Abstract
Epigenetics refers to functionally relevant modifications of the genome that do not involve a change in the nucleotide sequence. Examples of such modifications are DNA methylation and histone modifications. Both modifications serve to regulate gene expression without altering the underlying DNA sequence. The epigenome encodes critical information to regulate gene expression. The cellular epigenome is established during development and differentiation and maintained during cell division. These instructions are different in each cell type; therefore the epigenome is cell type specific. Nutrient availability and other environmental factors cause changes in the epigenome. Recent research suggests the critical contribution of the epigenome to the development of complex gene-environmental diseases including chronic kidney diseases.
What is Epigenetics?
Epigenetics, meaning ‘above (epi-) genetics’, is the study of gene expression regulation that cannot be directly attributed to changes in the DNA sequence. Among the 3 billion nucleotides in our genome, less than 2% are responsible for coding proteins. The rest of the sequences were long thought to be non-functional (‘junk’), possibly only there as a buffering zone for mutations. Recent reports, especially those published by the ENCODE consortium, indicate that, by describing and annotating the cellular epigenomes, functional roles could possibly be assigned to as much as 80% of the genome [1]. It seems that the instruction manual that governs gene expression is hidden somewhere in the ‘junk DNA’.
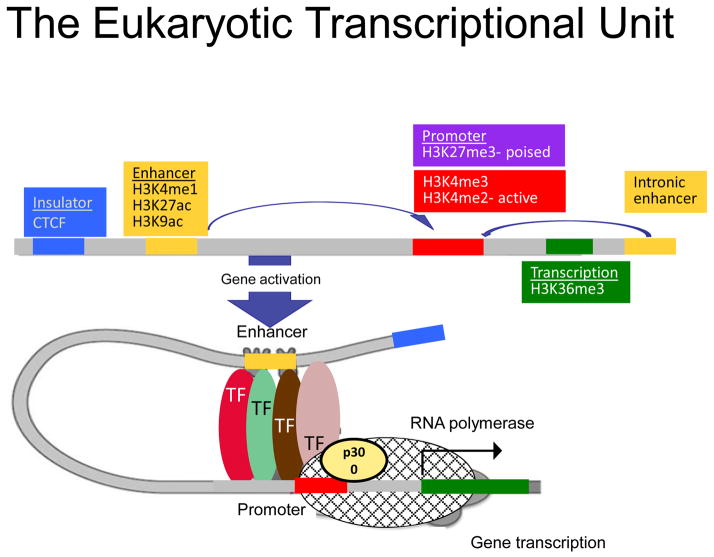
The epigenome involves ‘marking’ (i.e. chemical modification) of the DNA or associated proteins. Epigenetic marks include cytosine modification (mainly methylation) and histone tail modification. The epigenome is inherited during cell division to maintain cell identity. However, nutrition and other environmental factors change these chemical tags. Therefore these chemical tags (i.e. the epigenome) serve as the intersection between the stably inherited genome and the changing environment. Characterization of the epigenome has helped tremendously to define cell-type specific gene expression. A typical transcription unit in a multicellular eukaryote contains both clusters of proximal promoter elements and five types of cis-acting regulatory sequences (insulators, promoters, enhancers, silencers, and locus control regions) (Fig. 1). Insulator areas border and separate transcriptional units. Promoters are usually located at the 5′ end of transcription start site and contain elements called TATA box, which is bound by either a TATA-binding protein or a cluster of cytosine-guanine nucleotide pairs within the linear sequence, known as CpG islands (CGI). For target gene transcription to take place, not only promoters but also long- and short-range regulatory regions are needed. These cis-type gene regulatory regions are highly cell type specific and are critical for cell-type specific transcription (Figure 1). Simultaneous binding of transcription factors to each other and to the long-and short-range regulatory regions, results in genomic DNA loops that join distant regulatory DNA sequences together [2, 3].
Figure 1. The Eukaryotic transcription unit.
The eukaryotic transcription complex is characterized by a combination of transcription factors binding to promoters and one or multiple looping enhancers. Enhancers can be upstream or downstream (intronic). Enhancers are cell type specific. Both enhancer looping and TF binding are essential for mRNA transcription. Different chromatin regions are characterized by different histone tail marks.
