CP110 at centrioles through de-ubiquitylation. USP33 positively regulates centrosome duplication by regulating CP110 abundance. Together with the finding that SCFcyclin F promotes the ubiquitylation and degradation of CP1103, our study has uncovered an underlying cycle in which the equilibrium between ubiquitylation and de-ubiquitylation elaborately governs levels of a critical centrosome protein during the cell cycle, thereby ensuring centrosome homeostasis. Disruption of either the de-ubiquitylating or ubiquitylating enzymes could upset this equilibrium and lead to centrosome aberrations. Ubiquitylating and de-ubiquitylating enzymes have been the subject of intense scrutiny in cancer research, and inhibitors of the enzymes that promote or remove ubiquitin, including USP33, are targets for drug development12,19,20. Since many human tumors are associated with centrosome amplification and genomic instability, our findings suggest the possibility of therapeutic intervention in cancers with this hallmark.
METHODS
Cell culture and cell cycle synchronization
HeLa, U2OS, RPE1-hTERT, HEK-293T, COS-1 and COS-7 were obtained from ATCC and maintained according to its instruction. Pancreatic cell lines were kindly provided by Jonathan Pollack (Stanford University, Department of Pathology). Early passages of cells were used and they were not constitutively cultured for more than two months. For cell cycle synchronization, HeLa or U2OS cells were cultured in the presence of 2 mM thymidine (Sigma) for 16 h, washed twice with medium, and released into fresh medium without thymidine for 8 h. After another 16 h block in thymidine, cells were washed twice, released to fresh medium, and collected at indicated time points (5 hours, S phase; 10 hours, G2 phase). Nocodazole (100 ng/ml) was added 5h after release from the thymidine block to block cells in pro-metaphase.
Transient transfections
siRNAs were transfected into HeLa, U2OS, or RPE-1 cells using siIMPORTER (Millipore), HiPerFect (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plasmids were transfected into HeLa and U2OS cells using FuGene6 (Roche) or Lipofectamine 2000 (invitrogen) according to the manufacturer’s instructions. HEK-293T, COS-1 and COS-7 cells were transfected using the calcium phosphate method, as described1-3.
Antibodies
Rabbit polyclonal antibodies against CP1101, Neurl44, and Cep972 have been described. IHC staining of CP110 was preformed with CP110 antibody from Proteintech (12780-1-AP). Antibodies against USP33 (A300-925A; Bethyl Laboratories; 1:100 dilution for IF), USP20 (A301-189A; Bethyl Laboratories), Centrin 2 (20H5; millipore), Cyclin F (C-20; Santa Cruz Biotechnology), FLAG (M2; Sigma), α-tubulin (T5168; Sigma), γ-tubulin (mouse, T5326; Sigma), γ-tubulin (rabbit, T5192; Sigma), β-actin (AC-15; abcam), ubiquitin (P4D1, Enzo life sciences and FK2, Biomol International), GFP (rabbit, G1544; Sigma), GFP (mouse, Roche), Cyclin A (C-19; Santa Cruz Biotechnology), Cep290 (A301-659A; Bethyl Laboratories), phosphorylated Histone H3 (phospho-Ser10; Millipore), phosphorylated Cdc2 (phospho-Tyr15; Santa Cruz Biotechnology) were also used.
siRNA sequence
The 21-nucleotide siRNA sequence for the non-specific control was AATTCTCCGAACGTGTCACGT 2. The sequences for USP33 oligonucleotides 1 and 2 were GAUCAUGUGGCGAAGCAUAdTdT and GGCUUGGAUCUUCAGCCAUdTdT 11, respectively. The siRNA for CP110 was AAGCAGCATGAGTATGCCAGT 2. The siRNA sequence for USP20 was GGACAAUGAUGCUCACCUAUU 11. The siRNA sequence for Neurl4 was CCAUCAUGCAAGACGGUAAUU 4.
