Abstract
DNA double-strand breaks (DSBs) represent a threat to the genome because they can lead to loss of genetic information and chromosome rearrangements. The DNA repair protein p53 binding protein 1 (53BP1) protects the genome by limiting nucleolytic processing of DSBs by a mechanism that requires its phosphorylation, but whether it does so directly is not known. Here we identify Rapl-interacting factor 1 (Rif1) as an Ataxia-Telangiectasia Mutated (ATM) phosphorylation-dependent interactor of 53BP1, and show that absence of Rif1 results in 5′-3′ DNA end resection in mice. Consistent with enhanced DNA resection, Rif1 deficiency impairs DNA repair in the G1 and S phases of the cell cycle, interferes with class switch recombination (CSR) in B lymphocytes, and leads to accumulation of chromosome DSBs.
53BP1 is a multi-domain DNA damage response factor containing a chromatin-binding tudor domain, an oligomerization domain, tandem BRCA1 C-terminal (BRCT) domains, and an N-terminal domain with 28 SQ/TQ potential phosphorylation sites for phosphatidylinositol 3-kinase-related kinases (PIKKs, ATM/ATM and Rad3-related (ATR)/DNA-dependent protein kinase catalytic subunit (DNA-PKcs)) (1–3). 53BP1 contributes to DNA repair in several ways: it facilitates joining between intrachromosomal DSBs at a distance (synapsis) (4–7); it enables heterochromatic DNA repair through relaxation of nucleosome compaction (2, 3), and it protects DNA ends from resection and thereby favors repair of DSBs that occur in G1 by non-homologous end joining (NHEJ) (4, 5, 8). Consistent with its role in DNA end protection, 53BP1 is essential for CSR in B lymphocytes (9, 10).
Structure-function studies indicate that, besides its recruitment to DNA ends, protection requires 53BP1 phosphorylation (4), but how this protective effect is mediated is unknown. To identify phosphorylation-dependent interactors of 53BP1, we applied SILAC (Stable Isotope Labeling by Amino acids in Cell culture). Trp53bp1−/− (Trp53bp1 encodes 53BP1) B cells were infected with retroviruses encoding a C-terminal deleted version of 53BP1 (53BP1DB) or a phosphomutant in which all 28 N-terminal potential PIKK phosphorylation sites were mutated to alanine (53BP1DB28A) (4), in media containing isotopically heavy (53BP1DB) or light (53BP1DB28A) lysine and arginine (Fig. S1, A-C; (11)).
Most proteins co-precipitating with 53BP1DB and 53BP1DB28A displayed a H/(H+L) ratio of ~0.5, which is characteristic of phospho-independent association (average of 0.57 ± 0.09, peptide count ≥ 4; Fig. 1 and Table S1). Many of these proteins are non-specific contaminants, but others such as KRAB-associated protein 1 (KAP-1), dynein light chain LC8-type 1 (Dynll1), Nijmegen breakage syndrome 1 (Nbs1), and H2AX, represent authentic phospho-independent 53BP1 interacting proteins (Fig. S1D). Three proteins displayed an abundance ratio that was more than four standard deviations above the mean indicating that they interact specifically with phosphorylated 53BP1: Pax interaction with transcription-activation domain protein-1 (Paxip1, or PTIP; 0.95), PTIP associated protein 1 (Pa1; 0.97), and Rif1 (0.96; Fig. 1, S1D, and S2). PTIP was known to interact with 53BP1 in a phospho-dependent manner (12) whereas Pa1 and Rif1 were not.
Fig. 1. Identification of phospho-dependent 53BP1 interactors.
Graph shows the H/(H+L) ratio distribution of proteins identified by SILAC. Error bars represent the standard deviation of the H/(H+L) mean value for all the peptides identified for each individual protein (only proteins with 4 peptides were included). “H/(H+L)” and “σ” are the mean (0.57) and SD (0.09) of the distribution, respectively. H: heavy; L: light.
Rif1 was originally identified in budding yeast as a protein with a key role in telomere length maintenance (13). However, in mammalian cells, Rif1 is not essential for telomere homeostasis, but has been assigned a number of different roles in maintaining genome stability including participation in the DNA damage response (14–16), repair of S-phase DNA damage (17, 18), and regulation of origin firing during DNA replication (19, 20). However, the mechanism by which Rif1 might contribute to DNA repair and maintaining genome stability is not known.