Enhancers are also of critical importance (Figure 1). Enhancers can be located from a few kilobases (short-range), up to hundreds of kilobases (long-range) away from the regulated gene [4, 5]. They can be found on the opposite DNA strand, downstream of the regulated gene, or on intronic regions. Enhancers usually show significant enrichment for the binding of the cell type specific transcription factors [6]. Binding (or the presence) of multiple enhancers is critical for regulating the maximal transcription strength following an external stimulus. However, sometimes the loss of even one enhancer can result in loss of transcript. While the critical importance of enhancers is increasingly appreciated, genome-wide identification of enhancers has been exceedingly difficult. Recent advances in epigenetics, sequencing and computational methods have helped to define these important gene regulatory regions and will be discussed in detail later in this review.
Different histone modifications
The DNA in the nucleus is wrapped around small basic proteins called histones to form a larger organized structure called chromatin. When 147 base pairs of DNA are wrapped around histones, it forms a compact structure called a nucleosome. In addition to DNA, nucleosomes are comprised of a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4. Nucleosomes play important role in compacting DNA and the DNA wrapped on the nucleosome is not accessible to transcription factors. Amino acid modifications of histone tails appear to be central to the regulation of transcription. The “histone code” hypothesis proposes that chromatin-DNA interactions (i.e. the gene regulatory complex) are guided by histone tail modifications. There are molecular histone code ‘writers’ that establish these modifications and ‘readers’, enzymes that use these histone tail modifications to assign gene regulatory function to the DNA. More than 60 different histone modifications have been described. It seems that histone modifications are redundant and can be simplified into 10–15 different patterns, each with specific gene regulatory functions (Table I). Functional genomic elements are identified through sequence-association with histone modification, transcription factor binding and DNaseI hypersensitivity [7]. Cell type specific gene regulation can be understood by defining the cellular histone code (the epigenome). The pioneering ENCODE project has resulted in the ongoing characterization of cell type-specific annotations of gene regulatory regions through the use of cultured human cell lines [1].
Table I.
The most commonly analyzed histone modifications and their relationships to gene regulatory regions.
| Chromatin mark | Chromatin state |
|---|---|
| CTCF | Insulator/Repetitive/CNV |
| H3K27me3 | Poised promoter/repressed promoter |
| H3K36me3 | Transcription transition and elongation |
| H4K20me1 | Transcription transition |
| H3K4me1 | Enhancer |
| H3K4me3 | Active promoter |
| H3K4me2 | Promoter/enhancer |
| H3K27ac | Enhancer≫promoter |
| H3K9ac | Enhancer≫ promoter |
Transcription factor binding is a key characteristic of a regulatory region therefore; gene regulatory regions are nucleosome free (the DNA has to be open in these regions). Nucleosome free DNA is sensitive to DNaseI digestion, thus allowing the for identification of gene regulatory regions by the “DNaseI Hypersensitivity Method”. Nucleosomes that are directly adjacent to DNaseI hypersensitive sites are characterized by histone proteins that have specific tail modifications. For example, H3K4me3 (read as trimethylation of the 4th lysine:K in the H3 histone) in combination with H3K4me2 and H3K4me1 usually decorate active promoter areas. On the other hand, H3K37me3 and H3K9me3 usually marks inactive promoters. H3K4me1, H3K27ac and H3K9ac marks together are associated with enhancer activity [8]. H3K36me3 and H3K20me1 mark transcribed regions both for coding and non-coding transcripts (Table I). The binding of the transcriptional repressor, CTCF, defines insulator areas that separate different transcriptional units. Combinations of histone marks further tune gene regulatory regions within the genome. In particular, H3K4me1 and H3K27ac together have been shown to be very effective in determining the location of cis-regulatory active enhancers. H3K9me3 and H3K20me3 histone tail modifications locate satellite, telomeric and repeat regions.
In embryonic stem cells, large genomic regions have histone modifications with opposite characteristics to those seen in differentiated cell types. For example, many developmentally important transcription factor-encoding regions in embryonic stem cells contain the active mark of H3K4me3 and H3K27me3 functions as a transcriptional repressor. These regions are called ‘bivalent domains’ [9–11]. Promoters of key developmental transcription factors that control differentiation typically have bivalent domains [12, 13]. It is believed that during differentiation cells will take on either the active or the repressive mark depending on which pathway they follow.