In vivo deubiquitylation assay
The methods to detect ubiquitylated protein were modified from W. Tansey21. HEK293 cells were transiently transfected with pcB6-His Ubiquitin (gift from Richard Baer), HA- or Flag- tagged CP110, cherry-cyclin F, GFP, GFP-USP33 wild type, GFP-USP33 catalytic mutant, or GFP-USP1 (a kind gift from T. Huang) as indicated. After 36 h, cells were treated with 10 μm MG132 (Peptides International) for 4 h, cells were lysed and briefly sonicated in Buffer A (100 mM NaH2PO4, 10 mM Tris-Cl, 6M guanidine-HCl, 10 mM imidazole pH 8.0). The cell lysates were incubated with nickel-NTA resin (Qiagen) to precipitate His-tagged ubiquitylated proteins for three hours at room temperature. Ni-NTA beads were subjected to two washes with Buffer A, two washes with Buffer A/TI (1 volume Buffer A and 1 volume TI Buffer) (TI buffer, 25 mM Tris-Cl pH6.8 and 20 mM imidazole), and then one wash with TI Buffer. Lastly, Ni-NTA beads were eluted in SDS loading buffer containing 200 mM imidazole, separated by SDS-PAGE, and detected by immunoblotting.
In vitro deubiquitylation assay
Flag-tagged USP33 and CP110 and the SCFCyclin F complex (comprised of GST-Skp1, His-tagged Cul1, Roc1, and His-tagged cyclin F) were individually affinity purified using anti-Flag (CP110 and USP33) or glutathioneagarose (SCF Cyclin F) chromatography from extracts of baculovirus-infected insect cells10. CP110 was ubiquitylated in vitro by purified SCFCyclinF complex in ubiquitylation reaction buffer (50 mM Tris-HCl pH 7.6, 5 mM MgCl2, 1 μM Okadaic acid and 0.6 mM DTT) containing 2 mM ATP, 0.1 μM E1 (Boston Biochem),10 ng/μl Ubch3, 10 μg/ml Ubch5c, and 2.5 mg/ml ubiquitin wild type or ubiquitin-k0 (Boston Biochem). Ubiquitylated CP110 was then diluted 10-fold with deubiquitylation reaction buffer (20mM Hepes pH7.6, 50mM NaCl, 1mM EDTA, 0.2 μg/μl BSA, 1mM DTT, 100 μM MG132, 1μM AEBSF, 2μg/ml Aprotinin). Diluted ubiquitylated CP110 was incubated with 10ng/μl USP33 protein at 30°C for indicated times (0-1.5 hours). For the reactions with ubiquitin-aldehyde, 8 μM ubiquitin-aldehyde (Boston Biochem) was added. Samples were separated by SDS-PAGE, and detected by immunoblotting.
Correlative Light-Electron Microscopy
U2OS cells were plated at low confluence on gridded glass bottom dishes (MatTek) and transfected with GFP-USP33/RFP-centrin-2 or GFP along/RFP-centrin-2 using FuGene 6. After 48 hours, cells were examined by live-cell imaging to monitor green and red fluorescence and to obtain phase contrast images. Cells with both GFP and RFP expression were documented, and their relative positions on the grids were recorded. Cells were then fixed with 0.1M sodium cacodylate buffer (pH7.4) supplemented with 2.5% glutaraldehyde, and 1mM CaCl2, and then processed in a standard manner for transmission electron microscopy, as was described4.
Tissue Microarray
Pancreatic tissue arrays were purchased from US Biomax (PA1001 and PA207). IHC staining was performed as described22 and scored by two independent pathologists using a four-tier grading system (0=negative, 1=weak, 2=moderate and 3=strong staining intensity). Immunofluorescent staining was performed as described23.
Statistical analyses
All data represent the average from at least three independent experiments with at least 100 cells counted per experiment unless otherwise indicated. Significance was calculated by two-tail student t-test or ANOVA (Fig. S6e) using GraphPad software. Differences were considered significant when p was less than 0.05.