To confirm that Rif1 interaction with 53BP1 is phosphorylation dependent, we performed Western blot analysis of Flag-immunoprecipitates from lysates of irradiated Trp53bp1−/−B cells infected with retroviruses encoding 53BP1DB or 53BP1DB28A. Whereas Dynll1, a phospho-independent 53BP1 interactor (SILAC ratio: 0.55; Fig. S1D), co-immunoprecipitated with 53BP1DB and 53BP1DB28A to a similar extent (Fig. 2A), only 53BP1DB co-immunoprecipitated with Rif1. We conclude that the interaction between 53BP1 and Rif1 is dependent on phosphorylation of 53BP1.
Fig. 2. Rif1 interaction with 53BP1 is phospho-, damage- and ATM-dependent.
(A) Western blot analysis of anti-Flag immuno-precipitates from irradiated Trp53bp1−/− B lymphocytes infected with empty vector (vec), 53BP1DB, or 53BP1DB28A virus. Triangles indicate 3-fold dilution. Data are representative of two independent experiments. (B) Western blot analysis of anti-Flag immunoprecipitates from Trp53bp1−/− B cells infected with empty vector or 53BP1DB. Cells were either left untreated or irradiated (50 Gy, 45 min recovery) in the presence or absence of the ATM kinase inhibitor KU55933 (ATMi). Triangles indicate 3-fold dilution. Data are representative of two independent experiments. (C) Immunofluorescent staining for 53BP1 (Flag) and Rif1 in irradiated Trp53bp1−/− iMEFs reconstituted with 53BP1DB or 53BP1DB28A retroviruses (4). Magnification, 100X; Scale bars, 5 μm. Data are representative of two independent experiments.
ATM phosphorylates 53BP1 in response to irradiation-induced DSBs (1, 3). To determine whether ATM induces irradiation-dependent association between Rif1 and 53BP1, we compared irradiated and non-irradiated B cells in co-immunoprecipitation experiments. Although small amounts of Rif1 were detected in 53BP1DB immunoprecipitates from unirradiated cells, this was increased by a factor higher than 3 after irradiation, and the increase was abrogated by treatment with the ATM inhibitor KU55933 (Fig. 2B). We conclude that Rif1 preferentially interacts with phosphorylated 53BP1 in a DNA damage- and ATM-dependent manner.
Rif1 is recruited to DNA damage foci by 53BP1 (15). To determine whether 53BP1 phosphorylation is required for Rif1 focus formation, we tested Rif1 foci in irradiated Trp53bp1−/− immortalized mouse embryonic fibroblasts (iMEFs), which were stably transduced with either 53BP1DB or 53BP1DB28A. Rif1 foci were readily detected and co-localized with 53BP1DB (Fig. 2C). In contrast, although 53BP1DB28A formed normal appearing foci, there were only rare Rif1 foci that did not co-localize with 53BP1DB28A (Fig. 2C). Furthermore, Rif1 recruitment to ionizing radiation-induced foci (IRIF) and co-localization with 53BP1 was abrogated in ATM-deficient but not DNA-PKcs-deficient iMEFs (Fig. S3 and (15)). We conclude that Rif1 recruitment to DNA damage response foci is dependent on ATM-mediated 53BP1 phosphorylation.