The addition of methyl groups to histones is attributed to three families of enzymes that catalyze the addition of methyl groups donated from S-adenosylmethionine to histones. The SET-domain containing proteins and DOT1-like proteins have been shown to methylate lysines (K), and members of the Protein Arginine (R) N-methyltransferase (PRMT) family have been shown to methylate arginines. H3K27me3 is mostly catalyzed by the EZH subunit of the Polycomb repressive complex 2 (PRC2) [14–16]. Histone acetylation is catalyzed by histone acetyltransferases (HATs) and acetyl groups are removed by histone deacetylases (HDACs). These enzymes can be targeted by histone deacetylase inhibitors (HDACi) such as valproic acid and hydroxamic acids vorinostat (SAHA).
Cytosine Methylation
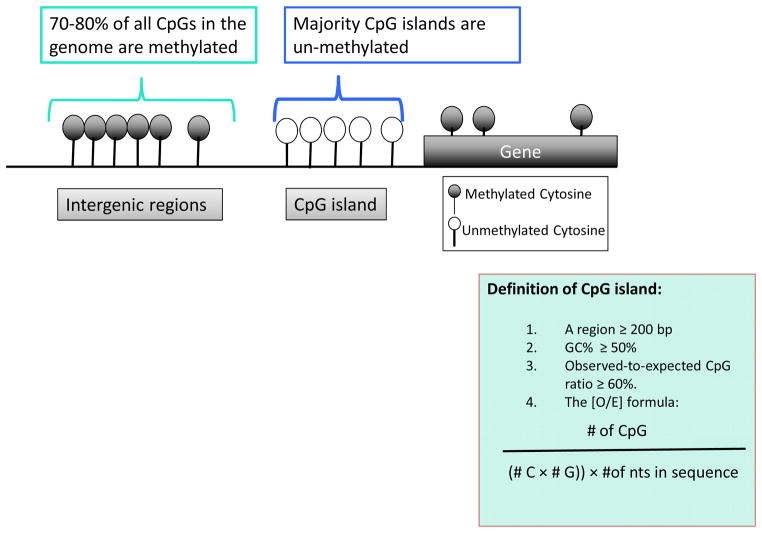
Methylation of cytosines on the 5th position (5-methylcytosine, m5C), also called ‘the fifth base’, is an epigenetic modification. The majority of the genome has a low cytosine/guanine content and these cytosines are usually methylated, including transposons and other repeated elements. Cytosine rich regions are organized into CpG islands (CGIs) in the genome; these are short stretches (about 300–3,000 bp) regions characterized by high cytosine content (more than 60%) [17]. These CGIs are enriched on gene promoter regions and cytosines in CGIs are usually unmethylated (Fig. 2). Methylation of promoters negatively correlates with transcript levels. There are two basic models for promoter hypermethylation induced transcriptional silencing: DNA methylation can directly repress transcription by blocking transcriptional activators from binding to cognate DNA sequences; or methyl binding proteins recognize methylated DNA and recruit co-repressors to silence gene expression directly. Genome-wide studies also indicate that methylation of regions adjacent to CGIs, so-called ‘CpG shore regions’, play a key role in diverse biological processes. Gene body regions can also be methylated (Fig. 2). The functional effect of gene body methylation is hotly debated, however, it may enhance transcription through inhibition of cryptic transcription initiation.
Figure 2. Cytosine methylation.
Cytosine methylation of CpG islands that are localized to promoter regions is usually low. Promoter methylation is usually associated with gene silencing. Cytosine methylation of regions outside the CpG islands is usually high.
Cytosine methylation levels are usually the highest in fully differentiated cells, however, cytosine methylation is erased after fertilization, with only a handful of imprinted regions remaining methylated in zygotes. During cell type specific differentiation, cytosine methylation is established by DNA (cytosine-5)-methyltransferases (DNMTs) [18–21]. DNMT3A and 3B participate in de novo DNA methylation of unmethylated cytosines. DNMT3a and DNMT3b appear to exhibit non-overlapping functions in development. DNMT3b is specifically required for methylation of centromeric minor satellite repeats. Mutations of human DNMT3B are found in ICF syndrome (Immunodeficiency, Centromeric instability and Facial anomalies), a developmental defect characterized by hypomethylation of pericentromeric repeats [22, 23]. DNMT1 is responsible for maintaining the already established DNA methylation levels during cell division. Homozygous Dnmt1 embryos are stunted, delayed in development, and do not survive past mid-gestation [24]. In addition, to cytosine methylation, hydroxymethylation of cytosines have been recently identified. Hydroxymethylated cytosine content is the highest in stem cells. Cytosines are hydroxymethylated by the ten-eleven translocation TET (TET1-3) proteins. Mutations in TET2 have been associated with leukemia [25].