Supplementary Material
Acknowledgements
We thank C. Hajdu, G. Miller, L. Chiriboga, H. Court, C. Zambirinis, E. Bekes, L. Taylor, S. Krauter, M. Philips, and D. Bar-Sagi for generous help with staining and analysis of pancreatic cancer samples. We thank F. Liang, C. Petzold, and K. Dancel for help with correlative EM. We thank J. Jastrab and S. Vijayakumar for initial experiments pertaining to USP20/CP110 interaction. We thank J. Pollack (Stanford University, Department of Pathology) for providing pancreatic cell lines. We thank G. Wu, S. Shenoy, R. Baer, and T. Huang for providing plasmids. We thank members of the Dynlacht lab and T. Huang for helpful advice. B.D.D. was supported by NIH grant 5R01HD069647-02 and March of Dimes grant FY11-432, and M.P. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.L., V.D., S.K., T.K., E.C., M.P. and B.D. performed all biochemical experiments and analyses of deubiquitylation and centrosome phenotypes. E.S., W.F., and J.L. performed pancreatic tissue studies. J. L. and B.D. wrote the manuscript. B.D. coordinated the study and oversaw all experiments. All authors discussed the results and commented on and assisted in revising the manuscript.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 2.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. doi:10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 3.D’Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. doi:10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, et al. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO reports. 2012 doi: 10.1038/embor.2012.40. doi:10.1038/embor.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. doi:10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, et al. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. doi:10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, et al. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. doi:10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 8.Curcio-Morelli C, et al. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Invest. 2003;112:189–196. doi: 10.1172/JCI18348. doi:10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorne C, Eccles RL, Coulson JM, Urbe S, Clague MJ. Isoform-specific localization of the deubiquitinase USP33 to the Golgi apparatus. Traffic. 2011;12:1563–1574. doi: 10.1111/j.1600-0854.2011.01261.x. doi:10.1111/j.1600-0854.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 10.D’Angiolella V, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. doi:10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthouze M, Venkataramanan V, Li Y, Shenoy SK. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. Embo J. 2009;28:1684–1696. doi: 10.1038/emboj.2009.128. doi:10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7:1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. doi:10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchholz M, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. doi:10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 15.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 16.Sato N, et al. Centrosome abnormalities in pancreatic ductal carcinoma. Clin Cancer Res. 1999;5:963–970. [PubMed] [Google Scholar]
- 17.Sato N, et al. Correlation between centrosome abnormalities and chromosomal instability in human pancreatic cancer cells. Cancer Genet Cytogenet. 2001;126:13–19. doi: 10.1016/s0165-4608(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 18.Shono M, et al. Stepwise progression of centrosome defects associated with local tumor growth and metastatic process of human pancreatic carcinoma cells transplanted orthotopically into nude mice. Laboratory investigation; a journal of technical methods and pathology. 2001;81:945–952. doi: 10.1038/labinvest.3780306. [DOI] [PubMed] [Google Scholar]
- 19.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. doi:10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Sippl W, Collura V, Colland F. Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 2011;7:619–632. doi: 10.2217/fon.11.39. doi:10.2217/fon.11.39. [DOI] [PubMed] [Google Scholar]
- 21.Tansey WP. Detection of Ubiquitylated Proteins in Mammalian Cells. Cold Spring Harbor Protocols. 2006;2006 doi: 10.1101/pdb.prot4616. pdb.prot4616, doi:10.1101/pdb.prot4616. [DOI] [PubMed] [Google Scholar]
- 22.Sabbaghian MS, et al. Levels of elevated circulating endothelial cell decline after tumor resection in patients with pancreatic ductal adenocarcinoma. Anticancer research. 2010;30:2911–2917. [PubMed] [Google Scholar]
- 23.Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer research. 2009;69:422–430. doi: 10.1158/0008-5472.CAN-08-1290. doi:10.1158/0008-5472.CAN-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.