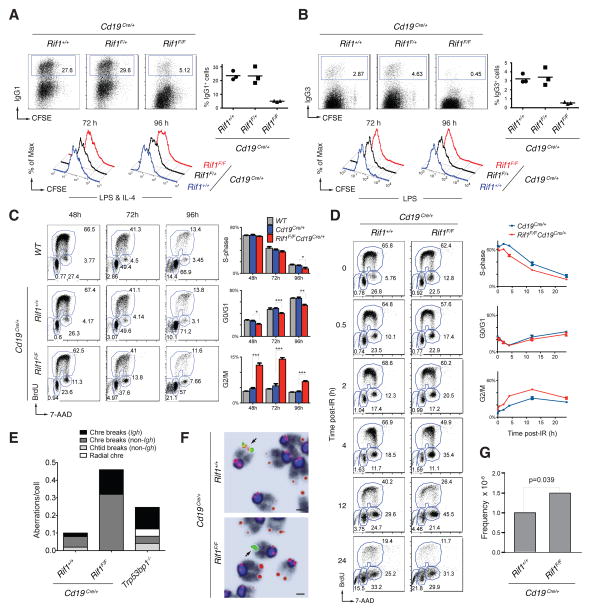
53BP1 phosphorylation is essential for CSR (4). To examine the role of Rif1 in joining DSBs during CSR, we conditionally ablated Rif1 in B cells using CD19Cre, which is expressed specifically in B cells (Rif1F/FCd19Cre/+ mice, Fig. S4, A, B and C). To induce CSR, B cells were activated with lipopolysaccharide (LPS) and interleukin (IL)-4 in vitro, and switching to IgG1 or IgG3 was measured by flow cytometry. CSR to IgG1 and IgG3 was markedly reduced in Rif1F/FCd19Cre/+ B cells, but less so than Trp53bp1−/− controls (Fig. 3, A and B and S5). Switch junctions from Rif1F/FCd19Cre/+ B cells were comparable to Trp53bp1−/− and wild type controls ((7) and Fig. S6), which indicates that, similar to 53BP1 deficiency, absence of Rif1 does not alter the nature of productive CSR joining events. A similar CSR defect was also obtained by conditionally deleting Rif1 with 4-hydroxy-tamoxifen (4HT) in Rif1F/FROSA26Cre-ERT2/+ B cells (Fig. S7). Finally, shRNA-mediated partial down-regulation of CtBP-interacting protein (CtIP), which interacts with Rif1 (Fig. S8C), and has been implicated in processing of DNA ends (21, 22), resulted in a very small but reproducible increase in CSR (Fig. S8, A and B). Thus, Rif1 is essential for normal CSR, and CtIP may not be the only factor that contributes to end processing in Rif1-deficient B cells.
Fig. 3. Rif1 deficiency impairs class switch recombination, and causes Igh and genome instability in primary B cells.
(A) Left: CSR to IgG1 96 hr post-stimulation of B lymphocytes with LPS and IL-4. Right: summary dot plot for three independent experiments (n = 3 mice per genotype). Mean values are: 23.6% for Cd19Cre/+, 23.4% for Rif1F/+Cd19Cre/+, and 5.0% for Rif1F/FCd19Cre/+ (P <0.008 with the paired student’s t-test). Bottom: B cell proliferation by carboxyfluorescein succinimidyl ester (CFSE) dilution. Data are representative of three independent experiments. (B) Same as in (A) but for CSR to IgG3 after stimulation with LPS alone. Mean values are: 3.2% for Cd19Cre/+, 3.4% for Rif1F/+Cd19Cre/+, and 0.5% for Rif1F/FCd19Cre/+ (P <0.008). (C) Left: Cell cycle analysis of primary B cells after stimulation with LPS and IL-4. Right: Summary histograms for S-phase, G0/G1 and G2/M cells from two independent experiments (n = 4 mice per genotype). Error bars indicate SEM. * 0.01 < P < 0.05, ** 0.001 < P < 0.01, *** P < 0.001. (D) Left: Cell cycle analysis of LPS- and IL-4-stimulated splenocytes at the indicated times post-irradiation (6 Gy). Right: Summary graphs for S-phase, G0/G1 and G2/M cells from two independent experiments (n = 3 mice per genotype). Error bars indicate SD. (E) Analysis of genomic instability in metaphases from B cell cultures. Chtid: chromatid; Chre: chromosome. Data are representative of two independent experiments (n = 50 metaphases analyzed per genotype per experiment). (F) Examples of Igh-associated aberrations in Rif1F/FCd19Cre/+ B cells. Chromosomes were hybridized with an Igh Cα probe (green; centromeric of Cγ1) and a telomere sequence-specific probe (red), and counterstained with DAPI (dark blue/black). Magnification, 63×; Scale bars, 1 μm. (G) Frequency of c-myc/Igh translocations in activated B cells. Graph shows combined results from 3 mice per genotype.