The epigenome as the molecular footprint of the environment
The cellular epigenome is established during development and differentiation. Cells constantly adjust to their environment and one key feature of the epigenome is its dynamic characteristics as it changes under environmental pressure. These changes are most prominent in yeast, where nutrient availability is the key determinant of the phenotype. How does nutrient availability change the epigenome? Most chromatin-modifying enzymes require substrates or cofactors that are intermediates of cell metabolism. For example, acetylation by histone acetyltransferases (HATs) depends on the local subcellular acetyl-CoA concentration [26]. H3 Lys4 trimethylation appears to be especially sensitive to changes in threonine metabolism. In vitro evidence supports that fluctuations of metabolite levels modulate activities of chromatin-modifying enzymes, thereby influencing chromatin dynamics. It is predicted that cells are likely to be even more sensitive to this “epigenetic shift” during development and differentiation when the epigenome is more plastic and gene regulatory regions are being established. Epigenetic changes established during development might even play important role in phenotype development later on by modulating how cells later respond to stimuli. For example, functionally competent podocytes develop even in the absence of a key histone modification enzyme (PTIP), although the cells have an abnormal response to environmental stressors for example during aging [27]. Recent reports suggest that we may even be able to determine cellular aging based on the epigenetic signature [28–30], indicating that the epigenome could provide a tractable link between the genome and the environment, with the epigenome emerging as a biochemical record of relevant life events.
Why should we study epigenetics in Chronic Kidney Disease?
It is well established that epigenetic silencing of tumor suppressors play key role in malignant transformation. In addition, mutations in many of the key chromatin modification enzymes (including TET and PRMD2 etc.) have been detected in various cancer types, indicating that genetic-epigenetic interaction drives carcinogenesis. However, very little information is available on the role and contribution of epigenetics to non-cancerous common diseases, such as chronic kidney disease (CKD).
CKD is a common complex gene-environmental disease. Although the genetics of CKD remains largely unexplored, lately multiple genes with small effects on serum creatinine levels have been identified [31–33]. In addition, several environmental factors have been shown to have a long lasting effect on CKD development. Environmentally induced epigenetic modifications could represent a plausible mediator to explain the impact of the environment on CKD development. Intrauterine malnutrition has been associated with metabolic syndrome, hypertension and renal disease development. This phenomenon became known as “fetal programming’ [19, 34]. Brenner and colleagues showed that low birth weight (LBW), a measure of intrauterine malnutrition, is associated with a reduced nephron number. A reduction in the number of nephrons results in increased glomerular pressure of the remaining nephrons. The increased work-load can then induce microalbuminuria and glomerulosclerosis, resulting in nephron loss and renal injury [35]. The most prominent observations came from long-term follow-ups of children born in the Netherlands during the winter of 1944–1945 (known as “The Hunger Winter”), when food was rationed and calorie count was reduced. Children born during this period had a greatly increased risk developing diabetes, cardiovascular disease, and obesity later in life [36–38]. In the Pi-ma Indians, a much-studied population of Native Americans in the southwestern United States, LBW correlates with type II diabetes and the progression of CKD [39]. As well, in the general population, a direct correlation of LBW to microalbuminuria in type I diabetes has been found [40]. Rodent models of intrauterine growth retardation indicate that lack of nutrient or oxygen availability will program the development of salt-sensitive hypertension and microalbuminuria. Together, these results indicate that the epigenome could be an important mediator of this long lasting intrauterine environmental effect [41], particularly as the epigenome is plastic during development.
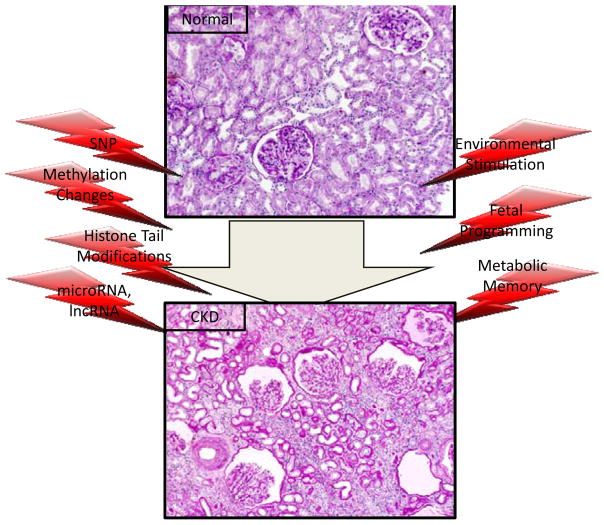
It was believed that differentiated cells have a stable epigenome, however lately a memory or programming effect of the environment has been described beyond development as well (Fig. 3). The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications studies (DCCT/EDIC), established a beneficial effect of intensive control of blood glucose and blood pressure for patients with diabetes [42]. Recent reports, including from the follow up ED-IC studies, indicate that an episode of hyperglycemia even when it is followed by more than 20 years of intensive glycemic control is associated with an increased incidence of progressive renal function decline; a finding interpreted as ‘hyperglycemic memory’ (Fig. 3) [43]. Metabolite fluctuation in diabetes could easily induce a shift in the cellular epigenome. The molecular mechanism of fetal programming and hyperglycemic memory are yet to be determined, but epigenetic adaptation could be involved. These observations provide a strong rationale to study the contribution of epigenetic changes to CKD development (Fig. 3).
Figure 3.
Chronic kidney disease is a complex disease with both genetic and environmental components
How should we study the epigenome?
Analyzing condition-relevant cell types is critical to understand the role of epigenetics to disease development (e.g., renal epithelial cells to understand kidney disease). As the cell type is the key determinant of the epigenome, it is less likely that much mechanistic understanding is gained from analyzing cells that are not functionally linked to disease development. Cytosine methylation is one of the most stable epigenetic mark; it can be studied even in archived samples as the genomic DNA is stable.
DNA Methylation
There are three major approaches to study cytosine methylation; restriction enzyme digestion, affinity enrichment, and sodium bisulfite conversion based. Each method has its strengths and caveats. Currently, all of these methods are available for both locus specific and genome-wide studies, when coupled with whole genome covering microarrays or sequencing technologies.
The restriction enzyme digestion method is based on isoschizomer enzymes that recognize the same nucleotides, but differentially digest DNA based on the cytosine methylation status. The most frequently used isoschizomer for DNA methylation studies are HpaII, and MspI, which recognize the same sequences. MspI is insensitive to methylation while Hpall will only digest the site when the cytosines are methylated. By comparing the fragments after restriction enzyme treatments, site-specific methylation differences can be determined. The digested DNA can be subjected either to PCR for locus-specific study or to microarray/sequencing for genome-wide screening. SmaI and its neoschizomer XmaI is another favored enzyme pair. Xmal and Smal both recognize the sequence, CCCGGG, but they digest it at different sites. Smal is sensitive to cytosine methylation while Xmal is methylation insensitive. The advantage of enzyme digestion is their sensitivity to hypomethylated regions (for examples CGIs). The drawback of this method is that the coverage and resolution is restricted by the regions where restriction enzyme sites are present. One of the most commonly used methods to take advantage of these restriction enzymes is the HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) assay developed by Greally et al [44, 45].
Affinity enrichment methods utilize antibodies against m5C, or methyl group binding protein domains (MBD) that target m5C, such as MeCP2, or MBC. There are five proteins with mCpG-binding motifs that have been identified in the MBD family: MBD1, MBD2, MBD3, MBD4, and MeCP2 [46]. To capture m5C with antibodies and proteins, the genomic DNA is randomly sheared into small fragments by sonication. The fragments are then subjected to immunoprecipitation with antibodies (above) targeting methylated cytosines in DNA (called MeDIP) [47, 48]. The advantage of MeDIP method is a true genome-wide coverage as it is not limited by restriction enzyme sites. However, a key disadvantage of MeDIP is a limited resolution. The binding of the antibody is determined by the cumulative methylation level of a region and the resolution is limited by the sonication.