CSR requires cell division, activation-induced cytidine deaminase (AID) expression and Igh germline transcription (23). There are conflicting reports that Rif1 is required for proliferation in MEFs, but not DT40 B cells (17, 18). We found that cell division profiles of Rif1F/FCd19Cre/+ and 4HT-treated Rif1F/FROSA26Cre-ERT2/+ B cells were indistinguishable from controls (Fig. 3, A and B; and Fig. S7, A, C, E and G), indicating that Rif1 is dispensable for B cell proliferation in vitro. Finally, AID mRNA and protein expression, and Igh germline transcription were unaffected by Rif1 deletion (Fig. S4, B and D).
We next examined the role of Rif1 in cell cycle progression in primary B cells. We found no major differences in the percentage of cells in G0/G1 and S-phases (Fig. 3C). However, the number of cells in G2/M was increased approximately twofold in the absence of Rif1 (2.64, 2.56 and 1.91 fold at 48, 72, and 96 hours respectively) (Fig. 3C). Similar results were also obtained using Rif1F/FROSA26Cre-ERT2/+ B cells treated with 4HT (Fig. S7, H and I). Furthermore, irradiation increases the accumulation of Rif1F/FCd19Cre/+ B cells in G2/M (Fig. 3D). In addition, Trp53bp1−/− iMEFs expressing 53BP1DB28A, which did not recruit Rif1 to IRIF (Fig. 2C), exhibited delayed progression through S-phase following DNA damage with accumulation of cells in G2 after irradiation (Fig. S9).
Accumulation of cells in G2/M may reflect the persistence of unrepaired DNA damage in a fraction of Rif1-deficient cells. To investigate this possibility, we analyzed metaphase spreads from B cells dividing in response to LPS and IL-4 in vitro. These cells express AID, which produces DSBs in Igh, and less frequently at off-target sites throughout the genome, in the G1 phase of the cell cycle (24–26). Chromosomal aberrations were increased in Rif1F/FCd19Cre/+ B cells compared to controls (Fig. 3E), with many localized to the Igh locus (Fig. 3E). Consistent with the observation that Igh is targeted by AID in the G1 phase of the cell cycle, all of the Igh breaks were chromosome breaks (Fig. 3, E and F). Interestingly, the frequency of c-myc/Igh translocations is moderately increased in Rif1F/FCd19Cre/+ B cells, however, the breakpoint distribution was similar to Cd19Cre/+ control (1.5 × 10−6 versus 1.0 × 10−6 in control, P = 0.039; Fig. 3G and Fig. S10). We conclude that in the absence of Rif1, DSBs fail to be resolved efficiently in the G1, S or G2 phases leading to increased levels of genomic instability including chromosome breaks at Igh and translocations in dividing B cells.
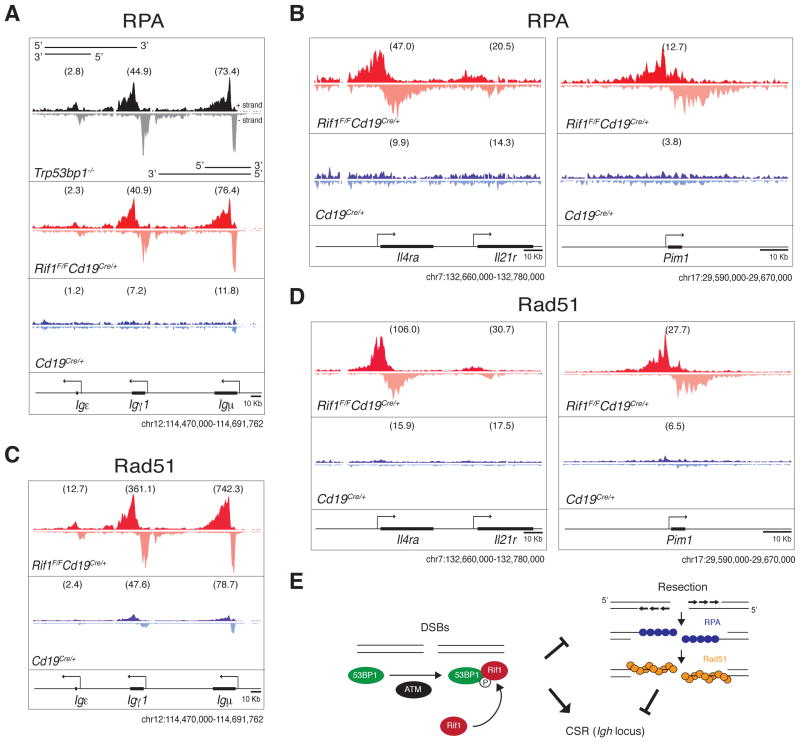
In the absence of 53BP1, DSBs produced by AID at the Igh locus accumulate the single-stranded DNA-binding replication protein A complex (RPA) as a result of increased DNA end resection (24). To determine if Rif1 is required for DNA end protection by 53BP1, we performed RPA-ChIP-seq (chromatin immunoprecipitation followed by massive parallel sequencing) experiments on Rif1F/FCd19Cre/+ and control B cells. Ablation of Rif1 was indistinguishable from loss of 53BP1 in that in its absence RPA decorates the Igh locus asymmetrically, in a manner consistent with 5′-3′ resection (Fig. 4A and (27)). In addition, absence of Rif1 also results in RPA accumulation at non-Igh genes like Il4ra and Pim1 that are damaged by AID in G1 (Fig. 4B) (24, 25). Rad51 is the recombinase that mediates repair of DSBs by homologous recombination in S/G2/M (22). To confirm that Rif1 prevents resection that takes place in S-phase, we monitored Rad51 accumulation in activated B cells by ChIP-Seq. Loss of Rif1 was indistinguishable from loss of 53BP1 (27), in that it led to asymmetric Rad51 accumulation at sites of AID-inflicted DNA damage (Fig. 4, C and D). We conclude that in the absence of Rif1, AID-induced DSBs incurred in G1 persist and undergo extensive 5′-3′ DNA end resection in S/G2/M, as measured by RPA and Rad51 accumulation.
Fig. 4. Rif1 prevents resection of DNA ends at sites of AID-induced DNA damage.
(A) to (D) RPA and Rad51 occupancy at the Igh locus (A and C) and at non-Igh AID targets genes (B and D) in B cells activated to undergo class switching. ChIP-seq libraries were resolved into upper (+) and lower (-) DNA strands to show RPA and Rad51 association with sense and antisense strands. Within a specified genomic window, graphs have the same scale and show tag density. Deep-sequencing samples were normalized per library size, and TPM (Tags Per Million) values were calculated for each genic region as indicated in Materials and Methods and shown in parenthesis. Data are representative of two independent experiments for RPA ChIP-seq and one for Rad51. (E) Model of Rif1 recruitment and DNA end protection at DSBs. DNA damage activates ATM, which phosphorylates many targets, including 53BP1. This event recruits Rif1 to 53BP1 at the DSB, where it inhibits DNA resection. The extensive resection in the absence of Rif1 impairs CSR at the Igh locus.
A role for Rif1 in maintenance of genome stability and protection of DNA ends against resection is consistent with its phosphorylation-dependent recruitment to the N-terminal domain of 53BP1 (4). 53BP1 facilitates DNA repair and prevents DNA end resection during CSR. In the absence of 53BP1, AID-induced DSBs are resolved inefficiently in G1 leading to chromosome breaks, Igh instability, and resolution by alternative-NHEJ or HR instead of classical-NHEJ (4, 8, 27). Our experiments show that in the absence of Rif1, 53BP1 is insufficient to promote genomic stability, mediate efficient Igh repair, DNA end protection or CSR. Thus, these 53BP1 activities require Rif1 recruitment to the phosphorylated N-terminus of 53BP1. Rif1 is likely to have additional functions beyond 53BP1, CSR and DNA end protection because whereas Trp53bp1−/− mice are viable, Rif1 deletion is lethal (17). Indeed, Rif1 is believed to play a role in the repair of S-phase DNA damage (17, 18), and in the regulation of replication timing (19, 20, 28). Analogously, additional CSR factor(s) may exist downstream of 53BP1, as class switching in Rif1-deficienct B cells is significantly higher than in Trp53bp1−/−.
In summary our data is consistent with a model whereby ATM-mediated phosphorylation of 53BP1 recruits Rif1 to sites of DNA damage, where it facilitates DNA repair in part by protecting DNA ends from resection (Fig. 4E). In the absence of Rif1, DNA breaks incurred in G1 fail to be repaired by NHEJ and undergo extensive 5′-3′ end resection resulting in accumulation of chromosome breaks and genome instability.