The sodium bisulfite conversion based cytosine methylation determination is an unbiased and sensitive method, which is currently the gold standard for determining cytosine methylation. The method is based on the principle that unmethylated cytosine will be converted to uracil after bisulfite treatment while methylated cytosines will not. Fragments can later be either subjected to cloning and sequencing or to mass spectrometry (for example using the Sequenom EpyTYPER-type equipment) to interrogate locus specific cytosine methylation. Genome-wide quantification of bisulfite-converted cytosines can be performed by either microarray or sequencing-based platforms. The Infinium HumanMethylation 450BeadChips arrays (bisulfate conversion-based assays) are gaining popularity as an easy and cost-effective methylation analysis. These arrays appear to perform quite well for epigenome-wide association studies. One key disadvantage is that they only analyze the methylation of 480,000 preselected loci, representing less then 1% of the whole genome. The ‘reduced representation bisulfite sequencing’ is another bisulfite-conversion based method that combines restriction enzyme based size selection followed by next-generation sequencing. This method provides excellent coverage for CGI and promoter regions. Genome-wide bisulfite sequencing provides the best, and highest resolution method to examine cytosine methylation status. The main drawback of this method is the cost associated with whole-genome sequencing and the data analysis.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation, followed by either microarrays or next generation sequencing (ChIP-chip/ChIP-Seq), are the most frequently used methods to map transcription factor binding and histone modifications. Here genomic DNA and proteins in either live cells or fresh tissues are cross-linked with formaldehyde, then the chromatin is fragmented into 200 base pair regions. The sheared DNA/protein complex is then subjected to immunoprecipitation with an antibody against the selected transcription factor or histone tail modification. Once the genomic DNA is reverse cross-linked and released from the protein complex, it can be analyzed using quantitative PCR, microarrays or sequencing. While ChIP has revolutionized our understanding of the epigenome, the method is not without limitations. First the method does not provide an epigenetic map with base pair resolution as the resolution is limited by the size of the sheared chromatin. To circumvent this issue, an enzyme digestion based method, the exo-ChIP has been developed [49]. The key limitation of the ChIP-seq relates mainly to the specificity of the antibodies used for the studies. To address this issue, the most commonly used antibodies are constantly evaluated with their quality posted on antibody validation databases (http://compbio.med.harvard.edu/antibodies/).
Next-Generation Sequencing Platforms and Data Analysis
For genome-wide bisulfite sequencing, high (usually at least 30x) genome coverage and long reads are needed. Therefore, 100–150 bp paired-end sequencing is used. For ChIP-seq, generally short, 50-bp single end sequencing is used. The SOLiD and the HiSeq (Illumina, Inc.) machines are excellent choices for these experiments. Multiple ChIP samples can be run on a single HiSeq lane, providing sufficient coverage for analysis. ChIP-Seq analysis typically begins by aligning each read to the genome and then determining the number of reads that aligns within each genomic region. As the accessibility of each genomic region is slightly different, an enrichment score is developed to show the enrichment fraction is normalized to input DNA (that is sonicated DNA without antibody enrichment). Both the degree of enrichment, and the size of the region that is marked by an individual histone mark, are important. Multiple different modifications can be present on an individual region. To address this challenge, several algorithms have been developed using computational techniques that incorporate histone mark information to identify functional elements from high-throughput genomic data sets. Recently, Ernst and Kellis developed a new method that is based on the Hidden Markov Model, which can be used to generate gene regulatory region annotation, based on a panel of histone ChIP-seq experiments performed on the same sample [50, 51]. The method takes into consideration of the presence of multiple different histone marks and their reference sequence based location [50]. This method will be highly valuable to compare multiple different histone marks either between different cell types, or within the same cell type but expressing different phenotypic characteristics, to highlight key similarities and differences in datasets.
What do we know about epigenetics and chronic kidney disease development? What do the mouse models tell us?
The most convincing result indicating the role of epigenetics in chronic kidney disease development comes from studies performed in the Zeisberg lab. In an elegant paper published in 2010, they showed differences in the cytosine methylation profiles of fibroblasts isolated from either control or CKD kidneys. Using a folic acid induced kidney fibrosis model, the authors specifically demonstrated that hypermethylation of a GTPase activating protein, RASAL1, causes increased Ras activation in fibroblasts, resulting in fibroblast proliferation and fibrosis development [52]. They also showed that genetic deletion or chemical inhibition of the DNMT1 is able to reduce fibrosis development in this model. Future studies using DNMT inhibitors in other progressive kidney disease models, as well as cell type specific Dnmt1 deletion in mice, will aide in elucidating how changes in cytosine methylation specifically contribute to kidney fibrosis development.
The contribution of HDACs has also been described in different mouse models of acute and chronic kidney injury. The HDAC inhibitor, TSA, ameliorates proliferative glomerulonephritis, long-term glomerulosclerosis, and proteinuria in a nephrotoxic serum nephritis model. Pre-treating mice with valproic acid, a class I-selective HDAC inhibitor, was highly effective reducing proteinuria as well as sclerosis in the adria-mycin-induced focal segmental glomerulosclerosis model [53]. Two separate studies have demonstrated that HDAC inhibitors can also attenuate diabetes induced renal hypertrophy and renal damage in rodents. Together, these chemical inhibitor based studies strongly suggest that histone deacetylases play a role in chronic kidney disease development. The proposed mechanism being that they are involved in reactivating major developmental pathways and promoting tissue repair following injury. Unfortunately, HDAC inhibitors like many inhibitors, have specificity issues. Therefore, follow-up studies using genetically engineered site-specific knock-out animals will be essential to determine the role and contribution of histone acetylation in kidney fibrosis.
Cell culture based studies from the Natarajan and El-Osta laboratories indicate that cells that are grown in high-glucose containing medium develop rapid and persistent changes in their histone tail modification patterns [43, 54]. In cultured aortic endothelial cells, even transient hyperglycemia caused changes in H3K4me1 on the promoter of the p65 subunit of NFkB and subsequently was associated with increased p65 transcript levels. The expression of p65 was increased in endothelial cells even several days after they were returned to normal glucose medium. A similar mechanism has also been shown in diabetic mice [55]. The sustained proinflammatory phenotype observed in vascular smooth muscle cells cultured from type 2 diabetic db/db mice, was associated with reduced levels of the repressive mark, H3K9me3, on the affected inflammatory gene promoters [56]. Treatment of glomerular mesangial cells with TGF-beta1 or high glucose could lead to changes in key active and repressive histone modifications at the promoters of inflammatory and fibrotic genes [57, 58]. These interesting observations link episodes of hyperglycemia with a persistent increase of inflammation and fibrosis, which are important characteristics of diabetic kidney disease.
Human Studies
Large-scale human studies showing epigenetic differences in chronic kidney disease have not been published. As the epigenome is cell type specific, human kidney cells will be necessary for such experiments. There are few human epigenome-wide-association studies that used peripheral blood mononuclear cells or other surrogate cell types. Sapienza et al. described differences in cytosine methylation profiles of diabetic patients with end stage renal disease (ESRD) when compared to diabetic patients without ESRD. They used the Illumina 27KBeadArrays to determine cytosine methylation changes. While there was significant patient heterogeneity, they have identified close to 100 loci with statistically significant methylation differences in ESRD samples as compared to those without. Most loci showed lower methylation levels in ESRD cases [59]. Many of the differentially methylated regions were in close proximity to genes that had previously been shown to be differentially expressed in kidney disease patients. While this is an important first step to understand the epigenome of chronic kidney disease, it will be interesting to see whether markers found in saliva correlate to epigenetic changes in renal epithelial cells.
A very nice study from the Falk group examined the potential contribution of epigenome to antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis. The AN-CA disease is associated with spontaneous development of autoantigens against myeloperoxidase (MPO) and proteinase 3 (PR3) proteins. They found that H3K27me3, a chromatin modification associated with silenced promoters, was decreased on the PR3 and MPO promoter areas in ANCA patients compared with healthy controls. This was also associated with decreased cytosine methylation of PR3 and MPO CGI promoter areas. The authors propose that normally increased RUNX3 levels are responsible for recruiting the H3K27 methyltransferase, a subunit of enhancer of zeste homolog 2 (EZH2) to PR3 and MPO loci. Loss of RUNX3 expression is the likely cause for the loss of H3K27methylation and the expression of MPO and PR3 in ANCA patients [60]. This is a very elegant illustration that epigenetic modifications associated with gene silencing are perturbed at ANCA autoantigen-encoding genes, potentially contributing to autoimmunity development. Future studies shall determine whether similar mechanisms might play role in other autoimmune diseases as well. Our lab been investigating the regulation of gene expression in the pathogenesis of DKD. The alterations in both gene expression and cytosine methylation in the TGFβ pathway (TGFBR3, SMAD3, SMAD6) have been observed in the dataset. These genes are known to be key regulators of kidney fibrosis development and have important roles in CKD (unpublished observations).
Summary
Epigenomics is a new and emerging field that has experienced incredible growth during the last few years. It has improved our understanding of the human genome organization. The epigenome represents a plausible factor in mediating the long-term footprint of environmental alterations. Epigenetic alterations play a key role in cancer development and new epigenome-based therapies are on the horizon for clinical testing. Clinical observational studies and animal model experiments strongly support the role of long-term programming in kidney disease development and progression. As methods to perform epigenome-wide analysis have improved and patient samples are increasingly available, future studies shall address the role and contribution of the epigenetic changes to acute and chronic kidney disease development.