Supplementary Material
Acknowledgments
We thank all members of the Nussenzweig laboratory for discussion, D. Bosque and T. Eisenreich for help in managing mouse colonies, A. Gazumyan for assistance with Igh germline and AID transcript levels analysis, and K. Yao for help with genotyping. We thank T. de Lange (The Rockefeller University, New York) for Rif1F/F mice, S. Buonomo (EMBL Mouse Biology Unit, Monterotondo) for the anti–mouse Rif1 serum #1240, and G. Gutierrez (NIAMS, NIH, Bethesda) for Illumina sequencing. We are grateful to N. Zampieri (Columbia University, New York) for assistance with IF image processing, and to M. P. Rout, J. LaCava, S. Obado and L. Hough (The Rockefeller University, New York) for invaluable help, discussions and protocols for cryolysis and magnetic bead-mediated immuno-isolation. The data presented in the manuscript are tabulated in the main text and in the supplementary materials. Sequence data shown in Fig. 4 have been deposited in the Gene Expression Omnibus database at http://www.ncbi.nlm.nih.gov/geo/. M.D.V. was a Fellow of the American Italian Cancer Foundation. A.D.G. was supported by NIH MSTP grant GM007739. This work was supported in part by NIH grants AI037526 (M.C.N.), RR022220 (B.T.C.), RR00862 (B.T.C.), GM103314 (B.T.C.), and by the intramural program of NIAMS at the NIH (R.C.). M.C.N. is an HHMI Investigator.
Footnotes
Materials and Methods
References and Notes
- 1.Adams MM, Carpenter PB. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell division. 2006;1:19. doi: 10.1186/1747-1028-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011 Oct;13:1161. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 3.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA Repair (Amst) 2011 Oct 10;10:1071. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011 May 6;42:319. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008 Nov 27;456:529. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008 Nov 27;456:524. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur J Immunol. 2007 Jan;37:235. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 8.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010 Apr 12;207:855. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004 May;5:481. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 10.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004 May 24;165:459. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary material on Science Online.
- 12.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003 Oct 24;302:636. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 13.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992 May;6:801. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, et al. Role for Rif1 in the checkpoint response to damaged DNA in Xenopus egg extracts. Cell Cycle. 2012 Mar 15;11:1183. doi: 10.4161/cc.11.6.19636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004 Sep 1;18:2108. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004 Dec 6;167:819. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonomo SB, Wu Y, Ferguson D, de Lange T. Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol. 2009 Nov 2;187:385. doi: 10.1083/jcb.200902039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu D, et al. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010 Sep 15;29:3140. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornacchia D, et al. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012 Jul 31; doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, et al. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012 Jul 31; doi: 10.1038/emboj.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007 Nov 22;450:509. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 23.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 24.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012 Feb 7; doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001 Dec 6;414:660. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011 Jan;12:62. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamane A, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA end resection during the S-G2/M phase of the cell cycle. Cell Reports. 2013;1 doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayano M, et al. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012 Jan 15;26:137. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Luca C, et al. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest. 2005 Dec;115:3484. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997 Mar 15;25:1317. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008 Dec 12;135:1028. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cristea IM, et al. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol. 2010 Jul;84:6720. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krutchinsky AN, Kalkum M, Chait BT. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal Chem. 2001 Nov 1;73:5066. doi: 10.1021/ac010682o. [DOI] [PubMed] [Google Scholar]
- 34.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 35.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008 Dec;26:1367. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 36.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004 May 3;199:1235. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callen E, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009 May 15;34:285. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000 Sep 1;102:553. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 39.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010 Oct 1;143:122. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003 Jun 16;197:1767. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006 Mar 2;440:105. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nature methods. 2008 Sep;5:829. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callen E, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007 Jul 13;130:63. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Huen MS, et al. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010 Mar 26;37:854. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo KW, et al. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem. 2005 Mar 4;280:8172. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 46.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001 Apr 30;153:613. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010 Feb;12:177. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 48.Boisvert FM, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005 Dec;4:1834. doi: 10.4161/cc.4.12.2250. [DOI] [PubMed] [Google Scholar]
- 49.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002 Jan;22:555. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.