Acknowledgments
Financial Support: Research in the Susztak lab is supported by the NIH (NIDDK), ADA and JDRF
We would like to thank the NIH (R01DK087635) for supporting the work of the Susztak lab.
Footnotes
Financial disclosure and conflict of interest statements: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148(5):908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gheldof N, et al. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38(13):4325–36. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin F, et al. Enhancers: multi-dimensional signal integrators. Transcription. 2011;2(5):226–30. doi: 10.4161/trns.2.5.17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–9. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22(9):1798–812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 11.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 12.Bergsland M, et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25(23):2453–64. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liber D, et al. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7(1):114–26. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 15.Bracken AP, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20(9):1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem. 2002;277(37):34521–30. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- 19.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 20.Feldman N, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–94. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 21.Yeo S, et al. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359(3):536–42. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 22.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96(25):14412–7. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlich M, et al. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity. 2008;41(4):253–71. doi: 10.1080/08916930802024202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 25.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 27.Lefevre GM, et al. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010;6(10):e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch CM, et al. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell. 2012;11(2):366–9. doi: 10.1111/j.1474-9726.2011.00784.x. [DOI] [PubMed] [Google Scholar]
- 29.Bork S, et al. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9(1):54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schellenberg A, et al. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging (Albany NY) 2011;3(9):873–88. doi: 10.18632/aging.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada Y, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44(8):904–9. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–84. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kottgen A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–7. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravelli AC, et al. Cardiovascular disease in survivors of the Dutch famine. Nestle Nutr Workshop Ser Pediatr Program. 2005;55:183–91. doi: 10.1159/000082602. discussion 191–5. [DOI] [PubMed] [Google Scholar]
- 35.Hershkovitz D, et al. Fetal programming of adult kidney disease: cellular and molecular mechanisms. Clin J Am Soc Nephrol. 2007;2(2):334–42. doi: 10.2215/CJN.03291006. [DOI] [PubMed] [Google Scholar]
- 36.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumey LH, et al. Cohort profile: the Dutch Hunger Winter families study. Int J Epidemiol. 2007;36(6):1196–204. doi: 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- 38.Ravelli AC, et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351(9097):173–7. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 39.Nelson RG, Morgenstern H, Bennett PH. Birth weight and renal disease in Pima Indians with type 2 diabetes mellitus. Am J Epidemiol. 1998;148(7):650–6. doi: 10.1093/aje/148.7.650. [DOI] [PubMed] [Google Scholar]
- 40.Yudkin JS, Phillips DI, Stanner S. Proteinuria and progressive renal disease: birth weight and microalbuminuria. Nephrol Dial Transplant. 1997;12(Suppl 2):10–3. [PubMed] [Google Scholar]
- 41.Vom Saal FS, et al. The estrogenic endocrine disrupting chemical bisphenol A (BPA) and obesity. Mol Cell Endocrinol. 2012;354(1–2):74–84. doi: 10.1016/j.mce.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Boer IH, et al. Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med. 2011;365(25):2366–76. doi: 10.1056/NEJMoa1111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper ME, El-Osta A. Epigenetics: mechanisms and implications for diabetic complications. Circ Res. 2010;107(12):1403–13. doi: 10.1161/CIRCRESAHA.110.223552. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki M, Greally JM. DNA methylation profiling using HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) Methods. 2010;52(3):218–22. doi: 10.1016/j.ymeth.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson RF, et al. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24(9):1161–7. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou X, et al. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 2012;40(6):2747–58. doi: 10.1093/nar/gkr1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 48.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 49.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147(6):1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9(5):473–6. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bechtel W, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16(5):544–50. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pang M, et al. Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am J Physiol Renal Physiol. 2009;297(4):F996–F1005. doi: 10.1152/ajprenal.00282.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res. 2011;90(3):421–9. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Osta A, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205(10):2409–17. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villeneuve LM, et al. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105(26):9047–52. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan H, et al. Involvement of p300/CBP and Epigenetic Histone Acetylation in TGF-beta1 Mediated Gene Transcription in Mesangial Cells. Am J Physiol Renal Physiol. 2012 doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun G, et al. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21(12):2069–80. doi: 10.1681/ASN.2010060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapienza C, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6(1):20–8. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 60.Ciavatta DJ, et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest. 2010;120(9):3209–19. doi: 10.1172/JCI40034. [DOI] [PMC free article] [PubMed] [Google Scholar]